Abstract
The kinase cascade of the septation initiation network (SIN), first revealed in fission yeast, activates the contraction of the actomyosin ring, and plays an essential role in fungal septation. Mob1p, an evolutionarily conserved SIN protein, is associated with the most downstream kinase of this cascade in fission yeast. In this study, the mobA gene encoding a homologous protein was isolated from the filamentous fungus Aspergillus nidulans, whose mycelium is made of multinucleate cells. The MOBA protein was required for septation and conidiation, but was not essential for hyphal extension and colony formation. To identify genes that act antagonistically against the SIN, UV mutagenesis was carried out to isolate suppressor (smo) mutations that restored conidiation when MOBA was not expressed. Microscopic examination indicated that the restored conidiation was concomitant with restored septation in the absence of the MOBA protein. Eight recessive smo mutations in five complementation groups also bypassed the requirement of the SIN kinases SEPH and SIDB for septum formation and conidiation. However, none of these smo mutations affected the localization of MOBA. Among smo mutations, smoA and smoB mutations caused reduced hyphal growth and colony formation. They also rendered hypersensitivity to low doses of the microtubule-depolymerizing agent benomyl for conidiation. Therefore, in A. nidulans, proteins encoded by the smo genes likely have an antagonistic interaction against the SIN pathway to regulate septation and conidiation.
IN the cell cycle, cytokinesis is the final step, which physically separates segregated genomes into two daughter cells. Thus, its timing is of paramount significance. To ensure that two groups of segregated genetic material are enclosed in different cytoplasms, cytokinesis has to be coordinated with mitosis so that the onset of cytokinesis is triggered by the completion of mitosis. Although organisms of different kingdoms have developed unique mechanisms to execute cytokinesis, signals that trigger the onset of cytokinesis may be evolutionarily conserved (Guertin et al. 2002a; Wolfe and Gould 2005).
Haploid fungal organisms like the unicellular yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe and the filamentous fungus Aspergillus nidulans serve as model systems for dissecting fundamental mechanisms that bring about cytokinesis in eukaryotes (Harris 2001; Krapp et al. 2004). Cytokinesis in fungi, or septation, results in the formation of a cross wall known as the septum. Serendipitous studies in S. pombe have revealed the septation initiation network, or SIN, with a kinase cascade that triggers the contraction of the actomyosin ring and consequently the assembly of the septum (Krapp et al. 2004; Wolfe and Gould 2005). Downregulation of the kinase cascade would abolish septation, and hyperactivation would induce septation precociously (Krapp et al. 2004). A similar signaling network, known as the mitotic exit network, has been characterized in S. cerevisiae, and it functions in mitotic exit and septation (Bardin and Amon 2001; McCollum and Gould 2001).
In S. pombe, central players of SIN are cascaded kinases of Cdc7p, Sid1p, and Sid2p (Krapp et al. 2004). These kinases and their associated proteins exhibit dynamic localization patterns during the cell cycle. Septation is triggered by a spindle-pole-body-associated small G-protein known as Spg1p. The GTP–Spg1p recruits Cdc7p to the spindle pole body during mitosis. Consecutively, the Sid1p and Sid2p kinases are activated. Upon its activation, Sid2p and its associated protein Mob1p appear at the septation site. Consequently, the septation signal seems to be rallied from the spindle pole body to the septation site. Then, Sid2p and Mob1p directly or indirectly activate the formation of the septum. However, how the SIN activates septum formation is still unclear.
The mycelium of the filamentous fungus A. nidulans consists of multinucleate cells. When ample nutrients are provided, the mycelium would undergo asexual reproduction by producing an aerial hypha called the stalk, which results in the formation of the vesicle. Conidiophore development is then led by formation of the primary sterigma, termed the metula, and followed by the emergence of the secondary sterigma, known as the phialide. Uninucleate asexual spores of the conidium are produced by a budding process on the top of phialides. This conidiation process is genetically regulated by a flurry of genes (Adams et al. 1998). Mutations that spatially or temporally deregulate septation would abolish conidiation (Liu and Morris 2000; Liu et al. 2003)
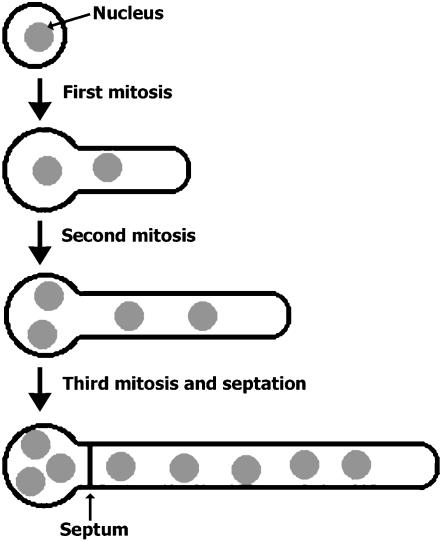
Upon the germination of the conidium, the first two rounds of mitosis are not coupled with septation (Harris 1997). The formation of the septum typically takes place after the third round of mitosis at the “neck” site near the region of what was once the conidium, or the spore body (Figure 1). The resulting five-nucleus apical cell retains robust tip growth, while the three-nucleus basal cell becomes dormant until a new tip growth event is initiated in this cell. Septation is regulated by cell cycle progression as well as by nuclear positioning in A. nidulans (Wolkow et al. 1996, 2000). Genetic studies have revealed several essential regulators of septation in this fungus (Harris et al. 1994, 1997; Harris and Hamer 1995). Among them, the SEPH protein is an ortholog of the yeast Cdc7p kinase (Bruno et al. 2001). But homologs of other SIN components have not been studied in A. nidulans. Moreover, it is not clear how septation is regulated in the multinucleate mycelium in this fungus.
Figure 1.
Diagram showing mitosis and septation after conidial germination. Note that the first septation event takes place after three rounds of mitosis and that the septum is assembled near the spore body.
In A. nidulans, it is intriguing how septation is regulated after conidial germination as the septation event is not coupled with the first and second mitosis. This is either because positive regulator(s) of septation like the SIN have not been activated after the first two mitoses or because negative regulator(s) need to be removed or inactivated before the completion of the third mitosis. The unusual cellularization feature endured by filamentous fungi like A. nidulans prompts us to postulate that unique regulatory mechanisms of septation may exist. Unfortunately, compared to organisms like yeasts, limited knowledge has been obtained in A. nidulans.
Another intriguing question is how cells respond to multinucleation in filamentous fungi. Mutations that lead to failures in septation typically confer lethality in organisms of uninucleate cells like yeasts (Gould and Simanis 1997). Although A. nidulans cells can tolerate cells with multiple nuclei, it is not known whether septation is dispensable for hyphal growth in this organism. Although little is known about the regulation of septation in other filamentous fungi, it is conceivable that common features in septation may exist among filamentous fungi. Thus, results garnered from studies in A. nidulans may unravel common mechanisms that regulate septation among filamentous fungi.
Because in S. pombe Mob1p and Sid2p translocate to the septation site upon activation, they may directly or indirectly interact with their substrates there. To gain further insights into regulatory mechanisms underlying septation in A. nidulans, we isolated the homologous mobA and sidB genes and analyzed their functions in septation. Furthermore, we attempted to address the question of whether septation can be restored when the SIN pathway is blocked. We report here the isolation and preliminary characterization of bypass suppressor mutations that can induce septation and conidiation processes in the absence of the MOBA protein and other SIN components. These suppressors illustrate the presence of a regulator pathway that antagonizes the SIN.
MATERIALS AND METHODS
A. nidulans strains and growth conditions:
A. nidulans strains used in this study are listed in Table 1. Growth conditions, transformation, crosses, germination of conidia, and induction conditions of alcA(p)-driven expressions were as described previously (Kafer 1977; Osmani et al. 1987; Liu and Morris 2000).
TABLE 1.
A. nidulans strains used in this study
| Strains | Genotype | Origin |
|---|---|---|
| AJM72 | sepH1 argB2; chaA1 veA1 | K. Bruno |
| GR5 | pyrG89; pyroA4; wA2 | FGSC |
| JKA8 | alcA(p)∷GFP-sidB∷pyrG; pyrG89; pabaA1 yA2 | This study |
| JKA12-12 | alcA(p)∷GFP-sidB∷pyrG; smoB101; pyrG89; pabaA1 yA2 | This study |
| JKA13-1 | alcA(p)∷GFP-sidB∷pyrG; smoA164; pyrG89; pabaA1 yA2 | This study |
| JKA19 | smoA164; pyrG89; pabaA1 yA2; pyroA4 | This study |
| JKA20 | smoB101; pyrG89; pabaA1 yA2; pyroA4 | This study |
| JKA26-5 | ΔmobA∷pyrG; smoA164; pyrG89; pyroA4; chaA1 | This study |
| JKA27-3 | ΔmobA∷pyrG; smoB101; pyrG89; pyroA4; chaA1 | This study |
| LB04 | pyroA4; chaA1 | This study |
| LBA49 | alcA(p)∷GFP-mobA∷pyrG; pyrG89; pyroA4; wA2 | This study |
| LBA53 | alcA(p)∷GFP-mobA∷pyrG; pyrG89; chaA1 | This study |
| LBA68 | ΔmobA∷pyrG; pyroA4; pyrG89; wA2 | This study |
| LLA1 | pyrG89; pabaA1 yA2 | This study |
| R21 | pabaA1 yA2 | FGSC |
| RSA6 | smoB101; chaA1; pyrG89 | This study |
| RSA7 | smoA137; chaA1; pyrG89 | This study |
| RSA8 | smoA152; chaA1; pyrG89 | This study |
| RSA9 | smoA164; chaA1; pyrG89 | This study |
| RSA10 | smoA302; chaA1; pyrG89 | This study |
| RSA46 | smoC265; chaA1; pyrG89 | This study |
| RSA47 | smoD299; chaA1; pyrG89 | This study |
| RSA48 | smoE306; chaA1; pyrG89 | This study |
| RSA49 | sepH1; smoB101; chaA1; pyrG89 | This study |
| RSA50 | sepH1; smoA137; chaA1; pyrG89 | This study |
| smo strains | alcA(p)∷GFP-mobA∷pyrG; pyrG89; chaA1; suppressor mutation | This study |
Standard DNA manipulation techniques:
The pBluescript KS+ plasmid (Stratagene, La Jolla, CA) was used as the cloning vector. The Vent DNA polymerase (New England Biolabs, Beverly, MA) was used for amplifying DNA fragments by polymerase chain reaction (PCR) from genomic DNA and from a λgt10 cDNA library (Osmani et al. 1988). The host strain for the recombinant DNA experiment was Escherichia coli DH5α (Invitrogen, Calsbad, CA), unless otherwise mentioned. Southern blotting hybridization was carried out as described before (Liu and Morris 2000).
Genetic manipulation of the mobA and sidB genes:
The genomic sequence of mobA was revealed by the A. nidulans genome project at the AN6288.2 locus (http://www.broad.mit.edu/annotation/fungi/aspergillus/). The cDNA sequence of mobA was isolated from the cDNA library using primers of mob11.5ra 5′-CCG AAT TCC CAT GGC TTC ATT CAT TAC C-3′ and mob11.3h 5′-GCC GAA GCT TGC TAC GTA ATA GCT GG-3′.
A mutant strain, in which mobA was expressed as a green fluorescent protein (GFP) fusion under the control of the alcA promoter, was generated according to the method described in our previous work except using the pLB04B plasmid (Liu and Morris 2000). At first, pLB04B was constructed using a strategy similar to that for the previously reported pLB01, except the pyr4 gene was replaced by pyrG. Briefly, the tandem alcA(p) and GFP-coding sequences were amplified using primers of ag_bst5a 5′-TAT ACC ACC GCG GTG GAA TTC CTG AAA AAG CTG-3′, and ag_not3b 5′-ATA TGC GGC CGC TCT GGA TCT CGG AGA TTT TGT ATA GTT CAT CC-3′. The pyrG fragment was isolated from the plasmid pXX1 using BamHI and EcoRV. The alcA(p)–GFP fragment was cloned into pBluescript KS+ at the BstXI and NotI sites, and the pyrG fragment was cloned into the derived plasmid at the BamHI and EcoRV sites to generate pLB04B. The 5′ truncated genomic sequence of mobA was amplified by PCR from wild-type genomic DNA using primers of mob15x 5′-CGC GTC TAG AAT GGC TTC ATT CAT TAC C-3′, and mob13b 5′-ATA GGA TCC TGC CAG AGG TAC TC-3′. The amplified genomic DNA was digested with XbaI and BamHI and cloned into pLB04B, previously digested with the same enzymes to give rise to pLB04B-mob1. This plasmid was then used to transform the GR5 strain.
A mobA deletion (ΔmobA) strain was constructed by replacing the mobA gene with the pyrG gene. The 5′ flanking sequence of mobA was amplified from genomic DNA using primers of dmob55na 5′-ATA AGA ATG CGG CCG CGT TCT CTA CCC TGT CAT CG-3′ and dmob53b 5′-CGC GGA TCC GTA TGG GAT ACA TGA ACG G-3′. After being digested with NotI and BamHI, the amplified fragment was cloned into pXX1 at NotI and BamHI sites to render pDMOB-5. The 3′ flanking region of the mobA gene was amplified using primers of dmob35p 5′-AAC TGC AGT ATC GCA CGG TTT CCT AG-3′ and dmob33sa 5′-TCC CCC GGG CAG CAC CGA GGT TGT TCA G-3′. The amplified fragment was digested with PstI and SmaI and then cloned into pDMOB-5 at the PstI and EcoRV sites to give rise to pDMOB-53. The fragment of the deletion construct was amplified from pDMOB-53 using the dmob55na and dmob33sa primers and introduced into the GR5 cells so that the mobA deletion strain was isolated.
An alcA(p)∷GFP-sidB strain was created using a strategy identical to that used for the alcA(p)∷GFP-mobA strain. The two primers GFP-sid2F 5′-ATA AGA ATG CGG CCG CAT GAC TCT GAA TCT CGA TTC TAA AGA CC-3′ and GFP-sid2R 5′-GGA CTA GTT CGC CCT GCC TAA GTC TGG T-3′ were used to amplify a 5′ truncated sequence of the A. nidulans sidB gene from the wild-type genome. The amplified DNA fragment was digested with NotI and SpeI and cloned into pLB04B at the corresponding sites. The resulting plasmid was used to transform the LLA1 strain to create the JKA8 strain.
Mutagenesis and isolation of septation revertants:
The strain LBA53 bearing the chaA1 mutation was used for mutagenesis. Approximately 2 × 108 conidia were suspended in 20 ml of sterile distilled water. They were irradiated with ultraviolet light at a dosage of 8000 μW/cm2 for 65 sec with agitation. The irradiation rendered ∼10–15% viability. Mutagenized spores were plated on the yeast extract, agar, glucose (YAG) medium in 100 × 50-mm petri dishes at a concentration of 1000 viable spores/plate. After being incubated at 30° for 3 days, plates were inspected by the naked eye. Colonies demonstrating the chartreuse color were picked as candidate conidiating revertants.
Microscopy and imaging acquisition/analysis:
For microscopic observations, conidia were inoculated onto precleaned glass coverslips on either the minimal medium plus glycerol for turning on the alcA(p) or the YAG medium for turning off the promoter. The GFP–MOBA signal was observed in live cells by placing the coverslips on a glass slide. DNA and chitin were stained using 4′,6-diamidino-2-phenylindole (DAPI) and calcofluor white (both from Sigma Aldrich, St. Louis), respectively, after the cells had been fixed with 4% paraformaldehyde (Polyscience, Warrington, PA).
Immunofluorescence was carried out according to a previous report using the same anti-γ-tubulin antibodies (Liu and Morris 2000). The γ-tubulin signal was observed using Texas red X-conjugated goat anti-rabbit IgG antibody (Molecular Probes, Eugene, OR).
Fluorescent microscopy was conducted using a Nikon Eclipse E600 microscope equipped with DIC and epifluorescence optics with a ×60 Plan-Apo or a ×100 Plan-Fluor objective (Nikon, Tokyo). The GFP signal was observed using a Chroma (Brattleboro, VT) GFP filter set (catalog no. 41017), and DAPI and calcofluor-stained signals were observed using a Chroma DAPI filter set (catalog no. 31000). Images were acquired using an Orca-100 CCD camera (Hamamatsu Photonics K.K., Hamamatsu City, Japan), driven by the ImagePro Plus 4.0 software package (Media Cybernetics, Silver Spring, MD). Colonies on solid media were scanned using an Epson 2450 scanner (Epson, Long Beach, CA). All images were assembled in the Adobe Photoshop 7.0 software package (Adobe, San Jose, CA).
RESULTS
Expression of a functional GFP–MOBA fusion under the alcA promoter:
In the fission yeast S. pombe, the Mob1p protein and its associated Sid2p kinase are two essential components of the SIN, which, upon activation, appear at the spindle pole body of daughter nuclei and then at the septation site (Salimova et al. 2000). Because the A. nidulans mycelium consists of multinucleate cells, we wondered how MOBA would behave in these cells. At first we determined the cDNA sequence of the mobA gene in A. nidulans. The cDNA sequence of mobA matched perfectly with the predicted sequence for the annotated AN6288.2 locus (accession no. XM_658800). The 218-aa MOBA protein showed 53% sequence identity at the amino acid level with its counterpart in fission yeast.
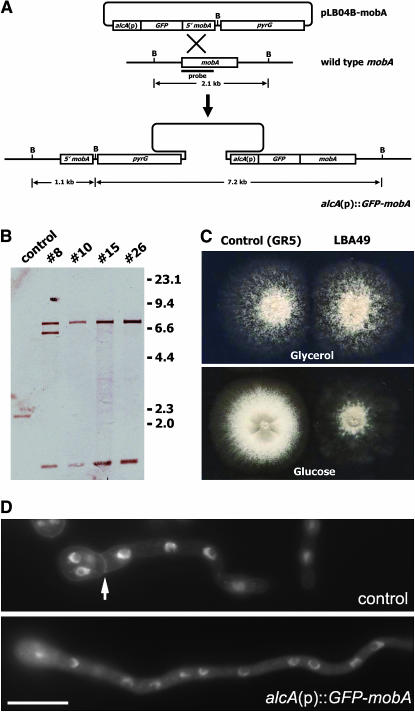
To test the intracellular localization pattern of MOBA and its function, the MOBA protein was tagged with GFP according to the strategy shown in Figure 2A. Southern blot analysis showed that, among four candidates, three had pLB04B-mobA integrated only once into the genome at the original mobA locus (Figure 2B). We chose transformant 10, designated as the LBA49 strain, for further analyses. In this mutant, there were only one full-length mobA gene, fused with the GFP-coding sequence, under the control of alcA(p), and a truncated 5′ mobA sequence.To test whether the resulting GFP–MOBA fusion was functional, the LBA49 strain was inoculated under both inducing and repressing conditions. On minimal medium containing glycerol, alcA(p) was moderately induced, and LBA49 demonstrated identical colony growth and conidiation pattern to the control GR5 strain (Figure 2C). Under the repressing condition on rich medium containing glucose (YAG), LBA49 produced a colony with a size comparable to that of GR5, if not larger (Figure 2C). However, the LBA49 strain formed fluffy-looking colonies. In areas where cells had direct contact with glucose (the medium), conidiation was abolished. Although conidia can be found in the middle of a well-grown colony, conidiation was triggered by leaky expression of GFP–MOBA when hyphae were air born on the top of the colony. The mobA deletion mutant (ΔmobA), however, showed no sign of conidiation (see below). Mycelial formation demonstrated by these strains indicates that mobA is not an essential gene in A. nidulans.
Figure 2.
Creation of the alcA(p)∷GFP-mobA strain. (A) Diagram showing the strategy of the creation of the conditional alcA(p)∷GFP-mobA strain. Note that pLB04B-mobA contains only a 5′ truncated sequence from the mobA gene. The integration of pLB04B-mob1 into the mobA locus in the wild-type genome renders a full-length mobA in fusion with GFP under the control of the alcA promoter. (B) Southern blot analysis showed the hybridization patterns of the control strain and four candidate transformants using the probe indicated in A. The control strain rendered a 2.1-kb band. If the plasmid was integrated once into the mobA locus, two bands of 1.1 and 7.2 kb should be detected. Thus, nos. 10, 15, and 26 transformants were such strains. The no. 10 strain was designated as LBA49 and used for further experiments. (C) The control (GR5) and the alcA(p)∷GFP-mobA (LBA49) strains were grown on rich medium containing glucose (bottom), and on minimal medium with glycerol as the sole carbon source (top). Under repressive conditions (with glucose), LBA49 failed to conidiate, while GR5 demonstrated robust conidiating capacity. Under inducing conditions (with glycerol), the two strains grew identically, indicating that the GFP–MOBA fusion protein was as functional as the wild-type MOBA. (D) Under repressive conditions (with glucose), a GR5 (control) hypha produced a septum (arrow). Under identical conditions, the LBA49 hypha failed to septate for an even longer time. Bar, 10 μm.
We further examined whether the colony growth phenotype on YAG was caused by the failure of septation. In the liquid YAG medium, control cells formed the first septum near the region of what was once the conidium (Figure 2D). In the same medium that turned off the expression of GFP–MOBA, however, all examined LBA49 cells failed to septate even after an extended growth time (Figure 2D). Thus, we concluded that in the LBA49 strain the GFP–MOBA fusion protein is the only functional form of the MOBA protein and that it is required for septation.
Intracellular localization of GFP–MOBA:
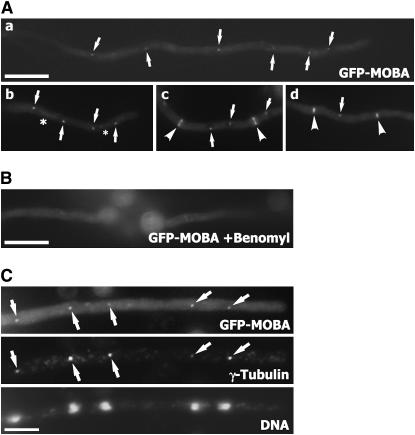
Because the GFP–MOBA fusion protein was functional, we used it to determine the intracellular localization pattern of the MOBA protein. In most cells, the GFP signal appeared to be in single dots distributed regularly along the hypha (Figure 3A,a). These GFP-labeled dots always appeared on top of dark objects, which represented the nuclei. We further examined GFP–MOBA in cells undergoing mitosis. In late mitotic cells, GFP–MOBA was predominantly localized to putative spindle pole bodies at the ends of the spindle (Figure 3A,b). An additional faint signal was also detected along the interzonal microtubules between the two spindle pole bodies (Figure 3A,b). During septation, GFP–MOBA signal appeared in a ring at the division site, and a fainter signal was still detected at the spindle pole body (Figure 3A,c). At later stages of septation, the GFP–MOBA signal at the septation site became confined in the central region of the septation site (Figure 3A,d). This reflected the contraction of the actomyosin ring toward the completion of septation. Thus, in A. nidulans, MOBA localizes to the spindle pole body and septation site as shown for its fission yeast ortholog (Hou et al. 2000). However, the fission yeast Mob1p does not contract with the developing septum.
Figure 3.
Intracellular localization of GFP–MOBA. (A,a) GFP–MOBA localized to putative spindle pole bodies (arrows) during interphase. (A,b) GFP–MOBA was detected at the spindle pole body (arrows) and along interpolar spindle microtubules (*). (A,c) During septation, GFP–MOBA decorated both the spindle pole body (arrows) and the septation site (arrowheads). The appearance at the septation site was in a ring. (A,d) At later stages of septation, the GFP–MOBA signal was restricted to the middle region (arrowheads), reflecting the contraction of the contractile rings. Signal was still detected at the spindle pole body (arrow). (B) Treatment of growing hyphae with benomyl led to the disappearance of GFP–MOBA at both the spindle pole body and the septation site. (C) Colocalization of GFP–MOBA with γ-tubulin. Arrows pointed at the spindle pole bodies decorated by both GFP–MOBA and anti-γ-tubulin signals. Bars, 10 μm in A and B; 5 μm in C.
Because GFP–MOBA localized to the microtubule-organizing center of the spindle pole body and along spindle microtubules, we tested whether the localization was dependent on microtubules. The microtubule-depolymerizing agent benomyl was used to depolymerize microtubules. After a 30-min exposure to 2.4 μg/ml benomyl, only diffuse GFP signal was detected in the cytoplasm (Figure 3B). Thus, our results showed that the localization of MOBA was dependent on the integrity of microtubules.
To determine whether GFP–MOBA decorated the spindle pole body, anti-γ-tubulin immunofluorescence was carried out as γ-tubulin has been shown as a bona fide spindle-pole-body-associated protein (Oakley et al. 1990). In hypha-containing nuclei at late anaphase, γ-tubulin decorated the spindle pole body associated with each nucleus (Figure 3C). The GFP–MOBA signal coincided with the γ-tubulin signal, indicating that MOBA was indeed associated with the spindle pole body.
Isolation of extragenic mutations that suppress loss of function of mobA:
On the YAG medium that shuts off the alcA promoter, strains harboring alcA(p)∷GFP-mobA failed to conidiate (Figure 2C). After spores were collected from the strain LBA53 bearing the chaA1 mutation for ease of visual screening, they were mutagenized by UV irradiation. In all examined A. nidulans mutants that cause septation defects, conidition is either abolished or significantly inhibited (Harris et al. 1994; Bruno et al. 2001; Liu et al. 2003). In addition, the conidium itself is the result of a specialized cell division. Hence, we used successful conidiation as an indicator of the recovery of the septation capability among the UV-mutagenized population. In the absence of MOBA, mutant colonies demonstrated light brown color (Figure 2C). After mutagenized spores were inoculated on the repressing YAG medium, tiny chartreuse colonies were spotted on the brown lawn of nonconidiating mycelia (data not shown). These individual colonies bearing conidia were streaked onto YAG to single colonies. Recovered mutant strains were subject to further characterization.
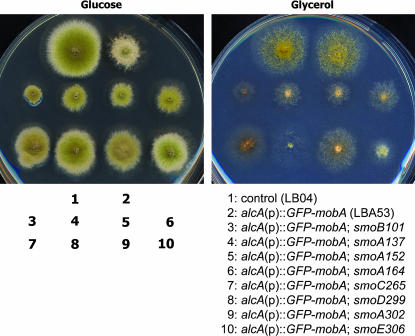
Among 110 independent mutations that restored conidiation for LBA53 on repressing medium, 8 demonstrated robust conidiation capacity (Figure 4). This report focuses on these 8 smo, or suppressor of mobA, mutations. The smo101, smo137, smo152, and smo164 strains showed significantly reduced colony size compared to LBA53 and the control LB04 strain (Figure 4). The smo299 and smo306 strains produced colonies very similar to those of the control strain. To determine whether the suppressor mutation was intragenic or extragenic, these smo strains were crossed to the control GR5 strain. All crosses resulted in progeny including those of wild-type (mobA+) and those of alcA(p)∷GFP-mobA phenotypes. Thus, all 8 mutations were extragenic mutations, which happened at genetic loci other than the mobA locus.
Figure 4.
Suppression of conidiation defect caused by downregulation of mobA expression. Colony growth phenotypes were shown on rich medium containing glucose (left) or on minimal medium with glycerol as the sole carbon source (right). Note that the LBA53 colony on rich medium failed to conidiate. The eight revertants restored conidiation at different degrees. On minimal medium, all mutants demonstrated a poor conidiation phenotype. The smo101, smo137, smo152, smo164, and smo306 mutants grew very poorly.
The strains containing smo mutations were also tested on inducing medium containing glycerol as the sole carbon source (Figure 4). Their growth phenotypes were tested side by side with control strains. Again, when the expression of GFP–MOBA was induced, the LBA53 strain produced a colony identical to that of a control strain. The suppressor mutations, however, all demonstrated different aspects of growth defects. Colonies produced by smo101, smo137, smo152, and smo164 strains were significantly smaller than those of control strains, indicating the inhibition of hyphal growth. To determine whether these mutations were at the same or different loci, genetic linkage tests were carried out among the strains containing these smo mutations alone. The results allowed us to place these eight mutations in five complementation groups, and the loci were designated as smoA–smoE.
To test whether smoA–smoE genes act in the same pathway, double-mutant analysis was carried out. Among smo mutations, smoB101 has a slightly stronger phenotype of retarded colony growth than smoA mutations. But smoA and smoB mutations have a far more severe phenotype than smoC265, smoD299, and smoE306 mutations. The smoA smoB double mutant behaved like smoB. Double mutants of smoA smoC, smoA smoD, and smoA smoE behaved like smoA (data not shown). Thus, we conclude that smoA, smoB, smoC, smoD, and smoE likely act in the same pathway.
smo mutations restored septation in the absence of MOBA:
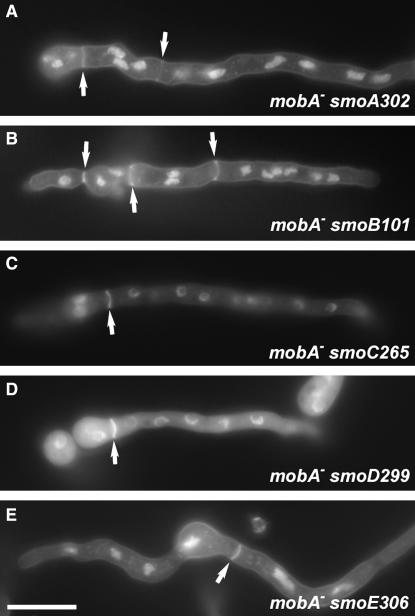
Because the aforementioned smo mutations rendered conidiation when MOBA expression was turned off, we wondered whether the restored conidiation was concomitant with the restoration of septation. When conidia of smo101-smo306 mutants were allowed to germinate in rich medium containing glucose, alcA(p)∷GFP-mobA was turned off, as demonstrated by the absence of the GFP–MOBA signal using fluorescent microscopy. No detectable signal was observed at the spindle pole body or at the septation site (data not shown). After germlings were fixed and stained with calcofluor white for visualizing chitin, septa were detected in all smo mutants (Figure 5). Thus, these smoA–smoE mutations had allowed the hyphae to septate in the absence of the MOBA protein.
Figure 5.
Restoration of septation in the absence of MOBA. (A–E) On rich medium containing glucose when the GFP–MOBA was not expressed, smoA302 (A), smoB101 (B), smoC265 (C), smoD299 (D), and smoE306 (E) restored septation. Bar, 10 μm.
To test whether the smo mutations acted directly on MOBA, we also examined the localization pattern of GFP–MOBA in these mutant backgrounds on glycerol medium. We detected a GFP–MOBA signal at the spindle pole body and at the septation site at intensities similar to those in control cells (supplemental Figure 1 at http://www.genetics.org/supplemental/). Thus, smoA–smoE mutations did not affect the activity of MOBA.
smo mutations suppressed the ΔmobA and sepH 1 mutations:
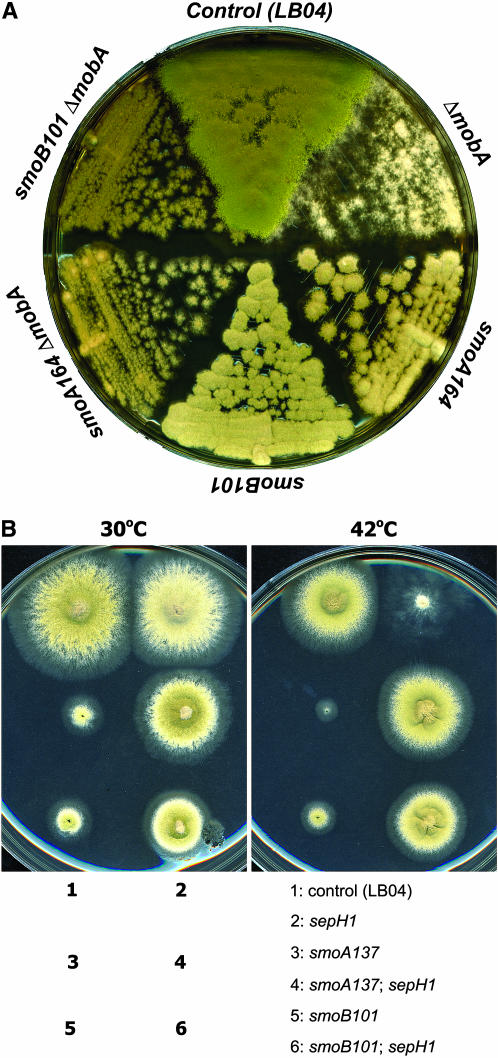
We further asked whether the smo mutations could suppress the deletion mutation at the mobA locus. The ΔmobA mutation showed a complete loss of conidiation (Figure 6A). Double mutants of smoA164 ΔmobA and smoB101 ΔmobA exhibited conidiation (Figure 6A), indicating that the smoA164 and smoB101 mutations suppressed the ΔmobA mutation.
Figure 6.
Suppression of the ΔmobA null mutation and the temperature-sensitive sepH 1 mutation by smo mutations. (A) The control (LB04), ΔmobA, smoA164, and smoB101 strains; ΔmobA smoA164 and smoB101ΔmobA double-mutant strains were grown on rich medium at 37°. Note that the ΔmobA strain formed fluffy colonies with no sign of conidiation. smoA164 and smoB101 mutations restored conidiation in the genetic background of mobA null mutation. (B) The control (LB04), sepH 1, smoA137, and smoB101 strains; sepH 1 smoA137 and sepH 1 smoB101 double-mutant strains were inoculated on duplicate plates and grew at either 30° or 42°. Note that the sepH 1 mutation abolished conidiation at 42° due to the failure of septation. The smoA137 and smoB101 mutations restored conidiation in the presence of the sepH 1 mutation at 42°. Note that the slow-growth phenotypes caused by smoA and smoB mutations were also suppressed by the sepH 1 mutation.
Since smoA–smoE mutations suppressed the loss-of-function mutation of mobA, we sought to determine whether these mutations could suppress mutations in upstream genes of mobA. An earlier work had indicated that the A. nidulans SEPH protein is an ortholog of the S. pombe Cdc7p—the most upstream kinase in the SIN pathway (Bruno et al. 2001). The sepH 1 mutation abolished septation and conidiation at the restrictive temperature of 42° (Harris et al. 1994; Bruno et al. 2001). At 42°, the sepH 1 mutant demonstrated fluffy colonies with loose hyphae similar to those of LBA49 (Figure 6B).
At 42°, the smoA137 sepH 1 and smoB101 sepH 1 double mutants formed colonies with sizes similar to that of the wild-type control strain, and the colonies conidiated robustly (Figure 6B). Thus, the failure of septation and conidiation caused by sepH 1 at restrictive temperature was suppressed by the smoA and smoB mutations. Similarly, smoC–smoE mutations also suppressed sepH 1 mutation (data not shown). Again, in these double mutants, the restored conidiation was concomitant with the restoration of septation at the restrictive temperature (data not shown).
As shown earlier, smoA137 and smoB101 significantly reduced growth rate at all tested temperatures. Interestingly, this slow-growth phenotype caused by the smoA and smoB mutations was also suppressed by the sepH 1 mutation at both 30° and 42°. Thus, we concluded that smoA/B and sepH genes act antagonistically on hyphal growth.
smoA and smoB mutations suppressed sidB null mutation:
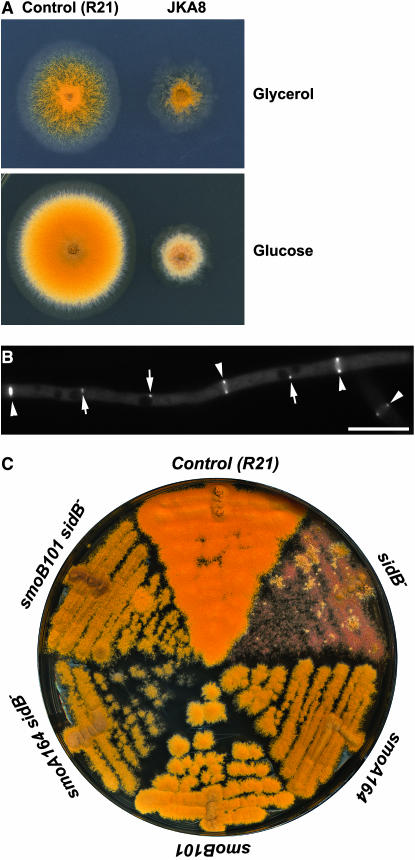
Because smoA–smoE mutations suppressed null mobA and sepH mutations, the restoration of septation could be the result of bypassing the requirement of the entire SIN pathway or the result of constitutively activating the Sid2p kinase homolog in A. nidulans. To distinguish these two possibilities, we identified the A. nidulans homolog of Sid2p encoded by the AN8751.2 locus sidB (accession no. XP_682020). Using an approach similar to that in Figure 2A, we created an A. nidulans strain, designated as JKA8, in which the only sidB gene was fused with the GFP-coding sequence and is under the control of the inducible/repressible alcA promoter (supplemental Figure 2A at http://www.genetics.org/supplemental/). When the GFP–SIDB fusion was induced on media containing glycerol as the sole carbon source, JKA8 produced a colony that conidiated robustly (Figure 7A). But the colony size was significantly smaller than that of the control R21 strain. Thus, the GFP–SIDB fusion was partially functional. When GFP–SIDB expression was turned off on the YAG medium, JKA8 completely failed to produce conidia, and it produced a fluffy colony with a much reduced size compared to the control strain (Figure 7A). This result suggested that, in addition to its essential function in septation and conidiation, SIDB also played a role in vegetative hyphal growth. Such a role was not shared by MOBA and SEPH as their loss-of-function mutations abolished only septation and conidiation, but did not affect the rate of hyphal growth and consequently the colony size.
Figure 7.
smo mutations suppressed the loss-of-function sidB mutation. (A). The control (R21) and the alcA(p)∷GFP-sidB (JKA8) strains were grown on minimal medium with glycerol as the sole carbon source (top) and on rich medium containing glucose (bottom). Under inducing conditions (with glycerol), JKA8 produced a colony with reduced size, and it conidiated robustly. Under repressive conditions (with glucose), JKA8 failed to conidiate, while R21 demonstrated healthy conidiating capacity. (B) GFP–SIDB localized to both the spindle pole bodies (arrows) and the septation site (arrowheads) during septation. (C) smoA164 and smoB101 mutations restored conidiation when GFP–SIDB expression was turned off on medium containing glucose. But the double mutants produced colonies with reduced sizes compared to the control (R21) strain. Bar, 10 μm.
We also examined the localization of the partially functional GFP–SIDB fusion protein. Similar to GFP–MOBA, GFP–SIDB also localized to the spindle pole bodies and septation sites (Figure 7B). Again, the GFP–SIDB signal coincided with the contraction of the actomyosin ring, which was different from its counterpart in fission yeast.
To test whether smoA and smoB mutations would suppress the loss-of-function sidB mutation, double mutants of alcA(p)∷GFP-sidB smoA137 and alcA(p)∷GFP-sidB smoB101 were created by standard genetic crosses. On rich medium containing glucose, these double mutants showed a restored conidiation phenotype (Figure 7C). However, they produced colonies of significantly reduced sizes compared to the control strain. Hence, smoA and smoB mutations could suppress the conidiation defect caused by the loss of SIDB, but not the slow-growth phenotype. This result again suggests that SIDB plays role(s) in hyphal growth in addition to its role in sepatation and conidiation.
In A. nidulans, in addition to sepH, mobA, and sidB, genes encoding other SIN components, except Sid4p, were cloned in silica on the basis of the genomic sequence released by the Broad Institute (J.-M. Kim and B. Liu, unpublished data). Thus, the data presented here as well as published work strongly suggested that A. nidulans also uses the SIN pathway to trigger the initiation of septation as seen in S. pombe. Since smoA and smoB mutations suppressed the loss-of-function sepH 1 and sidB mutations, we concluded that these mutations bypassed the requirement of the SIN for septation in A. nidulans.
The smoA and smoB mutations confer hypersensitivity to benomyl:
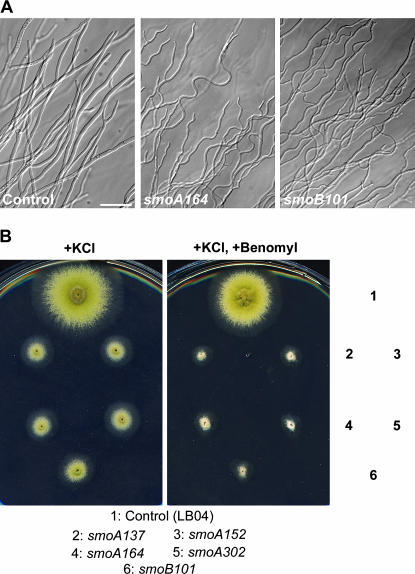
Because the smoA and smoB mutants showed reduced colony growth, we wondered whether hyphal growth in these mutants was also altered. As shown in Figure 5, smoA and smoB did not affect nuclear division in hyphae. Both smoA and smoB mutant hyphae, however, demonstrated wavy morphology (Figure 8A). Previous work indicates that the microtubule-depolymerizing agent benomyl inhibits hyphal growth and causes hyphae to appear in a highly wavy form (Riquelme et al. 2003). We reasoned whether the smoA and smoB mutations also impaired microtubules in the hyphae and consequently caused the distortion of hyphal morphology. To test this hypothesis, we grew the mutants in the presence of 0.4 μg/ml benomyl, a concentration that would still allow control strains to form conidiating colonies (Figure 8B). While colonies of the smoA and smoB mutants, although small, conidiated robustly, smoA and smoB showed no sign of conidiation on the medium containing benomyl (Figure 8B). The phenotype of growth inhibition and abolishment of conidiation by benomyl was linked to the suppression of loss of function of mobA (data not shown). Thus, smoA and smoB genes may encode proteins that play roles in microtubule stabilization.
Figure 8.
smoA and smoB mutations confer hypersensitivity to benomyl. (A) Compared to control (GR5) hyphae, smoA164 and smoB101 hyphae demonstrated a wavy appearance. (B) Growth phenotypes of smoA and smoB mutants on rich medium plus KCl and on rich medium plus KCl and benomyl. Compared to the control strain, smoA and smoB mutants showed a significantly retarded growth with smaller colonies that failed to conidiate. Bar, 50 μm.
It was intriguing to see whether simply destabilizing microtubules would suppress the loss of the SIN. We were able to destabilize microtubules by the application of low doses of benomyl or by introducing mutations in the ebA gene encoding a microtubule plus end-binding protein. Our results indicated that simply destabilizing microtubules did not restore septation and conidiation in the absence of the SIN (R. Shao and B. Liu, unpublished data).
DISCUSSION
In this study, we report the characterization of the SIN proteins of MOBA and SIDB in A. nidulans. Our results demonstrated that in this fungus, although the SIN was required for septation and conidiation during vegetative growth and asexual reproduction, it was not required for continuous mycelial growth and colony formation. We have also taken advantage of this system to isolate extragenic smo suppressor mutations that restored septation and conidiation in the absence of the SIN. The SMO proteins are predicted to antagonize the SIN for septation and conidiation in A. nidulans.
SIN and septation in A. nidulans:
The results presented here revealed that functions of both MOBA and SIDB for septation are conserved in A. nidulans as are their counterparts in S. pombe. Earlier work by Bruno et al. (2001) also has proved that the Cdc7p ortholog SEPH is essential for septation in A. nidulans. On the basis of the high degree of sequence identity, the A. nidulans genome also contains genes encoding homologous proteins of other SIN components in S. pombe, except the Sid4p protein (J.-M.. Kim and B. Liu, unpublished results). Sid4p, together with Cdc11p, acts as a scaffolding factor for anchoring the SIN components to the spindle pole body (Krapp et al. 2001; Tomlin et al. 2002). In S. pombe, these two stable spindle pole body components also organize regulators of SIN, such as the polo kinase Plo1p and the spindle checkpoint protein Dma1p, to the spindle pole body (Guertin et al. 2002,b; Morrell et al. 2004). It is believed that they assemble the SIN and its regulators to a close proximity so that robust signal transduction activities can take place to coordinate cell division with nuclear division (Morrell et al. 2004). It was surprising for us to realize the absence of an A. nidulans protein showing a high degree of primary sequence homology with S. pombe Sid4p. Sid4p contains an N-terminal spindle-pole-body-targeting domain followed by coiled-coil domains (Chang and Gould 2000). Its coiled coils are capable of self-dimerization (Chang and Gould 2000). It is believed that Sid4p is linked to the SIN through Cdc11p as Cdc11p directly interacts with SIN components like Sid2p (Morrell et al. 2004). In A. nidulans, other protein(s), such as the spindle-pole-body-associated SNAD protein (Liu and Morris 2000), may serve as a scaffolding factor for anchoring the SIN molecules.
As reported in S. pombe, sepH, mobA, and sidB genes are essential for septation in A. nidulans. Surprisingly, none of them is essential for hyphal growth and colony formation. This is different from the results in S. pombe as SIN genes are essential genes (Gould and Simanis 1997). Our results indicated that mycelia with aseptate hyphae formed normal-size colonies (Figure 2B). In organisms with uninucleate cells, failure of cytokinesis may cause the formation of multinucleate cells or activate signals that prevent nuclei from entering new rounds of mitosis, which triggers cells to undergo cell death. Many filamentous fungi like A. nidulans, however, form mycelia of multinucleate cells. In these organisms, special mechanisms that allow cells to tolerate the failure of septation may have evolved. While fungi of uninucleate cells see the failure of septation as a death signal, filamentous fungi may sense such a failure as a tolerable mistake.
It has been reported that in fission yeast the SIN is activated by the polo kinase Plo1p, which is required for septation (Ohkura et al. 1995; Tanaka et al. 2001). Moreover, overexpression of Plo1p induces the formation of multiple septa without mitosis (Ohkura et al. 1995). Recently, the A. nidulans ortholog of polo kinase, PLKA, has been identified (Bachewich et al. 2005). PLKA is an essential protein localized to the spindle pole body. Surprisingly, overexpression of PLKA inhibits septation in A. nidulans, suggesting that its roles do not totally overlap with those of polo kinases from fission yeast and other organisms (Bachewich et al. 2005). It will be interesting to isolate a conditional mutation at the plka locus and test how the SIN and septation would respond to the loss of plka function.
Proteins antagonizing components of the SIN:
Septation initiation in fission yeast is triggered by the small G-protein Spg1p when it is in a GTP-bound form (Schmidt et al. 1997). Two-component GTPase-activating protein of Byr4p and Cdc16p negatively regulates the septation initiation process by acting on Spg1p directly (McCollum and Gould 2001). The spindle checkpoint protein Dma1p acts as an inhibitor of the SIN, and it does so by preventing Plo1p from localizing to the spindle pole body (Guertin et al. 2002,b).
Genetic screens have revealed several other proteins that antagonize the SIN for septation in S. pombe. Protein phosphatases Par1p and Par2p all act negatively on septation as deletion of par1 and par2 genes leads to a phenotype with multiple septa formed between daughter cells (Jiang and Hallberg 2000; Tanabe et al. 2001). The par1 gene also was isolated as an extragenic suppressor of loss-of-function mutation in the spg1 gene (Le Goff et al. 2001). As a matter of fact, deletion of par1 and par2 also suppresses mutations in cdc11 and cdc7 genes, but not in other downstream SIN genes (Jiang and Hallberg 2001; Le Goff et al. 2001). Thus, Par1p and Par2p negatively regulate activities of Spg1p and Cdc7p for septation in fission yeast. Another suppressor gene of defects in SIN is scw1, which encodes a putative RNA-binding protein (Karagiannis et al. 2002). Null mutations in the scw1 gene suppress temperature-sensitive mutations, including a mob1 mutation, but not null mutations, of genes encoding the SIN components (Karagiannis et al. 2002; Jin and McCollum 2003). It is not clear how this RNA-binding protein, which localizes diffusely in the cytoplasm, affects septation by antagonizing the SIN. Nevertheless, the scw1 deletion mutation acts as a better suppressor of mutations of SIN genes than par1 and par2 deletions in S. pombe. But none of these mutations seem to bypass the SIN pathway. The AN7700.2 locus in the A. nidulans genome (accession no. XP 680969) encodes a 628-aa polypeptide with 28% sequence identity with the fission yeast Scw1p (J.-M. Kim and B. Liu, unpublished data). The genomic fragment of the A. nidulans homolog of scw1, including 2 kb each of its 5′ and 3′ flanking sequences, was amplified and used to transform the smoA and smoB mutants reported here. The DNA fragment did not complement the mutations. Thus, we concluded that smoA and smoB genes were not scw1 homologs. It would be interesting to test whether a loss-of-function mutation in the A. nidulans scw1 gene also could suppress mutations in the SIN pathway.
Unlike the above-mentioned fission yeast suppressor mutations, the smo mutations reported here suppress null mutations in three tested SIN genes in A. nidulans. The hypersensitivity of smoA and smoB mutants to benomyl provides another line of evidence that these genes do not function similarly to scw1. The scw1 deletion mutations increase the tolerance of yeast cells to a microtubule-depolymerizing drug (Jin and McCollum 2003). Thus, the smoA, smoB, and smoC–E genes encode factors that probably act in an antagonizing pathway against the SIN pathway. A better understanding of the mechanism of suppression awaits the cloning of these smo genes. While SIDB activates its substrates by phosphorylation to trigger septation, the smo genes may encode negative regulators of these substrates to prevent septation from taking place precociously. Phosphorylation of the substrates may lead to the dissociation of the substrates from inhibitors like SMO proteins. Inactivation of SMO proteins would release the inhibitory effect of SMO on these substrates. Consequently, septation could take place without the influence of the SIN. One can anticipate that the substrates could be proteins directly or indirectly involved in actomyosin ring formation preceding septum assembly. While the SIN activates the ring formation, SMO proteins act to prevent septation from taking place prior to receiving signals from the SIN.
Microtubules and septation in A. nidulans:
Although microtubules are not directly involved in the septum assembly in fungi, they play multifaceted regulatory roles in septation. Data obtained in fission yeast show that the γ-tubulin complex provides a link between the metaphase checkpoint and septation, probably by providing an inhibitory role in SIN activation (Vardy et al. 2002). More recently, studies in fission yeast demonstrate that cytoplasmic microtubules play a role in the positioning of the actomyosin medial ring during septation (Pardo and Nurse 2003). In A. nidulans, depolymerization of microtubules abolished the formation of the medial ring, and the later contraction of the ring requires intact microtubules (Momany and Hamer 1997; Kaminskyj 2000). Our own work indicates that in A. nidulans the microtubule minus end-directed motor cytoplasmic dynein plays a role in the determination of the septum position, but not the timing (Liu and Morris 2000; Liu et al. 2003).
In this report, we show that GFP–MOBA is transiently associated with the mitotic spindle (Figure 3). In fission yeast, full-length and truncated forms of the Sid2p protein decorate cytoplasmic and spindle microtubules (Hou et al. 2004). The significance of the microtubule association is not clear. One possibility is that the translocation of the SIDB/Sid2p–MOBA/Mob1p complex is carried out by an active transport mechanism instead of by passive diffusion. Thus, microtubules may act as tracks for the active transport of the complex from the spindle pole body to the septation site.
The connection between microtubules and septation also can be reflected by the phenotypes of the scw1 mutant in fission yeast and the smoA and smoB mutants reported here. The scw1 deletion mutation increases the resistance to the microtubule-depolymerizing agent methyl-2-benzimidazolecarbamate (Jin and McCollum 2003). smoA and smoB mutations, however, caused hypersensitivity to benomyl. Our preliminary results also indicated that simply destabilizing microtubules by pharmacological and genetic means could not suppress the loss-of-function mobA mutation. Thus, how microtubules cross-talk with signals regulating septation remains illusive.
Missing links between the SIN and the actomyosin ring:
One of the goals of the current genetic screen was to uncover the players that might link the SIN with the actomyosin ring contraction. While the SIN itself has been shown as one of the most elegant signaling cascades, its downstream target(s) remain elusive. Recently, a screen for an ethanol-dependent septation mutant has recovered a novel gene named etd1 in S. pombe (Daga et al. 2005). Upon activation by the SIN, the Etd1p translocates from the plasma membrane at cell tips to the cortical region at the septation area, and it plays a role in the constriction of the actomyosin ring. Thus, it is an ideal candidate linker between the SIN and the actomyosin ring. However, Etd1p seems to be a fission yeast-specific protein. Therefore, if it acts as a substrate of the SIN, it must be a unique substrate found only in S. pombe.
Because the SIN pathway seems to be evolutionarily conserved, its phosphorylation target(s) should demonstrate a certain degree of conservancy. Unraveling such universal substrates awaits further investigations.
Acknowledgments
We thank Kenneth Bruno and Chris Staiger for providing the sepH strains. The corrections and comments by two anonymous reviewers are greatly appreciated. This work was supported by a grant from the National Science Foundation (grant no. MCB-0235364).
References
- Adams, T. H., J. K. Wieser and J. H. Yu, 1998. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62: 35–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachewich, C., K. Masker and S. Osmani, 2005. The polo-like kinase PLKA is required for initiation and progression through mitosis in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 55: 572–587. [DOI] [PubMed] [Google Scholar]
- Bardin, A. J., and A. Amon, 2001. MEN and SIN: What's the difference? Nat. Rev. Mol. Cell Biol. 2: 815–826. [DOI] [PubMed] [Google Scholar]
- Bruno, K. S., J. L. Morrell, J. E. Hamer and C. J. Staiger, 2001. SEPH, a Cdc7p orthologue from Aspergillus nidulans, functions upstream of actin ring formation during cytokinesis. Mol. Microbiol. 42: 3–12. [DOI] [PubMed] [Google Scholar]
- Chang, L., and K. L. Gould, 2000. Sid4p is required to localize components of the septation initiation pathway to the spindle pole body in fission yeast. Proc. Natl. Acad. Sci. USA 97: 5249–5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daga, R. R., A. Lahoz, M. J. Munoz, S. Moreno and J. Jimenez, 2005. Etd1p is a novel protein that links the SIN cascade with cytokinesis. EMBO J. 24: 2436–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould, K. L., and V. Simanis, 1997. The control of septum formation in fission yeast. Genes Dev. 11: 2939–2951. [DOI] [PubMed] [Google Scholar]
- Guertin, D. A., S. Trautmann and D. McCollum, 2002. a Cytokinesis in eukaryotes. Microbiol. Mol. Biol. Rev. 66: 155–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin, D. A., S. Venkatram, K. L. Gould and D. McCollum, 2002. b Dma1 prevents mitotic exit and cytokinesis by inhibiting the septation initiation network (SIN). Dev. Cell 3: 779–790. [DOI] [PubMed] [Google Scholar]
- Harris, S. D., 1997. The duplication cycle in Aspergillus nidulans. Fungal Genet. Biol. 22: 1–12. [DOI] [PubMed] [Google Scholar]
- Harris, S. D., 2001. Septum formation in Aspergillus nidulans. Curr. Opin. Microbiol. 4: 736–739. [DOI] [PubMed] [Google Scholar]
- Harris, S. D., and J. E. Hamer, 1995. SepB: an Aspergillus nidulans gene involved in chromosome segregation and the initiation of cytokinesis. EMBO J. 14: 5244–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, S. D., J. L. Morrell and J. E. Hamer, 1994. Identification and characterization of Aspergillus nidulans mutants defective in cytokinesis. Genetics 136: 517–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, S. D., L. Hamer, K. E. Sharpless and J. E. Hamer, 1997. The Aspergillus nidulans sepA gene encodes an FH1/2 protein involved in cytokinesis and the maintenance of cellular polarity. EMBO J. 16: 3474–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, M. C., J. Salek and D. McCollum, 2000. Mob1p interacts with the Sid2p kinase and is required for cytokinesis in fission yeast. Curr. Biol. 10: 619–622. [DOI] [PubMed] [Google Scholar]
- Hou, M. C., D. A. Guertin and D. McCollum, 2004. Initiation of cytokinesis is controlled through multiple modes of regulation of the Sid2p-Mob1p kinase complex. Mol. Cell. Biol. 24: 3262–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W., and R. L. Hallberg, 2000. Isolation and characterization of par1(+) and par2(+): two Schizosaccharomyces pombe genes encoding B′ subunits of protein phosphatase 2A. Genetics 154: 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W., and R. L. Hallberg, 2001. Correct regulation of the septation initiation network in Schizosaccharomyces pombe requires the activities of par1 and par2. Genetics 158: 1413–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Q.-W., and D. McCollum, 2003. Scw1p antagonizes the septation initiation network to regulate septum formation and cell separation in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 2: 510–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafer, E., 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19: 33–131. [DOI] [PubMed] [Google Scholar]
- Kaminskyj, S. G. W., 2000. Septum position is marked at the tip of Aspergillus nidulans hyphae. Fungal Genet. Biol. 31: 105–113. [DOI] [PubMed] [Google Scholar]
- Karagiannis, J., R. Oulton and P. G. Young, 2002. The Scw1 RNA-binding domain protein regulates septation and cell-wall structure in fission yeast. Genetics 162: 45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp, A., S. Schmidt, E. Cano and V. Simanis, 2001. S. pombe cdc11p, together with sid4p, provides an anchor for septation initiation network proteins on the spindle pole body. Curr. Biol. 11: 1559–1568. [DOI] [PubMed] [Google Scholar]
- Krapp, A., M. P. Gulli and V. Simanis, 2004. SIN and the art of splitting the fission yeast cell. Curr. Biol. 14: R722–R730. [DOI] [PubMed] [Google Scholar]
- Le Goff, X., S. Buvelot, E. Salimova, F. Guerry, S. Schmidt et al., 2001. The protein phosphatase 2A B′-regulatory subunit par1p is implicated in regulation of the S. pombe septation initiation network. FEBS Lett. 508: 136–142. [DOI] [PubMed] [Google Scholar]
- Liu, B., and N. R. Morris, 2000. A spindle pole body-associated protein, SNAD, affects septation and conidiation in Aspergillus nidulans. Mol. Gen. Genet. 263: 375–387. [DOI] [PubMed] [Google Scholar]
- Liu, B., X. Xiang and Y. R. J. Lee, 2003. The requirement of the LC8 dynein light chain for nuclear migration and septum positioning is temperature dependent in Aspergillus nidulans. Mol. Microbiol. 47: 291–301. [DOI] [PubMed] [Google Scholar]
- McCollum, D., and K. L. Gould, 2001. Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell. Biol. 11: 89–95. [DOI] [PubMed] [Google Scholar]
- Momany, M., and J. E. Hamer, 1997. Relationship of actin, microtubules, and crosswall synthesis during septation in Aspergillus nidulans. Cell Motil. Cytoskeleton 38: 373–384. [DOI] [PubMed] [Google Scholar]
- Morrell, J. L., G. C. Tomlin, S. Rajagopalan, S. Venkatram, A. S. Feoktistova et al., 2004. Sid4p-Cdc11p assembles the septation initiation network and its regulators at the S. pombe SPB. Curr. Biol. 14: 579–584. [DOI] [PubMed] [Google Scholar]
- Oakley, B. R., C. E. Oakley, Y. Yoon and M. K. Jung, 1990. Gamma-tubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans. Cell Cycle 61: 1289–1301. [DOI] [PubMed] [Google Scholar]
- Ohkura, H., I. M. Hagan and D. M. Glover, 1995. The conserved Schizosaccharomyces pombe kinase plo1, required to form a bipolar spindle, the actin ring, and septum, can drive septum formation in G1 and G2 cells. Genes Dev. 9: 1059–1073. [DOI] [PubMed] [Google Scholar]
- Osmani, S. A., G. S. May and N. R. Morris, 1987. Regulation of the messenger RNA levels of nimA. A gene required for the G2-M transition In Aspergillus nidulans. J. Cell Biol. 104: 1495–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani, S. A., R. T. Pu and N. R. Morris, 1988. Mitotic induction and maintenance by overexpression of a G2-specific gene that encodes a potential protein kinase. Cell 53: 237–244. [DOI] [PubMed] [Google Scholar]
- Pardo, M., and P. Nurse, 2003. Equatorial retention of the contractile actin ring by microtubutes during cytokinesis. Science 300: 1569–1574. [DOI] [PubMed] [Google Scholar]
- Riquelme, M., R. Fischer and S. Bartnicki-Garcia, 2003. Apical growth and mitosis are independent processes in Aspergillus nidulans. Protoplasma 222: 211–215. [DOI] [PubMed] [Google Scholar]
- Salimova, E., M. Sohrmann, N. Fournier and V. Simanis, 2000. The S. pombe orthologue of the S. cerevisiae mob1 gene is essential and functions in signalling the onset of septum formation. J. Cell Sci. 113: 1695–1704. [DOI] [PubMed] [Google Scholar]
- Schmidt, S., M. Sohrmann, K. Hofmann, A. Woollard and V. Simanis, 1997. The Spg1p GTPase is an essential, dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev. 11: 1519–1534. [DOI] [PubMed] [Google Scholar]
- Tanabe, O., D. Hirata, H. Usui, Y. Nishito, T. Miyakawa et al., 2001. Fission yeast homologues of the B′ subunit of protein phosphatase 2A: multiple roles in mitotic cell division and functional interaction with calcineurin. Genes Cells 6: 455–473. [DOI] [PubMed] [Google Scholar]
- Tanaka, K., J. Petersen, F. Maclver, D. P. Mulvihill, D. M. Glover et al., 2001. The role of Plo1 kinase in mitotic commitment and septation in Schizosaccharomyces pombe. EMBO J. 20: 1259–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlin, G. C., J. L. Morrell and K. L. Gould, 2002. The spindle pole body protein Cdc11p links Sid4p to the fission yeast septation initiation network. Mol. Biol. Cell 13: 1203–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardy, L., A. Fujita and T. Toda, 2002. The γ-tubulin complex protein Alp4 provides a link between the metaphase checkpoint and cytokinesis in fission yeast. Genes Cells 7: 365–373. [DOI] [PubMed] [Google Scholar]
- Wolfe, B. A., and K. L. Gould, 2005. Split decisions: coordinating cytokinesis in yeast. Trends Cell. Biol. 15: 10–18. [DOI] [PubMed] [Google Scholar]
- Wolkow, T. D., S. D. Harris and J. E. Hamer, 1996. Cytokinesis in Aspergillus nidulans is controlled by cell size, nuclear positioning and mitosis. J. Cell Sci. 109: 2179–2188. [DOI] [PubMed] [Google Scholar]
- Wolkow, T. D., P. M. Mirabito, S. Venkatram and J. E. Hamer, 2000. Hypomorphic bimA(APC3) alleles cause errors in chromosome metabolism that activate the DNA damage checkpoint blocking cytokinesis in Aspergillus nidulans. Genetics 154: 167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]