Abstract
14-3-3 proteins are highly conserved polypeptides that participate in many biological processes by binding phosphorylated target proteins. The Saccharomyces cerevisiae BMH1 and BMH2 genes, whose concomitant deletion is lethal, encode two functionally redundant 14-3-3 isoforms. To gain insights into the essential function(s) shared by these proteins, we searched for high-dosage suppressors of the growth defects of temperature-sensitive bmh mutants. Both the protein kinase C1 (Pkc1) and its upstream regulators Wsc2 and Mid2 were found to act as high dosage suppressors of bmh mutants' temperature sensitivity, indicating a functional interaction between 14-3-3 and Pkc1. Consistent with a role of 14-3-3 proteins in Pkc1-dependent cellular processes, shift to the restrictive temperature of bmh mutants severely impaired initiation of DNA replication, polarization of the actin cytoskeleton, and budding, as well as cell wall integrity. Because Pkc1 acts in concert with the Swi4-Swi6 (SBF) transcriptional activator to control all these processes, the defective G1/S transition of bmh mutants might be linked to impaired SBF activity. Indeed, the levels of the G1 cyclin CLN2 transcripts, which are positively regulated by SBF, were dramatically reduced in bmh mutants. Remarkably, budding and DNA replication defects of bmh mutants were suppressed by CLN2 expression from an SBF-independent promoter, suggesting that 14-3-3 proteins might contribute to regulating the late G1 transcriptional program.
THE 14-3-3 proteins are a large family of highly conserved, ubiquitously expressed acidic polypeptides of 28–33 kDa found in all eukaryotes. At least 7 isoforms are present in mammals and up to 15 isoforms are present in plants, while 2 isoforms have been identified in yeast, Drosophila melanogaster, and Caenorhabditis elegans (reviewed in Hermeking 2003; Dougherty and Morrison 2004). They form homo- and heterodimers able to bind protein ligands that are usually phosphorylated on serine or threonine residues of consensus binding motifs (Jones et al. 1995; Muslin et al. 1996; Yaffe et al. 1997; Chaudhri et al. 2003). By inducing conformational changes or steric hindrance in protein ligands, 14-3-3 proteins can activate/repress their enzymatic activity, prevent their degradation, modulate their localization, and/or facilitate/inhibit their modifications and interactions (reviewed in Hermeking 2003; Dougherty and Morrison 2004). Targets of 14-3-3 family members are found in all subcellular compartments and include transcription factors, biosynthetic enzymes, cytoskeletal proteins, signaling molecules, checkpoint and apoptosis factors, and tumor suppressors. This plethora of interacting proteins allows 14-3-3 to play important roles in a wide range of regulatory processes such as cell cycle control, mitogenic signal transduction, and apoptotic cell death and to be implicated in carcinogenesis and some human diseases (reviewed in Dougherty and Morrison 2004). However, because multiple 14-3-3 isoforms are present in mammals and 14-3-3 proteins have several binding targets, the mechanisms underlying 14-3-3 functions are not fully understood.
The two Saccharomyces cerevisiae members of the 14-3-3 family, sharing 93% amino acid identity, are encoded by the BMH1 and BMH2 genes. While single bmh1Δ and bmh2Δ mutants do not show detectable growth defects compared to wild type, the bmh1 bmh2 double disruption is lethal in most laboratory strains (van Heusden et al. 1992, 1995; Gelperin et al. 1995; Roberts et al. 1997).
Although their essential functions are not well understood, budding yeast Bmh proteins appear to be involved in many cellular processes. For example, they modulate the activity of several transcription factors. In fact, loss-of-function mutations impairing the SIN4 or the RTG3 genes, encoding a global transcriptional regulator and a basic helix–loop–helix transcription factor, respectively, suppress the temperature-sensitive phenotype of a bmh1 bmh2 mutant (van Heusden and Steensma 2001). Moreover, Bmh1 physically interacts with phosphorylated Rtg3, suggesting that 14-3-3 proteins inhibit Rtg3 transcriptional activation function by binding its phosphorylated form (van Heusden and Steensma 2001). Finally, Bmh1 physically interacts with Msn2 and Msn4, two transcription factors required to activate a large number of stress-related genes, and retains their phosphorylated forms in the cytoplasm (Beck and Hall 1999).
Vesicular transport and cortical actin network organization also likely involve 14-3-3 proteins (Gelperin et al. 1995; Roth et al. 1999). In fact, S. cerevisiae cells overproducing the carboxy-terminal region of Bmh2 fail to polarize vesicular transport and show a disrupted actin cytoskeleton (Roth et al. 1999). Moreover, 14-3-3 proteins interact with many proteins involved in cytoskeletal regulation in both yeast and mammals (Jin et al. 2004). In particular, two-hybrid interactions have been reported for Bmh2 with Msb3 (Mayordomo and Sanz 2002), which is involved in actin cytoskeleton organization (Bach et al. 2000; Bi et al. 2000), and with Gic2 (Mayordomo and Sanz 2002), which is required together with Gic1 for cytoskeleton polarization during bud emergence (Brown et al. 1997; Chen et al. 1997). Both Bmh1 and Bmh2 interact also with the p21-activated kinase (PAK) Ste20. This interaction appears to be specifically required for Ras/MAPK cascade signaling during pseudohyphal development (Roberts et al. 1997). Finally, mammalian 14-3-3 proteins regulate actin dynamics by stabilizing phosphorylated cofilin, a family of proteins essential for high rates of actin filament turnover through regulation of the actin polymerization/depolymerization cycles (Gohla and Bokoch 2002). By interacting with various regulatory proteins, 14-3-3 proteins participate in diverse signal-transduction pathways. In fact, cell lethality caused by Bmh depletion can be suppressed by hyperactivating the Ras/cAMP-dependent protein kinase A (PKA) pathway through overproduction of Tpk1, the PKA catalytic subunit (Gelperin et al. 1995). Consistent with a link between 14-3-3 and PKA, Bmh proteins are dispensable for yeast cell viability in Σ1278b background (Roberts et al. 1997), where the Ras/cAMP signaling pathway is hyperactivated (Stanhill et al. 1999). However, bmh1Δ bmh2Δ Σ1278b derivative cells exhibit osmoremediable temperature sensitivity and sensitivity to high osmolarity (Roberts et al. 1997), suggesting that some functions of Bmh proteins are still required at 37° even in this background. Moreover, 14-3-3 proteins have been implicated also in Ras/MAPK cascade signaling in vertebrates (Fantl et al. 1994; Li et al. 1995) and during pseudohyphal development in S. cerevisiae (Roberts et al. 1997). Finally, vertebrate 14-3-3 proteins were shown to inhibit or activate protein kinase C (PKC), which is involved in many signaling processes (Toker et al. 1990; Isobe et al. 1992; Tanji et al. 1994), and to stimulate the interaction between PKC and the mitogen-stimulated Raf1 kinase that controls cell growth (Van Der Hoeven et al. 2000).
In a previous study we isolated four bmh1 alleles, whose presence in the cell as the sole 14-3-3 source caused temperature-sensitive growth (Lottersberger et al. 2003). To provide new insights into the essential functions of the S. cerevisiae 14-3-3 proteins, we carried out a detailed phenotypic characterization of these mutants and searched for high-dosage suppressors of their temperature sensitivity. We provide evidence that hyperactivation of the protein kinase C1 (Pkc1)-dependent pathways suppresses the growth defects of these bmh mutants, suggesting that 14-3-3 proteins functionally interact with Pkc1. Accordingly, bmh mutants are impaired in Pkc1-regulated processes at the G1/S transition, such as budding, initiation of DNA replication, actin cytoskeleton polarization, and cell wall integrity. Our data suggest that both the temperature sensitivity and the G1/S transition defects of our bmh mutants might be ascribed to an impaired activity of the Swi6/Swi4 (SBF) transcription factor, which is known to act in concert with Pkc1 to control all the processes described above.
MATERIALS AND METHODS
Yeast strains and media:
The relevant genotypes of all the yeast strains are listed in Table 1. All the strains used during this study were derivatives of W303 (MATa or MATα, ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 ssd1).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Relevant genotype | Reference/Source |
|---|---|---|
| YLL1081 | MATabmh2Δ∷KanMX4 bmh1-221∷LEU2∷bmh1Δ∷HIS3 | Lottersberger et al. (2003) |
| YLL1082 | MATabmh2Δ∷KanMX4 bmh1-103∷LEU2: bmh1Δ∷HIS3 | Lottersberger et al. (2003) |
| YLL1092 | MATabmh2Δ∷KanMX4 bmh1-342∷LEU2: bmh1Δ∷HIS3 | Lottersberger et al. (2003) |
| YLL1120 | MATabmh2Δ∷KanMX4 bmh1-266∷LEU2: bmh1Δ∷HIS3 | Lottersberger et al. (2003) |
| DMP4436/3C | MATabmh2Δ∷KanMX4 bmh1-103∷LEU2∷bmh1Δ∷HIS3 cdc24-1 | This study |
| DMP4430/7A | MATabmh2Δ∷KanMX4 bmh1-103∷LEU2∷bmh1Δ∷HIS3 cdc42-1 | This study |
| DMP4439/6B | MATabmh2Δ∷KanMX4 bmh1-221∷LEU2∷bmh1Δ∷HIS3 cdc24-1 | This study |
| DMP4433/10B | MATabmh2Δ∷KanMX4 bmh1-221∷LEU2∷bmh1Δ∷HIS3 cdc42-1 | This study |
| DMP4440/5C | MATabmh2Δ∷KanMX4 bmh1-266∷LEU2∷bmh1Δ∷HIS3 cdc24-1 | This study |
| DMP4434/7B | MATabmh2Δ∷KanMX4 bmh1-266∷LEU2∷bmh1Δ∷HIS3 cdc42-1 | This study |
| DMP4441/5A | MATabmh2Δ∷KanMX4 bmh1-342∷LEU2∷bmh1Δ∷HIS3 cdc24-1 | This study |
| DMP4435/4B | MATabmh2Δ∷KanMX4 bmh1-342∷LEU2∷bmh1Δ∷HIS3 cdc42-1 | This study |
| L96 | MATa/α CLN2/cln2∷GAL-CLN2∷URA3 | L. Dirick |
| DMP4357/1B | MATacln2∷GAL-CLN2∷URA3 | This study |
| DMP4370/2D | MATabmh2Δ∷KanMX4 bmh1-103∷LEU2∷bmh1Δ∷HIS3 cln2∷GAL-CLN2∷URA3 | This study |
| DMP4372/3B | MATabmh2Δ∷KanMX4 bmh1-221∷LEU2∷bmh1Δ∷HIS3 cln2∷GAL-CLN2∷URA3 | This study |
| DMP4373/7C | MATabmh2Δ∷KanMX4 bmh1-266∷LEU2∷bmh1Δ∷HIS3 cln2∷GAL-CLN2∷URA3 | This study |
| DMP4465/3B | MATabmh2Δ∷KanMX4 bmh1-342∷LEU2∷bmh1Δ∷HIS3 cln2∷GAL-CLN2∷URA3 | This study |
Strains YLL1082, YLL1081, YLL1120, and YLL1092 were previously described (Lottersberger et al. 2003). Wild-type, bmh1-103 bmh2Δ, bmh1-221 bmh2Δ, bmh1-266 bmh2Δ, and bmh1-342 bmh2Δ strains carrying either the 2μ vector or 2μ WSC2 or 2μ MID2 or 2μ GIC1 plasmids were constructed by transforming strains W303, YLL1082, YLL1081, YLL1120, and YLL1092 with plasmids YEplac195 (2μ URA3), pML489 (2μ WSC2 URA3), pML490 (2μ MID2 URA3), and pML493 (2μ GIC1 URA3), respectively.
A MATα strain, carrying the GAL-CLN2 construct integrated at the CLN2 chromosomal locus and obtained after sporulation of the diploid L96 kindly provided by L. Dirick (Montpellier, France), was crossed to strains YLL1082, YLL1081, YLL1120, and YLL1092 to obtain DMP4370/2D, DMP4372/3B, DMP4373/7C, and DMP4465/3B strains, respectively. Strain DMP4357/1B was obtained after sporulation of the diploid L96. Strain YLL1906, carrying the deletion of the PKC1 gene, was kindly provided by R. Tisi (University of Milano-Bicocca, Italy).
Strains DMP4436/3C, DMP4439/6B, DMP4440/5C, and DMP4441/5A were meiotic segregants from crosses of strains YLL1082, YLL1081, YLL1120, and YLL1092, respectively, with a MATα cdc24-1 strain. Strains DMP4430/7A, DMP4433/10B, DMP4434/7B, and DMP4435/4B strains were meiotic segregants from crosses of strains YLL1082, YLL1081, YLL1120, and YLL1092, respectively, with a MATα cdc42-1 strain.
The accuracy of all gene replacements and integrations was verified by Southern blot analysis or PCR. Standard yeast genetic techniques and media were according to Rose et al. (1990). Cells were grown in YEP medium (1% yeast extract, 2% bactopeptone, 50 mg/l adenine) supplemented with 2% glucose (YEPD) or 2% raffinose (YEP+raf) or 2% raffinose and 1% galactose (YEP+raf+gal). Transformants carrying the KANMX4 cassette were selected on YEPD plates containing 400 μg/ml G418 (US Biological).
Plasmids:
To obtain plasmid pML490, containing a MID2 fragment spanning from 663 bp upstream of the coding region start codon to 425 bp downstream of the stop codon, a 2219 bp MID2 fragment was amplified by PCR using yeast genomic DNA as template and the oligonucleotides PRP551 (5′-CGG GAT CCC GAT TGA GAG ATC TCA CGG AAA TG-3′) and PRP552 (5′-CGG GAT CCC GTC ACA GAA CTC GGT AAG TTT TC-3′) as primers. The PCR amplification product was then cloned into the BamHI site of plasmid YEplac195 (Gietz and Sugino 1988).
To obtain plasmid pML489, containing the WSC2 ORF flanked by 459 bp upstream of the start codon and 293 bp downstream of the stop codon, a 2264 bp WSC2 fragment was amplified by PCR using yeast genomic DNA as template and the oligonucleotides PRP543 (5′-CGG GAT CCC GCT ACG GTA AAC ATG CCT GAT GG-3′) and PRP544 (5′-CGG GAT CCC GTG TGA TCT AGC ACT TCT CCC AG-3′) as primers. The PCR amplification product was then cloned into the BamHI site of plasmid YEplac195.
To obtain plasmid pML493, containing the GIC1 ORF flanked by 343 bp upstream of the start codon and 282 bp downstream of the stop codon, a 1570-bp GIC1 fragment was amplified by PCR using yeast genomic DNA as template and the oligonucleotides PRP561 (5′-GGG GTA CCC CGT TGT CTG AGC AGG AAT AAA GAG-3′) and PRP562 (5′-GGG GTA CCC CGG GTA GTA GAC ATC GCT ATT ATC-3′) as primers. The PCR amplification product was then cloned into the KpnI site of plasmid YEplac195.
Plasmid YEplac112 (Gietz and Sugino 1988), carrying the PKC1 gene, was kindly provided by R. Tisi (University of Milano-Bicocca, Italy).
Search for high-dosage suppressors:
To search for high-dosage suppressors of the temperature sensitivity caused by the bmh1-266 mutation, strain YLL1120 was transformed with an S. cerevisiae genomic library on the basis of the multicopy 2μ vector YEp24 (Carlson and Botstein 1982). Ura+ transformants were tested for their ability to grow at 37° on YEPD plates, which inhibited the untransformed strain. Plasmids from transformants showing cosegregation of the thermo-resistance with the URA3 vector marker were recovered and introduced again into the YLL1120 strain, to confirm their ability to suppress bmh1-266 temperature sensitivity. Restriction analysis allowed us to identify several classes of plasmids containing different yeast genomic fragments. The nucleotide sequences of both ends of the smallest DNA insert of each plasmid class were determined and compared with the whole S. cerevisiae genomic sequence in the Saccharomyces Genome Database. Since most inserts contained several ORFs, the suppressor genes were identified by cloning subfragments of the inserts into the 2μ plasmid YEplac195 and testing the derivative plasmids for their ability to suppress the temperature sensitivity of the bmh1-266 bmh2Δ mutant strain.
Other techniques:
Synchronization experiments were performed as described in Lottersberger et al. (2003). Flow cytometric DNA analysis was determined on a Becton-Dickinson FACScan. To stain actin cytoskeleton, cells were treated 2 min with 20 units/ml rhodamine-phalloidin (Sigma-Aldrich) in PBS and then washed three times in PBS buffer. Digital images were taken with a CCD camera and software (CoolSNAP; Photometrics). For Western blot analysis, native protein extracts were prepared in 0.1% SDS, 1% Triton, 50 mm Tris pH 7.5, 1 mm sodium deoxycholate, 120 mm β-glycerophosphate, 1.72 mm sodium orthovanadate, 10 mm DTT, 1 mm AEBSF, 15 mm paranitrophenylphosphate, and a protease inhibitor cocktail (Boehringer Mannheim). To detect phosphorylated Mpk1 and Mpk1, polyclonal anti-phospho p42/p44 (Cell Signaling) and anti-Mpk1 (Santa Cruz Biotechnology) antibodies were used, respectively, after 1:1000 dilution in BSA–TBS. Secondary antibodies were purchased from Amersham and proteins were visualized by an enhanced chemiluminescence system according to the manufacturer.
RESULTS
MID2, WSC2, PKC1, and GIC1 act as high-dosage suppressors of bmh mutants' temperature sensitivity:
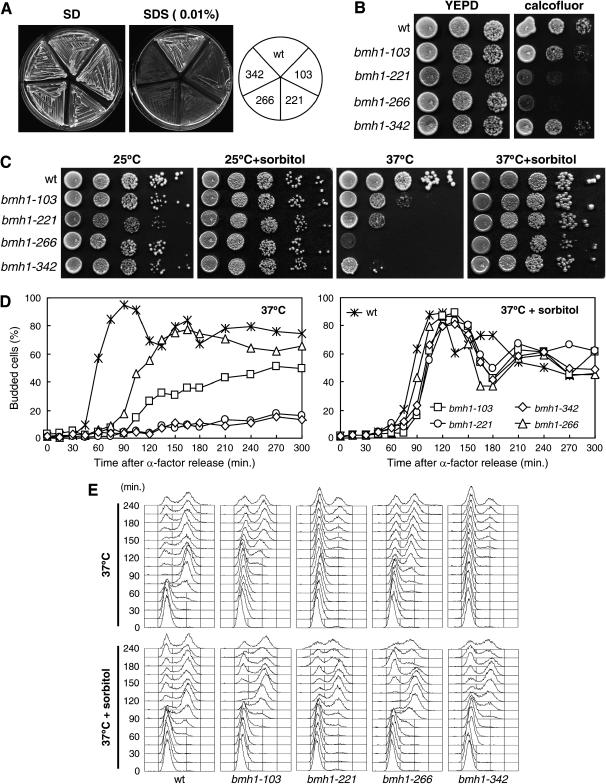
We previously generated bmh1-103, bmh1-221, bmh1-266, and bmh1-342 yeast temperature-sensitive mutants (Lottersberger et al. 2003 and Figure 1). In each of our mutant strains, the bmh1 mutant allele was the sole 14-3-3 source, because all strains carried a BMH2 deletion, which did not itself cause any of the phenotypes described during this study (data not shown). Importantly, overproduction of the Tpk1 catalytic subunit of PKA was unable to suppress the growth defects of these bmh mutants at 37° (data not shown), in contrast with its ability to suppress cell lethality caused by Bmh depletion at 25° (Gelperin et al. 1995). Therefore, our bmh mutants are not solely impaired in the activation of the Ras/cAMP-dependent PKA pathway.
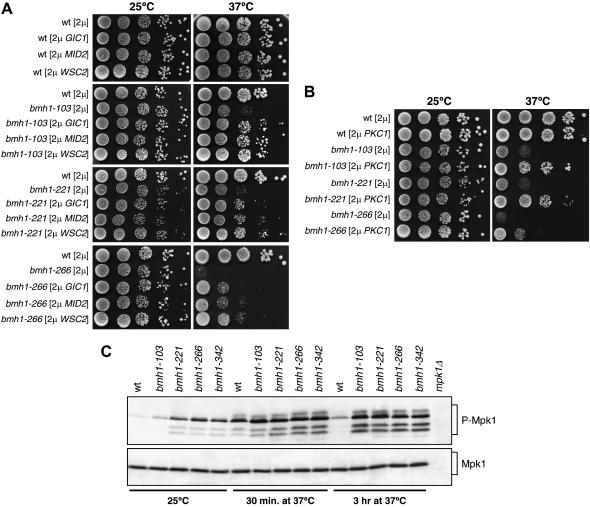
Figure 1.
High-dosage suppressors of the bmh temperature-sensitive growth defects. (A and B) Exponentially growing cell cultures (selective media at 25°) of wild-type (W303), bmh1-103 bmh2Δ (YLL1082), bmh1-221 bmh2Δ (YLL1081), and bmh1-266 bmh2Δ (YLL1120) strains transformed with 2μ plasmids, either empty or carrying the GIC1, MID2, WSC2 (A), or PKC1 (B) genes, were spotted on YEPD plates and incubated at 25° or at 37° for 3 days. (C) Cell cultures of wild type (K699), bmh1-103 bmh2Δ (YLL1082), bmh1-221 bmh2Δ (YLL1081), bmh1-266 bmh2Δ (YLL1120), and bmh1-342 bmh2Δ (YLL1092), exponentially growing in YEPD at 25°, were shifted to 37°. Aliquots were withdrawn at time zero (25°) and 30 min or 3 hr after shift at 37° to prepare protein extracts, which were subjected to Western blot analysis with anti-phospho-p44/p42 antibodies (Cell Signaling) to detect Mpk1 phosphorylation (top, P-Mpk1). The two faster migrating bands were likely P-Mpk1 degradation products, and they were not detected by polyclonal antibodies raised against a C-terminal Mpk1 peptide (Santa Cruz Biotechnology), which were used to measure total Mpk1 levels in the same samples (bottom, Mpk1). Specificity of the antibodies was checked by using protein extract prepared from an mpk1Δ (YLL1906) strain incubated 30 min at 37°.
To identify cellular partners for 14-3-3 proteins, we searched for high-dosage suppressors of the temperature sensitivity of the bmh1-266 mutant. To this purpose, bmh1-266 cells were transformed with an S. cerevisiae genomic library constructed in the YEp24 2μ vector (Carlson and Botstein 1982), and 40,000 Ura+ transformants were screened for the ability to form colonies on YEPD plates at 37° (see materials and methods). In addition to 190 BMH2- and 20 BMH1-bearing plasmids, the screen allowed the recovery of 10 plasmids carrying different ORFs. Subcloning of the several ORFs carried by 3 of these plasmids in the YEplac195 2μ vector revealed that high copy number of the GIC1, WSC2, or MID2 genes could partially suppress the temperature sensitivity of bmh1-266, bmh1-103, and bmh1-221 cells (Figure 1A). Unfortunately, we were unable to assess this suppressing ability in bmh1-342 cells due to their high frequency of 2μ plasmid loss.
While the GIC1 and GIC2 genes encode two homologous proteins required for actin polarization and bud formation (Brown et al. 1997; Chen et al. 1997), WSC2 and MID2 gene products are transmembrane cell surface sensors. They have been proposed to perform partially overlapping functions in cell wall remodeling during vegetative growth and under stress conditions (Verna et al. 1997; Marcoux et al. 1998; Ketela et al. 1999; Rajavel et al. 1999; Philip and Levin 2001) and to detect and signal the cell wall status to Pkc1. The latter is involved in a multiplicity of pathways, including those related to bud emergence, cell wall integrity, and organization of the actin cytoskeleton, in response to heat shock, pheromone, low osmolarity, nutrient starvation, and cell cycle progression (Heinisch et al. 1999). We therefore examined whether an excess of Pkc1 could also suppress the temperature sensitivity of our bmh mutants. Indeed, high copy number of PKC1 improved the ability of bmh1-103, bmh1-221, and bmh1-266 cells to form colonies at 37° (Figure 1B), indicating that hyperactivation of a Pkc1-dependent cascade may compensate for defects in 14-3-3 proteins.
Pkc1 is believed to possess multiple functions (Lee and Levin 1992; Verna et al. 1997; Delley and Hall 1999; Ketela et al. 1999; Andrews and Stark 2000; Zanelli and Valentini 2005), only one of which is to regulate the activity of the MAPK cascade that ultimately regulates cell wall integrity, bud emergence, response to hypotonic shock, and actin reorganization (reviewed in Levin and Errede 1995). Because high levels of Wsc2, Mid2, or Pkc1 (predicted to result in increased signaling through Pkc1) suppressed the temperature-sensitive growth defects of our bmh mutants, we asked whether the latter were defective in activating the Pkc1-dependent MAPK cascade. To this end, we monitored Mpk1 phosphorylation, which is an established marker for activation of the Pkc1–MAPK pathway (Lee et al. 1993; Zarzov et al. 1996; de Nobel et al. 2000). As shown in Figure 1C, the amount of phosphorylated Mpk1 in bmh mutants was higher than in wild type already at the permissive temperature. Moreover, heat shock, which is known to induce a rapid transient depolarization of the actin cytoskeleton and cell wall weakening (Delley and Hall 1999), increased the amount of Mpk1 phosphorylated forms in both wild-type and bmh mutants after a shift to 37° for 30 min (Figure 1C). However, phosphorylated Mpk1 level significantly decreased in wild-type cells within 3 hr at 37°, due to adaptation to the high temperature, whereas it remained high in bmh cells under the same conditions (Figure 1C). Thus, the growth defects of our bmh mutants are unlikely due to faulty MAPK signaling, as the latter appears to be hyperactivated instead in these mutants. Rather, these mutants could be deficient in some Pkc1-regulated pathway that parallels the one involving MAP kinases. In this view, the MAPK cascade could be hyperactivated in bmh mutants as a compensatory mechanism to maintain cell viability in the absence of 14-3-3 function. Accordingly, we found that MPK1 deletion was lethal for the bmh1-103, bmh1-221, bmh1-266, and bmh1-342 mutants (data not shown). Therefore, an excess of Wsc2, Mid2, or Pkc1 may suppress temperature sensitivity of bmh mutants by acting through a Pkc1-dependent MAPK-independent pathway.
Temperature-sensitive bmh mutants are defective in the G1/S transition and actin polarization:
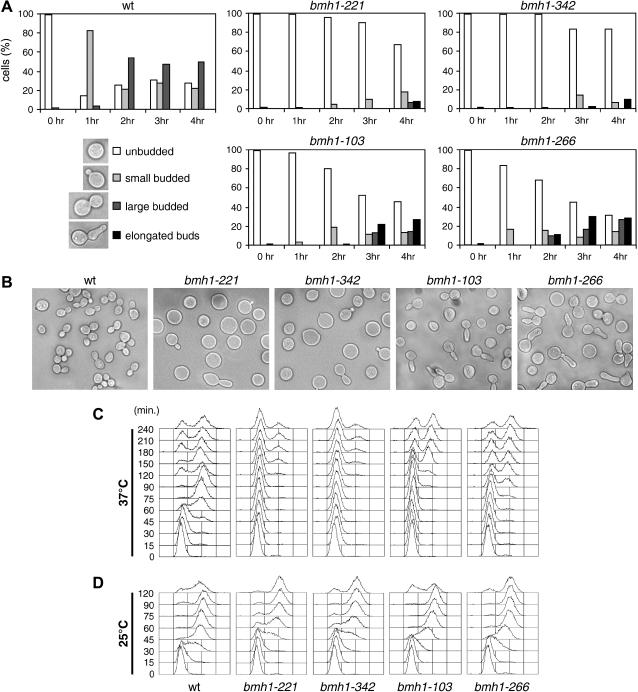
Because enhanced Pkc1 signaling contributes to cell viability in the absence of 14-3-3 function, we asked whether bmh mutants were impaired in Pkc1-regulated cellular processes, such as bud formation, actin reorganization, cell wall remodeling, and cell cycle progression (Heinisch et al. 1999). To investigate whether defects in 14-3-3 functions may affect bud formation at the G1/S transition, exponentially growing cultures of wild-type, bmh1-103, bmh1-221, bmh1-266, and bmh1-342 cells were arrested in G1 with α-factor at 25° and then released into fresh medium at 37°. As shown in Figure 2A, most of bmh1-103, bmh1-221, bmh1-266, and bmh1-342 cells were still largely unbudded after 1 hr at 37°, when bud emergence had already occurred in 90% of similarly treated wild-type cells. After 3 hr at 37°, most bmh1-221 and bmh1-342 mutant cells were still unbudded, while ∼50% of bmh1-103 and bmh1-266 cells managed to bud (Figure 2, A and B). However, their buds appeared mostly misshapen and, upon further incubation at 37°, continued to elongate (Figure 2, A and B). Moreover, some elongated budded cells appeared also in bmh1-221 and bmh1-342 mutants at later time points. Thus, bmh mutants might be impaired in the switch between apical to isotropic growth.
Figure 2.
Temperature-sensitive bmh mutants are defective in bud emergence and initiation of DNA replication. Cell cultures of wild-type (W303), bmh1-103 bmh2Δ (YLL1082), bmh1-221 bmh2Δ (YLL1081), bmh1-266 bmh2Δ (YLL1120), and bmh1-342 bmh2Δ (YLL1092) strains, exponentially growing at 25° in YEPD, were arrested in G1 with α-factor for 2 hr and released at time zero in YEPD at 25° or 37°. (A) A total of 200 cells for each strain were analyzed to determine the frequency of cells with no, small, large, or elongated buds at 25° (0 hr) and at the indicated time points after shift at 37°. (B) Photographs were taken 3 hr after shift at 37°. (C and D) To determine DNA contents by fluorescence-activated cell sorting (FACS) analysis, samples were withdrawn at the indicated times after release in YEPD at 37° (C) or at 25° (D).
Delayed bud formation in bmh mutants was parallel with defects in DNA synthesis initiation. In fact, all bmh mutants severely delayed initiation of DNA replication, although to different extents after a shift to 37° (Figure 2C). In fact, the onset of DNA replication took place in bmh1-221 and bmh1-342 cells only ∼150 min after release at 37° from the G1 block, while wild-type cells initiated DNA replication after 45–60 min under the same conditions (Figure 2C). In addition, a major fraction of cells in both mutants were unable to replicate DNA by 240 min (Figure 2C). The bmh1-103 and bmh1-266 cells started DNA replication ∼120 and 75 min after release at 37°, respectively, and again only a fraction of these mutant cells managed to complete DNA replication by 240 min at 37° (Figure 2C). As shown in Figure 2D, initiation of DNA replication upon release at 25° of the same G1-arrested cell cultures was delayed by 15–30 min in bmh mutant cell cultures compared to wild type. Altogether, these data indicate that 14-3-3 proteins are required for a timely G1/S transition.
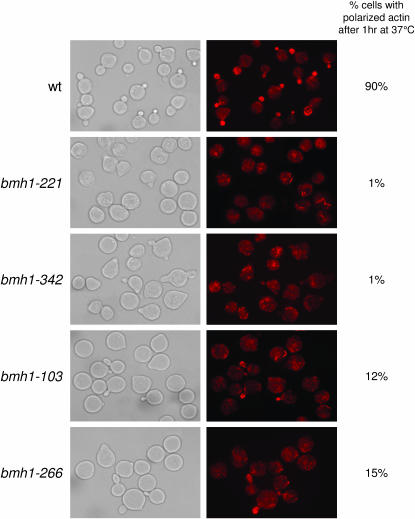
Both bud emergence and its subsequent surface growth require the polarization of the actin cytoskeleton, such that cortical patches and actin cables converge at the bud site (reviewed in Pruyne and Bretscher 2000). Since 14-3-3 proteins have been previously linked to actin cytoskeleton organization (Gelperin et al. 1995; Roth et al. 1999), impaired bud formation in the above bmh mutants might be related to defects in this process. To address this issue, we analyzed actin polarization upon Alexa-Fluor 546 phalloidin staining of wild-type and bmh mutant cells arrested in G1 by α-factor and then released at 37° for 1 hr. As shown in Figure 3, actin cortical patches, which normally clustered at the bud tips of wild-type cells, were completely missing in bmh1-221 and bmh1-342 cells and appeared only in a small fraction of bmh1-103 and bmh1-266 cells. Therefore, organization of the actin cytoskeleton at the future bud emergence sites is perturbed in bmh mutants, thus affecting bud formation at the G1/S transition.
Figure 3.
Actin organization in the temperature-sensitive bmh mutants. Cell cultures of wild-type (W303), bmh1-103 bmh2Δ (YLL1082), bmh1-221 bmh2Δ (YLL1081), bmh1-266 bmh2Δ (YLL1120), and bmh1-342 bmh2Δ (YLL1092) strains, exponentially growing at 25° in YEPD, were synchronized in G1 with α-factor and released at time zero in YEPD at 37°. Cells were fixed 1 hr after the release at 37°, stained with fluorochrome-conjugated phalloidin, and scored for the presence of cells with polarized actin by fluorescence microscopy. Differential interference contrast (left) and epifluorescence (right) images are shown as examples.
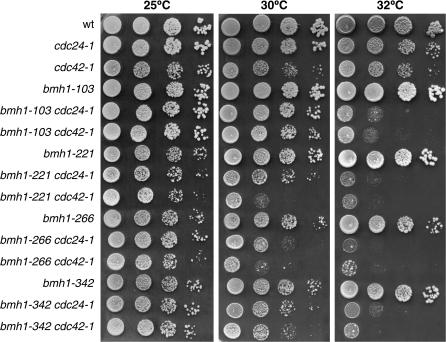
We then combined the different bmh1 alleles, together with the BMH2 deletion, with the temperature-sensitive cdc42-1 or cdc24-1 alleles, altering the essential Cdc42 GTPase and its guanine-nucleotide-exchange factor (GEF) Cdc24 (Adams et al. 1990; Johnson and Pringle 1990; Van Aelst and D'Souza-Schorey 1997), which are both required to establish actin cytoskeleton polarity (reviewed in Pruyne and Bretscher 2000). As shown in Figure 4, the ability of all bmh1 bmh2Δ cdc24-1 and bmh1 bmh2Δ cdc42-1 triple mutants to form colonies at 32° was severely impaired compared to that of the parental mutants. This synthetic effect between bmh and cdc24 or cdc42 mutant alleles further supports a role for budding yeast 14-3-3 proteins in actin polarization and bud formation.
Figure 4.
Synthetic effects between bmh and polarization mutant alleles. The following strains were used: wild type (W303), cdc24-1, cdc42-1, bmh1-103 bmh2Δ (YLL1082), bmh1-103 cdc24-1 bmh2Δ (DMP4436/3C), bmh1-103 cdc42-1 bmh2Δ (DMP4430/7A), bmh1-221 bmh2Δ (YLL1081), bmh1-221 cdc24-1 bmh2Δ (DMP4439/6B), bmh1-221 cdc42-1 bmh2Δ (DMP4433/10B), bmh1-266 bmh2Δ (YLL1120), bmh1-266 cdc24-1 bmh2Δ (DMP4440/5C), bmh1-266 cdc42-1 bmh2Δ (DMP4434/7B), bmh1-342 bmh2Δ (YLL1092), bmh1-342 cdc24-1 bmh2Δ (DMP4441/5A), and bmh1-342 cdc42-1 bmh2Δ (DMP4435/4B). Serial dilutions of cell cultures, exponentially growing in YEPD at 25°, were spotted on YEPD plates and incubated at the indicated temperatures for 3 days.
Bmh defects cause sensitivity to cell wall stress and their effects on the G1/S transition can be relieved by osmotic support:
Pkc1 controls cell wall metabolism by regulating both β-glucan synthesis at the site of wall remodeling and expression of cell wall biosynthesis genes necessary for maintaining cellular integrity during bud formation and in response to heat shock, pheromone, and nutrient starvation (reviewed in Levin 2005). We therefore asked whether defects in 14-3-3 functions might result in impaired cell wall integrity. We analyzed the ability of bmh mutants to grow at permissive temperature in the presence of compounds such as the chitin antagonist calcofluor white and SDS. In fact, both compounds are powerful tools for revealing yeast cell wall defects (Ram et al. 1994). As shown in Figure 5A, bmh1-103, bmh1-221, bmh1-266, and bmh1-342 cells were unable to grow on YEPD plates supplemented with 0.01% SDS, which did not affect wild-type cell growth. Moreover, growth of all bmh mutants on YEPD was compromised, although to different degrees, by addition of 0.01 mg/ml calcofluor white (Figure 5B). Finally, microscopic examination of the bmh mutant cell cultures revealed accumulation of cell debris at 37° (data not shown), suggesting that cell lysis frequently occurred. Thus, 14-3-3 proteins appear to be required for a stable cell wall structure.
Figure 5.
Temperature-sensitive bmh mutants are sensitive to cell wall stress. (A) Serial dilutions of wild-type (W303), bmh1-103 bmh2Δ (YLL1082), bmh1-221 bmh2Δ (YLL1081), bmh1-266 bmh2Δ (YLL1120), and bmh1-342 bmh2Δ (YLL1092) cell cultures, exponentially growing in YEPD at 25°, were streaked on SD plates with or without SDS (0.01%). (B) The same cultures in A were spotted on YEPD plates with or without Calcofluor (0.01 mg/ml). Plates were incubated at 25° for 4 days. (C) Serial dilution of wild-type (W303), bmh1-103 bmh2Δ (YLL1082), bmh1-221 bmh2Δ (YLL1081), bmh1-266 bmh2Δ (YLL1120), and bmh1-342 bmh2Δ (YLL1092) cell cultures, exponentially growing in YEPD at 25°, were spotted on YEPD plates in the absence or presence of 1 m sorbitol and incubated at the indicated temperatures for 3 days. (D and E) Cell cultures of wild-type (W303), bmh1-103 bmh2Δ (YLL1082), bmh1-221 bmh2Δ (YLL1081), bmh1-266 bmh2Δ (YLL1120), and bmh1-342 bmh2Δ (YLL1092) strains, exponentially growing at 25° in YEPD, were arrested in G1 with α-factor for 2 hr and released at time zero in YEPD at 37° in the absence or presence of 1 m sorbitol. Samples were withdrawn at the indicated times after α-factor release to analyze the kinetics of bud emergence (D) and DNA contents by FACS analysis (E).
We therefore examined whether osmotic stabilization of the medium might relieve the temperature sensitivity and the G1/S transition defects of our bmh mutants. As shown in Figure 5C, addition of the osmotic stabilizer sorbitol restored the ability of bmh1-103, bmh1-221, bmh1-266, and bmh1-342 cells to grow on YEPD plates at 37°. Moreover, the presence of sorbitol in the medium largely rescued the defects in bud emergence (Figure 5D) and initiation of DNA replication (Figure 5E) displayed by bmh mutants upon G1 release at 37°. Thus, slow growth and delayed G1/S transition that are caused by defective 14-3-3 proteins are osmoremediable, suggesting a primary defect of bmh mutants in cell wall biogenesis.
Wsc2, Mid2, or Pkc1 high dosage can partially suppress the G1/S transition defects of bmh mutants:
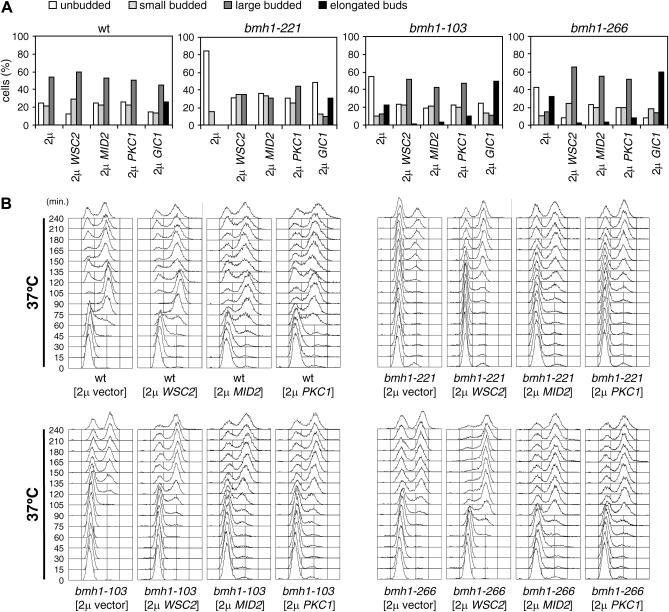
Because enhanced Pkc1-dependent signaling by high copy number WSC2, MID2, or PKC1 suppressed the temperature sensitivity of bmh mutants, we asked whether it also suppressed their G1/S transition defects. As shown in Figure 6A, bmh1-103, bmh1-221, and bmh1-266 cells carrying WSC2, MID2, or PKC1 on a 2μ plasmid and released from α-factor at 37° underwent budding more efficiently than the same mutant cells carrying the empty vector. Moreover, an excess of Wsc2, Mid2, or Pkc1 attenuated the abnormal bud morphology of bmh1-103 and bmh1-266 cells after 3 hr at 37° (Figure 6A). Similarly, bud emergence took place more efficiently in bmh1-221, bmh1-103, and bmh1-266 cells containing an excess of Gic1, although this, as expected, caused bud elongation even in wild-type cells due to sustained polarized growth (Figure 6A) (Brown et al. 1997; Chen et al., 1997).
Figure 6.
WSC2, MID2, and PKC1 overexpression can suppress the G1/S transition delay of bmh mutants. Exponentially growing (selective media at 25°) cell cultures of wild-type (W303), bmh1-103 bmh2Δ (YLL1082), bmh1-221 bmh2Δ (YLL1081), and bmh1-266 bmh2Δ (YLL1120) strains transformed with 2μ plasmids, either empty or carrying the WSC2, MID2, PKC1, or GIC1 genes, were arrested in G1 with α-factor and released at time zero in YEPD at 25° or 37°. (A) A total of 200 cells for each strain were analyzed to determine the frequency of cells with no, small, large, or elongated buds after 3 hr at 37°. (B) Samples were withdrawn at the indicated times after α-factor release to analyze DNA contents by FACS analysis.
Suppression of bmh defects in DNA replication initiation was also apparent upon WSC2, MID2, or PKC1 increased dosage. In fact, bmh1-103, bmh1-221, and bmh1-266 cells carrying high copy number WSC2-, MID2-, or PKC1-bearing plasmids initiated DNA replication at 37° earlier and more efficiently than the same mutants with the empty vector (Figure 6B). Thus, an excess of Wsc2, Mid2, or Pkc1 can partially suppress the G1/S transition defects of bmh mutants.
Low G1 cyclin-Cdk1 levels may account for the G1/S transition defects of bmh mutants:
During the G1/S transition, Pkc1 acts in concert with the SBF transcription factor to control the actin cytoskeleton, cell cycle progression, and transcription of cell wall biosynthesis genes (reviewed in Levin 2005). SBF is composed of the Swi6 and Swi4 subunits and is responsible for transcriptional activation of the CLN1 and CLN2 cyclin genes, whose products associate with the cyclin-dependent kinase 1 (Cdk1) to promote bud morphogenesis and DNA replication (reviewed in Levin et al. 1995; Nasmyth 1996).
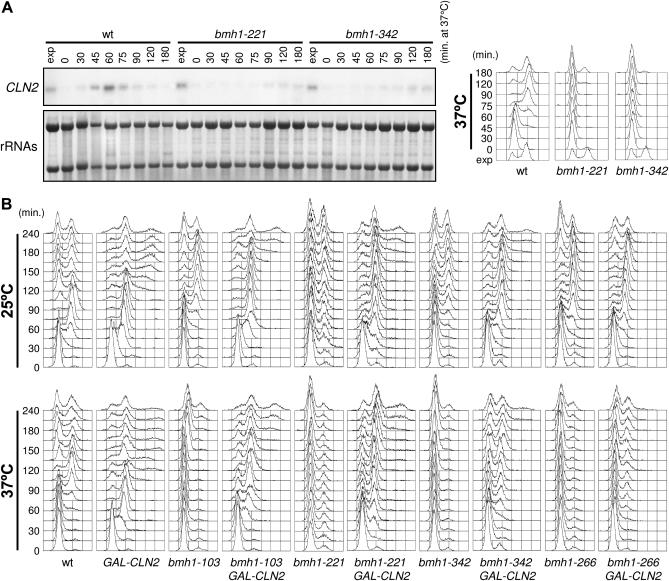
Since Pkc1 hyperactivation was shown to partially compensate for the lack of SBF activity (Gray et al. 1997; Igual et al. 1996), we asked whether the G1/S transition defects of bmh mutants might be related to impaired formation of G1 cyclin/Cdk1 complexes. We therefore measured the levels of CLN2 mRNA in the bmh1-221 and bmh1-342 mutants, which showed the most severe G1/S transition defects at 37° when compared to the other bmh mutants (Figure 2). Exponentially growing cultures of wild-type, bmh1-221, and bmh1-342 cells were arrested in G1 with α-factor and released into the cell cycle at 37°. Total RNA was prepared at different time points after release and subjected to Northern blot analysis with a CLN2 probe. As shown in Figure 7A, CLN2 mRNA was detectable in wild-type cells starting 30–45 min after release, just before bud emergence (data not shown) and initiation of DNA replication. Conversely, its amount was dramatically reduced in both bmh1-221 and bmh1-342 mutant cells that remained arrested with 1C DNA contents for at least 180 min after release at 37° (Figure 7A). If the G1/S transition defects of our bmh mutants were due to low amounts of G1 cyclin/Cdk1 complexes caused by the reduced CLN1 and CLN2 mRNA levels, CLN2 expression from an ectopic promoter might suppress the G1/S transition defects of our bmh mutants. To test this hypothesis, cultures of wild-type, bmh1-103, bmh1-221, bmh1-266, and bmh1-342 strains, carrying or lacking a galactose-inducible GAL1-CLN2 construct, were grown in YEP+raffinose at 25°, arrested in G1 with α-factor, and then released at 25° or 37° in galactose-containing medium to induce CLN2 expression. GAL1-CLN2 induction significantly rescued the G1/S defects of most bmh mutants. In fact, both bud emergence (data not shown) and initiation of DNA replication (Figure 7B) were advanced upon galactose induction in all bmh GAL1-CLN2 strains compared to the isogenic bmh strains, at both 25° and 37°. In particular, S phase entry took place in GAL-CLN2, bmh1-103 GAL-CLN2, bmh1-221 GAL-CLN2, and bmh1-342 GAL-CLN2 strains at 30, 45, 30, and 60 min, respectively, after release at 37° in galactose-containing medium. Similarly treated bmh1-103, bmh1-221, and bmh1-342 cells neither budded (data not shown) nor initiated DNA replication up to 4 hr after release (Figure 7B). Conversely, ectopic CLN2 expression had only a marginal effect on bmh1-266 cells, allowing only 30% of them to initiate DNA replication by 4 hr at 37° (Figure 7B). This suggests that functions other than activation of Cln1, 2/Cdk1 might be affected in this mutant. It is worth noting that expression of CLN2 from the GAL1 promoter caused cytokinesis defects at late time points in most strains, leading to accumulation of cells with elongated buds (data not shown) and more than 2C DNA contents (Figure 7B), as previously reported (Lew and Reed 1993). Altogether, these data indicate that reduced amounts of G1 cyclin/Cdk1 complexes may partially account for the G1/S transition defects of our bmh mutants.
Figure 7.
CLN2 mRNA levels and CLN2 ectopic expression in bmh mutants. (A) Cell cultures of wild-type (W303), bmh1-221 bmh2Δ (YLL1081), and bmh1-342 bmh2Δ (YLL1092) strains, exponentially growing in YEPD at 25°, were synchronized in G1 with α-factor and released at time zero into YEPD at 37°. Samples were taken at the indicated times after the release into the cell cycle to analyze CLN2 mRNA by Northern analysis (left) and to determine DNA contents by FACS analysis (right). Loading control of the Northern blot is a methylene blue-stained filter of ribosomal RNAs (rRNAs). (B) Cell cultures of wild-type (W303), GAL-CLN2 (DMP4357/1B), bmh1-103 bmh2Δ (YLL1082), bmh1-103 bmh2Δ GAL-CLN2 (DMP4370/2D), bmh1-221 bmh2Δ (YLL1081), bmh1-221 bmh2Δ GAL-CLN2 (DMP4372/3B), bmh1-266 bmh2Δ (YLL1120), bmh1-266 bmh2Δ GAL-CLN2 (DMP4373/7C), bmh1-342 bmh2Δ (YLL1092), and bmh1-342 bmh2Δ GAL-CLN2 (DMP4465/3B) strains, exponentially growing in YEP+raffinose at 25°, were synchronized in G1 with α-factor for 2 hr. Galactose was added 30 min before release. Synchronized cells were then released at time zero into YEP+raf+gal at 25° (top) or at 37° (bottom). Samples were taken at the indicated times after release to determine DNA contents by FACS analysis.
DISCUSSION
To understand the essential function(s) of S. cerevisiae 14-3-3 proteins, we searched for high dosage suppressors of the temperature sensitivity of bmh mutants, carrying bmh1 mutant alleles as the sole 14-3-3 sources (Lottersberger et al. 2003). We found that the growth defects of bmh1-103, bmh1-221, and bmh1-266 at 37° can be rescued by overproducing Pkc1 or its transmembrane cell surface sensors Wsc2 and Mid2. The latter have been proposed to perform partially overlapping functions in cell wall remodeling during vegetative growth and under stress conditions by detecting and transmitting cell wall status to Pkc1 (Verna et al. 1997; Ketela et al. 1999; Rajavel et al. 1999; Philip and Levin 2001). Pkc1 is believed to possess multiple functions (Lee and Levin 1992; Verna et al. 1997; Delley and Hall 1999; Ketela et al. 1999; Andrews and Stark 2000; Zanelli and Valentini 2005), one of which is to regulate the MAPK cascade involved in cell wall construction and polarized growth (reviewed in Levin and Errede 1995). On the basis of Mpk1 phosphorylation, the Pkc1-dependent MAPK cascade appears to be hyperactivated at both 25° and 37° in our bmh1 mutants, suggesting that defects in 14-3-3 proteins affect a pathway that is regulated by Pkc1 but does not involve Mpk1. Thus, Wsc2, Mid2, or Pkc1 may act as high-dosage suppressors by stimulating the former pathway, whereas the hyperactivation of the MAPK cascade in our mutants could be the result of a compensatory mechanism that contributes to their cell viability at the permissive temperature. Accordingly, deletion of MPK1 was lethal for our bmh mutants, indicating that 14-3-3 proteins and Mpk1 act in different branches of the Pkc1 pathway to sustain cell viability.
Pkc1, together with its upstream regulators Wsc1-3 and Mid2, controls actin cytoskeleton reorganization, cell cycle progression, and transcription of cell wall biosynthesis genes involved in synthesis and assembly of cell wall components at the bud (reviewed in Levin 2005). Since enhanced Pkc1-dependent signaling can partially suppress the temperature sensitivity of bmh mutants, some of the above Pkc1-regulated processes might be impaired in these mutants. Indeed, we found that all our temperature-sensitive bmh1 alleles cause defects in G1/S transition, actin polarization at the pre-bud site, and cell wall integrity. In fact, a shift to the restrictive temperature severely impairs bud formation and initiation of DNA replication in bmh1-221 and bmh1-342 mutants and significantly slows the same processes in bmh1-103 and bmh1-266 cells. When the entry into S phase and bud emergence take place in some of the latter mutant cells, buds are elongated, suggesting a defective apical–isotropic switch in bud growth. Consistent with a function for 14-3-3 proteins in bud formation and actin polarization, bmh mutant alleles also cause synthetic effects at semipermissive temperature when combined with the cdc42-1 and cdc24-1 temperature-sensitive alleles. These alleles alter the Rho-family GTPase Cdc42 and its GEF, respectively, which are essential for polarizing the actin cytoskeleton (reviewed in Pruyne and Bretscher 2000). Moreover, high levels of the Cdc42 effector Gic1, which binds to the activated GTP-bound form of Cdc42 and is required for cytoskeletal polarization during bud emergence (Chen et al. 1997; Brown et al. 1997), can partially suppress the temperature sensitivity of bmh1-103, bmh1-221, and bmh1-266 mutants. Finally, our bmh mutants undergo cell lysis at the restrictive temperature and are hypersensitive to calcofluor and SDS at the permissive temperature, suggesting that they are impaired in cell wall integrity. In agreement with a 14-3-3 role in cell wall biogenesis, both the growth defects and the G1/S transition delay at 37° of our bmh mutants can be rescued by the addition of the osmostabilizer sorbitol.
Both initiation of DNA replication and bud morphogenesis require activation of G1 cyclin/Cdk1 complexes (reviewed in Nasmyth 1996). In particular, when cells reach a critical size, Cln3-Cdk1 activates the SBF transcription factor that induces transcription of the CLN1 and CLN2 genes (Nasmyth and Dirick 1991; Ogas et al. 1991; Cross et al. 1994; Dirick et al. 1995). The Pkc1-dependent cascade acts in concert with SBF to control the actin cytoskeleton and transcription of cell wall biosynthesis genes involved in maintaining cellular integrity during bud formation (Levin and Bartlett-Heubusch 1992; Lew and Reed 1993; Mazzoni et al. 1993; Igual et al. 1996; Marini et al. 1996; Zarzov et al. 1996; Gray et al. 1997; Madden et al. 1997; Delley and Hall 1999). Accordingly, swi4Δ and swi6Δ mutants are sensitive to cell wall stresses and the growth defects of swi4Δ cells can be partially relieved by osmotic stabilization, supporting a role for SBF in cell wall biogenesis (Igual et al. 1996; Gray et al. 1997). Moreover, swi4 and pkc1 mutations are synthetically lethal (Madden et al. 1997), whereas the temperature-sensitive growth of swi4Δ cells can be suppressed by overproduction of Pkc1 or Wsc1, the latter belonging to the Wsc1-3 family of transmembrane proteins required for heat stress activation of the Pkc1-MAPK cascade (Igual et al. 1996; Gray et al. 1997). Finally, Pkc1 seems to play redundant functions with G1 cyclins since deletion of PKC1 causes cell death in a cln1Δ cln2Δ double mutant (Gray et al. 1997).
The partially redundant function of Pkc1 and SBF-dependent pathways and the similarities in the behavior of bmh and swi4 or swi6 mutants raise the possibility that the phenotypes of bmh mutants might arise from defective SBF activity. In agreement with this hypothesis, Cln1, 2/Cdk1 complexes appear to be limiting for execution of the G1/S transition in bmh mutants. In fact, the amount of CLN2 mRNA is dramatically reduced in both bmh1-103 and bmh1-342 mutants at 37° compared to wild type. Moreover, expression of CLN2 from an SBF-independent promoter can partially suppress bmh mutant defects in budding and DNA replication. Consistent with the possibility that defective 14-3-3 proteins may impair SBF-dependent accumulation of Cln1, 2/Cdk1 complexes, a mutation in the SIN4 gene, whose lack of function bypasses the requirement for Swi4 and Swi6 to transcribe the HO-LacZ reporter gene (Nasmyth et al. 1987; Lycan et al. 1994; Li et al. 2005), was shown to suppress the temperature sensitivity of a bmh2 bmh1Δ mutant (van Heusden and Steensma 2001). Since SBF-dependent induction of Cln1, 2/Cdk1 triggers both entry into S phase by turning on proteolysis of the cyclin B-Cdk1 inhibitor Sic1 and cytoskeleton polarization for bud formation (Lew and Reed 1993; Schwob et al. 1994; Tyers 1996), impaired SBF-dependent Cln1, 2/Cdk1 complex formation may account for both budding and DNA replication defects of our bmh mutants. It should in fact be noted that, unlike in most of the other laboratory strains, simultaneous deletion of CLN1 and CLN2 is lethal in the W303 genetic background that we used for all our experiments (Cvrckova et al. 1995). Therefore, low levels of Cln1, 2/Cdk1 complexes in bmh mutants may be the cause of their G1/S transition defects at high temperatures.
In any case, since SBF and Pkc1 play a partially redundant role in allowing bud formation and cell integrity (Igual et al. 1996; Gray et al. 1997; Madden et al. 1997), hyperactivation of the Pkc1-dependent cascades by high levels of Wsc2, Mid2, or Pkc1 might suppress the growth and G1/S transition defects of bmh mutants by partially compensating defects in SBF activity and G1 cyclin/Cdk1 complex accumulation.
Taken together, these data indicate that S. cerevisiae 14-3-3 proteins play essential functions in regulating processes that occur at the G1/S transition. Since 14-3-3 proteins are highly conserved in evolution, our studies may also help to elucidate their essential functions in higher eukaryotes.
Acknowledgments
We thank L. Dirick and R. Tisi for providing yeast strains and plasmids and M. Vai and all the members of our laboratory for useful discussions and critiques. This work was supported by grants from Associazione Italiana per la Ricerca sul Cancro and Fondazione Cassa di Risparmio delle Provincie Lombarde to M.P.L. and S.P., Cofinanziamento 2005 Ministero dell'Istruzione dell'Università e della Ricerca/Università di Milano-Bicocca to M.P.L. and Fondo per gli Investimenti della Ricerca di Base (FIRB) to G.L. F.L. was supported by a fellowship from Fondazione Italiana per la Ricerca sul Cancro.
References
- Adams, A. E., D. I. Johnson, R. M. Longnecker, B. F. Sloat and J. R. Pringle, 1990. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J. Cell Biol. 111: 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, P. D., and M. J. Stark, 2000. Type I protein phosphatase is required for maintenance of cell wall integrity, morphogenesis and cell cycle progression in Saccharomyces cerevisiae. J. Cell Sci. 113: 507–520. [DOI] [PubMed] [Google Scholar]
- Bach, S., O. Bouchat, D. Portetelle and M. Vandenbol, 2000. Co-deletion of the MSB3 and MSB4 coding regions affects bipolar budding and perturbs the organization of the actin cytoskeleton. Yeast 16: 1015–1023. [DOI] [PubMed] [Google Scholar]
- Beck, T., and M. N. Hall, 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402: 689–692. [DOI] [PubMed] [Google Scholar]
- Bi, E., J. B. Chiavetta, H. Chen, G. C. Chen, C. S. Chan et al., 2000. Identification of novel, evolutionarily conserved Cdc42p-interacting proteins and of redundant pathways linking Cdc24p and Cdc42p to actin polarization in yeast. Mol. Biol. Cell 11: 773–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J. L., M. Jaquenoud, M. P. Gulli, J. Chant and M. Peter, 1997. Novel Cdc42-binding proteins Gic1 and Gic2 control cell polarity in yeast. Genes Dev. 11: 2972–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, M., and D. Botstein, 1982. Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell 28: 145–154. [DOI] [PubMed] [Google Scholar]
- Chaudhri, M., M. Scarabel and A. Aitken, 2003. Mammalian and yeast 14–3-3 isoforms form distinct patterns of dimers in vivo. Biochem. Biophys. Res. Commun. 300: 679–685. [DOI] [PubMed] [Google Scholar]
- Chen, G. C., Y. J. Kim and C. S. Chan, 1997. The Cdc42 GTPase-associated proteins Gic1 and Gic2 are required for polarized cell growth in Saccharomyces cerevisiae. Genes Dev. 11: 2958–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, F. R., M. Hoek, J. D. Mckinney and A. H. Tinkelenberg, 1994. Role of Swi4 in cell cycle regulation of CLN2 expression. Mol. Cell. Biol. 14: 4779–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvrckova, F., C. De Virgilio, E. Manser, J. R. Pringle and K. Nasmyth, 1995. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 9: 1817–1830. [DOI] [PubMed] [Google Scholar]
- De Nobel, H., C. Ruiz, H. Martin, W. Morris, S. Brul et al., 2000. Cell wall perturbation in yeast results in dual phosphorylation of the Slt2/Mpk1 MAP kinase and in an Slt2-mediated increase in FKS2-lacZ expression, glucanase resistance and thermotolerance. Microbiology 146: 2121–2132. [DOI] [PubMed] [Google Scholar]
- Delley, P. A., and M. N. Hall, 1999. Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J. Cell Biol. 147: 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirick, L., T. Bohm and K. Nasmyth, 1995. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 14: 4803–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty, M. K., and D. K. Morrison, 2004. Unlocking the code of 14–3-3. J. Cell Sci. 117: 1875–1884. [DOI] [PubMed] [Google Scholar]
- Fantl, W. J., A. J. Muslin, A. Kikuchi, J. A. Martin, A. M. MacNicol et al., 1994. Activation of Raf-1 by 14–3-3 proteins. Nature 371: 612–614. [DOI] [PubMed] [Google Scholar]
- Gelperin, D., J. Weigle, K. Nelson, P. Roseboom, K. Irie et al., 1995. 14–3-3 proteins: potential roles in vesicular transport and Ras signaling in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 92: 11539–11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R. D., and A. Sugino, 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527–534. [DOI] [PubMed] [Google Scholar]
- Gohla, A., and G. M. Bokoch, 2002. 14–3-3 regulates actin dynamics by stabilizing phosphorylated cofilin. Curr. Biol. 12: 1704–1710. [DOI] [PubMed] [Google Scholar]
- Gray, J. V., J. P. Ogas, Y. Kamada, M. Stone, D. E. Levin et al., 1997. A role for the Pkc1 MAP kinase pathway of Saccharomyces cerevisiae in bud emergence and identification of a putative upstream regulator. EMBO J. 16: 4924–4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinisch, J. J., A. Lorberg, H. P. Schmitz and J. J. Jacoby, 1999. The protein kinase C-mediated MAP kinase pathway involved in the maintenance of cellular integrity in Saccharomyces cerevisiae. Mol. Microbiol. 32: 671–680. [DOI] [PubMed] [Google Scholar]
- Hermeking, H., 2003. The 14–3-3 cancer connection. Nat. Rev. Cancer 3: 931–943. [DOI] [PubMed] [Google Scholar]
- Igual, J. C., A. L. Johnson and L. H. Johnston, 1996. Coordinated regulation of gene expression by the cell cycle transcription factor Swi4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 15: 5001–5013. [PMC free article] [PubMed] [Google Scholar]
- Isobe, T., Y. Hiyane, T. Ichimura, T. Okuyama, N. Takahashi et al., 1992. Activation of protein kinase C by the 14–3-3 proteins homologous with Exo1 protein that stimulates calcium-dependent exocytosis. FEBS Lett. 308: 121–124. [DOI] [PubMed] [Google Scholar]
- Jin, J., F. D. Smith, C. Stark, C. D. Wells, J. P. Fawcett et al., 2004. Proteomic, functional, and domain-based analysis of in vivo 14–3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr. Biol. 14: 1436–1450. [DOI] [PubMed] [Google Scholar]
- Johnson, D. I., and J. R. Pringle, 1990. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J. Cell Biol. 111: 143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D. H., S. Ley and A. Aitken, 1995. Isoforms of 14–3-3 protein can form homo- and heterodimers in vivo and in vitro: implications for function as adapter proteins. FEBS Lett. 368: 55–58. [DOI] [PubMed] [Google Scholar]
- Ketela, T., R. Green and H. Bussey, 1999. Saccharomyces cerevisiae Mid2p is a potential cell wall stress sensor and upstream activator of the PKC1–MPK1 cell integrity pathway. J. Bacteriol. 181: 3330–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K. S., and D. E. Levin, 1992. Dominant mutations in a gene encoding a putative protein kinase (BCK1) bypass the requirement for a Saccharomyces cerevisiae protein kinase C homolog. Mol. Cell. Biol. 12: 172–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K. S., K. Irie, Y. Gotoh, Y. Watanabe, H. Araki et al., 1993. A yeast mitogen-activated protein kinase homolog (Mpk1p) mediates signalling by protein kinase C. Mol. Cell. Biol. 13: 3067–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, D. E., 2005. Cell wall integrity signalling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69: 262–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, D. E., and E. Bartlett-Heubusch, 1992. Mutants in the S. cerevisiae PKC1 gene display a cell cycle-specific osmotic stability defect. J. Cell Biol. 116: 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, D. E., and B. Errede, 1995. The proliferation of MAP kinase signaling pathways in yeast. Curr. Opin. Cell Biol. 7: 197–202. [DOI] [PubMed] [Google Scholar]
- Levin, K., A. H. Tinkelenberg and F. Cross, 1995. The CLN gene family: central regulators of cell cycle Start in budding yeast. Prog. Cell Cycle Res. 1: 101–114. [DOI] [PubMed] [Google Scholar]
- Lew, D. J., and S. I. Reed, 1993. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol. 120: 1305–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., T. Quinton, S. Miles and L. L. Breeden, 2005. Genetic interactions between mediator and the late G1-specific transcription factor Swi6 in Saccharomyces cerevisiae. Genetics 171: 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., P. Janosch, M. Tanji, G. C. Rosenfeld, J. C. Waymire et al., 1995. Regulation of Raf-1 kinase activity by the 14–3-3 family of proteins. EMBO J. 14: 685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lottersberger, F., F. Rubert, V. Baldo, G. Lucchini and M. P. Longhese, 2003. Functions of Saccharomyces cerevisiae 14–3-3 proteins in response to DNA damage and to DNA replication stress. Genetics 165: 1717–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lycan, D., G. Mikesell, M. Bunger and L. Breeden, 1994. Differential effects of Cdc68 on cell cycle-regulated promoters in Saccharomyces cerevisiae. Mol. Cell. Biol. 14: 7455–7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden, K., Y. J. Sheu, K. Baetz, B. Andrews and M. Snyder, 1997. SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science 275: 1781–1784. [DOI] [PubMed] [Google Scholar]
- Marcoux, N., Y. Bourbonnais, P. M. Charest and D. Pallotta, 1998. Overexpression of MID2 suppresses the profilin-deficient phenotype of yeast cells. Mol. Microbiol. 29: 515–526. [DOI] [PubMed] [Google Scholar]
- Marini, N. J., E. Meldrum, B. Buehrer, A. V. Hubberstey, D. E. Stone et al., 1996. A pathway in the yeast cell division cycle linking protein kinase C (Pkc1) to activation of Cdc28 at START. EMBO J. 15: 3040–3052. [PMC free article] [PubMed] [Google Scholar]
- Mayordomo, I., and P. Sanz, 2002. The Saccharomyces cerevisiae 14–3-3 protein Bmh2 is required for regulation of the phosphorylation status of Fin1, a novel intermediate filament protein. Biochem. J. 365: 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni, C., P. Zarov, A. Rambourg and C. Mann, 1993. The SLT2 (MPK1) MAP kinase homolog is involved in polarized cell growth in Saccharomyces cerevisiae. J. Cell Biol. 123: 1821–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslin, A. J., J. W. Tanner, P. M. Allen and A. S. Shaw, 1996. Interaction of 14–3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84: 889–897. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K., 1996. Viewpoint: putting the cell cycle in order. Science 274: 1643–1645. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K., and L. Dirick, 1991. The role of SWI4 and SWI6 in the activity of G1 cyclins in yeast. Cell 66: 995–1013. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K., D. Stillman and D. Kipling, 1987. Both positive and negative regulators of HO transcription are required for mother-cell-specific mating-type switching in yeast. Cell 48: 579–587. [DOI] [PubMed] [Google Scholar]
- Ogas, J., B. J. Andrews and I. Herskowitz, 1991. Transcriptional activation of CLN1, CLN2, and a putative new G1 cyclin (HCS26) by SWI4, a positive regulator of G1-specific transcription. Cell 66: 1015–1026. [DOI] [PubMed] [Google Scholar]
- Philip, B., and D. E. Levin, 2001. Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a guanine nucleotide exchange factor for Rho1. Mol. Cell. Biol. 21: 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne, D., and A. Bretscher, 2000. Polarization of cell growth in yeast. J. Cell Sci. 113: 571–585. [DOI] [PubMed] [Google Scholar]
- Rajavel, M., B. Philip, B. M. Buehrer, B. Errede and D. E. Levin, 1999. Mid2 is a putative sensor for cell integrity signaling in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 3969–3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram, A. F., A. Wolters, R. Ten Hoopen and F. M. Klis, 1994. A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitivity to calcofluor white. Yeast 10: 1019–1030. [DOI] [PubMed] [Google Scholar]
- Roberts, R. L., H. U. Mosch and G. R. Fink, 1997. 14–3-3 proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell 89: 1055–1065. [DOI] [PubMed] [Google Scholar]
- Rose, M. D., F. Winston and P. Hieter, 1990. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Roth, D., J. Birkenfeld and H. Betz, 1999. Dominant-negative alleles of 14–3-3 proteins cause defects in actin organization and vesicle targeting in the yeast Saccharomyces cerevisiae. FEBS Lett. 460: 411–416. [DOI] [PubMed] [Google Scholar]
- Schwob, E., T. Bohm, M. D. Mendenhall and K. Nasmyth, 1994. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79: 233–244. [DOI] [PubMed] [Google Scholar]
- Stanhill, A., N. Schick and D. Engelberg, 1999. The yeast ras/cyclic AMP pathway induces invasive growth by suppressing the cellular stress response. Mol. Cell. Biol. 19: 7529–7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji, M., R. Horwitz, G. Rosenfeld and J. C. Waymire, 1994. Activation of protein kinase C by purified bovine brain 14–3-3: comparison with tyrosine hydroxylase activation. J. Neurochem. 63: 1908–1916. [DOI] [PubMed] [Google Scholar]
- Toker, A., C. A. Ellis, L. A. Sellers and A. Aitken, 1990. Protein kinase C inhibitor proteins. Purification from sheep brain and sequence similarity to lipocortins and 14–3-3 protein. Eur. J. Biochem. 191: 421–429. [DOI] [PubMed] [Google Scholar]
- Tyers, M., 1996. The cyclin-dependent kinase inhibitor p40SIC1 imposes the requirement for Cln G1 cyclin function at Start. Proc. Natl. Acad. Sci. USA 93: 7772–7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aelst, L., and C. D'Souza-Schorey, 1997. Rho GTPases and signaling networks. Genes Dev. 11: 2295–2322. [DOI] [PubMed] [Google Scholar]
- Van Der Hoeven, P.C., J. C. Van Der Wal, P. Ruurs and W. J. Van Blitterswijk, 2000. Protein kinase C activation by acidic proteins including 14–3-3. Biochem. J. 347: 781–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heusden, G. P., and H. Y. Steensma, 2001. 14–3-3 Proteins are essential for regulation of RTG3-dependent transcription in Saccharomyces cerevisiae. Yeast 18: 1479–1491. [DOI] [PubMed] [Google Scholar]
- Van Heusden, G. P., T. J. Wenzel, E. L. Lagendijk, H. Y. De Steensma and J. A. Van Den Berg, 1992. Characterization of the yeast BMH1 gene encoding a putative protein homologous to mammalian protein kinase II activators and protein kinase C inhibitors. FEBS Lett. 302: 145–150. [DOI] [PubMed] [Google Scholar]
- Van Heusden, G. P., D. J. Griffiths, J. C. Ford, T. F. Chin-A-Woeng, P. A. Schrader, et al., 1995. The 14–3-3 proteins encoded by the BMH1 and BMH2 genes are essential in the yeast Saccharomyces cerevisiae and can be replaced by a plant homologue. Eur. J. Biochem. 229: 45–53. [PubMed] [Google Scholar]
- Verna, J., A. Lodder, K. Lee, A. Vagts and R. Ballester, 1997. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94: 13804–13809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe, M. B., K. Rittinger, S. Volinia, P. R. Caron, A. Aitken et al., 1997. The structural basis for 14–3-3:phosphopeptide binding specificity. Cell 91: 961–971. [DOI] [PubMed] [Google Scholar]
- Zanelli, C. F., and S. R. Valentini, 2005. Pkc1 acts through Zds1 and Gic1 to suppress growth and cell polarity defects of a yeast eIF5A mutant. Genetics 171: 1571–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarzov, P., C. Mazzoni and C. Mann, 1996. The SLT2(MPK1) MAP kinase is activated during periods of polarized cell growth in yeast. EMBO J. 15: 83–91. [PMC free article] [PubMed] [Google Scholar]