Abstract
In the developing Drosophila eye, the morphogenetic furrow is a developmental organizing center for patterning and cell proliferation. The furrow acts both to limit eye size and to coordinate the number of cells to the number of facets. Here we report the molecular and functional characterization of Drosophila mini-me (mnm), a potential regulator of cell proliferation and survival in the developing eye. We first identified mnm as a dominant modifier of hedgehog loss-of-function in the developing eye. We report that mnm encodes a conserved protein with zinc knuckle and RING finger domains. We show that mnm is dispensable for patterning of the eye disc, but required in the eye for normal cell proliferation and survival. We also show that mnm null mutant cells exhibit altered cell cycle profiles and contain excess nucleic acid. Moreover, mnm overexpression can induce cells to proliferate and incorporate BrdU. Thus, our data implicate mnm as a regulator of mitotic progression during the proliferative phase of eye development, possibly through the control of nucleic acid metabolism.
CELL proliferation and growth in the developing Drosophila compound eye are regulated in two distinct phases, separated by the morphogenetic furrow (Thomas et al. 1994; Ready et al. 1976; Baker 2001). During embryogenesis, ∼20 cells are set aside to form the eye imaginal disc and grow by unpatterned proliferation. In the third instar, a wave of differentiation and patterning called the morphogenetic furrow passes across the eye field from posterior to anterior (Ready et al. 1976). In the furrow, cells are held in G1 arrest and a process of Delta/Notch-mediated lateral inhibition initiates pattern formation by specifying ommatidial founder cells (the future R8 photoreceptors, Baker 2001; Frankfort and Mardon 2002). Posterior to the furrow, cells surrounding the R8 are recruited to specific fates by successive rounds of Ras pathway signaling, modulated by further Notch-mediated signals (Nagaraj and Banerjee 2004; Voas and Rebay 2004).
The first five ommatidial cells remain in G1 arrest posterior to the furrow, but the surrounding cells reenter the cell cycle for one more round of cell division, the “second mitotic wave.” The remaining 15 ommatidial cells are derived from the daughters of this division (Ready et al. 1976; Tomlinson 1988; Baker 2001). Later, in pupal life, excess cells are removed by programmed cell death (Cagan and Ready 1989; Wolff and Ready 1991; Baker 2001) and the end result is a precisely constructed eye with 20 cells per facet. The regulation of cell cycle progression in the second mitotic wave has been shown to depend on Egfr, Notch, Hedgehog, and Decapentaplegic signaling, acting through Cyclins A and E, as well as on RBF, E2F, and Dacapo (de Nooij et al. 1996, 2000; Baker and Yu 2001; Duman-Scheel et al. 2002; Tseng and Hariharan 2002; Baonza and Freeman 2005; Firth and Baker 2005).
Thus, in normal development the different ommatidial cell types are derived from two different proliferative generations. However, this generational difference is not required. When the second mitotic wave is abolished (by the ectopic expression of a cyclin kinase inhibitor), all the retinal cells are derived from cell divisions that occur anterior to the furrow (de Nooij and Hariharan 1995). Under these circumstances, the eye lacks sufficient cells and some terminal fates are left unfilled; yet, most cells differentiate normally.
The morphogenetic furrow acts both to limit eye size (by ending the first mitotic wave) and to coordinate the number of cells to the number of facets (the second mitotic wave). The key regulator of the furrow is Hedgehog, which is expressed posterior to the furrow and activates downstream genes anterior to the furrow, via the regulation of Smoothened and Patched (Heberlein and Moses 1995; Lum and Beachy 2004). In addition, Hedgehog induces Decapentaplegic expression in the furrow (Heberlein and Moses 1995). Decapentaplegic is thought to act redundantly with Hedgehog anterior to the furrow (Greenwood and Struhl 1999; Fu and Baker 2003) and independently of Hedgehog at the margins of the disc (Pignoni and Zipursky 1997). Some genetic regulators of the cell cycle differ in the first and second mitotic waves. For example, mosaic clones lacking the Hedgehog receptor Smoothened are as large as their wild-type twin spots anterior to the furrow (Strutt and Mlodzik 1996) but do not synthesize DNA in the second mitotic wave (Duman-Scheel et al. 2002). In contrast, the size of Egfr clones (and other Ras pathway gene clones) is stunted on both sides of the furrow (Xu and Rubin 1993).
Hedgehog signaling has been implicated as a direct regulator of cell proliferation in the developing eye disc and the Hedgehog pathway element patched was recovered in a screen for genes that interact with RBF (Duman-Scheel et al. 2002). Hedgehog can regulate the transcription of Cyclins E and D, and ectopic Hedgehog signaling can activate Cyclin E reporter expression in the furrow (Duman-Scheel et al. 2002). Hedgehog also may act redundantly with Decapentaplegic to regulate G1 cell cycle arrest in the furrow (Penton et al. 1997; Horsfield et al. 1998; Duman-Scheel et al. 2002; Firth and Baker 2005). Cells lacking the Hedgehog pathway transcription factor gene cubitus interruptus (ci) arrest prematurely in G1 (Firth and Baker 2005). Furthermore, Ci overexpression in the furrow causes cells normally arrested in G1 to enter S-phase and incorporate Bromo-deoxy Uridine (BrdU) (Duman-Scheel et al. 2002). Also, cells that are doubly mutant for Hedgehog and Decapentaplegic pathway signaling show the strongest effects on G1 arrest by retaining Cyclin B expression and BrdU incorporation (Firth and Baker 2005).
Here we report the identification and genetic, molecular, and phenotypic characterization of a Drosophila gene, mini-me (mnm). We first identified an allele of mnm as a dominant modifier of hedgehog loss-of-function. We find that mnm encodes a conserved protein and that mnm transcription is regulated by Hedgehog signaling, on both sides of the furrow. mnm is required for normal cell proliferation and for cell survival anterior to the furrow, but not posterior to it. Mnm appears to function in the regulation of cell nucleic acid metabolism. Thus mnm may provide new insight into the control of cell proliferation and survival in the developing eye disc.
MATERIALS AND METHODS
Drosophila stocks, mutagenesis screen, and germline excision:
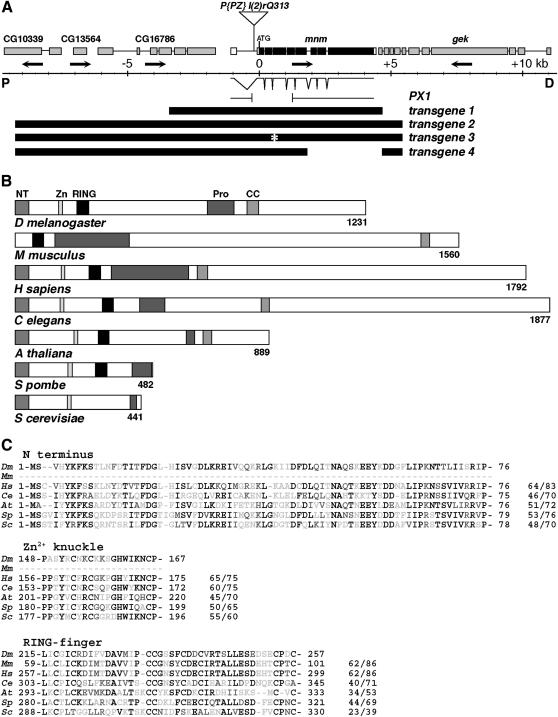
Wild-type stocks were w1118 and ry506. For the screen, autosomally isogenic w1118; cn1; es P((w, ry)D)3 hhbar3 males were treated with 25 mm EMS and crossed to autosomally isogenic w1118; hh8/TM6B females. Mutations were recovered from the F1 male progeny. For the excision screen, p(ry+(t7.2) = Delta2–3)99B (Laski and Rubin 1989) was used to mobilize the p(PZ) element in l(2)rQ313. Five hundred thirty rosy− lines were tested by genomic DNA gel blot (probe, transgene 2, Figure 2A). mnmPX1 deletes 1488 bp rightward from l(2)rQ313, removing the translation start site and three conserved protein domains (N-terminal, Zn knuckle, and RING). Three precise excision revertant alleles restored viability, confirming that the lethality associated with the l(2)rQ313 chromosome was solely due to the P-element insertion in mnm.
Figure 2.
Molecular-genetic characterization of the mnm locus. (A) Map of the mnm region, in polytene region 60B8–11: proximal (“P”) is left; distal (“D”) is right; center is a genomic scale, marked in kilobases. Orientations of transcription units are indicated by arrows: CG10339, CG13564 and CG16786 (deduced from genomic sequence), mnm (CG3231), and genghis khan (gek). The position of the insertion of P(PZ)l(2)rQ313 is indicated. Exon/intron structure of the mnm cDNA (clone LD21643) is shown. The structure of the 1488-bp deletion in mnmPX1 is indicated. Below this are the structures of four transgenes, as described in the text. (B) A comparison between the deduced Drosophila Mini-me protein domain structure and the closest homologs in other species: M. musculus (mP2P-R), H. sapiens (hRBBP6), C. elegans, A. thaliana, S. pombe, and S. cerevisiae (Mpe1). Conserved domains: NT, N-terminal domain; Zn, Zinc knuckle; RING, RING finger, Pro, proline rich; and CC, coiled coil. (C) Amino acid sequence alignments of three of the conserved domains, as indicated. Species are as in B. Residue numbers are indicated, and the percentages of identity/similarity to Mnm are given. Solid residues are identical, residues with dark shading are conservative changes, and residues with light shading are unconserved changes.
Mutant/transgenic stocks:
w1118; hhts2/TM6B
hh:GAL4/TM6B and hh:GAL4 UAS:GFP/TM6B (gifts from T. Tabata)
en:GAL4 (gift from Ruth Palmer)
From the Bloomington Stock Center:
l(2)rQ313rQ313/CyO
w1118; P(ry+=neoFRT)42D P(w+mC=Ubi:GFP.nls)2R1 P(Ubi:GFP.nls)2R2
w1118; P(ry+=neoFRT)42D
w1118; P(ry+=neoFRT)42D P(w+,ry+)47A
w1118/GMR:p35
sp/CyO; UAS-P35/TM6B
ey:FLP (Newsome et al. 2000)
hs:FLP (Xu and Rubin 1993)
Phage library screen, DNA constructs, and transgenic lines:
Library: 17–23 kb, Sau3a partially digested, genomic DNA from the autosomally isogenic screen parent line was inserted into the XhoI site of λFIX (Stratagene, La Jolla, CA).
Probe: PCR fragment flanking the l(2)rQ313 site [Roche (Indianapolis) High Prime DNA-labeling kit]. Thirty-nine unique phage isolates and transgenic constructs were confirmed by restriction mapping and end sequencing. Germline transformations were performed as previously described (Rubin and Spradling 1982).
Transgene 1: 8023-bp XbaI genomic fragment (2308 bp left of mnm transcript to 331 bp after the 3′ end) in pCaSpeR-4 (Thummel and Pirotta 1992).
Transgene 2: 14,701-bp EagI genomic fragment in the NotI site of pCaSpeR-3 (Thummel and Pirotta 1992).
Transgene 3: From transgene 2 by Acc65I digest and then Klenow fill-in to disrupt the splice acceptor site at the start of exon 4 for a frameshift at residue 135.
Transgene 4: From transgene 2 by SpeI digest and religation to produce a 2888-bp deletion, eliminating mnm exons 7–9, and terminating the protein after residue 491. Tests of rescue were for adult viability of all homozygous and trans-heterozygous combinations of mnm1, mnmP, and mnmPX1.
mnm overexpression construct: cDNA LD21643 (3937 bp; Research Genetics, Birmingham, AL) between the NotI and XhoI of pUAST (Brand and Perrimon 1993). Germline transformations were performed as described above. mnm overexpression was driven by en:GAL4 and flies were raised at 18°, 25°, or 29°. Ten transgenic lines that were obtained exhibited similar phenotypes.
Gel blots:
Poly(A)+ RNA from w1118 embryos, larvae, and adults was analyzed by gel blot (Sambrook et al. 1989). The probe was 32P-labeled cDNA LD21643 (Roche High Prime DNA-labeling kit).
RT–PCR:
Single mnm heterozygote or mutant embryos were identified using GFP balancer chromosomes and confirmed by PCR. RNA was isolated from single embryos [QIAGEN (Valencia, CA) RNeasy kit]. The RNA preparation contained contaminating genomic DNA, which was included as a loading control. The RT–PCR reactions were performed according to the QIAGEN One-Step RT–PCR protocol. RT–PCR products were resolved by agarose gel electrophoresis. The primers used to detect the mnm transcript were primers that amplified a portion of exons 8 and 9. mnm primer sequences were 5′-GCTGCTTTGTGATGCTTCCG-3′ and 5′-CAACTCCAGGGATAATCTCAAGGAC-3′.
Microscopy, in situ hybridization, and immunohistochemistry:
Scanning electron microscopy was performed as previously described (Tio and Moses 1997). The statistical analyses of ommatidium numbers were by paired Student's t-tests. Facet counts were (a) hh8/+, n = 3, mean = 674.67, SD = 32.52; (b) hhbar3, n = 4, mean = 228.25, SD = 27.68; (c) hh8/hhbar3, n = 6, mean = 316.67, SD = 8.59; (d) mnm1/+;hh8/hhbar3, n = 6, mean = 241.33, SD = 20.19; (e) mnmP/+;hh8/hhbar3, n = 9, mean = 274.33, SD = 21.44; and (f) mnmPX1/+;hh8/hhbar3, n = 4, mean = 195.25, SD = 13.60. Adult eye sections were prepared as previously described (Tomlinson 1985). Whole-mount in situ hybridizations were performed as previously described (Wolff 2000). The probes for the in situ hybridization were single-stranded digoxygenin (DIG)-labeled DNA by PCR from cDNA LD21643 and glass cDNA 5A6 (Roche PCR DIG probe synthesis kit; Moses et al. 1989). Eye disc immunohistochemistry was performed as previously described (Kumar et al. 1998). BrdU was performed as described (Tapon et al. 2001). F-actin was detected with Rhodamine-phalloidin [1:50; Molecular Probes (Eugene, OR) A-12380]. DNA was stained with Hoechst 33342 for fluorescence-activated cell sorting (FACS) (1:500; Sigma, St. Louis). The primary antisera used were rabbit anti-Ato (1:1000; Jarman et al. 1993), mouse anti-BrdU (1:100; BD Biosciences 33281A), mouse anti-Cyclin E (1:5, gift of B. Edgar; Richardson et al. 1995), rabbit activated Caspase-3 (1:200; BD Biosciences 551150; Srinivasan et al. 1998), rat anti-Elav (1:500, 7E8A10 from Developmental Studies Hybridoma Bank (DSHB); O'Neill et al. 1994), mouse anti-BarH1 (1:10, gift of K. Saigu; Higashijima et al. 1992), mouse anti-Cut (1:10, mAb 2B10 from DSHB; Blochinger et al. 1990), guinea-pig anti-Senseless (1:1000, gift of G. Mardon; Frankfort et al. 2001), mouse anti-Pros (1:100, mAb MR1A DSHB; Campbell et al. 1994), mouse anti-Boss (1:1000, gift from S. L. Zipursky; Cagan et al. 1992), mouse anti-Cyclin A (1:10, A12 from DSHB, a gift of I. Hariharan; Knoblich and Lehner 1993), mouse anti-Cyclin B (1:50 F2F4 from DSHB, gift of I. Hariharan; Knoblich and Lehner 1993), mouse anti-Cyclin D [1:10, gift of K. Moberg (unpublished data)], rabbit anti-phospho histone H3 (1:1000, Cell Signaling Technologies 9701), rat anti-Ci155 (1:1, 2A1, gift of R. Holmgren; see Motzny and Holmgren 1995), rabbit anti-Hedgehog (1:625, gift of I. Guererro), rabbit anti-pMad (1:500, gift of T. Tabata; Persson et al. 1998), mouse anti-Notch intracellular domain (1:200, from DSHB, gift of K. Moberg), guinea pig anti-Eyg (1:200, gift of K. Moberg) and rabbit anti-Lamin (1:1000, gift of D. Kiehardt). The secondary antibodies were from Jackson ImmunoResearch (West Grove, PA) and were goat anti-mouse Cy5 (1:500, 115-175-003), goat anti-rabbit TRITC (1:250, 111-025-003), goat anti-rabbit HRP (1:100, 111-035-003), goat anti-mouse HRP (1:40, 115-035-003), and goat anti-rat TRITC (1:200, 112-025-003).
Mosaic clones and flow cytometry:
mnmP and mnmPX1 clones were generated using ey:FLP (Newsome et al. 2000) or hs:FLP (Xu and Rubin 1993). For heat-shock experiments, clones were induced 24, 48, 72, or 96 hr before dissection, by one incubation at 37° for 1 hr. Discs were dissected from wandering third instar larvae. Wing discs were obtained 24 hr after heat shock and flow cytometry was performed as previously described (Tapon et al. 2001). The following genotypes were derived for mosaics and/or flow cytometry:
w1118/y1 w1118 ey:FLP; P(ry+=neoFRT)42D P(w+,ry+)47A/P(ry+=neoFRT)42D
w1118/y1 w1118 ey:FLP; P(ry+=neoFRT)42D P(w+,ry+)47A/P(ry+=neoFRT)42D mnmP
w1118/y1 w1118 ey:FLP; P(ry+=neoFRT)42D P(w+,ry+)47A/P(ry+=neoFRT)42D mnmPX1
w1118/w1118 hs:FLP; P(ry+=neoFRT)42D P(w+mC=Ubi:GFP.nls)2R1 P(Ubi:GFP.nls)2R2/P(ry+=neoFRT)42D
w1118/w1118 hs:FLP; P(ry+=neoFRT)42D P(w+mC=Ubi:GFP.nls)2R1 P(Ubi:GFP.nls)2R2/P(ry+=neoFRT)42D mnmPX1
w1118/y1 w1118 ey:FLP; P(ry+=neoFRT)42D P(w+mC=Ubi:GFP.nls)2R1 P(Ubi:GFP.nls)2R2/P(ry+=neoFRT)42D
w1118/y1 w1118 ey:FLP; P(ry+=neoFRT)42D P(w+mC=Ubi:GFP.nls)2R1 P(Ubi:GFP.nls)2R2/P(ry+=neoFRT)42D mnmPX1
w1118/y1 w1118 ey:FLP; P(ry+=neoFRT)42D P(w+mC=Ubi:GFP.nls)2R1 P(Ubi:GFP.nls)2R2/P(ry+=neoFRT)42D mnmPX1; P(w+mC=mnm transgene)
w1118 GMR:P35/y1 w1118 ey:FLP; P(ry+=neoFRT)42D P(w+mC=Ubi:GFP.nls)2R1 P(Ubi:GFP.nls)2R2/P(ry+=neoFRT)42D mnmPX1
w1118/w1118 hs:FLP; P(ry+=neoFRT)42D P(w+mC=Ubi:GFP.nls)2R1 P(Ubi:GFP.nls)2R2/FRT42D mnmPX1;hh:GAL4/UAS:P35.
NCBI accession numbers:
Drosophila melanogaster (Mnm): AAD34765.1
Homo sapiens (RBBP6): NP_008841.2
Mus musculus (PACT/P2P-R): AAC72432.1
Caenorhabditis elegans: T21861
Arabidopsis thaliana: NP_199554.1
Schizosaccharomyces pombe: NP_596522.1
Saccharomyces cerevisiae (Mpe1): NP_012864.1
RESULTS
mini-me is a dominant genetic enhancer of hedgehog in the developing eye:
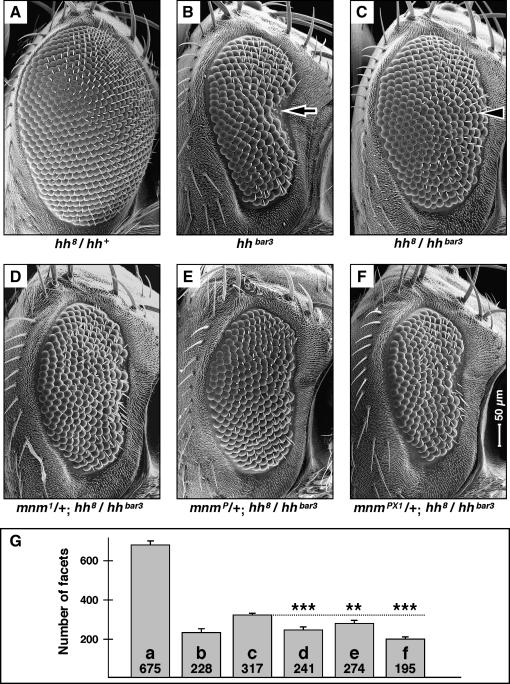
We undertook a genetic screen to discover genes that interact with hedgehog (hh) in the developing Drosophila eye, using a viable heteroallelic genotype. hh8 (also known as hh13C) is a homozygous lethal allele (Mohler 1988; Porter et al. 1995) and hh8 heterozygous eyes are phenotypically indistinguishable from wild type (Figure 1A). hhbar3 is a homozygous viable allele (Ives 1950; Mohler 1988) and has a strong recessive eye phenotype with an indented anterior side (arrow in Figure 1B and Lee et al. 1992). The hh8/hhbar3 heterozygote has an intermediate eye phenotype, with no anterior indentation (arrowhead in Figure 1C). We used the hh8/hhbar3 heterozygote as the basis for the genetic screen.
Figure 1.
mnm loss-of-function is a dominant enhancer of hedgehog loss-of-function in the compound eye. (A–F) Scanning electron micrographs of adult female compound eyes: dorsal, up; anterior, right and to the same scale (indicated in F). (A) hh8/hh+, phenotypically indistinguishable from wild type; (B) hhbar3/hhbar3, has a rough and reduced eye; (C) hh8/hhbar3, has a slightly rough and reduced eye; (D) mnm1/+; hh8/hhbar3; (E) mnmP/+; hh8/hhbar3; (F) mnmPX1/+; hh8/hhbar3. Note that the hedgehog eye phenotype is enhanced by mnm loss-of-function. (G) Quantification of facet counts; also see materials and methods. The number in each bar is the mean number of facets and the letter identifies which genotype and section (as above) it refers to. Error bars are standard deviation. ***P < 0.005; **P < 0.01.
We treated isogenized hhbar3 homozygous males with the chemical mutagen EMS and crossed them to isogenized hh8 balanced females. We screened ∼10,000 F1 males for modified eye phenotypes and recovered 62 mutations in 49 autosomal loci (by noncomplementation for lethality). Mutations were recovered in nine known genes with effects on embryonic development: Egfr, even skipped, gooseberry, huckebein, odd paired, patched, smoothened, thickveins, and tramtrack (Nüsslein-Volhard and Wieschaus 1980). These included members of the hedgehog pathway, the receptor component genes patched and smoothened, (Lum and Beachy 2004) as well as genes known to act in pathways also associated with eye development, the decapentaplegic and Egfr pathways (Heberlein and Moses 1995; Freeman 1997; Voas and Rebay 2004). In addition, we recovered mutations in 40 loci that we could not identify by complementation testing to known mutations. This article is focused on one of these: En(hh)2A. For reasons explained below, we named this EMS-induced allele mini-me1 (mnm1, Figure 1D).
Molecular characterization of mnm:
mnm1 fails to complement the lethality of l(2)rQ313rQ313, which is a P(PZ) insertion recovered in a screen for P-induced lethals (Spradling et al. 1999). The P-element lies in the first intron of a cytogene (CG3231, Figure 2A), immediately to the left of genghis khan (gek) (Luo et al. 1997). We renamed l(2)rQ313rQ313 mnmP. We obtained an embryonic 3937-bp cDNA from Research Genetics (LD21643). There are nine exons containing an open reading frame that encodes a 1231-residue protein (Figure 2, A and B).
We found that mnm1 is a late pupal lethal, while mnmP dies in stage 16 of embryogenesis. We suspected that one or both alleles might not be a null and there was no large deletion available for the region, so we excised the P element to generate mnmPX1, which removes the start of the open reading frame and three of the most conserved protein domains (Figure 2A). mnmPX1 is an embryonic lethal and we take it to be a null. mnmP and mnmPX1 are also dominant enhancers of the hh8/hhbar3 eye phenotype (Figure 1, E and F) and this enhancement is statistically significant (Figure 1G).
The phenotypic effects of all three mnm lesions (the EMS, P, and excision deletion) could be through a cis effect on one of the flanking genes. We were able to exclude gek by complementation, but could not likewise eliminate CG16786, the gene to the left (as no mutations are known for it). We attempted to rescue mnm function using an 8-kb genomic fragment (transgene 1, Figure 2A), as this includes both ends of the cDNA, but it does not rescue mnm lethality. A longer 14.7-kb genomic fragment (transgene 2, Figure 2A) does rescue the lethality associated with the mnmP and mnmPX1 chromosomes, but not mnm1. It may be that the mnm1 chromosome contains a second lethal lesion, but we could not rescue any heteroallelic combination containing mnm1. RT–PCR analyses of mnm embryos suggest that mnm1 may be a hypermorphic or neomorphic allele (Figure 3B, see below). Transgene 2 also rescued the eye development defects of mnmP and mnmPX1 (see below). On the basis of the argument that mnmPX1 is a single-gene null, we suggest that transgene 2 contains all the genomic sequences required for mnm genetic function.
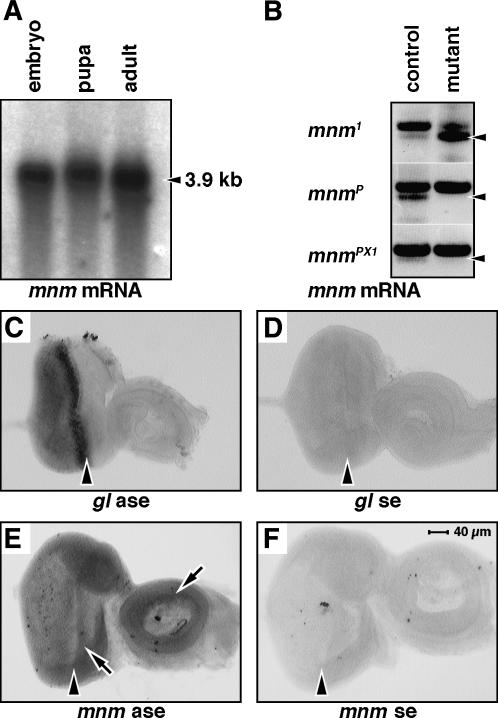
Figure 3.
mnm expression. (A) RNA gel blot, stages as indicated. mnm mRNA is indicated by an arrowhead. (B) RT gel. RNA from single embryos was isolated and analyzed by RT–PCR using gene-specific mnm primers. The arrowheads mark the predicted RT product in all sections. The top band in all sections is the predicted product from genomic DNA remaining in the reaction. Genotypes are mnm1/CyO and mnm1 (top), mnmP/CyO and mnmP (middle), and mnmPX1/CyO and mnmPX1 (bottom). There was no detectable transcript in mnmP or mnmPX1 homozygote embryos compared to the heterozygote controls (middle and bottom sections). The mnm transcript appeared to be overexpressed in the mnm1 homozygote embryos compared to controls (top), suggesting that mnm1 may be a hypermorphic or neomorphic allele. (C–F) RNA in situ hybridization experiments: (C) glass antisense, (D) glass sense, (E) mnm antisense, and (F) mnm sense. Third instar eye-imaginal discs: anterior right, to the same scale indicated in F. The morphogenetic furrow is indicated by an arrowhead. Note the elevated expression of mnm mRNA in the eye field and in parts of the antenna (arrows in E).
However, transgene 2 includes the entire predicted coding sequences of both CG3231 and CG16786 and thus does not eliminate either gene as a candidate for mnm. So we tested two derivatives of transgene 2 that selectively knocked out CG3231 function: transgene 3 includes an engineered 4-base mutation (a predicted frameshift in CG3231, Figure 2A), and transgene 4 includes a deletion that terminates CG3231 after amino acid 491 (Figure 2A). While transgene 2 rescues the lethality of the mnm null mutation, transgenes 3 and 4 do not. Thus, CG3231 is required for mnm function and we henceforth refer to CG3231 as mnm (Figure 2A).
The deduced Mnm protein contains several conserved domains (Figure 2, B and C). The first 76 amino acids form a previously uncharacterized but conserved N-terminal domain (NT). The closest human homolog, retinoblastoma binding protein 6 (RBBP6), contains the NT domain (Figure 2C and Sakai et al. 1995). The closest murine homolog is PACT/P2P-R (Simons et al. 1997; Witte and Scott 1997) and reported partial cDNAs do not encode the NT domain, but this domain is encoded in the genomic sequence (although we have not shown it in Figure 2, B and C). Following the NT domain is a conserved “zinc-knuckle” domain, thought to be involved in nucleic acid binding (Summers 1991). A “RING finger” domain often associated with Ubiquitin ligases is conserved in the human and mouse homologs (Figure 2, B and C; Freemont 2000). There are also protein-interaction motifs: proline-rich (Pro) and coiled-coil (CC) domains (Mason and Arndt 2004).
S. cerevisiae has a genetically characterized homolog of mnm called Mpe1 (Vo et al. 2001), shown to function in polyadenylation. Uncharacterized mnm homologs are present in the published genome sequences of C. elegans, A. thaliana, and S. pombe. All of the homologs, except ScMpe1, share all the conserved domains (Figure 2B).
Expression of mnm:
To characterize the temporal expression of mnm, we prepared poly(A)+ RNA from embryos, larvae, and adults and probed a gel blot, using the mnm cDNA as the probe. There is a single major transcript at ∼3.9 kb that persists throughout development, confirming the predicted size of the transcript (Figure 3A). In very long exposures, we can also detect an uncharacterized 1.8-kb minor transcript (not shown).
RNA was isolated from single mnm heterozygote or homozygote mutant embryos (genotyped using a GFP balancer chromosome) and used as templates for RT–PCR. RT–PCR products were resolved by agarose gel electrophoresis (Figure 3B). While mnm-specific primers were able to detect the predicted product in an mnmP heterozygote embryo (Figure 3B, arrowhead, middle, left lane), no transcript was detected in mnmP homozygote embryos (Figure 3B, middle, right lane). There was also no detectable mnm transcript in mnmPX1 homozygote embryos compared to the heterozygote control (Figure 3B; bottom, left and right lanes). The mnm transcript appeared to be overexpressed in the mnm1 homozygote embryos compared to controls, suggesting that mnm1 may be a hypermorphic or neomorphic allele (Figure 3B; top, left and right lanes).
As a positive control, we used RNA in situ hybridization to visualize the expression of a known gene in the developing eye (glass, Figure 3C; Moses et al. 1989). We find that mnm mRNA is expressed across the entire eye field as well as a ring in the antennal disc (arrows, Figure 3E); this is clearly above the background level (sense strand control, Figure 3F). The level of mnm mRNA appears slightly elevated anterior to the furrow.
We attempted to generate specific antibodies to the Mnm protein by two approaches. Neither rabbit polyclonal antisera raised against two Mnm peptides nor antisera against Mnm–GST fusion proteins showed any specificity for the Mnm protein by mosaic clonal analyses (not shown). We conclude that the mRNA in situ hybridization experiments reveal the true expression pattern of mnm mRNA, because we have controlled for nonspecific expression through the sense strand control.
mnm mRNA expression is regulated by hedgehog signaling:
While we originally identified mnm as a dominant enhancer of hedgehog, the genetic and regulatory relationships between the two genes are not at all clear. As hedgehog functions to induce many events in the furrow and influences the activities of several other signaling pathways (including Decapentaplegic, Notch, and Egfr) any regulatory relationship may be indirect.
To characterize the relationship between mnm and hedgehog, we used a conditional, temperature-sensitive allele of hedgehog, hhts2 (Ma et al. 1993) to remove hedgehog function. In hhts2/+ (phenotypically wild-type) discs, no signal was detected with a mnm sense strand control (Figure 4A) and normal signal was seen in hhts2/+ discs with the mnm antisense probe (Figure 4B). However, in hhts2/hhts2 homozygous siblings taken from the same vial after 4 hr at 29° (the nonpermissive temperature) the mnm signal was absent (Figure 4C). Thus the level of mnm transcript is sensitive to hedgehog function. While loss-of-hedgehog function clearly does affect mnm expression, the effect is global (the entire eye and antennal discs) and not limited to the territories in which Hedgehog signals are known to be received (e.g., anterior to the furrow). This may argue for an indirect mechanism for hedgehog regulation of mnm through the activation of other pathways (see below).
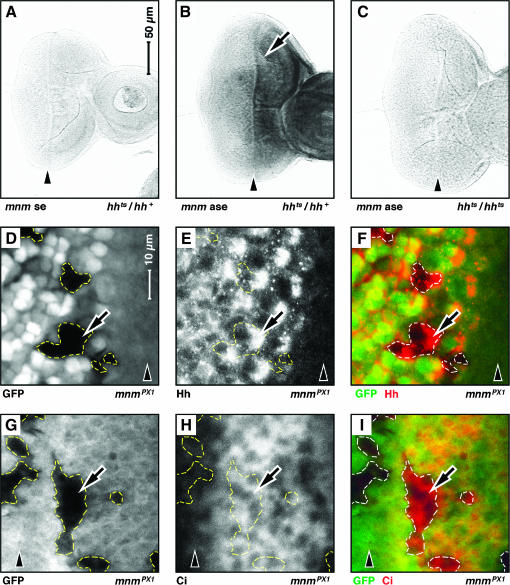
Figure 4.
mnm is genetically downstream of hedgehog signaling. Third instar eye-imaginal discs: anterior, right. A–C and D–I are to the same scale; see bars in A and D. Arrowheads indicate the position of the morphogenetic furrow. (A–C) RNA in situ hybridization experiments: (A) hhts2/+, mnm sense (se) strand control; (B) hhts2 heterozygote, mnm anti-sense (ase) strand [note elevated level of mnm mRNA anterior to the morphogenetic furrow (arrow)]; (C) hhts2/hhts2, mnm anti-sense strand (mnm signal is lost in hhts2/hhts2 animals raised at the nonpermissive temperature). (D–I) Mosaic clones of mnmPX1 cells. Clones are negatively marked with GFP and are outlined (white in D and G and green in F and I). E (in white) and F (in red) show the expression of Hedgehog antigen. Note that Hedgehog is not lost from mnmPX1 mutant cells (arrow). H (in white) and I (in red) show the expression of activated Ci antigen. Note that activated Ci is not lost from mnmPX1 mutant cells (arrow).
It was possible that mnm acts upstream of Hedgehog protein expression or downstream of Hedgehog activation. To test these possibilities, we induced GFP-negatively marked, mnmPX1 null homozygous clones in the developing eye, which have normal levels and localization of Hedgehog protein (arrows in Figure 4, D–F). Similarly, we stained retinal mnmPX1 clones for the Hedgehog signaling-activated transcription factor, Cubitus interruptus (Ci) but see no changes (arrows in Figure 4, G and H). Taken together these data strongly suggest that mnm is not genetically upstream but rather may be downstream of Hedgehog signaling and is controlled at the transcriptional level.
mnm is required for cell proliferation and survival in the developing eye:
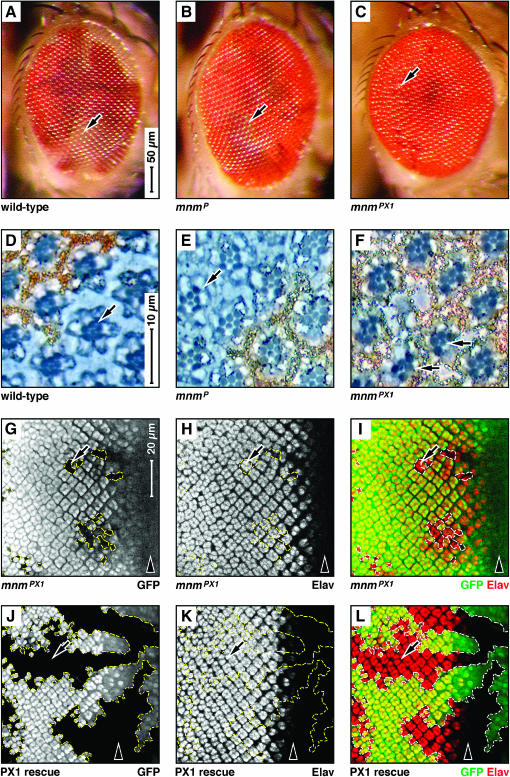
We used ey:FLP (Newsome et al. 2000) to induce mnm homozygous mutant clones, marked by white−, in the developing eye. These were negatively marked in the adult by white+. Wild-type control clones were large and occupied roughly half of the eye (arrow in Figure 5A), as did clones homozygous for mnmP (arrow in Figure 5B). However, clones for the deletion allele mnmPX1 were largely absent in the adult retina, leaving only scars (arrow in Figure 5C). In retinal sections, wild-type, mnmP, and mnmPX1 clones all contained white− photoreceptor cells (arrows in Figure 5, D–F), although there were very few in mnmPX1 clones (Figure 5F). These data suggest that mnm null cells either do not proliferate or die in the developing eye. However, the detection of some persistent mnm null cells suggests that mnm null cells may have differential requirements for survival.
Figure 5.
mnm loss-of-function affects eye development. (A–C) Scanning electron micrographs of female adult compound eyes containing ey:Flp-induced mosaic clones marked by white: dorsal, up; anterior, right; scale indicated in A. Genotypes: (A) wild type, (B) mnmP, and (C) mnmPX1. Note that the weak allele mnmP clones are as large as wild type (arrows in A and B) but that the null allele mnmPX1 leaves scars (arrow in C). (D–F) Sections of adult compound eyes containing eyeless:Flp-induced mosaic clones marked by white: dorsal, up; anterior, right; scale indicated in D. Genotypes: (D) wild type, (E) mnmP, and (F) mnmPX1. Note that the wild-type and mnm clones contain mutant (white−) photoreceptor cells (arrows in D–F). (G–L) Third instar eye-imaginal discs containing eyeless:Flp-induced mnmPX1 mosaic clones (outlined), negatively marked by GFP; anterior, right and to the same scale (bar in G). Arrowheads indicate the position of the morphogenetic furrow. (G and J) GFP in white; (H and K) Elav antigen in white; (I and L) GFP in green and Elav in red. Note that mnmPX1 clones are small, but express the differentiation marker Elav (arrow in G–I). (J–L) mnm rescuing transgene (see text). Note that the size of the mnmPX1 clones is entirely rescued (arrows).
To investigate whether mnm null cells can differentiate normally, we derived ey:FLP-induced mnmPX1 homozygous mutant clones that were negatively marked by GFP. In late larval discs we observed small mnmPX1 clones that were positive for the neural-specific protein Elav (arrows in Figure 5, G–I), suggesting that there is a strong effect of the mnm null mutation on cell number, but perhaps no effect on differentiation (see below). We also found that transgene 2 (see above) can fully rescue this cell number defect (arrows in Figure 5, J–L).
mnm null cells are underrepresented and die in proliferative tissue, but have no detectable defects in nonproliferative tissue:
The small size of the ey:FLP-induced mnm null clones in larval discs could be due to defects in cell proliferation, growth, or survival. To distinguish between these possibilities, we used hs:FLP to induce clones and then examined their progeny, in the retina, at a series of times after induction.
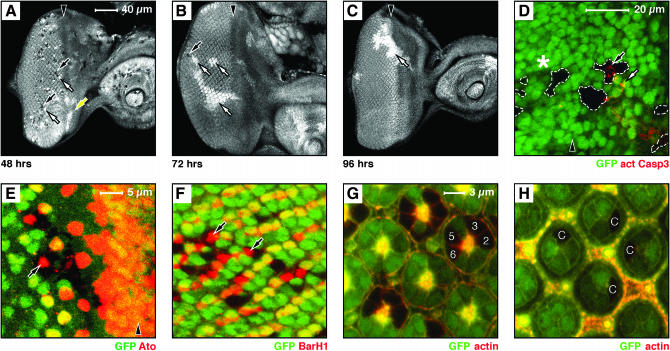
mnm null clones induced 24 hr before dissection are frequent, small, and similar in size (about two to four cells) to their adjacent wild-type twin spots (not shown). By 48 hr after induction, mnm null clones and wild-type twin spots are seen posterior to the furrow (postmitotic territory, Figure 6A: mutant, black arrows; twin spot, white arrow), but in the region anterior to the furrow, the twin spots grow larger while the mutant clones are lost (a proliferative territory, yellow arrow in Figure 6A). A day later (72 hr after induction) the mnm null clones are rare, small, and found only posterior to the furrow (postmitotic territory, black arrow in Figure 6B). The twin spots are large (white arrows in Figure 6B) and often not associated with a null clone. By 96 hr, only large twin spots are seen (white arrow in Figure 6C) and mnm null clones are not found. These data suggest that the defects seen in the hs:FLP clones take ∼48 hr to begin to develop; thus, mnm clones can survive one or two cycles of cell division, perhaps through perdurance of the Mnm protein. It should be noted that 72 hr before dissection, all the cells were in a proliferative territory (the furrow had not initiated yet), so the lack of remaining posterior null clones may be due to their deaths between 48 and 72 hr after induction.
Figure 6.
mnm is required for the survival of proliferating cells in the developing eye. (A–H) Eye-imaginal discs containing mnmPX1 mosaic clones, negatively marked by GFP; anterior, right. (A–F) Late third instar eye discs; (G–H) 48-hr pupal eye discs. A–C are to the same scale (bar in A), D is at higher magnification (see bar), E and F are to the same scale (bar in E), and G and H are to the same scale (bar in G). Arrowheads indicate the position of the morphogenetic furrow in A–E. (A–C) hs:FLP-induced clones, with the induction time before dissection indicated below: (A) 48 hr, (B) 72 hr, and (C) 96 hr. (A–C) GFP is shown as white. Note that 48 hr after induction (A) many small mnmPX1 homozygous clones are seen (black clones and black arrows) together with their homozygous wild-type twin spots (white twin spots and white arrow) posterior to the furrow. Note that the twin spots immediately anterior to the furrow lack clones (white twin spots and yellow arrow). At 72 hr (B) only very rare and small mutant clones are seen (black arrow) and the twin spots are far larger than the clones (white arrows). By 96 hr (C) only twin spots are seen (white arrow). (D) Activated Caspase3 antigen within (black arrow) or near (white arrow) mnmPX1 clones, anterior to the furrow. Note that there is no activated Caspase3 staining associated with mnm clones posterior to the furrow (asterisk). (E) Atonal antigen (red) and (F) BarH1 antigen (red) expressed within mnmPX1 clones (arrows). G and H show that mnmPX1 mutant cells can persist into pupal life and can differentiate normally [numbers indicate examples of R cell types (G) and cone cells (H)].
If the primary effect of mnm loss-of-function is to direct cells to apoptotic death, we might expect to observe a marker of cell death in the clones. We used an activated Caspase3 stain to detect apoptotic cell death and found staining close to, but not always contained within, mnm clones that lie anterior to the furrow (proliferative territory, arrows in Figure 6D). Dying cells delaminate from epithelia in the developing wing (Gibson and Perrimon 2005; Shen and Dahmann 2005), so we suggest that this stain is associated with both dying cells and debris remaining from dead cells that have been extruded. We never see such stain posterior to the furrow (asterisk in Figure 6D), suggesting that mnm apoptotic death is limited to the anterior, proliferative domain.
In addition, if the primary effect of mnm loss-of-function is to direct cells to apoptotic death, then inhibiting apoptosis might rescue the size of the clones. Thus we induced mnm clones in the eye disc in the presence of the baculovirus P35 protein (a potent inhibitor of cell death), driven by the GMR enhancer posterior to the furrow in the developing eye, but the size of mnm mutant clones was unchanged (data not shown and see Hay et al. 1994). These data are consistent with our observations that the apoptotic marker, activated Caspase3, is associated only with mnm clones that are anterior to the furrow (Figure 6D). However, this P35 experiment does not test for the effects of cell death anterior to the furrow (because GMR:P35 is expressed only on the posterior side).
Similarly, mnm mutant cells in the developing wing disc are small and rarely detectable beyond 48 hr of clone induction, as in the eye (not shown). We never observe activated Caspase3 in these wing clones, nor is their size rescued by the local expression of P35 (expressed in the posterior compartment using hh:Gal4, data not shown). In this wing experiment, we express P35 in the same territory in which the clones develop their size defect. Thus we suggest that the primary defect that produces the small clone size is not apoptotic cell death.
If mnm clones are small due to competition effects, then conferring a growth advantage on mnm mutant clones might overcome the small clone phenotype. We derived eye imaginal discs with mnm clones surrounded by Minute heterozygous cells. However, we did not recover any mnm mutant tissue surviving beyond 48 hr (not shown).
If mnm were to affect cell growth, then we might expect to find that mnm cells grow more slowly than wild-type cells. Because we could not inhibit cell death in mnm cells by overexpressing P35 (GMR∷P35 or hh∷P35, see above), we could not accurately determine the growth rate of mnm cells. We examined the sizes of persisting mnm null cells to determine if mnm loss-of-function had any effect on cell size. mnm null cells do not appear any smaller than their neighbors (Figure 6, G and H). Furthermore, we counted cells in mutant clones (by marking their nuclei through anti-Lamin D staining) and find that the number of cells per unit area is also not affected (not shown). It is possible that mnm cells can compensate for a slower growth rate by slowing their cell cycle time, and thus we are not able to use cell size as a measure of cell growth. However, we cannot conclude that mnm has any role in regulating cell growth.
Taken together these data suggest that it is unlikely that the mnm clones posterior to the furrow are small due to local apoptosis or competitive effects. Rather, we propose that mnm is required for cell survival in the proliferating cells anterior to the furrow in the eye and more generally in the wing (see below).
Surviving mnm null cells posterior to the furrow can differentiate and persist into later life:
We stained for several cell-type markers in surviving induced mnm homozygous clones and these appear normal (Atonal for R8 photoreceptors in Figure 6E; BarH1 for R1 and R6 in Figure 6F; and Boss, Sevenless, Prospero, and Cut, data not shown). Moreover, mnm null cells are found as morphologically normal photoreceptors (Figure 6G) and accessory cells (Figure 6H) 48 hr after puparium formation. These data are consistent with our observations that mnm cells are present in the adult eye and stain positively for neural-specific Elav in ey:FLP-induced clones in the eye disc (Figure 5, G–I). Thus, we conclude that mnm does not control cell-type specification or differentiation.
mnm affects nucleic acid content:
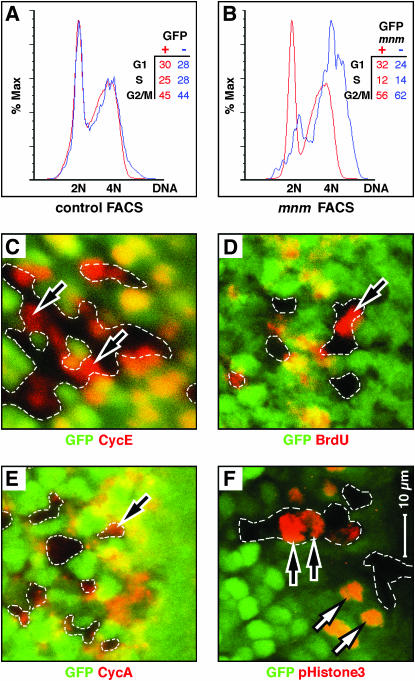
Our data are consistent with a function for mnm in proliferative cells in the eye and wing, with a secondary effect on survival. To examine the cell cycle profiles of mnm mutant cells, we used the developing wing disc as a rich source of proliferating cells. The use of wing rather than eye disc cells for FACS analysis also has the benefit of avoiding artifacts associated with the specific morphologies of the differentiating retinal cells posterior to the morphogenetic furrow. We used hs:FLP to induce negatively GFP-marked mnm null clones in the wing disc and 24 hr later (at least 24 hr before we observe small mnm clones in the wing disc), we dissected and dissociated wing imaginal discs for FACS, as described by Tapon et al. (2001). In control discs (in which both the GFP+ and GFP− cells are mnm+), we detect well-superimposed nucleic acid content profiles, which represent the G1, S, and G2/M phases of the cell cycle (Figure 7A). However, mnm null cells (blue curve, Figure 7B) are strongly shifted to the right, toward higher nucleic acid content compared to GFP+ mnm+ cells in the same discs. Moreover, mnm cells appear to accumulate DNA beyond 4N (blue curve, Figure 7B). Forward scatter profiles of mnm mutant cells are similar to those of wild-type cells (not shown), consistent with our observation that cell size is not affected in mnm mutant cells.
Figure 7.
mnm loss-of-function affects cell cycle progression. (A and B) Fluorescence-activated cell sorting (FACS) traces. Red curves are GFP+-expressing cells and blue are GFP− cells. DNA content (x-axes) is plotted against the percentage of maximal absorbance (y-axes). Insets show the percentages of cells in each genotype distributed between the phases G1, S, and G2/M. Cells were prepared from third larval wing imaginal discs containing hs:FLP-induced, GFP-marked mosaic clones, 24 hr after clone induction. (A) Control discs in which both the GFP+ and GFP− cell populations are mnm+. Note that the red and blue curves are superimposed with major peaks interpreted as 2N and 4N DNA content. (B) Cells prepared from imaginal discs in which the GFP+ cells are either mnm+ homozygotes or mnm+/mnmPX1. As mnm is recessive, all the GFP+ cells are phenotypically wild type. The GFP− cells are mnmPX1 homozygotes. Note the rightward shift of the GFP− mnm mutant cells, suggesting elevated DNA content, and an increased percentage of cells are scored as G2/M. (C–F) Third instar eye-imaginal discs, with fields shown posterior to the morphogenetic furrow, containing GFP-marked mnmPX1 homozygous clones. Anterior is to the right and C–F are to the same scale (bar in F). GFP is green and the antigens are in red: (C) Cyclin E (G1 phase), (D) BrdU (S phase), (E) Cyclin A (G2 phase), and (F) phosphorylated Histone H3 (pH3, M phase). Note that mnmPX1 mutant cells can express all four of these markers (black arrows), indicating that no cell cycle phase is absent.
We stained mnm null clones in the developing eye for cell cycle markers to test for a specific stage defect: Cyclin E for G1 (Figure 7C, as well as Cyclin D, data not shown), BrdU incorporation for S (Figure 7D), Cyclin A for G2 (Figure 7E, as well as Cyclin B, data not shown), and phosphorylated Histone H3 (pH3) for mitosis (Figure 7F). In all cases, we could find some cells expressing these markers in the clones and in roughly the normal frequencies.
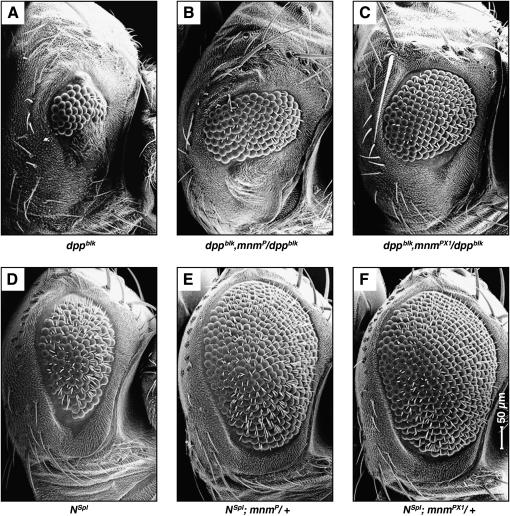
mnm is a dominant suppressor of dpp loss-of-function and Notch gain-of-function in the eye:
We examined whether mnm loss-of-function could genetically interact with mutations in other signaling pathways that caused adult eye phenotypes, by removing one copy of mnm using either the mnmP or mnmPX1 allele. While we observed no effect by either allele on the eye phenotypes of wggla/+, ellipseB1/+, roughD/+, or rolledSEM/+ flies (not shown), we did observe strong suppressive effects on a loss-of-function mutation in dpp, dppblk (Figure 8, A–C;Treisman and Rubin 1995), and a gain-of-function mutation in Notch, Notchspl (Nspl, Figure 8, D–F; Nagel and Preiss 1999), by both alleles. To determine whether Dpp or Notch signaling is affected in cells lacking mnm, we stained mnm null clones in the eye for pMad (target of Dpp signaling; Wiersdorff et al. 1996), the Notch intracellular domain, or eye gone (target of Notch signaling; Chao et al. 2004; Dominguez et al. 2004). However, we saw no effect in mnm null clones posterior or anterior to the furrow (not shown). This result was not surprising since we observe no effects on patterning in mnm null clones posterior to the furrow; and activated cell death in mnm null clones ahead of the furrow leads to cell loss. Altogether, our data suggest that mnm may be interacting with some component of these pathways in opposite ways, to antagonize Dpp signaling and enhance Notch signaling functions. Both the Dpp and the Notch pathways interact with Hh signaling during eye development (Curtiss and Mlodzik 2000; Fu and Baker 2003) and might explain the indirect regulation of mnm by hedgehog.
Figure 8.
mnm dominantly suppresses dpp loss-of-function and notch gain-of-function in the eye. (A–F) Scanning electron micrographs of adult compound eyes: dorsal, up; anterior, right and to the same scale (indicated in F). (A) dppblk, small, rough eye with very few facets; (B) dppblk, mnmP/dppblk, loss of one copy of mnmP can suppress the small eye size and the eye contains more facets; (C) dppblk, mnmPX1/dppblk, the mnmPX1 null allele strongly suppresses the small eye phenotype; (D) Notchspl (Nspl), reduced, rough eye with missing or double bristles; (E) Nspl; mnmP/+ and (F) Nspl; mnmPX1/+, both the mnmP and the mnmPX1 strongly suppress the small-eye phenotype, the null allele to a greater degree.
Excess mnm causes overproliferation and melanotic mass formation:
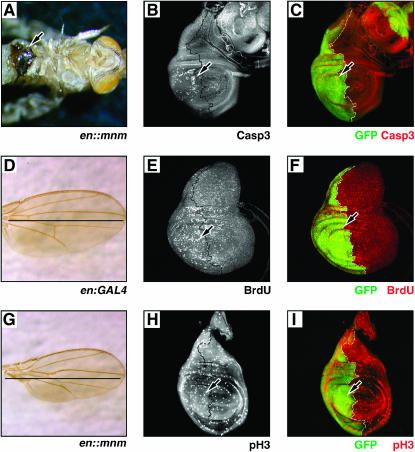
We derived transgenic flies that expressed the mnm cDNA under the control of the UAS-activating element. mnm overexpression was driven by en:GAL4 in the posterior parasegment compartments; flies were raised at 18°, 25°, or 29° and monitored throughout their lifetime. mnm overexpression led to reduced viability in all 10 lines at 18° and larval or pupal lethality at 25° or 29°. Six of the 10 lines exhibited small black melanotic masses in their larval epidermis compared to their sibling controls at 18°, similar to the metastatic masses seen by others (not shown; Pagliarini and Xu 2003). Adult escapers exhibited gross patterning defects in the engrailed-expressing dorsal abdomen (not shown) and also often contained melanotic masses in the ventral epidermis (arrowhead, Figure 9A). The wings of adult escapers were patterned normally, but were reduced in size in the posterior compartment compared to control (Figure 9, D and G).
Figure 9.
Excess mnm causes overproliferation and melanotic mass formation. (A) w1118; en:Gal4/UAS:mnm pharate adult raised at 18°, ventral side up. Legs have been removed. Arrow marks the melanotic mass in the ventral epidermis. (B, C, E, F, H, and I) en:Gal4/UAS:GFP, UAS:mnm wing discs obtained from third instar larvae raised at 25°. Arrows mark mnm-overexpressing cells in the posterior compartment, black and white dotted lines mark the A–P boundaries; anterior is right. (B and C) Caspase3 (white in B, red in C) and BrdU (white in E, red in F) staining are increased, while phosphorylated Histone H3 (pH3, white in H, red in I) is unaffected in the en∷mnm posterior compartments. (D) en:Gal4 control and (E) en∷mnm adult wings of flies raised at 18°. Black lines mark the A–P boundaries. Note that patterning is unaffected in en∷mnm wings but the size of the posterior compartment is reduced compared to the control wing.
One explanation for the small posterior wing phenotype of en∷mnm flies is increased cell death in the posterior compartment of the developing wing disc. We dissected wing discs from en∷mnm wandering larvae that were raised at 25° and stained for activated Caspase3. These larvae also contained UAS:GFP to help identify the mnm overexpressing domain (Figure 9, C, F, and I). Indeed, there was increased activated Caspase3 staining in the posterior compartment of the en∷mnm wing discs (Figure 9, B and C).
To determine if mnm overexpression could induce proliferation, we also stained en∷mnm wing discs for BrdU (Figure 9, E and F) and pH3 (Figure 9, H and I). While we observed no appreciable differences in pH3 staining, we did observe more BrdU-positive cells in the posterior compartment in these discs, suggesting that mnm overexpression is sufficient to induce cells to proliferate, and this overproliferation likely leads to the activation of programmed cell death.
DISCUSSION
In this study we report the identification of a potential regulator of cell proliferation and survival. The Drosophila mnm gene encodes a conserved protein with a novel N terminus and Zinc knuckle, RING finger, and proline-rich and coiled coil domains. mnm is expressed everywhere in the developing eye disc and is enriched ahead of the morphogenetic furrow. The expression of mnm is dependent upon Hedgehog signaling (perhaps indirectly), as loss of Hedgehog signaling through an inactivating mutation in hedgehog greatly reduces its expression.
From our timed analysis of mutant clones, it appears that mnm null cells in proliferative regions of the developing eye (and wing) can replicate for two or three times over 48 hr, but between 48 and 72 hr after clone induction they suffer some crisis and die. It may be that this delayed defect is due to perdurance of the Mnm protein. If during that time window they receive developmental signals to cease proliferation and differentiate, they can then survive. The morphogenetic furrow and subsequent events do provide such differentiation signals so that mnm null clones can persist in the retina, if they are induced late enough. Our data show that if the furrow passes over mnm null cells in the first 24–48 hr after they become homozygous, they can persist to the adult eye, and many differentiate as morphologically normal photoreceptors and accessory cells. Taken together, these data suggest that Mnm is required for some function in proliferative cells, but not in postmitotic cells. Because the mnm mutant clones posterior to the furrow can survive and differentiate as apparently perfect, yet tiny copies of their wild-type twin spots, we named the gene “mini-me” (Myers and McCullers 1999).
Our FACS analysis of mnm null cells suggests that mnm null cells have abnormal nucleic acid content. This could reflect changes in nuclear DNA, mitochondrial DNA, and/or RNA content. It could be that the mnm mutant cells have lost the correct coupling of DNA synthesis to cell division and accumulate DNA beyond 4N. It may be that these cells overreplicate DNA during S phase, missegregate DNA during mitosis, or fail to divide and become aneuploid. mnm overexpression is sufficient to induce proliferation; and this excessive proliferation is toxic and leads to cell death. An increase in nucleic acid content associated with mnm loss-of-function and the overproliferation of mnm-overexpressing cells are consistent with a role for mnm as a regulator of mitotic progression, although whether Mnm plays a role in DNA replication, the DNA damage checkpoints, or mitotic entry/exit is not clear.
The closest human and murine homologs of Drosophila Mnm are RBBP6 and PACT/P2P-R. These proteins have been shown to associate with Retinoblastoma (Rb) protein and p53 proteins in vitro, which are potent regulators of the cell cycle, including regulating entry into S phase and the monitoring of DNA integrity (Sakai et al. 1995; Simons et al. 1997). This could be consistent with our suggestion that loss of Mnm may lead to aberrant DNA metabolism. Furthermore P2P-R is downregulated in differentiating cells (Witte and Scott 1997), consistent with our observation of a lack of Mnm function in postmitotic territories in the developing eye. RNAi knockdown of P2P-R in mouse 3T3 cells affects nocodazole-induced arrest and UV-induced apoptosis, also possibly consistent with a disturbance in DNA metabolism (Gao et al. 2002; Scott and Gao 2002).
Hedgehog signaling has been implicated in cell cycle regulation in both flies and vertebrates (Forbes et al. 1996; Duman-Scheel et al. 2002; Roy and Ingham 2002). The link between hedgehog and mnm may be a new mechanism for this control. However, the interaction between hedgehog and mnm could be indirect: the small phenotype of mnm clones is quite dissimilar to that of smoothened clones, which are not small (lacking the Hedgehog receptor; Strutt and Mlodzik 1996). We also observe phenotypic effects of mnm loss-of-function outside of the territories where the Hedgehog signal is received. Thus we suggest that while mnm may be controlled in part by hedgehog, it has much more general functions and is likely, also, to be regulated by other pathways.
It is interesting that loss of mnm function strongly interacts genetically with the Dpp and Notch pathways in opposite ways. Both pathways have recently been characterized to have significant roles in regulating cell cycle progression in the developing eye. Dpp signaling promotes G1 arrest, while Notch signaling regulates S-phase entry in the second mitotic wave (Baonza and Freeman 2005; Firth and Baker 2005). It could be that mnm is interacting directly with these pathways to regulate cell cycle progression. However, the precise mechanism remains to be resolved.
Acknowledgments
We thank Philip Beachy for suggesting the hedgehog screen genotype; Ken Moberg, Maureen Powers, and the Moses lab for their helpful comments; Summer Cook for technical assistance; B. Edgar, I. Guererro, I. Hariharan, A. Jarman, D. Kiehardt, L. Luo, G. Mardon, K. Moberg, K. Saigu, T. Tabata, R. Holmgren, and S. L. Zipursky for their gifts of reagents; and C. Commisso and G. L. Boulianne for their biochemical help. This work was supported in the Moses lab by a grant from the National Eye Institute (EY09299), R. Reifegerste was supported in part by a Deutsche Forschungsgemeinschaft/German fellowship (RE 1089/1-1), and C. Jones was supported in part by training grants T32GM008367 and T32EY007092 and a supplement to National Eye Institute grant R01EY012537.
References
- Baker, N. E., 2001. Cell proliferation, survival, and death in the Drosophila eye. Semin. Cell. Dev. Biol. 12: 499–507. [DOI] [PubMed] [Google Scholar]
- Baker, N. E., and S. Y. Yu, 2001. The EGF receptor defines domains of cell cycle progression and survival to regulate cell number in the developing Drosophila eye. Cell 104: 699–708. [DOI] [PubMed] [Google Scholar]
- Baonza, A., and M. Freeman, 2005. Control of cell proliferation in the Drosophila eye by Notch signaling. Dev. Cell 8: 529–539. [DOI] [PubMed] [Google Scholar]
- Blochinger, K., R. Bodmer, L. Y. Jan and Y. N. Jan, 1990. Patterns of expression of Cut, a protein required for external sensory organ development in wild-type and cut mutant Drosophila embryos. Genes Dev. 4: 1322–1331. [DOI] [PubMed] [Google Scholar]
- Brand, A. H., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Cagan, R. L., and D. F. Ready, 1989. The emergence of order in the Drosophila pupal retina. Dev. Biol. 136: 346–362. [DOI] [PubMed] [Google Scholar]
- Cagan, R. L., H. Krämer, A. C. Hart and S. L. Zipursky, 1992. The Bride of Sevenless and Sevenless interaction: internalization of a transmembrane ligand. Cell 69: 393–399. [DOI] [PubMed] [Google Scholar]
- Campbell, G., H. Göring, T. Lin, E. Spana, S. Andersson et al., 1994. RK2, a glial-specific homeodomain protein required for embryonic nerve cord condensation and viability in Drosophila. Development 120: 2957–2966. [DOI] [PubMed] [Google Scholar]
- Chao, J. L., Y. C. Tsai, S. J. Chiu and Y. H. Sun, 2004. Localized Notch signal acts through eyg and upd to promote global growth in Drosophila eye. Development 131: 3839–3847. [DOI] [PubMed] [Google Scholar]
- Curtiss, J., and M. Mlodzik, 2000. Morphogenetic furrow initiation and progression during eye development in Drosophila: the roles of decapentaplegic, hedgehog and eyes absent. Development 127: 1325–1336. [DOI] [PubMed] [Google Scholar]
- de Nooij, J. C., and I. K. Hariharan, 1995. Uncoupling cell fate determination from patterned cell division in the Drosophila eye. Science 270: 983–985. [DOI] [PubMed] [Google Scholar]
- de Nooij, J. C., M. A. Letendre and I. K. Hariharan, 1996. A cyclin-dependent kinase inhibitor, Dacapo, is necessary for timely exit from the cell cycle during Drosophila embryogenesis. Cell 87: 1237–1247. [DOI] [PubMed] [Google Scholar]
- de Nooij, J. C., K. H. Graber and I. K. Hariharan, 2000. Expression of the cyclin-dependent kinase inhibitor Dacapo is regulated by cyclin E. Mech. Dev. 97: 73–83. [DOI] [PubMed] [Google Scholar]
- Dominguez, M., D. Ferres-Marco, F. J. Gutierrez-Avino, S. A. Speicher and M. Beneyto, 2004. Growth and specification of the eye are controlled independently by Eyegone and Eyeless in Drosophila melanogaster. Nat. Genet. 36: 31–39. [DOI] [PubMed] [Google Scholar]
- Duman-Scheel, M., L. Weng, S. Xin and W. Du, 2002. Hedgehog regulates cell growth and proliferation by inducing Cyclin D and Cyclin E. Nature 417: 299–304. [DOI] [PubMed] [Google Scholar]
- Firth, L. C., and N. E. Baker, 2005. Extracellular signals responsible for spatially regulated proliferation in the differentiating Drosophila eye. Dev. Cell 8: 541–551. [DOI] [PubMed] [Google Scholar]
- Forbes, A. J., H. Lin, P. W. Ingham and A. C. Spradling, 1996. hedgehog is required for the proliferation and specification of ovarian somatic cells prior to egg chamber formation in Drosophila. Development 122: 1125–1135. [DOI] [PubMed] [Google Scholar]
- Frankfort, B. J., and G. Mardon, 2002. R8 development in the Drosophila eye: a paradigm for neural selection and differentiation. Development 129: 1295–1306. [DOI] [PubMed] [Google Scholar]
- Frankfort, B. J., R. Nolo, Z. Zhang, H. Bellen and G. Mardon, 2001. senseless repression of rough is required for R8 photoreceptor differentiation in the developing Drosophila eye. Neuron 32: 403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman, M., 1997. Cell determination strategies in the Drosophila eye. Development 124: 261–270. [DOI] [PubMed] [Google Scholar]
- Freemont, P. S., 2000. RING for destruction? Curr. Biol. 10: R84–R87. [DOI] [PubMed] [Google Scholar]
- Fu, W., and N. E. Baker, 2003. Deciphering synergistic and redundant roles of Hedgehog, Decapentaplegic and Delta that drive the wave of differentiation in Drosophila eye development. Development 130: 5229–5239. [DOI] [PubMed] [Google Scholar]
- Gao, S., M. M. Witte and R. E. Scott, 2002. P2P-R protein localizes to the nucleolus of interphase cells and the periphery of chromosomes in mitotic cells which show maximum P2P-R immunoreactivity. J. Cell Physiol. 191: 145–154. [DOI] [PubMed] [Google Scholar]
- Gibson, M. C., and N. Perrimon, 2005. Extrusion and death of DPP/BMP-compromised epithelial cells in the developing Drosophila wing. Science 307: 1785–1789. [DOI] [PubMed] [Google Scholar]
- Greenwood, S., and G. Struhl, 1999. Progression of the morphogenetic furrow in the Drosophila eye: the roles of Hedgehog, Decapentaplegic and the Raf pathway. Development 126: 5795–5808. [DOI] [PubMed] [Google Scholar]
- Hay, B. A., T. Wolff and G. M. Rubin, 1994. Expression of baculovirus P35 prevents cell death in Drosophila. Development 120: 2121–2129. [DOI] [PubMed] [Google Scholar]
- Heberlein, U., and K. Moses, 1995. Mechanisms of Drosophila retinal morphogenesis: the virtues of being progressive. Cell 81: 987–990. [DOI] [PubMed] [Google Scholar]
- Higashijima, S.-I., T. Kojima, T. Michiue, S. Ishimaru, Y. Emori et al., 1992. Dual Bar homeo box genes of Drosophila required in two photoreceptor cells, R1 and R6, and primary pigment cells for normal eye development. Genes Dev. 6: 50–60. [DOI] [PubMed] [Google Scholar]
- Horsfield, J., A. Penton, J. Secombe, F. M. Hoffmann and H. Richardson, 1998. decapentaplegic is required for arrest in G1 phase during Drosophila eye development. Development 125: 5069–5078. [DOI] [PubMed] [Google Scholar]
- Ives, P., 1950. New mutants report: bar-3. Dros. Inf. Serv. 24: 58. [Google Scholar]
- Jarman, A. P., Y. Grau, L. Y. Jan and Y. N. Jan, 1993. atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell 73: 1307–1321. [DOI] [PubMed] [Google Scholar]
- Knoblich, J. A., and C. F. Lehner, 1993. Synergistic action of Drosophila cyclins A and B during the G2-M transition. EMBO J. 12: 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, J. P., M. Tio, F. Hsiung, S. Akopyan, L. Gabay et al., 1998. Dissecting the roles of the Drosophila EGF receptor in eye development and MAP kinase activation. Development 125: 3875–3885. [DOI] [PubMed] [Google Scholar]
- Laski, F. A., and G. M. Rubin, 1989. Analysis of the cis-acting requirements for germ-line-specific splicing of the P-element ORF2–ORF3 intron. Genes Dev. 3: 720–728. [DOI] [PubMed] [Google Scholar]
- Lee, J. J., D. P. von Kessler, S. Parks and P. A. Beachy, 1992. Secretion and localized transcription suggest a role in positional signaling for products of the segmentation gene hedgehog. Cell 71: 33–50. [DOI] [PubMed] [Google Scholar]
- Lum, L., and P. A. Beachy, 2004. The Hedgehog response network: sensors, switches, and routers. Science 304: 1755–1759. [DOI] [PubMed] [Google Scholar]
- Luo, L., T. Lee, L. Tsai, G. Tang, L. Y. Jan et al., 1997. Genghis Khan (Gek) as a putative effector for Drosophila Cdc42 and regulator of actin polymerization. Proc. Natl. Acad. Sci. USA 94: 12963–12968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, C., Y. Zhou, P. A. Beachy and K. Moses, 1993. The segment polarity gene hedgehog is required for progression of the morphogenetic furrow in the developing Drosophila eye. Cell 75: 927–938. [DOI] [PubMed] [Google Scholar]
- Mason, J. M., and K. M. Arndt, 2004. Coiled coil domains: stability, specificity, and biological implications. Chembiochem 5: 170–176. [DOI] [PubMed] [Google Scholar]
- Mohler, J., 1988. Requirements for hedgehog, a segmental polarity gene, in patterning larval and adult cuticle of Drosophila. Genetics 120: 1061–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses, K., M. C. Ellis and G. M. Rubin, 1989. The glass gene encodes a zinc-finger protein required by Drosophila photoreceptor cells. Nature 340: 531–536. [DOI] [PubMed] [Google Scholar]
- Motzny, C. K., and R. Holmgren, 1995. The Drosophila cubitus interruptus protein and its role in the wingless and hedgehog signal transduction pathways. Mech. Dev. 52: 137–150. [DOI] [PubMed] [Google Scholar]
- Myers, M., and M. McCullers, 1999. Austin Powers: The Spy Who Shagged Me. New Line Cinema, Los Angeles.
- Nagaraj, R., and U. Banerjee, 2004. The little R cell that could. Int. J. Dev. Biol. 48: 755–760. [DOI] [PubMed] [Google Scholar]
- Nagel, A. C., and A. Preiss, 1999. Notchspl is deficient for inductive processes in the eye, and E(spl)D enhances split by interfering with proneural activity. Dev. Biol. 208: 406–415. [DOI] [PubMed] [Google Scholar]
- Newsome, T. P., B. Asling and B. J. Dickson, 2000. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development 127: 851–860. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard, C., and E. Wieschaus, 1980. Mutations affecting segment number and polarity in Drosophila. Nature 287: 795–801. [DOI] [PubMed] [Google Scholar]
- O'Neill, E. M., I. Rebay, R. Tjian and G. M. Rubin, 1994. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78: 137–147. [DOI] [PubMed] [Google Scholar]
- Pagliarini, R. A., and T. Xu, 2003. A genetic screen in Drosophila for metastatic behavior. Science 302: 1227–1231. [DOI] [PubMed] [Google Scholar]
- Penton, A., S. B. Selleck and F. M. Hoffmann, 1997. Regulation of cell cycle synchronization by decapentaplegic during Drosophila eye development. Science 275: 203–206. [DOI] [PubMed] [Google Scholar]
- Persson, U., H. Izumi, S. Souchelnytskyi, S. Itoh, S. Grimsby et al., 1998. The L45 loop in type I receptors for TGF-beta family members is a critical determinant in specifying Smad isoform activation. FEBS Lett. 434: 83–87. [DOI] [PubMed] [Google Scholar]
- Pignoni, F., and S. L. Zipursky, 1997. Induction of Drosophila eye development by Decapentaplegic. Development 124: 271–278. [DOI] [PubMed] [Google Scholar]
- Porter, J. A., D. P. von Kessler, S. C. Ekker, K. E. Young, J. J. Lee et al., 1995. The product of Hedgehog autoproteolytic cleavage active in local and long-range signaling. Nature 374: 363–366. [DOI] [PubMed] [Google Scholar]
- Ready, D. F., T. E. Hanson and S. Benzer, 1976. Development of the Drosophila retina, a neurocrystalline lattice. Dev. Biol. 53: 217–240. [DOI] [PubMed] [Google Scholar]
- Richardson, H., L. V. O'Keefe, T. Marty and R. Saint, 1995. Ectopic cyclin E expression induces premature entry into S phase and disrupts pattern formation in the Drosophila eye imaginal disc. Development 121: 3371–3379. [DOI] [PubMed] [Google Scholar]
- Roy, S., and P. W. Ingham, 2002. Hedgehogs tryst with the cell cycle. J. Cell Sci. 115: 4393–4397. [DOI] [PubMed] [Google Scholar]
- Rubin, G. M., and A. C. Spradling, 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218: 348–353. [DOI] [PubMed] [Google Scholar]
- Sakai, Y., M. Saijo, K. Coelho, T. Kishino, N. Niikawa et al., 1995. cDNA sequence and chromosomal localization of a novel human protein, RBQ-1 (RBBP6), that binds to the retinoblastoma gene product. Genomics 30: 98–101. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Scott, R. E., and S. Gao, 2002. P2P-R deficiency modifies nocodazole-induced mitotic arrest and UV-induced apoptosis. Anticancer Res. 22: 3837–3842. [PubMed] [Google Scholar]
- Shen, J., and C. Dahmann, 2005. Extrusion of cells with inappropriate Dpp signaling from Drosophila wing disc epithelia. Science 307: 1789–1790. [DOI] [PubMed] [Google Scholar]
- Simons, A., C. Melamed-Bessudo, R. Wolkowicz, J. Sperling, R. Sperling et al., 1997. PACT: cloning and characterization of a cellular p53 binding protein that interacts with Rb. Oncogene 14: 145–155. [DOI] [PubMed] [Google Scholar]
- Spradling, A. C., D. Stern, A. Beaton, E. J. Rhem, T. Laverty et al., 1999. The Berkeley Drosophila Genome Project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153: 135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan, A., K. A. Roth, R. O. Sayers, K. S. Shindler, A. M. Wong et al., 1998. In situ immunodetection of activated caspase-3 in apoptotic neurons in the developing nervous system. Cell Death Differ. 5: 1004–1016. [DOI] [PubMed] [Google Scholar]
- Strutt, D. I., and M. Mlodzik, 1996. The regulation of hedgehog and decapentaplegic during Drosophila eye imaginal disc development. Mech. Dev. 58: 39–50. [DOI] [PubMed] [Google Scholar]
- Summers, M. F., 1991. Zinc finger motif for single-stranded nucleic acids? Investigations by nuclear magnetic resonance. J. Cell Biochem. 45: 41–48. [DOI] [PubMed] [Google Scholar]
- Tapon, N., N. Ito, B. J. Dickson, J. E. Treisman and I. K. Hariharan, 2001. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell 105: 345–355. [DOI] [PubMed] [Google Scholar]
- Thomas, B. J., D. A. Gunning, J. Cho and S. L. Zipursky, 1994. Cell cycle progression in the developing Drosophila eye: roughex encodes a novel protein required for the establishment of G1. Cell 77: 1003–1014. [DOI] [PubMed] [Google Scholar]
- Thummel, C. S., and V. Pirotta, 1992. Technical notes: new pCaSpeR P-element vectors. Dros. Inf. Serv. 71: 150. [Google Scholar]
- Tio, M., and K. Moses, 1997. The Drosophila TGFα homolog Spitz acts in photoreceptor recruitment in the developing retina. Development 124: 343–351. [DOI] [PubMed] [Google Scholar]
- Tomlinson, A., 1985. The cellular dynamics of pattern formation in the eye of Drosophila. J. Embryol. Exp. Morphol. 89: 313–331. [PubMed] [Google Scholar]
- Tomlinson, A., 1988. Cellular interactions in the developing Drosophila eye. Development 104: 183–193. [DOI] [PubMed] [Google Scholar]
- Treisman, J. E., and G. M. Rubin, 1995. wingless inhibits morphogenetic furrow movement in the Drosophila eye disc. Development 121: 3519–3527. [DOI] [PubMed] [Google Scholar]
- Tseng, A. S., and I. K. Hariharan, 2002. An overexpression screen in Drosophila for genes that restrict growth or cell-cycle progression in the developing eye. Genetics 162: 229–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo, L. T., M. Minet, J. M. Schmitter, F. Lacroute and F. Wyers, 2001. Mpe1, a zinc knuckle protein, is an essential component of yeast cleavage and polyadenylation factor required for the cleavage and polyadenylation of mRNA. Mol. Cell. Biol. 21: 8346–8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voas, M. G., and I. Rebay, 2004. Signal integration during development: insights from the Drosophila eye. Dev. Dyn. 229: 162–175. [DOI] [PubMed] [Google Scholar]
- Wiersdorff, V., T. Lecuit, S. M. Cohen and M. Mlodzik, 1996. Mad acts downstream of Dpp receptors, revealing a differential requirement for dpp signaling in initiation and propagation of morphogenesis in the Drosophila eye. Development 122: 2153–2162. [DOI] [PubMed] [Google Scholar]
- Witte, M. M., and R. E. Scott, 1997. The proliferation potential protein-related (P2P-R) gene with domains encoding heterogeneous nuclear ribonucleoprotein association and Rb1 binding shows repressed expression during terminal differentiation. Proc. Natl. Acad. Sci. USA 94: 1212–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff, T., 2000. Histological techniques for the Drosophila eye part 1: larva and pupa, pp. 200–227 in Drosophila Protocols, edited by W. Sullivan, M. Ashburner and R. S. Hawley. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Wolff, T., and D. F. Ready, 1991. Cell death in normal and rough eye mutants of Drosophila. Development 113: 825–839. [DOI] [PubMed] [Google Scholar]
- Xu, T., and G. M. Rubin, 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117: 1223–1237. [DOI] [PubMed] [Google Scholar]