Abstract
Amplification and eventual elimination of dispersed repeats, especially those of the retroelement origin, account for most of the profound size variability observed among plant genomes. In most higher plants investigated so far, differential accumulation of various families of elements contributes to these differences. Here we report the identification of giant Ty3/gypsy-like retrotransposons from the legume plant Vicia pannonica, which alone make up ∼38% of the genome of this species. These retrotransposons have structural features of the Ogre elements previously identified in the genomes of pea and Medicago. These features include extreme size (25 kb), the presence of an extra ORF upstream of the gag–pol region, and a putative intron dividing the prot and rt coding sequences. The Ogre elements are evenly dispersed on V. pannonica chromosomes except for terminal regions containing satellite repeats, their individual copies show extraordinary sequence similarity, and at least part of them are transcriptionally active, which suggests their recent amplification. Similar elements were also detected in several other Vicia species but in most cases in significantly lower numbers. However, there was no obvious correlation of the abundance of Ogre sequences with the genome size of these species.
NUCLEAR genomes of higher plants differ considerably in their size, ranging from 0.1 pg (98 Mbp) in Fragaria viridis to 89.5 pg (87,686 Mbp) in Fritillaria davisii (Bennett and Leitch 2004). Even closely related species belonging to the same genus can display 5- to 10-fold differences in their haploid genome size, as it is, for example, in Phalenopsis, Scilla, or Vicia (Bennett and Leitch 2005). First investigations of this phenomenon using DNA reassociation kinetics (Chooi 1971; Flavell et al. 1974) revealed that genome size variation is mainly caused by differences in the proportion of repetitive DNA sequences. This was later confirmed by finding many families of repetitive sequences from a number of species. Among these, satellite repeats and retroelements have the most significant impact on genome size. Satellite repeats are organized as long arrays of tandemly repeated units (monomers). Although the monomer sequences are usually only tens to hundreds of nucleotides long (Macas et al. 2002), they can be amplified up to millions of copies (Kato et al. 1984; Ingham et al. 1993; Irifune et al. 1995; Macas et al. 2000), making up to 20% of the genome (Ingham et al. 1993). However, in most plant species investigated so far, the majority of repetitive DNA is composed of various families of retroelements (reviewed in Kumar and Bennetzen 1999; Feschotte et al. 2002). This high proportion of retroelements within plant genomes is a consequence of their replicative (copy-and-paste) mode of transposition (retrotransposition), which generates a new copy of the element each time it is retrotransposed. Although the retroelements do not attain as high copy numbers as the satellite repeats, their impact on the genome size is more pronounced due to their considerable length, ranging from a few up to 14 kb (Hirochika et al. 1992; Martinez-Izquierdo et al. 1997; Neumann et al. 2005). The recent discovery of a new group of giant retrotransposons, named Ogre elements, showed that the upper length limit of retrotransposons could be even longer. These Ty3/gypsy-like elements identified in pea (Pisum sativum) and Medicago truncatula are up to 22 kb long, and they occur at ∼10,000 copies in the pea genome, corresponding to at least 5% of its nuclear DNA (Neumann et al. 2003).
The genus Vicia (Fabaceae) includes >160 species differing considerably in their haploid nuclear DNA content (1.9–14.4 pg, corresponding to 1862–14,112 Mbp) (Bennett and Leitch 2005). Several studies revealed that there is a number of differentially amplified repeats of retroelement origin that significantly contribute to these differences (Pearce et al. 1996; Kumar et al. 1997; Nouzová et al. 2001; Hill et al. 2005). However, these studies described only partial retroelement sequences, which do not allow precise evaluation of the contribution that specific element families give to the evolution of the Vicia genome. In this work, we show that many of these partial sequences belong to a retrotransposon family closely related to the giant Ogre elements previously identified in pea and Medicago. We describe full-length Ogre-like elements isolated from the genome of Vicia pannonica and present data suggesting that significant expansion of the genome size in some Vicia species was caused by recent amplification of these elements.
MATERIALS AND METHODS
Plant material:
Seeds of V. pannonica (cv. Dětěnická panonská) and other species used in this study were obtained from the seed bank of the Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany; Agritec Šumperk, Czech Republic; and Plant Breeding Station, Boršov, Czech Republic. Total genomic DNA was extracted from leaves as described by Dellaporta et al. (1983). All DNA concentration measurements were done using the PicoGreen dye (Molecular Probes, Eugene, OR) according to manufacturer's recommendations. The measurements were performed in microwell plates and PicoGreen fluorescence was evaluated using a fluoroimager (Typhoon 9410, Amersham, Buckinghamshire, UK). The whole set of genomic DNAs and control fragments used for dot blotting was measured simultaneously, using the same DNA concentration standards.
Cloning procedures and sequence analysis:
Restriction fragments appearing as bands on ethidium-bromide-stained agarose gels of V. pannonica DNA digested with BglII or KpnI were cut out of the gel, purified, and cloned into pBluescript II SK+ (Stratagene, La Jolla, CA) digested with the corresponding enzyme. The fragments were sequenced using the dideoxy-mediated chain-termination method (Sanger et al. 1977).
A cosmid library was prepared by partial digestion of high-molecular-weight genomic DNA of V. pannonica with MboI, followed by its dephosphorylation and cloning into BamHI-digested vector SuperCos1 (Stratagene). The library was screened with the cloned BglII and KpnI fragments as probes using AlkPhos direct hybridization and detection kit (Amersham). A clone selected for sequencing (VP-cosC6) was subcloned and the sequencing templates from individual subclones were prepared using the GeneJumper primer insertion kit (Invitrogen, San Diego). Sequence assembly and basic analysis was done with Staden Package software (Staden 1996). As the cosmid clone contained several highly similar elements, the sequence assembly of the whole clone was verified by its restriction analysis and by PCR with primers specific for its individual parts. Computer analysis of the resulting sequence was performed using the Dotter program (Sonnhammer and Durbin 1995), program tools implemented at the Biology WorkBench website (http://workbench.sdsc.edu/), and EMBOSS (Rice et al. 2000). Multiple-sequence comparisons were done using Clustal W (Thompson et al. 1994). BLAST and FASTA (Pearson and Lipman 1988; Altschul et al. 1997) were employed for homology searches and the RPS-Blast (Marchler-Bauer et al. 2003) was used to search for conserved protein domains. Phylogenetic analyses were done using Clustal W and a phylogenetic tree was reconstructed using the Phylojava client/server tool. Sequences of reverse transcriptase domains were taken mostly from the alignment ALIGN_000602 (Vicient et al. 2001). Sequences of tRNAs used for identification of the primer binding site (pbs) were obtained from the Arabidopsis thaliana tRNA database (Lowe and Eddy 1997). Splice-site analysis was performed at the NetGene2 server (Hebsgaard et al. 1996).
Copy-number estimation:
To estimate the copy number of Ogre elements in the genomes of selected species, serial dilutions of their genomic DNAs were quantitatively dot blotted on Hybond N+ membrane (Amersham) together with fragments of the Ogre element as hybridization standards. The quantity of spotted genomic DNA corresponding to 50–105 copies of the haploid genomes (1C) was compared to 5 × 106–1 × 1010 copies of the hybridization standards. Dot blots were hybridized with the probes specific for different regions of the Ogre elements (LTR, ORF1, -2, and -3) prepared by PCR using VP-cosC6 DNA as a template and employing the following primers: VP1 5′-AAC TTT TAG TCA TTT ACT TTC AAT AAA CA-3′ and MT-pbs 5′-TCC CCA GTG AAG TCG CCA-3′; VP17 5′-TGG GAA GAA GAA ACA CCA AG-3′ and VP18 5′-CAT CTT CAT TTG ACG AGC AA-3′; VP19 5′-AAC GAG CTT CGT GGT ACA AT-3′ and VP20 5′-CTC GAG GAT TGT TGT GAC AG-3′; VP23 5′-CGA AGA GGA TGA AGA AGA GG-3′; and VP24 5′-TTT CTT GAC TGC ATC AGC AT-3′. Probe positions in the Ogre-VP1 element are shown in the Figure 2A. Probe labeling and hybridization were done using the AlkPhos kit following the manufacturer's instructions. Hybridization and washing temperatures were 65°. Probe detection was performed using CDP-Star substrate (Amersham) and the signals were captured on X-ray film. The signals were also captured and quantified using Typhoon 9410 scanner and ImageQuant TL software (Amersham).
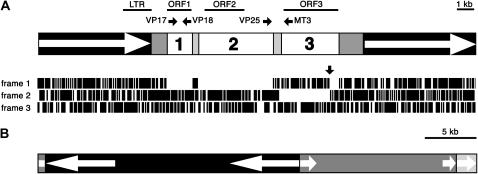
Figure 2.
Ogre elements identified in V. pannonica. (A) Structure of the full-length element Ogre-VP1. LTRs are depicted as open arrows in solid boxes. The open reading frames ORF1, ORF2, and ORF3 are represented by open boxes marked with corresponding numbers. Lightly shaded boxes indicate regions separating individual ORFs and darkly shaded boxes represent putative 5′- and 3′-UTRs. Lines above the scheme show positions of probes used for hybridizations. The horizontal solid arrows mark positions and directions (5′ → 3′) of primers used for RT–PCR experiments. Positions of stop codons in three reading frames of the Ogre-VP1 sequence are shown as vertical lines below the scheme. The vertical arrow indicates the frameshift in the ORF3. (B) Structure of the clone VP-cosC6 (GenBank accession no. AY936172) showing positions and orientation of Ogre-VP1 (solid), Ogre-VP2 (dark shading), and Ogre-VP3 (light shading) elements. Open arrows represent LTRs.
Alternatively, estimation of Ogre copy numbers in the V. pannonica genome was based on the number of positive clones observed after screening a short-insert shotgun genomic library with the probes described above. The shotgun library was prepared from V. pannonica DNA subjected to sonication, mung bean nuclease treatment, and size fractionation on an agarose gel. The 600- to 800-bp fragments were purified from the gel, treated with polynucleotide kinase in the presence of ATP, and cloned into dephosphorylated SmaI-cut plasmid vector (pBluescript II SK+). Copy number (CN) was calculated for individual probes using the formula CN = GS × PG/Lp, where GS is the genome size of V. pannonica (6.51 × 109 bp), PG is the proportion of the probe sequence in the genome, and Lp is the probe length. The PG-value was defined as PG = Lh/Lt, where Lh is the length of hybridizing sequences within the library and Lt is the sum of insert lengths of all screened clones. As it was supposed that many of positive clones did not hybridize over their whole length, the Lh-value could not be calculated by simply multiplying the number of positive clones by the average insert length. Instead, the Lh-value was calculated considering the theoretical frequency of clones hybridizing over their whole length (F1), the theoretical frequency of clones hybridizing with only part of their sequences (F2), the number of positively hybridizing clones (Np), the average insert size (Li = 700 bp), the minimum length of sequence capable of efficient hybridization (Lmin = 100 bp), and the average length of partially hybridizing sequences [Lavg = (Li − Lmin − 1)/2], using the formula Lh = Np × F1 × Li + Np × F2 × Lavg. The F1- and F2-values were calculated using the formulas F1 = (Lp − Li)/((Lp − Li) + 2 × (Li − Lmin − 1)) and F2 = 2 × (Li − Lmin − 1)/((Lp − Li) + 2 × (Li − Lmin − 1)).
RNA isolation and RT–PCR:
The tissues used for RNA isolation (leaf, root, flower) were harvested from plants cultivated in pots under a 15 hr light/9 hr dark photoperiod at 22° and 18°, respectively. Total RNA was isolated using the total RNA isolation kit (Ambion, Austin, TX). All RNA samples were treated with RNase-free DNase (Invitrogen) to remove any contaminant DNA. Reverse transcription was carried out using the SuperScript II reverse transcription kit (Invitrogen) by a random priming method according to the manufacturer's recommendations, using 0.5 μg of template RNA. The RT–PCR reaction mix (25 μl) consisted of 1 × PCR buffer, 0.2 mm dNTPs, 0.2 μm primers, 1.5 mm MgCl2, 1 unit of platinum Taq polymerase (Invitrogen), and 2.5 ng of reversely transcribed RNA or an equal amount of reverse-transcriptase-untreated RNA as a negative control. The following primers were used in the RT–PCR experiments: VP17 and VP18; VP25 5′-ACG TTC TCT TTC ATC GAT GC-3′ and MT3 5′-CGG TAG TCA ACA CAC ATT CTG AC-3′. The reaction profile included 35 cycles of 30 sec at 94°, 50 sec at 55°, and 1–3 min at 72°, preceded by initial denaturation (3 min at 94°) and followed by a final extension step (10 min at 72°). Reaction products were resolved on agarose gel electrophoresis.
Fluorescence in situ hybridization:
Fluorescence in situ hybridization (FISH) was performed on isolated chromosomes prepared as described by Gualberti et al. (1996) and centrifuged onto slides using a Hettich centrifuge equipped with cytospin chambers. The Ogre probe was derived from the longest EcoRV restriction fragment of the clone VP-cosC6 containing LTR, 5′-untranslated region (5′-UTR), ORF1, and ORF2 regions. The probe was labeled with biotin-16-dUTP (Boehringer Mannheim, Indianapolis) using random priming and detected using streptavidin-Alexa Fluor-568 (Molecular Probes) as described by Leitch et al. (1994). Treatment of slides before hybridization, composition of hybridization mix, and hybridization conditions of the Ogre probe were as described by Neumann et al. (2001). Following hybridization of the Ogre probe, the slides were dehydrated using the ethanol series, air dried, and used for the second round of hybridization with fluorescein-labeled oligonucleotide probe (5′-AAG ATT RTC TTG TGY TAT AST ACA TAA AAK TCA CGA AGT-3′) specific for satellite repeats VicTR-A (Macas et al. 2000), which produce labeling patterns allowing discrimination of all chromosome types within the V. pannonica karyotype (Navrátilová et al. 2003). Hybridization of the VicTR-A probe (0.5 ng/μl) was performed at 37° for 16 hr in the hybridization mix consisting of 2× SSC, 100 ng/μl sheared calf thymus DNA, and 0.125% SDS, and posthybridization washes were done in 2× SSC at 42° for 10 min. Chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and examined using a Nikon Eclipse 600 microscope equipped with appropriate filter sets. The images were captured with a CCD camera and analyzed using Lucia software (Laboratory Imaging).
RESULTS
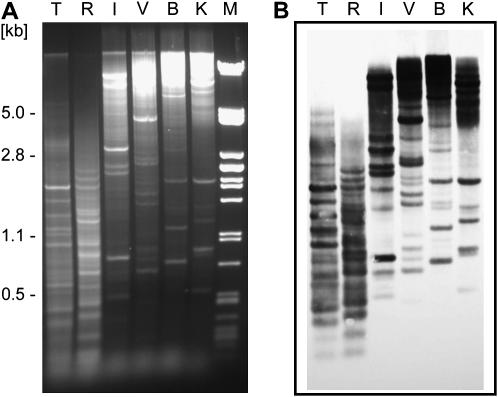
In our preliminary experiments aimed at comparative analysis of Vicia genomes, we noted that V. pannonica produces characteristic restriction patterns when its DNA is digested with restriction endonucleases and resolved on agarose gels. Contrary to the majority of species used for the experiments, which produced smears with a few distinct bands, these patterns consisted of a series of well-distinguished bands in most enzymes tested (Figure 1A). This observation suggests that there is a prominent and conserved DNA repeat making up a large portion of the V. pannonica genome. To isolate and characterize this repeat, we cloned four of the distinct bands from the DNA digested with BglII (850 and 1250 bp) or KpnI (560 and 1000 bp). Sequence analysis of the cloned fragments revealed their similarity to different parts of the giant retroelement Ogre previously described in pea (Neumann et al. 2003). To isolate the corresponding full-length element from V. pannonica, a cosmid library was constructed and screened with the cloned fragments as probes. Thirty randomly picked clones hybridizing to all four probes were subjected to restriction analysis to check for the presence of fragments corresponding to the conserved genomic bands. These fragments were found in most of the clones, 13 of which (43%) produced bands corresponding to all four fragments used as probes. One such clone (VP-cosC6) was selected for complete sequencing and further analysis.
Figure 1.
V. pannonica Ogre sequences are highly conserved in restriction sites. (A) Genomic DNA of V. pannonica digested with TaqI (T), RsaI (R), EcoRI (I), EcoRV (V), BglII (B), and KpnI (K). λDNA digested with PstI was used as a size marker (M). (B) Southern blot hybridization of the gel shown in A using the whole VP-cosC6 insert sequence as a probe.
Sequence characterization and transcriptional activity of Ogre elements in V. pannonica:
The insert cloned in VP-cosC6 (GenBank accession no. AY936172) was >42 kb in length and entirely composed of Ogre sequences belonging to three different elements (Figure 2). There was one complete element, designated Ogre-VP1, which contained intact LTRs surrounding internal coding regions and was flanked by a 5-bp target-site duplication (5′-ATGCC/ATGCC-3′). This element was inserted into another Ogre element (Ogre-VP2), whose sequence was truncated at its left LTR due to cloning and at its right LTR due to insertion of the third element, Ogre-VP3. Most of the Ogre-VP3 sequence was lost through cloning except for a part of its left LTR. All three elements shared high mutual similarities. The overall similarity between Ogre-VP1 and Ogre-VP2 was 92.3%. The Ogre-VP3 LTR shared 92.1% similarity with the LTR sequence of Ogre-VP1 and 81.5% similarity with LTR of Ogre-VP2. Similarity of these elements to the partial sequences cloned as conserved BglII and KpnI fragments was 86–99%.
The only full-length element, Ogre-VP1, was 25,049 bp long, which makes it the largest plant retroelement described so far. This extreme size was mainly due to exceptionally long LTRs, each of which spans 6438 bp. Sequence similarity between the LTRs was 99.9% (only six nucleotide substitutions over their entire length), which indicates that Ogre-VP1 represents a very recent insertion. The overall arrangement of Ogre-VP1 coding and structural regions is identical to that of the Ogre elements identified in the P. sativum and M. truncatula genomes (Neumann et al. 2003) and further confirms its assignment into this group of Ty3/gypsy-like retrotransposons. Close relationships of Ogre sequences from all three species were also apparent from a phylogenetic analysis of the reverse transcriptase protein domains (supplementary Figure S1 at http://www.genetics.org/supplemental/). The coding region of Ogre-VP1 is divided into three reading frames (ORF1–3, Figure 2A) separated by short regions containing several stop codons in all three frames. These ORFs could be directly translated into protein sequences, with the exception of ORF3, containing a +1 frameshift due to an insertion of one nucleotide, which had to be removed to allow its conceptual translation.
Similarly to the pea elements, ORF1 of Ogre-VP1 encoded a protein with unknown function. This ORF was 1527 bp in length and was separated from ORF2 by a 237-bp-long noncoding region containing stop codons. The partial Ogre-VP2 sequence contained an ORF1 of the same length with 97% sequence similarity to ORF1 in Ogre-VP1. All of the protein domains typical for plant retroelements could be recognized within two ORFs: ORF2 and ORF3. Gag and protease (prot) domains were encoded by ORF2, whereas reverse transcriptase (rt), ribonuclease H (rh), and integrase (int) were encoded by ORF3 (Table 1). These ORFs were separated by a region of 317 bp, which contained multiple stop codons. The fact that the position of this region corresponded to that of an intron within the pea and M. truncatula Ogre sequences (Neumann et al. 2003 and our unpublished data), in addition to further computer analysis using NetGene2 server (Hebsgaard et al. 1996), strongly suggested that this region could also represent an intron sequence. Moreover, the eventual removal of this predicted intron sequence by splicing would result in joining of ORF2 (+2 frame; 4403 bp) with ORF3 (+1 frame; 3464 bp) into one frame encoding a polyprotein of 2622 amino acids.
TABLE 1.
Conserved domains found in the putative polyprotein encoded by Ogre-VP1
| CDa | E-value | Positionb | Active-site motif | Note |
|---|---|---|---|---|
| gag (pfam03732) | 3e−09 | 264–350 | The domain is followed by putative zinc-finger motif CX2HX5HX4C (513–527)f | |
| prot | — | 986–1054 | DTGc | The domain was identified based on similarity to pea Ogre element (accession no. AY299398) |
| rt (pfam00078) | 7e−24 | 1624–1793 | YVDDd | |
| rh (pfam00075) | 1e−08 | 2053–2173 | DEDDe | |
| int (pfam00665) | 2e−21 | 2334–2488 | DDEg | The domain is preceded by zinc-binding motif HHCC (2280–2314)g |
Domains typical for LTR retrotransposon polyproteins were identified by searching the conserved domain (CD) database using RPS-Blast (Marchler-Bauer et al. 2003) and by noting similarity to the putative protein encoded by the pea Ogre element identified in the clone Ps-cos16 (accession no. AY299398).
The putative polyprotein sequence was deduced from ORF2 and ORF3, which were joined after the removal of predicted intron.
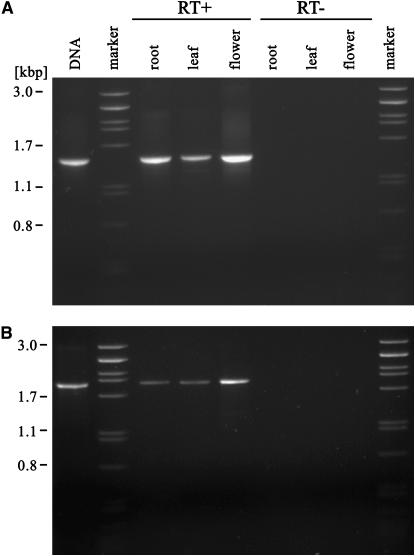
Transcriptional activity of V. pannonica Ogre elements was tested using RT–PCR with total RNA isolated from leaves, roots, and flowers. Primers were designed to amplify the ORF1 sequence as this region is specific for Ogre elements and is absent in all other types of retrotransposons described so far. The products of expected length were detected in all three organs tested (Figure 3A). To test the splicing of the predicted intron between ORF2 and ORF3, primers directed toward this region were also used. Although the corresponding RNA was again detected in all samples, the size of the amplified fragments did not correspond to the spliced sequence (Figure 3B). Cloning and sequence analysis of these fragments revealed 92–97% similarity to the corresponding region in Ogre-VP1. Comparison of the RT–PCR-amplified sequences with genomic sequences confirmed that none of them was spliced.
Figure 3.
Transcriptional activity of Ogre retrotransposons in V. pannonica analyzed by RT–PCR. The reactions were performed on total RNA isolated from roots, leaves, or flowers (RT+). Negative controls with omitted reverse transcriptase step were included to check for false-positive results (RT−). (A) Amplification of the ORF1 sequence using primers VP17 and VP18. (B) Amplification of the region containing putative intron using primers VP25 and MT3.
Abundance of Ogre-like sequences in Vicia and other legume species:
The copy number of Ogre elements in the V. pannonica genome was measured using two different approaches. The first estimate was based on quantitative dot-blot hybridization of serial dilutions of genomic DNA and control fragments (cloned Ogre sequences) with probes derived from four regions of the element (see Figure 2A for the probe positions). Resulting signals corresponded to 1–5 × 105 copies/haploid genome (1C) for the LTR probe and to 1–2 × 105 copies/1C for each of the remaining probes (ORF1, ORF2, and ORF3). The second method employed hybridization of probes for LTR, ORF1, or ORF2 to a shotgun, short-insert library of V. pannonica genomic DNA. Of 6530 clones screened, 409, 169, or 237 clones, respectively, displayed positive hybridization signal. Considering the length of individual probes (1615, 1518, and 1999 bp) and using formulas described in materials and methods, the estimated copy number per 1C was 2 × 105 for LTR, 8 × 104 for ORF1, and 9 × 104 for ORF2. Thus, the copy-number calculations based on these two principally different methods were in good accordance and a copy number of 1 × 105 copies/1C can be considered a reliable estimation. Taking into account the genome size of V. pannonica (6.75 pg; Raina and Rees 1983) and assuming that all element copies are full length, the copy number of 105/1C corresponds to 38% of the genome being made up by Ogre sequences.
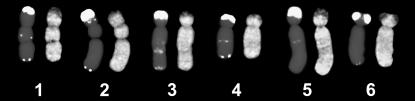
To test if such a high amplification of Ogre elements resulted in their accumulation in specific genomic regions or if it was accompanied by an overall increase of the element copies throughout the entire genome, we performed detection of the Ogre repeats on mitotic chromosomes using in situ hybridization. These experiments showed that the elements were dispersed over the entire genome, spanning all of the chromosomes (Figure 4). The only exception to this homogenous chromosome labeling was the subtelomeric regions of the short chromosome arms, which produced weaker signals due to the presence of the highly amplified satellite repeat VicTR-A (Macas et al. 2000).
Figure 4.
FISH on metaphase chromosomes of V. pannonica. Two images of each chromosome type are shown. (Left of chromosome pair) DAPI-stained chromosomes (shaded) hybridized with the VicTR-A repeat (open signals). (Right of chromosome pair) The same chromosome hybridized with the probe specific for Ogre-VP1.
The quantitative dot-blot hybridization was also used to estimate the abundance of Ogre-like sequences in 12 Vicia species and in several other legumes (Table 2). Two of these species, V. melanops and V. hybrida, were found to contain highly amplified Ogre elements, which were estimated to reach up to 1 × 105 and 1–5 × 104 copies/1C, respectively. These findings were in agreement with the results of Southern blot experiments in which only these two species showed prominent signals in addition to V. pannonica (Figure 5). Other Vicia species produced hybridization signals corresponding to lower numbers of Ogre sequences; however, the numbers estimated for individual species varied, depending on the probe used (Table 2). This observation most likely reflects sequence divergence of the corresponding regions (e.g., ORF1) among species or eventual cross-hybridization of probes derived from regions conserved among various groups of retroelements (ORF3). This is even more evident for the estimates made for P. sativum, which were considerably lower than those made using Pisum-derived Ogre probes (Neumann et al. 2003 and Table 2). The species from other genera, including Vigna unquiculata, Cicer arietinum, Glycine max, Lotus angustifolius, Phaseolus vulgaris, and M. truncatula, produced no or very weak hybridization, not exceeding signals corresponding to <100–500 copies/1C (our unpublished data).
TABLE 2.
Estimated copy numbers of Ogre sequences per haploid genomes (1C) of selected Vicia species and P. sativum
| 1C(Mbp) | 1C(pg) | Dot-blot probes
|
Other estimates | % in the genomed | |||||
|---|---|---|---|---|---|---|---|---|---|
| Section | Species | LTR | ORF1 | ORF2 | ORF3 | ||||
| Hypechusa | V. pannonica | 6,615 | 6.75 | 1–5 × 105 | 1 × 105 | 1 × 105 | 1 × 105 | 1 × 105 a | 38 |
| V. hybrida | 6,640 | 6.78 | 1 × 105 | 1 × 104 | 1 × 104 | 1–5 × 104 | — | 4 | |
| V. lutea | 7,252 | 7.40 | 0.5–1 × 104 | 0.5–1 × 103 | 5 × 102 | 1–5 × 103 | — | 0.2 | |
| V. melanops | 9,800 | 10.00 | 5 × 105 | 1 × 105 | 1 × 105 | 1 × 105 | 1 × 105 b | 26 (26)b | |
| Cracca | V. villosa | 2,230 | 2.28 | 5 × 103 | 0.5–1 × 103 | 1 × 103 | 5 × 103 | — | 1 |
| Attosa | V. sepium | 4,582 | 4.68 | 1 × 104 | 1 × 103 | 5 × 103 | 1–5 × 104 | — | 3 |
| Vicia | V. sativa | 2,205 | 2.25 | 5 × 103 | 1–5 × 103 | 1–5 × 103 | 1–5 × 104 | 1 × 104 b | 6 (12)b |
| V. grandiflora | 3,283 | 3.35 | 0.5–1 × 104 | 1 × 103 | 5 × 103 | 1–5 × 104 | — | 4 | |
| Wiggersia | V. lathyroides | 2,573 | 2.63 | 1 × 102 | 0–50 | 0.5–1 × 102 | 5 × 103 | — | 0.1 |
| Narbonensis | V. narbonensis | 7,130 | 7.28 | 1–5 × 103 | 1 × 103 | 1–5 × 103 | 1–5 × 104 | — | 2 |
| Faba | V. faba | 13,255 | 13.50 | 1 × 103 | 0.5–1 × 102 | 1–5 × 102 | 1–5 × 104 | — | 0.1 |
| Peregrina | V. peregrina | 9,286 | 9.48 | 1–5 × 103 | 1–5 × 102 | 1 × 103 | 1–5 × 104 | — | 0.3 |
| V. michauxii | 8,134 | 8.30 | 1 × 103 | 1 × 102 | 1 × 103 | 1 × 104 | — | 0.3 | |
| P. sativum | 4,337 | 4.43 | 1–5 × 102 | 5 × 102 | 1–5 × 103 | 1–5 × 104 | 1 × 104 c | 3 (6)c | |
Based on the number of positively hybridizing clones in the V. pannonica library.
According to Nouzová et al. (2001).
According to Neumann et al. (2003).
Calculations were based on estimated copy numbers of the ORF2 probe. Values in parentheses show genome proportions calculated using estimates obtained with other species-specific Ogre probes (as shown in the column “Other estimates”).
Figure 5.
Agarose gel electrophoresis (A) and corresponding Southern blot (B) of genomic DNAs of selected Vicia species digested with TaqI and probed with the whole VP-cosC6 insert sequence.
In addition to previously described Ogre sequences (Neumann et al. 2003), a number of partial clones of repetitive elements from V. melanops, V. sativa, and V. narbonensis were identified as most similar to Ogre-VP elements in BLAST and FASTA homology searches (Table 3). The high-sequence similarities strongly suggest that these clones correspond to various regions of Ogre-like elements, including ORF1.
TABLE 3.
Ogre-like sequences identified in Vicia species by FASTA sequence similarity searches using Ogre-VP1 as a query
| Species | Accession no. | Similarity (%/length) | E-value | Region | Copy nos. (copies/1C)a |
|---|---|---|---|---|---|
| V. melanops | AJ391781 | 88/666 | 1.6 × 10−70 | LTR | 105–106 |
| AJ391800 | 75/982 | 1.7 × 10−86 | LTR | 104–105 | |
| AJ391797 | 78/493 | 1.1 × 10−37 | ORF1/spacer | 105–106 | |
| AJ391792 | 80/479 | 1.2 × 10−94 | ORF2 | 105–106 | |
| AJ391793 | 86/222 | 2.6 × 10−46 | ORF2 | 104–105 | |
| AJ391794 | 90/337 | 4.5 × 10−84 | ORF2 | 104–105 | |
| AJ391802 | 75/211 | 2.2 × 10−30 | ORF2 | 104–105 | |
| AJ391799 | 85/332 | 6.9 × 10−62 | ORF2/intron | 105–106 | |
| AJ391754, AJ391755, AJ391759, AJ391760, AJ391761, AJ391764, AJ391765 | 87/490–92/757 | 2.3 × 10−105–4.1 × 10−187 | ORF3 | Unknown | |
| AJ391774 | 91/308 | 7.3 × 10−49 | ORF3/3′-UTR | Unknown | |
| V. sativa | AJ391782 | 69/781 | 4.8 × 10−30 | LTR | 104–105 |
| AJ391804 | 64/118 | 8.1 | 5′-UTR | 104 | |
| AJ391758, AJ391762, AJ391763, AJ391766, AJ391767, AJ391769 | 68/275–87/421 | 9.1 × 10−27–1.4 × 10−88 | ORF3 | Unknown | |
| AJ391776, AJ391778 | 82/65, 85/65 | 6 × 10−6, 7.8 × 10−5 | 3′-UTR | Unknown | |
| V. narbonensis | AJ391771 | 62/320 | 1.9 × 10−13 | ORF1 | Unknown |
| AJ391756, AJ391757, AJ391768, AJ391770 | 70/382–68/664 | 7.3 × 10−71 | ORF3 | Unknown | |
| AJ391777 | 76/107 | 0.0029 | 3′-UTR | Unknown |
Copy numbers are according to Nouzová et al. (2001).
DISCUSSION
A crucial role of the amplification and eventual elimination of retroelement sequences in plant genome evolution has become evident through the accumulation of sequence data and subsequent comparative analysis of species with different genome sizes. For example, a comparison of the Adh1-containing orthologous regions in maize and sorghum demonstrated that while >60% of this region in maize is composed of LTR retrotransposons, no retrotransposons were detected in the orthologous region in sorghum (SanMiguel et al. 1996; SanMiguel and Bennetzen 1998). These LTR retrotransposons constitute 49–78% of the maize genome, suggesting that the threefold greater genome size of maize compared to sorghum was a result of retrotransposon amplification (SanMiguel and Bennetzen 1998). Similar role of retroelements in genome-size evolution has also been revealed in other species (Shirasu et al. 2000; Wicker et al. 2001, 2005). In general, all high-copy-number retrotransposons were identified in species with large genomes (Pearce et al. 1996, 1997; Meyers et al. 2001; Muniz et al. 2001; Kentner et al. 2003) while species with small genomes contain these elements at relatively low copy numbers. The latter is the case of well-characterized genomes of A. thaliana (130 Mb) and Oryza sativa (430 Mb) in which retrotransposons make up only 4 and 22% of the genome (Ma et al. 2004; Peterson-Burch et al. 2004; Zhang and Wessler 2004). All plant genomes characterized so far are composed of many diverse families of LTR retrotransposons with different degrees of amplification. The copy number of elements belonging to one family can vary greatly from a few to ∼105 copies/genome (Kumar and Bennetzen 1999; Feschotte et al. 2002). The most abundant plant LTR retrotransposons, such as PREM-2, Opie, or Huck-2 in Zea mays and IRRE in Iris, can make up to 10% of the genome (SanMiguel and Bennetzen 1998; Meyers et al. 2001; Kentner et al. 2003; Chantret et al. 2005). Thus, concurrent amplification of elements belonging to several families can account for considerable increases in genome size as demonstrated for Zea species (SanMiguel and Bennetzen 1998; Meyers et al. 2001). Our results described here demonstrate that even the amplification of a single retroelement family can increase the genome size by >50% (based on the calculation that this element makes up 38% of the V. pannonica genome). Although the Ogre sequences were detected in all Vicia species tested, their abundance differed by several orders of magnitude (Table 2). Whereas the copy numbers in some species could be underestimated due to sequence divergence of the elements, this great variability in copy numbers was evident even among closely related species, each belonging to the taxonomic section Hypechusa. Among the four species tested, the Ogre sequences were ∼10-fold less abundant in V. lutea than in V. hybrida and 100-fold less abundant than in V. pannonica and V. melanops. Interestingly, the genome size of V. lutea is similar to those of the other three species, suggesting that other repetitive elements may have been amplified there instead of Ogre. The amplification of other elements probably accompanied the expansion of the genome size in V. melanops, as the copy number of the Ogre family in this species is the same as in V. pannonica, but its genome is larger (Table 2). Therefore, although it is evident from the differences in Ogre copy numbers in individual species that these elements have played a crucial role in the increase of genome size for at least some Vicia species, there is no simple correlation between abundance of Ogre sequences and the genome size in this genus.
The differences in copy numbers observed among Vicia species could be in part explained by a loss of Ogre sequences in some genomes due to unequal homologous recombination and illegitimate recombination, which were shown to be responsible for deletions of retrotransposon sequences from genomes in several plant species (Shirasu et al. 2000; Devos et al. 2002; Vitte and Panaud 2003; Wicker et al. 2003; Ma et al. 2004). An indicator of unequal recombination is a presence of solo LTRs, which remain in a genome after a recombination event. The copy number estimated for LTR sequences in several species was considerably higher compared to inside regions (e.g., V. lutea and V. hybrida, Table 2), although the LTR sequences are believed to be less conserved and thus less likely to hybridize with a probe from another species than the internal coding sequences. Thus, it is possible that in these species the reduction of the genome size through the elimination of Ogre sequences took place, leaving an excess of solo LTRs.
Molecular mechanisms facilitating the eventual regulation of retroelement amplification in plant genomes are only poorly understood. Mechanisms that are likely to play a role in the suppression of mobile elements include transcriptional silencing through DNA methylation (Hirochika et al. 2000; Wright and Voytas 2002; Liu et al. 2004), chromatin modifications (Jackson et al. 2002), and post-transcriptional silencing by RNA interference mechanism (Timmons 2002). The high amplification of Ogre in a limited number of species and its occurrence in only moderate copy numbers in others can be explained either by an insufficient function of mechanisms suppressing transposition of retroelements or by the escape of Ogre elements from such mechanisms. Since virtually all prominent bands visible on digested genomic DNA of V. pannonica hybridized to Ogre, it seems that only this family of retrotransposons was amplified to high copy numbers, whereas other retroelements remained suppressed. Considering the high sequence similarity among Ogre sequences isolated from V. pannonica, the high level of conservation in restriction sites, and the recent insertion of Ogre-VP1 element (inferred from the similarity of its LTRs), it is likely that the burst of amplification of the Ogre family happened quite recently. As these elements are still transcribed in V. pannonica, the amplification of the Ogre family in this species may not be over yet. In this respect, it is interesting to mention that the transcription of Ogre elements also was detected in P. sativum (Neumann et al. 2003) containing one order of magnitude fewer copies compared to V. pannonica and in M. truncatula having a small genome (0.48 pg or 466 Mbp; Arumuganathan and Earle 1991), which, consequently, can be occupied by relatively few Ogre elements. This could imply that transcription itself is not sufficient for transpositional activity and that post-transcriptional regulation plays an important role in the suppression of transposition.
From the results described here, as well as from our previous findings (Neumann et al. 2003), it is evident that Ogre elements represent a group of retrotransposons capable of inducing significant changes in the genome size of some plant species. Up to now, full-length Ogre elements have been detected in only a relatively narrow range of legume taxa, including the genera of Pisum, Medicago, and Vicia. However, except for the hybridization-based detection described here no attempts were performed to identify related elements in other plants. Moreover, no or very weak hybridization signals obtained in species from more distant genera (Vigna, Cicer, Glycine, Lotus, and Phaseolus) do not necessarily indicate the absence of Ogre elements but could rather be caused by their further sequence divergence. Thus, the alternative approaches on the basis of sequence analysis of regions typical for the Ogre family, including mainly ORF1 and pbs, should be employed to find Ogre-like elements in other species. Future experiments should also be directed to structural and functional analysis of the extremely long Ogre LTRs harboring regulatory regions controlling the element replication cycle, which could provide clues for explaining the high amplification rate of Ogre elements in some species.
Acknowledgments
We thank H. Štěpančíková for excellent technical assistance, M. Nouzová for help with DNA sequencing, and S. M. Rafelski and Lara Colton for assistance in preparation of the manuscript. This work was supported by grants GACR521/00/0655 and GACR521/02/P007 from the Czech Science Foundation, grant LC06004 from the Ministry of Education, Youth and Sports of the Czech Republic, and grant AVOZ50510513 from the Academy of Sciences of the Czech Republic.
Sequence data from this article have been deposited with the EMBL/GenBank Data libraries under accession no. AY936172.
References
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumuganathan, K., and E. D. Earle, 1991. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9: 208–218. [Google Scholar]
- AsanteAppiah, E., and A. M. Skalka, 1997. A metal-induced conformational change and activation of HIV-1 integrase. J. Biol. Chem. 272: 16196–16205. [DOI] [PubMed] [Google Scholar]
- Bennett, M. D., and I. J. Leitch, 2004. Plant DNA C-Values Database (release 3.0). http://www.rbgkew.org.uk/cval/homepage.html.
- Bennett, M. D., and I. J. Leitch, 2005. Nuclear DNA amounts in angiosperms: progress, problems and prospects. Ann. Bot. 95: 45–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd, C. G., and G. Dreyfuss, 1994. Conserved structures and diversity of functions of RNA-binding proteins. Science 265: 615–621. [DOI] [PubMed] [Google Scholar]
- Chantret, N., J. Salse, F. Sabot, S. Rahman, A. Bellec et al., 2005. Molecular basis of evolutionary events that shaped the hardness locus in diploid and polyploid wheat species (Triticum and Aegilops). Plant Cell 17: 1033–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chooi, W. Y., 1971. Comparison of DNA of six Vicia species by method of DNA-DNA hybridization. Genetics 68: 213–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, J. F., Z. Hostomska, Z. Hostomsky, S. R. Jordan and D. A. Matthews, 1991. Crystal-structure of the ribonuclease-H domain of HIV-1 reverse-transcriptase. Science 252: 88–95. [DOI] [PubMed] [Google Scholar]
- Dellaporta, S. L., J. Wood and J. B. Hicks, 1983. A plant DNA minipreparation: version II. Plant Mol. Biol. Rep. 1: 19–21. [Google Scholar]
- Devos, K. M., J. K. M. Brown and J. L. Bennetzen, 2002. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12: 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, J. P., K. Das, Y. Hsiou, S. G. Sarafianos, A. D. Clark et al., 1998. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 angström resolution. J. Mol. Biol. 284: 1095–1111. [DOI] [PubMed] [Google Scholar]
- Feschotte, C., N. Jiang and S. R. Wessler, 2002. Plant transposable elements: where genetics meets genomics. Nat. Rev. Genet. 3: 329–341. [DOI] [PubMed] [Google Scholar]
- Flavell, R. B., M. D. Bennett, J. B. Smith and D. B. Smith, 1974. Genome size and proportion of repeated nucleotide-sequence DNA in plants. Biochem. Genet. 12: 257–269. [DOI] [PubMed] [Google Scholar]
- Gualberti, G., J. Doležel, J. Macas and S. Lucretti, 1996. Preparation of pea (Pisum sativum L.) chromosome and nucleus suspensions from single root tips. Theor. Appl. Genet. 92: 744–751. [DOI] [PubMed] [Google Scholar]
- Hebsgaard, S. M., P. G. Korning, N. Tolstrup, J. Engelbrecht, P. Rouze et al., 1996. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 24: 3439–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, P., D. Burford, D. M. A. Martin and A. J. Flavell, 2005. Retrotransposon populations of Vicia species with varying genome size. Mol. Genet. Genomics 273: 371–381. [DOI] [PubMed] [Google Scholar]
- Hirochika, H., A. Fukuchi and F. Kikuchi, 1992. Retrotransposon families in rice. Mol. Gen. Genet. 233: 209–216. [DOI] [PubMed] [Google Scholar]
- Hirochika, H., H. Okamoto and T. Kakutani, 2000. Silencing of retrotransposons in Arabidopsis and reactivation by the ddm1 mutation. Plant Cell 12: 357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham, L. D., W. W. Hanna, J. W. Baier and L. C. Hannah, 1993. Origin of the main class of repetitive DNA within selected Pennisetum species. Mol. Gen. Genet. 238: 350–356. [DOI] [PubMed] [Google Scholar]
- Irifune, K., K. Hirai, J. Zheng, R. Tanaka and H. Morikawa, 1995. Nucleotide-sequence of a highly repeated DNA-sequence and its chromosomal localization in Allium fistulosum. Theor. Appl. Genet. 90: 312–316. [DOI] [PubMed] [Google Scholar]
- Jackson, J. P., A. M. Lindroth, X. F. Cao and S. E. Jacobsen, 2002. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556–560. [DOI] [PubMed] [Google Scholar]
- Kato, A., K. Yakura and S. Tanifuji, 1984. Sequence analysis of Vicia faba repeated DNA, the FokI repeat element. Nucleic Acids Res. 12: 6415–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentner, E. K., M. L. Arnold and S. R. Wessler, 2003. Characterization of high-copy-number retrotransposons from the large genomes of the Louisiana Iris species and their use as molecular markers. Genetics 164: 685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A., and J. L. Bennetzen, 1999. Plant retrotransposons. Annu. Rev. Genet 33: 479–532. [DOI] [PubMed] [Google Scholar]
- Kumar, A., S. R. Pearce, K. McLean, G. Harrison, J. S. Heslop-Harrison et al., 1997. The Ty1-copia group of retrotransposons in plants: genomic organisation, evolution, and use as molecular markers. Genetica 100: 205–217. [PubMed] [Google Scholar]
- Leitch, A. R., T. Schwarzacher, D. Jackson and I. J. Leitch, 1994. In situ Hybridization. BIOS Scientific, Oxford.
- Liu, Z. L., F. P. Han, M. Tan, X. H. Shan, Y. Z. Dong et al., 2004. Activation of a rice endogenous retrotransposon Tos17 in tissue culture is accompanied by cytosine demethylation and causes heritable alteration in methylation pattern of flanking genomic regions. Theor. Appl. Genet. 109: 200–209. [DOI] [PubMed] [Google Scholar]
- Lowe, T. M., and S. R. Eddy, 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J. X., K. M. Devos and J. L. Bennetzen, 2004. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 14: 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macas, J., D. Požárková, A. Navrátilová, M. Nouzová and P. Neumann, 2000. Two new families of tandem repeats isolated from genus Vicia using genomic self-priming PCR. Mol. Gen. Genet. 263: 741–751. [DOI] [PubMed] [Google Scholar]
- Macas, J., T. Mészáros and M. Nouzová, 2002. PlantSat: a specialized database for plant satellite repeats. Bioinformatics 18: 28–35. [DOI] [PubMed] [Google Scholar]
- Maignan, S., J. P. Guilloteau, Q. Zhou-Liu, C. Clement-Mella and V. Mikol, 1998. Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases. J. Mol. Biol. 282: 359–368. [DOI] [PubMed] [Google Scholar]
- Malik, H. S., and T. H. Eickbush, 2001. Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res. 11: 1187–1197. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer, A., J. B. Anderson, C. DeWeese-Scott, N. D. Fedorova, L. Y. Geer et al., 2003. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31: 383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Izquierdo, J. A., J. Garcia-Martinez and C. M. Vicient, 1997. What makes Grande1 retrotransposon different? Genetica 100: 15–28. [PubMed] [Google Scholar]
- Meyers, B. C., S. V. Tingley and M. Morgante, 2001. Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res. 11: 1660–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniz, L. M., A. Cuadrado, N. Jouve and J. M. Gonzalez, 2001. The detection, cloning, and characterisation of WIS 2–1A retrotransposon-like sequences in Triticum aestivum L. and × Triticosecale Wittmack and an examination of their evolution in related Triticeae. Genome 44: 979–989. [PubMed] [Google Scholar]
- Navrátilová, A., P. Neumann and J. Macas, 2003. Karyotype analysis of four Vicia species using in situ hybridization with repetitive sequences. Ann. Bot. 91: 921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann, P., M. Nouzová and J. Macas, 2001. Molecular and cytogenetic analysis of repetitive DNA in pea (Pisum sativum L.). Genome 44: 716–728. [PubMed] [Google Scholar]
- Neumann, P., D. Požárková and J. Macas, 2003. Highly abundant pea LTR retrotransposon Ogre is constitutively transcribed and partially spliced. Plant Mol. Biol. 53: 399–410. [DOI] [PubMed] [Google Scholar]
- Neumann, P., D. Požárková, A. Koblížková and J. Macas, 2005. PIGY, a new plant envelope-class LTR retrotransposon. Mol. Genet. Genomics 273: 43–53. [DOI] [PubMed] [Google Scholar]
- Nouzová, M., P. Neumann, A. Navrátilová, D. W. Galbraith and J. Macas, 2001. Microarray-based survey of repetitive genomic sequences in Vicia spp. Plant Mol. Biol. 45: 229–244. [DOI] [PubMed] [Google Scholar]
- Pearce, S. R., G. Harrison, P. J. S. Heslop-Harrison, A. J. Flavell and A. Kumar, 1997. Characterization and genomic organization of Ty1-copia group retrotransposons in rye (Secale cereale). Genome 40: 617–625. [DOI] [PubMed] [Google Scholar]
- Pearce, S. R., G. Harrison, D. T. Li, J. S. Heslop-Harrison, A. Kumar et al., 1996. The Ty1-copia group retrotransposons in Vicia species: copy number, sequence heterogeneity and chromosomal localisation. Mol. Gen. Genet. 250: 305–315. [DOI] [PubMed] [Google Scholar]
- Pearson, W. R., and D. J. Lipman, 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85: 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson-Burch, B. D., D. Nettleton and D. F. Voytas, 2004. Genomic neighborhoods for Arabidopsis retrotransposons: a role for targeted integration in the distribution of the Metaviridae. Genome Biol. 5: R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina, S. N., and H. Rees, 1983. DNA variation between and within chromosome complements of Vicia species. Heredity 51: 335–346. [Google Scholar]
- Rice, P., I. Longden and A. Bleasby, 2000. EMBOSS: the European Molecular Biology open software suite. Trends Genet. 16: 276–277. [DOI] [PubMed] [Google Scholar]
- Sanger, F., D. Nicklen and A. R. Coulson, 1977. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. USA 74: 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel, P., and J. L. Bennetzen, 1998. Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann. Bot. 82: 37–44. [Google Scholar]
- SanMiguel, P., A. Tikhonov, Y. K. Jin, N. Motchoulskaia, D. Zakharov et al., 1996. Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 765–768. [DOI] [PubMed] [Google Scholar]
- Shirasu, K., A. H. Schulman, T. Lahaye and P. Schulze-Lefert, 2000. A contiguous 66-kb barley DNA sequence provides evidence for reversible genome expansion. Genome Res. 10: 908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalka, A. M., 1989. Retroviral proteases:first glimpses at the anatomy of a processing machine. Cell 56: 911–913. [DOI] [PubMed] [Google Scholar]
- Sonnhammer, E. L. L., and R. Durbin, 1995. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167: GC1–GC10. [DOI] [PubMed] [Google Scholar]
- Staden, R., 1996. The Staden sequence analysis package. Mol. Biotechnol. 5: 233–241. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. Clustal-W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons, L., 2002. The long and short of siRNAs. Mol. Cell 10: 435–437. [DOI] [PubMed] [Google Scholar]
- Vicient, C. M., R. Kalendar and A. H. Schulman, 2001. Envelope-class retrovirus-like elements are widespread, transcribed and spliced, and insertionally polymorphic in plants. Genome Res. 11: 2041–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitte, C., and O. Panaud, 2003. Formation of solo-LTRs through unequal homologous recombination counterbalances amplifications of LTR retrotransposons in rice Oryza sativa L. Mol. Biol. Evol. 20: 528–540. [DOI] [PubMed] [Google Scholar]
- Wicker, T., N. Stein, L. Albar, C. Feuillet, E. Schlagenhauf et al., 2001. Analysis of a contiguous 211 kb sequence in diploid wheat (Triticum monococcum L.) reveals multiple mechanisms of genome evolution. Plant J. 26: 307–316. [DOI] [PubMed] [Google Scholar]
- Wicker, T., N. Yahiaoui, R. Guyot, E. Schlagenhauf, Z. D. Liu et al., 2003. Rapid genome divergence at orthologous low molecular weight glutenin loci of the A and A(m) genomes of wheat. Plant Cell 15: 1186–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker, T., W. Zimmermann, D. Perovic, A. H. Paterson, M. Ganal et al., 2005. A detailed look at 7 million years of genome evolution in a 439 kb contiguous sequence at the barley Hv-eIF4E locus: recombination, rearrangements and repeats. Plant J. 41: 184–194. [DOI] [PubMed] [Google Scholar]
- Wright, D. A., and D. F. Voytas, 2002. Athila4 of Arabidopsis and Calypso of soybean define a lineage of endogenous plant retroviruses. Genome Res. 12: 122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, Y., and T. H. Eickbush, 1990. Origin and evolution of retroelements based upon their reverse-transcriptase sequences. EMBO J. 9: 3353–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. Y., and S. R. Wessler, 2004. Genome-wide comparative analysis of the transposable elements in the related species Arabidopsis thaliana and Brassica oleracea. Proc. Natl. Acad. Sci. USA 101: 5589–5594. [DOI] [PMC free article] [PubMed] [Google Scholar]