Abstract
African populations of Drosophila simulans are thought to be ancestral in this model species and are increasingly used for testing general hypotheses in evolutionary genetics. It is often assumed that African populations are more likely to be at a neutral mutation drift equilibrium than other populations. Here we examine population structuring and the demographic profile in nine populations of D. simulans. We surveyed sequence variation in four X-linked genes (runt, sevenless, Sex-lethal, and vermilion) that have been used in a parallel study in the closely related species D. melanogaster. We found that an eastern group of populations from continental Africa and Indian Ocean islands (Kenya, Tanzania, Madagascar, and Mayotte Island) is widespread, shows little differentiation, and has probably undergone demographic expansion. The other two African populations surveyed (Cameroon and Zimbabwe) show no evidence of population expansion and are markedly differentiated from each other as well as from the populations from the eastern group. Two other populations, Europe and Antilles, are probably recent invaders to these areas. The Antilles population is probably derived from Europe through a substantial bottleneck. The history of these populations should be taken into account when drawing general conclusions from variation patterns.
DROSOPHILA melanogaster and D. simulans are two cosmopolitan sibling species widely used in evolutionary studies. Despite their recent common ancestry, they show surprising differences in genomic parameters. Sequence variation parameters differ across chromosomes, across species, and between African and European populations. A number of hypotheses involving differences in history, in effective population size, in life history traits, and in chromosome inversion frequencies have been alleged for explaining these contrasts (see, e.g., Begun 1996; Moriyama and Powell 1996; Irvin et al. 1998; Andolfatto and Przeworski 2000; Andolfatto 2001; Charlesworth 2001; Capy and Gibert 2004; Gravot et al. 2004). A long and established history of studies has made these two species excellent models for evolutionary analyses. They have independently undergone a similar evolution and are thus considered useful systems for studying the critical factors involved in genetic evolution. Despite these strengths as model systems, population histories can add unique, contingent factors to the patterns of variation that can obscure patterns of change if they are not accounted for in data interpretation.
Biometrical studies conducted in D. melanogaster by Teissier (1957) showed long ago that this species is geographically differentiated. More recent studies based on molecular markers have confirmed that African D. melanogaster populations are genetically differentiated (Bénassi and Veuille 1995; Michalakis and Veuille 1996; Veuille et al. 1998; Baudry et al. 2004). In D. simulans, Hamblin and Veuille (1999) studied two genes, vermilion (v) and Glucose-6 phosphate dehydrogenase (G6pd), in several populations from the Old and the New World. While G6pd was subject to selection and thus was not a good marker of population history, vermilion appeared to show neutral patterns of distribution. African populations appeared to be split into three regions (Cameroon, Zimbabwe, and Kenya–Tanzania) that did not individually significantly depart from a neutral mutation-drift equilibrium but were significantly differentiated from each other. Compared with these populations, those from Europe and Antilles, along with a U. S. population previously studied by Begun and Aquadro (1995), had a lower nucleotide variation and showed evidence of excess linkage disequilibrium, two features that could result from recent bottlenecks. This study supported Lachaise et al.'s (1988) hypothesis that African populations are the probable ancestors of non-African populations. However, it did not indicate the migration routes followed by D. simulans in its spread out of Africa. This study also lacked populations from the Indian Ocean, where biogeographical data suggest that D. simulans ancestors have been present for a long time (Lachaise et al. 1988).
In this study, we analyze DNA sequence variation at several genes in nine populations of D. simulans from continental Africa, Indian Ocean islands, and Antilles. We recorded DNA sequence variation at four X-linked genes that have been used in a parallel study in D. melanogaster (Baudry et al. 2004): runt (run), sevenless (sev), Sex-lethal (Sxl), and vermilion (v). We use these data for checking demographic hypotheses and for comparing historical processes between populations.
MATERIALS AND METHODS
Sample collection:
Sample sites for Antilles, Cameroon, Kenya, Tanzania, and Zimbabwe were previously described (Hamblin and Veuille 1999). Samples from La Saline (Reunion) and Dzaoudzi (Mayotte) were collected by C. Montchamp-Moreau in 1996 and 1999, respectively. The sample from Antananarivo (Madagascar) was collected by M. Veuille in November 2000. The samples from Madagascar and Reunion were previously used by Derome et al. (2004) for the study of a selective sweep caused by the sex-ratio factor of D. simulans, and variation in vermilion was recorded for these two populations. For Europe, our previous work on vermilion involved a sample from Italy (Hamblin and Veuille 1999). Since the DNA was of poor quality, it was replaced for the other three genes by a sample from Banyuls (France) collected by M. Veuille in July 2001. Isochromosomal lines were started in the laboratory by crossing single wild-caught males to virgin attached-X females. Stocks were frozen and stored at −80° until subsequent use. DNA extraction, PCR amplification, and purification of gene fragments were carried out using classical protocols. For v, the sequenced region overlaps positions 654–1350 of GenBank accession no. U27204. For the other three genes, it overlaps the same ∼0.6-kb fragment as was used in D. melanogaster (Baudry et al. 2004). The three genes are located in distant regions of the X chromosome (Sex lethal: 6F5; vermilion: 9F11; sevenless: 10A4; runt: 19E2) that all correspond to highly recombining parts of the chromosome. The recombination rate in females ranges from 2.7 to 4.9 × 10−8 event/generation/bp in D. melanogaster.
Sequence analysis:
DNA sequencing was performed using an ABI-310 automated sequencer. Sequences were aligned using the Bioedit program, and the analysis of molecular variation was carried out using Dnasp 4.0 (Rozas et al. 2003). Fifteen chromosomes were sequenced in all new samples except Kenya (14 for all genes), Cameroon (12 for all genes), and Mayotte (14 for sevenless). We aligned 581 bp in runt, 800 bp in sevenless, 643 in Sex-lethal, and 698 bp in vermilion, covering most of the 728-bp vermilion sequence formerly used by Hamblin and Veuille (1999). Polymorphic indels ranging from 1 to 33 bp were observed in introns at three of four loci: Sxl, sev, and runt. They were excluded from all analyses.
Several tests have been developed to detect departure from neutrality in sequence data. Since no single test has a high power for all kinds of departure from neutrality (Depaulis et al. 2001), it has become a systematic practice to run several tests on the same data set, which raises the issue of the significance of multiple tests. We used five neutrality tests. The nine populations of D. simulans analyzed shared many polymorphic sites (see results), suggesting a long common history, and tests run on them are not statistically independent. We therefore did not use a Bonferroni correction. Since the four loci used in this study were genetically independent, our conclusions relied on consistency among neutrality tests over the four loci.
Among neutrality tests, Tajima's D (Tajima 1989a) summarizes the frequency spectrum. Fu's FS (1997) and the H-haplotype test (Depaulis and Veuille 1998; Depaulis et al. 2001), respectively, compare the observed number of haplotypes and the haplotype diversity in a sample of those expected under neutrality. The McDonald and Kreitman (1991) test compares synonymous and nonsynonymous variation within and between species. We also used the CV test (Mousset et al. 2004) for examining the deviation from neutrality resulting from population expansion. The test is based on theoretical work from Tajima (1989b), Slatkin and Hudson (1991), and Rogers and Harpending (1992) on properties of the mismatch distribution that were subsequently applied, e.g., to estimating human population expansion parameters from mitochondrial DNA variation (Excoffier and Schneider 1999; Schneider and Excoffier 1999). The test is performed by comparing the value of the coefficient of variation of pairwise differences observed among sequences to the value expected under the neutral model (Mousset et al. 2004).
A test on a recombining region must include an independently derived estimate of the recombination rate. The recombination rate estimated from laboratory experiments in D. melanogaster was used as a reference, since its value in these four chromosome X regions does not differ between D. melanogaster and D. simulans (True et al. 1996). The recombination rate varies between 2.7 and 4.9 × 10−8 event/generation/bp in the four genes. To run the tests conservatively (Wall 1999), we used a lower value of recombination rate (c = 5.10−9/event/generation/pb) and tested only a decrease in haplotype number or diversity for Fu's FS and the H-test. To be able to detect possible population expansion, the CV test was performed two tailed (Mousset et al. 2004). We therefore used the estimated recombination rates to run this test.
Genetic differentiation among populations was analyzed by means of the FST (Hudson et al. 1992a,b) statistic. It was calculated for the four genes simultaneously by treating each polymorphic site as an independent locus (Hudson et al. 1992a,b). We also used the nearest-neighbor statistic, Snn, of Hudson (2000), which is a measure of how often the “nearest neighbors” (in sequence space) of sequences are from the same locality in geographic space. We used 10,000 permutations to determine whether the observed values of the two statistics were statistically significant (Hudson et al. 1992a,b).
RESULTS
DNA sequence variation:
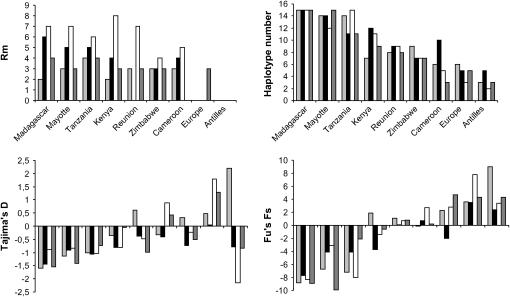
Summary statistics for molecular variation are shown in Table 1 and some of them are illustrated in Figure 1. Populations are ranked in decreasing order of number of polymorphic sites over all genes (from 154 in Madagascar to 59 in Antilles). This broadly corresponds to a decreasing order of variation over all genes. Values for θw vary from a twofold range (2.30–5.33% in vermilion) to a threefold range (0.90–2.99% in runt, 1.05–2.76 in Sex-lethal) to a fourfold range (0.52–2.02% in sevenless). Values for θπ vary in the same way, although in a somewhat smaller range (from 1.41 to 1.88% in Sex-lethal to 0.71–1.92% in runt), indicating that differences in variation level owe more to the number of polymorphic sites (S) than to their frequency. Since θπ incorporates more information on polymorphic-site frequency than θw does, the difference between the two estimators means that population differences in sequence variation mostly rest on low-frequency polymorphisms. For all genes, the different samples actually differ more in the number of singletons (Table 2, Se), than in the number of other polymorphic sites (Si). Singletons are at a high frequency in Mayotte, Madagascar, and Tanzania. In these three populations, the number of haplotypes is also high, since it is equal to the number of chromosomes in Madagascar for all genes, and almost equal to it for the other two populations (Figure 1). At the other extreme, Europe and Antilles tend to show few haplotypes and small θ values with some fluctuation over genes. This can be illustrated by pooling the number of polymorphic sites found in each population over loci (supplemental Table S1 at http://www.genetics.org/supplemental/). Even though the number of sequences obtained is slightly different, populations do not vary much in the number of internal polymorphic sites (range 54–72, when setting Antilles apart) whereas the difference in the number of singletons is drastic (range 22–98).
TABLE 1.
Polymorphism data at four X-linked loci in nine populations of D. simulans
| Population | n | K | S | Si | Se | Rm | θw (%) | θπ (%) |
|---|---|---|---|---|---|---|---|---|
| Sex-lethal (643 bp; 403.0 silent sitesa) | ||||||||
| Madagascar | 15 | 15 | 36 | 10 | 26 | 2 | 2.76 | 1.74 |
| Mayotte | 15 | 14 | 31 | 13 | 18 | 3 | 2.35 | 1.72 |
| Tanzania | 15 | 14 | 27 | 12 | 15 | 4 | 1.82 | 1.41 |
| Kenya | 14 | 7 | 28 | 20 | 8 | 2 | 2.05 | 1.88 |
| Reunion | 15 | 8 | 22 | 16 | 6 | 3 | 1.48 | 1.70 |
| Zimbabwe | 15 | 9 | 24 | 16 | 8 | 3 | 1.72 | 1.59 |
| Cameroon | 12 | 6 | 22 | 18 | 4 | 3 | 1.59 | 1.72 |
| Europe | 15 | 6 | 24 | 13 | 11 | 0 | 1.61 | 1.80 |
| Antilles | 15 | 3 | 16 | 16 | 0 | 0 | 1.07 | 1.66 |
| vermilion (698 bp; 288.3 silent sitesa) | ||||||||
| Madagascar | 15 | 15 | 46 | 18 | 28 | 6 | 4.70 | 3.20 |
| Mayotte | 15 | 14 | 48 | 22 | 26 | 5 | 5.33 | 4.23 |
| Tanzania | 11 | 11 | 38 | 12 | 26 | 5 | 4.61 | 3.57 |
| Kenya | 13 | 12 | 34 | 14 | 20 | 4 | 4.02 | 3.30 |
| Reunion | 15 | 9 | 27 | 20 | 7 | 0 | 2.88 | 2.62 |
| Zimbabwe | 10 | 7 | 28 | 14 | 14 | 3 | 3.56 | 3.26 |
| Cameroon | 12 | 10 | 29 | 14 | 15 | 4 | 3.44 | 2.88 |
| Europe | 12 | 5 | 22 | 12 | 10 | 0 | 2.53 | 2.55 |
| Antilles | 12 | 5 | 20 | 10 | 10 | 0 | 2.30 | 1.88 |
| sevenless (800 bp; 566.8 silent sitesa) | ||||||||
| Madagascar | 15 | 15 | 35 | 15 | 20 | 7 | 1.80 | 1.44 |
| Mayotte | 14 | 12 | 35 | 15 | 20 | 7 | 1.85 | 1.50 |
| Tanzania | 15 | 15 | 38 | 17 | 21 | 6 | 2.02 | 1.54 |
| Kenya | 14 | 11 | 38 | 22 | 16 | 8 | 1.94 | 1.62 |
| Reunion | 15 | 9 | 33 | 19 | 14 | 7 | 1.64 | 1.52 |
| Zimbabwe | 15 | 7 | 23 | 21 | 2 | 4 | 1.22 | 1.50 |
| Cameroon | 12 | 5 | 19 | 14 | 5 | 5 | 0.98 | 0.92 |
| Europe | 15 | 3 | 14 | 14 | 0 | 0 | 0.73 | 1.05 |
| Antilles | 15 | 2 | 10 | 0 | 10 | 0 | 0.52 | 0.23 |
| runt (581 bp; 325.8 silents sitesa) | ||||||||
| Madagascar | 15 | 15 | 37 | 13 | 24 | 4 | 2.99 | 1.92 |
| Mayotte | 15 | 15 | 33 | 10 | 23 | 3 | 2.40 | 1.61 |
| Tanzania | 15 | 11 | 25 | 13 | 12 | 4 | 1.96 | 1.63 |
| Kenya | 14 | 9 | 20 | 16 | 4 | 3 | 1.57 | 1.59 |
| Reunion | 15 | 8 | 28 | 13 | 15 | 3 | 2.26 | 1.74 |
| Zimbabwe | 15 | 7 | 16 | 15 | 1 | 3 | 1.42 | 1.24 |
| Cameroon | 12 | 3 | 13 | 10 | 3 | 0 | 1.10 | 0.98 |
| Europe | 15 | 5 | 17 | 16 | 1 | 3 | 1.19 | 1.58 |
| Antilles | 15 | 3 | 13 | 6 | 7 | 0 | 0.90 | 0.71 |
n, number of sequences; K, number of haplotypes; S, number of mutations; Si, number of internal polymorphic sites. Se, number of external polymorphic sites (singletons); Rm, minimum number of recombination events estimated using the four-gamete rule (Hudson and Kaplan 1985), θw, Watterson's (1975) estimates of the mutation parameter calculated on silent sites; θπ, nucleotide diversity calculated on silent sites.
The number of silent sites may vary among populations due to the presence of polymorphic indels in introns.
Figure 1.
Variation statistics (minimum number of recombination events, number of haplotypes) and neutral distribution tests (Tajima's D and Fu's FS) at four loci (from left to right: Sex-lethal, vermilion, sevenless, runt) in nine D. simulans populations in the same ranking order from left to right.
TABLE 2.
FST (top right) and Snn (bottom left) values at four X-linked loci in nine populations of D. simulans
| Madagascar | Mayotte | Tanzania | Kenya | Reunion | Zimbabwe | Cameroon | Europe | Antilles | |
|---|---|---|---|---|---|---|---|---|---|
| Madagascar | 0.009 | 0.017 | 0.017 | 0.044** | 0.093** | 0.194** | 0.117** | 0.272** | |
| Mayotte | 0.600 | 0.031* | 0.040* | 0.049** | 0.096** | 0.212** | 0.134** | 0.252** | |
| Tanzania | 0.755* | 0.817* | −0.004 | 0.058** | 0.089** | 0.156** | 0.098** | 0.258** | |
| Kenya | 0.695 | 0.724 | 0.655 | 0.058** | 0.118** | 0.174** | 0.101** | 0.239** | |
| Reunion | 0.6333 | 0.683 | 0.917** | 0.879** | 0.154** | 0.251** | 0.159** | 0.312** | |
| Zimbabwe | 0.875** | 0.933** | 0.956** | 0.931** | 1.000** | 0.200** | 0.089** | 0.264** | |
| Cameroon | 0.926** | 0.926** | 0.796* | 0.923** | 0.987** | 0.889** | 0.274** | 0.412** | |
| Europe | 0.917** | 0.967** | 0.733* | 0.873* | 0.967** | 0.794* | 1.000** | 0.141** | |
| Antilles | 0.950** | 1.000** | 0.928** | 0.937** | 0.967** | 0.933** | 0.963** | 0.697 |
*P < 0.05; **P < 0.01.
Very few amino acid polymorphisms were observed. None of the four loci presented fixed nonsynonymous differences between D. simulans and D. melanogaster. The sevenless and vermilion genes showed six and four intraspecific low-frequency replacement polymorphisms, respectively, whereas runt and Sex-lethal showed none. These observations are consistent with the action of purifying selection on most nonsynonymous sites in the four gene regions.
The minimum number of recombination events was estimated using the four-gamete rule (Hudson and Kaplan 1985). It was high in all African populations, and zero in Europe and Antilles, except in one case (runt in Europe). Overall, variation statistics suggest some ranking between populations. There are three highly variable African populations (Mayotte, Madagascar, and Tanzania). Kenya, Zimbabwe, and Cameroon (in this order) are intermediate between these and the non-African populations (Europe and Antilles). Reunion also shows intermediate values.
Tests of neutral distribution:
The results of the tests of neutral distribution are illustrated in Figure 1 (details of the tests are in supplemental Table S3 at http://www.genetics.org/supplemental/). Tajima's (1989) D was negative for most African populations. It was always negative in Mayotte, Madagascar, Kenya, and Tanzania, consistent with the high proportion of low-frequency variants. It was significantly negative for three of four genes in Madagascar. In Europe and Antilles, Tajima's D showed four significant values—three positive and one negative. Both types of values result from the marked deficit of haplotypes observed in these populations. Positive values were observed when the few haplotype classes were evenly represented (e.g., Europe at sev), whereas negative values result from the presence of a rare haplotype (e.g., Antilles at sev).
As with Tajima's D, Fu's FS (1997) was consistently negative in all four genes for Madagascar, Mayotte, and Tanzania, indicating an excess of haplotypes in these populations. Kenya, Zimbabwe, and Cameroon had both positive and negative Fs values. The Cameroon population showed a significant deficit of haplotype at three loci, as did Zimbabwe and Kenya at one locus each. They thus differed from the other African populations. Finally, the test was significantly positive at all four loci for Europe and Antilles. The H-test yielded similar results. The coefficient of variation of the mismatch distribution was generally smaller for Mayotte, Madagascar, and Tanzania than for the other samples. The CV values were significantly too low in Madagascar at three of four loci, and at one locus in Mayotte and Kenya. Conversely, CV values were significantly too high for Antilles at all loci, for Europe at three loci, for Cameroon at two loci, and for Reunion at one locus. Finally, the McDonald and Kreitman tests performed at the two loci with nonsynonymous polymorphisms, sev and v, did not detect departure from neutrality.
Mismatch distribution profiles:
The differences among the four eastern African populations (Mayotte, Madagascar, Kenya, and Tanzania) and the other ones appears clearly in a diagram showing the distribution of the pairwise differences (supplemental Figure S1 at http://www.genetics.org/supplemental/). The distributions from East Africa populations were unimodal for all genes, whereas those of the other populations were multimodal for all genes. Unimodal distributions are observed when the genealogy of the sample resembles a star phylogeny, which in turn is observed during demographic expansions (Slatkin and Hudson 1991) or recovery phases following a selective sweep (Depaulis et al. 2003).
Pairwise comparisons between chromosomes bearing the same haplotype increase the score of the no-difference class, whereas comparisons between chromosomes from two different haplotypes increase the frequency of higher-scoring classes. A small number of haplotypes thus tends to generate a small number of peaks and to produce a dog-toothed pattern, as found for Cameroon, Zimbabwe, Reunion, Europe, and especially for Antilles. This pattern is expected after a recent bottleneck or after a recent partial selective sweep (Depaulis et al. 2003).
Genetic differentiation among populations:
We estimated genetic differentiation among populations using FST (Hudson et al. 1992a,b) and the Snn statistics (Hudson 2000). Table 4 summarizes the values of these statistics for the pooled genes. Overall, similar results were observed with the two statistics, although a few pairwise comparisons have slightly different probability values. It appears that eastern African populations (Kenya, Tanzania, Madagascar, and Mayotte) make up the only close group, with little or no differentiation. The most extreme Indian Ocean population, Reunion, shows a moderate differentiation from these eastern populations. The other two continental African populations (Cameroon and Zimbabwe) are very different from each other as well as from the eastern populations. In Table 4, Europe does not appear to be closely related to any of the African populations, even though it differs less from Zimbabwe, Tanzania, and Kenya than from the other populations. The Antillean population seems closer to the European population than to the African and Indian Ocean populations. The Antillean and European populations are still highly significantly differentiated when the FST statistics are considered whereas this is not true with the Snn statistics. This discrepancy between the two statistics suggest that, while the two populations differ markedly in their polymorphic-site frequencies, they present similar haplotypes.
TABLE 4.
Number of sites unique to the populations in the rows, when compared to the populations in the columns
| Madagascar | Mayotte | Tanzania | Kenya | Reunion | Zimbabwe | Cameroon | Europe | Antilles | |
|---|---|---|---|---|---|---|---|---|---|
| Madagascar | 84 | 90 | 88 | 88 | 101 | 102 | 112 | 119 | |
| Mayotte | 75 | 81 | 76 | 78 | 88 | 93 | 101 | 108 | |
| Tanzania | 61 | 61 | 45 | 68 | 66 | 64 | 82 | 88 | |
| Kenya | 56 | 53 | 42 | 63 | 67 | 56 | 79 | 82 | |
| Reunion | 33 | 32 | 42 | 40 | 50 | 54 | 63 | 65 | |
| Zimbabwe | 38 | 34 | 32 | 36 | 42 | 35 | 33 | 47 | |
| Cameroon | 28 | 28 | 19 | 14 | 35 | 24 | 40 | 47 | |
| Europe | 32 | 30 | 31 | 31 | 38 | 16 | 34 | 21 | |
| Antilles | 20 | 18 | 18 | 15 | 21 | 11 | 22 | 2 |
Genetic relationships between populations:
FST and Snn values between a pair of populations depend on their genetic similarity, but they are also influenced by the level of variation of these populations (Charlesworth 1998). They are thus inappropriate for tracing lines of descent when new populations have been established through a bottleneck. Another way of exploring these relationships consists of examining shared and unique polymorphic sites among populations (Table 3). Comparing the number of shared and private sites across populations is possible in this study, since the sample sizes are very similar.
TABLE 3.
Shared polymorphic sites in pairwise comparisons among populations
| Mayotte | Tanzania | Kenya | Reunion | Zimbabwe | Cameroon | Europe | Antilles | |
|---|---|---|---|---|---|---|---|---|
| Madagascar | 65 | 59 | 61 | 61 | 48 | 47 | 37 | 30 |
| Mayotte | 59 | 64 | 62 | 52 | 47 | 39 | 32 | |
| Tanzania | 75 | 52 | 54 | 56 | 38 | 32 | ||
| Kenya | 54 | 50 | 61 | 38 | 35 | |||
| Reunion | 44 | 40 | 31 | 29 | ||||
| Zimbabwe | 51 | 53 | 39 | |||||
| Cameroon | 35 | 28 | ||||||
| Europe | 48 |
There is no obvious pattern in African populations (including continental and Indian ocean island populations), where the number of shared sites fluctuates in a narrow range (40–65). In contrast, the number of unique sites is highly variable but they mostly occur as singletons, which carry no information on shared origins. Some relatedness is suggested between Europe and Zimbabwe. They share 53 sites, whereas Europe shares only 31–39 sites with the other populations. Moreover, Europe shows only 16 unique sites with respect to Zimbabwe, compared to from 30 to 38 with the other populations. Similarly, of 19 haplotypes observed in the Europe population, 6 are also present in the Zimbabwe sample (supplemental Figure S2 at http://www.genetics.org/supplemental/), whereas only one or two are also present in the other African and Indian Ocean populations. Our data thus suggest that, among the African and Indian Ocean populations of our sampling, the European population is most closely related to the Zimbabwe population. However, a relatively large part of the European variation (in terms of polymorphic sites or haplotypes) is not found in the other populations of our sampling, suggesting that the European population did not simply originate through a bottleneck from one of these populations.
Some relatedness is also suggested between Antilles and Europe, since Antilles shares 48 sites with Europe, compared to 28–39 with the other populations. It shows only two unique sites with respect to Europe, compared to 11–22 with the other populations. The same pattern is observed when haplotypes are considered (supplemental Figure S2 at http://www.genetics.org/supplemental/): of 13 haplotypes observed in the Antilles population, 10 are also present in the Europe sample, whereas between 0 and 4 are also present in the other African and Indian Ocean populations. In the two cases, the asymmetry of the pattern suggests that the European population is derived from a Zimbabwe-related population and the Antilles population is derived from a Europe-related population.
DISCUSSION
Demographic vs. selective effects in D. simulans populations:
We observed numerous instances of departure from the expectations of the neutral model in D. simulans. In a group of eastern populations (Madagascar, Mayotte, Kenya, and Tanzania, hereafter the “eastern group”), an excess of singletons, an excess of haplotypes, and a bell-shaped mismatch distribution were observed at all four loci. Such features can theoretically been explained by two very different processes: recovery after a complete selective sweep or a demographic expansion. Demography is a force that affects the entire genome whereas natural selection has only a local effect around the target of selection. Given that we observed the same pattern at four unlinked loci, a demographic expansion is therefore a much more parsimonious hypothesis than selection. Moreover, at the four loci, the populations of the eastern group show very high levels of polymorphism, which is less compatible with recent complete selective sweeps.
Similarly, two populations in our sample (Europe and Antilles) showed consistent features at all four loci. They showed a relatively low level of polymorphism, a deficit of haplotypes, a quasi-absence of detectable recombination events, and a dog-toothed mismatch distribution. Again, this could theoretically be explained by a selective hypothesis, for example, by an incomplete selective sweep. Partial sweeps occur, for example, when recombination takes place during the selective stage between the region surveyed and the selected site. They tend to result in a deficit of haplotypes (Depaulis et al. 2003). However, this hypothesis would imply that four independent selective events recently occurred in the Europe and Antilles populations. It is more parsimonious to suppose that these populations have undergone a demographic bottleneck, probably during the expansion of D. simulans out of Africa. Alternatively, the Europe and Antilles populations may have formed by admixture of two or more divergent populations.
The three remaining populations show a more complex pattern. The populations of Zimbabwe, Reunion, and Cameroon, respectively, show significant departures from neutrality at one, two, and three loci. In these cases, selective and/or demographic explanations could explain the data.
Structuring and demographic history of D. simulans populations from Africa and the Indian Ocean:
This study extends Hamblin and Veuille's (1999) observation that D. simulans populations are highly structured in Africa when nuclear polymorphism is considered. Extending the analysis to populations from Indian Ocean islands delineates four groups of genetically differentiated populations. Several populations overlapping Central–East Africa and the closest Indian Ocean islands (Kenya, Tanzania, Mayotte, and Madagascar) make up the first group. Reunion, the second group, shows a moderate differentiation from these eastern populations. Zimbabwe and Cameroon are the last two groups.
Our finding of a widespread, little differentiated, expanding population centered around Madagascar is in good agreement with the postulated geographic origin of D. simulans. It has long been assumed from biogeographic data and coancestry with insular endemics that D. simulans originated from East Africa (Lachaise et al. 1988). This has been recently confirmed by Dean and Ballard (2004). Using polymorphism data from three unlinked loci, they showed that phylogenetic inferences strongly support Madagascar as the geographic origin of D. simulans. Similarly, data from mitochondrial DNA suggest that Madagascar or continental East Africa may be the geographic origin of D. simulans (Montchamp-Moreau et al. 1991; Ballard 2004; Solignac 2004). A demographic expansion coupled with a range expansion over a wide area could explain the near absence of genetic differentiation that we observed from Kenya to Madagascar. There is no obvious cause for such a range expansion, since this area is fractionated into a continental area and remote islands and covers highly divergent habitats and ecosystems (for instance, Madagascar is remarkable for its endemic flora and fauna). However, it is tempting to speculate that the suggested range and demographic expansion may be related to the wild-to-domestic habit shift of D. simulans (Lachaise and Silvain 2004). Interestingly, Glinka et al. (2003) also found evidence for a recent demographic expansion in an African population of D. melanogaster.
The Cameroon population of D. simulans is significantly differentiated from all the other African populations. The singularity of this population was not unexpected as individuals from this area were previously shown to differ from all other D. simulans flies in their cuticular hydrocarbon pattern (Luyten 1982; Rouault et al. 2000, 2004). The difference may be linked to sexual behavior, since cuticular hydrocarbons are involved in mate recognition (Ferveur et al. 1996). Zimbabwe is another genetically isolated population. This seems surprising, given the genetic continuity among Kenya, Tanzania, Mayotte, and Madagascar. We cannot say, however, whether an explanation is needed for the discontinuity between Zimbabwe and the eastern group or for the lack of differentiation within the eastern group. To conclude this part, although our observations are in general agreement with the admitted hypothesis that D. simulans originates from the Africa–Madagascar area, it shows a complex population pattern that can make it difficult to choose an adequate “ancestral African” population for use in comparative evolutionary studies.
Comparison between D. simulans and D. melanogaster:
One result of this study is that D. simulans and D. melanogaster show a different structuration pattern in Africa. Baudry et al. (2004) used the same set of four loci in a sample of 11 populations of D. melanogaster and found only slight population structuring in Africa (mean Fst of 0.047 within continental African populations). D. melanogaster populations from Niger, Kenya, and Zimbabwe did not significantly differ from each other and were only slightly, although significantly, differentiated from the Ivory Coast population. In contrast, D. simulans populations show a much stronger structuration within continental Africa (mean FST of 0.162). These conclusions stand in marked contrast to results of studies of allozyme variation, morphology, or chromosome arrangements, which generally show much more differentiation among African D. melanogaster populations than among African D. simulans populations (review in Capy and Gibert 2004).
We consider the D. simulans population from Europe and Antilles as examples of derived populations. They share several features: low nucleotide variation, a significant deficit in the number of haplotypes, and an absence or near absence of detectable recombination events. They thus appear to be invading populations, consistent with Lachaise et al.'s (1988) hypothesis. However, they differ from derived populations of D. melanogaster. The latter species was studied using the same loci in African and non-African populations (Baudry et al. 2004). The seven derived populations of D. melanogaster showed evidence of a strong bottleneck, resulting in only a few remaining haplotypes (typically one or two). There was a clear-cut change in variation parameters between African and non-African populations. In D. simulans, the European population is only slightly less polymorphic than the less variable African populations (Zimbabwe, Cameroon, and Reunion), suggesting that it suffered a relatively minor bottleneck. The Antilles population underwent a more severe bottleneck. The difference between the derived populations of the two species may be related to the different timing of their out-of-Africa colonization. It is commonly thought that D. melanogaster expanded its range out of Africa in conjunction with the rise of agriculture after the Neolithic revolution ∼10,000 years ago (Lachaise et al. 1988). On the basis of allozyme data, it has been suggested that D. simulans spread worldwide much more recently (Morton et al. 2004).
Conclusion: accounting for historical complexity in genomes:
It should be noted that our conclusions on the demographic histories and structuring of D. simulans populations are based on X chromosome data. However, analyses based on sequence polymorphism (Begun and Whitley 2000; Andolfatto 2001) and a large number of microsatellite loci (Schöfl and Schlötterer 2004) have demonstrated that X chromosomes are about as variable as autosomes in African populations of D. simulans, whereas non-African populations show a markedly lower level of polymorphism on the X chromosome compared to the autosomes. Similarly, Dean and Ballard (2004) have shown that support for a Madagascar origin of D. simulans seems to differ between X- and autosome-located loci. A comparable discrepancy between African and non-African X chromosomes and autosomes is observed in the closely related D. melanogaster (Andolfatto 2001; Kauer et al. 2002). These differences between the X chromosome and the autosomes suggest that their selective and/or demographic histories differ. For example, the effects of background selection and selective sweeps may be different between the two types of chromosomes (Begun and Whitley 2000). Alternatively, since the effective population size is lower for the X chromosome than for the autosomes, the effects of bottlenecks are expected to be more severe for the X (Wall et al. 2002). In conclusion, it will be necessary to gather more polymorphism data, particularly from the autosomes of African and Indian Ocean populations, to confirm the present findings.
Natural selection and demographic events are two forces that shape DNA variation patterns and cause them to depart from theoretical expectations under neutrality. African samples of D. simulans have been used for avoiding demographic disturbances due to commensalism (e.g., Begun and Aquadro 1993; Rozas et al. 2001; Andolfatto and Wall 2003: Quesada et al. 2003; Schlenke and Begun 2004). Our study, however, reveals a substantial complexity in Africa. “African” populations of D. simulans are not a homogeneous category. The properties of these populations must be accounted for in designing genetic studies. For instance, an African population may be of interest in comparative studies either because it is ancestral to European ones or because it is thought to be at neutral mutation-drift equilibrium. Assuming that D. simulans is of African origin, we do not know whether its ancestral population is still alive in Africa or is at neutral mutation-drift equilibrium, as we did not observe any such population in our study.
Acknowledgments
We thank Matthew Cobb, Catherine Montchamp-Moreau, Laura Rose, and two anonymous reviewers for helpful comments on the manuscript. This research was supported by the Groupe de Recherche GDR-1928 “Evolution des génomes dans les populations” of the Centre National de la Recherche Scientifique, by the Unité Mixte de Recherche 7625 (Laboratoire d'Ecologie) and by the Programme Pluri-Formations “Populations fractionnées et insulaires” of the Ecole Pratique des Hautes Etudes.
References
- Andolfatto, P., 2001. Contrasting patterns of X–Linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol. Biol. Evol. 18: 279–290. [DOI] [PubMed] [Google Scholar]
- Andolfatto, P., and M. A. Przeworski, 2000. Genome-wide departure from the standard neutral model in natural populations of Drosophila. Genetics 156: 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto, P., and J. D. Wall, 2003. Linkage disequilibrium patterns across a recombination gradient in African Drosophila melanogaster. Genetics 165: 1289–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard, J. W., 2004. Sequential evolution of a symbiont inferred from the host: Wolbachia and Drosophila simulans. Mol. Biol. Evol. 21: 428–442. [DOI] [PubMed] [Google Scholar]
- Baudry, E., B. Viginier and M. Veuille, 2004. Non-African populations of Drosophila melanogaster have a unique origin. Mol. Biol. Evol. 21: 1482–1491. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., 1996. Population genetics of silent and remplacement variation in Drosophila simulans and D. melanogaster: X/autosome differences? Mol. Biol. Evol. 13: 1405–1407. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., and C. F. Aquadro, 1993. African and North American populations of Drosophila melanogaster are very different at the DNA level. Nature 365: 548–550. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., and C. F. Aquadro, 1995. Molecular variation at the vermilion locus in geographically diverse populations of Drosophila melanogaster and D. simulans. Genetics 140: 1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun, D. J., and P. Whitley, 2000. Reduced X-linked nucleotide polymorphism in Drosophila simulans. Proc. Natl. Acad. Sci. USA 97: 5960–5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bénassi, V., and M. Veuille, 1995. Comparative population structuring of molecular and allozyme variation of Drosophila melanogaster Adh between Europe, West Africa and East Africa. Genet. Res. 65: 95–103. [DOI] [PubMed] [Google Scholar]
- Capy, P., and P. Gibert, 2004. Drosophila melanogaster, Drosophila simulans: so similar yet so different. Genetica 120: 5–16. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., 1998. Measures of divergence between populations and the effect of forces that reduce variability. Mol. Biol. Evol. 15: 538–543. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., 2001. The effect of life-history and mode of inheritance on neutral genetic variability. Genet. Res. 77: 153–166. [DOI] [PubMed] [Google Scholar]
- Dean, M. D., and J. W. Ballard, 2004. Linking phylogenetics with population genetics to reconstruct the geographic origin of a species. Mol. Phylogenet. Evol. 32: 998–1009. [DOI] [PubMed] [Google Scholar]
- Depaulis, F., and M. Veuille, 1998. Neutrality tests based on the distribution of haplotypes under an infinite site model. Mol. Biol. Evol. 15: 1788–1790. [DOI] [PubMed] [Google Scholar]
- Depaulis, F., S. Mousset and M. Veuille, 2001. Haplotype tests using coalescent simulations conditional on the number of segregating sites. Mol. Biol. Evol. 18: 1136–1138. [DOI] [PubMed] [Google Scholar]
- Depaulis, F., S. Mousset and M. Veuille, 2003. Power of neutrality tests to detect bottlenecks and hitchhiking. J. Mol. Evol. 57: S190–S200. [DOI] [PubMed] [Google Scholar]
- Derome, N., K. Métayer, C. Montchamp-Moreau and M. Veuille, 2004. Signature of selective sweep associated with the evolution of sex-ratio drive in Drosophila simulans. Genetics 166: 1357–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier, L., and S. Schneider, 1999. Why hunter-gatherer populations do not show signs of Pleistocene demographic expansions. Proc. Natl. Acad. Sci. USA 96: 10597–10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferveur, J. F., M. Cobb, H. Boukella and J. M. Jallon, 1996. World-wide variation in Drosophila melanogaster sex pheromone: behavioural effects, genetic bases and potential evolutionary consequences. Genetica 97: 73–80. [DOI] [PubMed] [Google Scholar]
- Fu, Y., 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147: 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinka, S., L. Ometto, S. Mousset, W. Stephan and De Lorenzo, 2003. Demography and natural selection have shaped genetic variation in Drosophila melanogaster: a multi-locus approach. Genetics 165: 1269–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravot, E., M. Huet and M. Veuille, 2004. Effect of breeding structure on population genetic parameters in Drosophila. Genetics 166: 779–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblin, M. T., and M. Veuille, 1999. Population structure among African and derived populations of D. simulans: evidence for ancient subdivision and recent admixture. Genetics 153: 305–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., 2000. A new statistic for detecting genetic differentiation. Genetics 155: 2011–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., and N. L. Kaplan, 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111: 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., D. D. Boos and N. L. Kaplan, 1992. a A statistical test for detecting geographic subdivision. Mol. Biol. Evol. 9: 138–151. [DOI] [PubMed] [Google Scholar]
- Hudson, R. R., M. Slatkin and W. P. Maddison, 1992. b Estimation of levels of gene flow from DNA sequence data. Genetics 132: 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvin, S. D., K. A. Wetterstrand, C. M. Hutter and C. F. Aquadro, 1998. Genetic variation and differentiation at microsatellite loci in Drosophila simulans: evidence for founder effects in New World populations. Genetics 150: 777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer, M., B. Zangerl, D. Dieringer and C. Schlotterer, 2002. Chromosomal patterns of microsatellite variability contrast sharply in African and non-African populations of Drosophila melanogaster. Genetics 160: 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaise, D., and J. F. Silvain, 2004. How two Afrotropical endemics made two cosmopolitan human commensals: the Drosophila melanogaster-D. simulans palaeogeographic riddle. Genetica 120: 17–39. [DOI] [PubMed] [Google Scholar]
- Lachaise, D., M.-L. Cariou, J. R. David, F. Lemeunier and L. Tsacas, 1988. Historical biogeography of the Drosophila melanogaster species subgroup. Evol. Biol. 22: 159–225. [Google Scholar]
- Luyten, I., 1982. Variation intraspécifique et interspécifique des hydrocarbures cuticulaires chez Drosophila simulans et des espèces affinés. C. R. Acad. Sci. Paris. Sci. Vie 295: 733–736. [Google Scholar]
- McDonald, J. H., and M. Kreitman, 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654. [DOI] [PubMed] [Google Scholar]
- Michalakis, Y., and M. Veuille, 1996. Length variation of CAG/CAA trinucleotide repeats in natural populations of Drosophila melanogaster and its relation to the recombination rate. Genetics 143: 1713–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montchamp-Moreau, C., J. F. Ferveur and M. Jacques, 1991. Geographic distribution and inheritance of three cytoplasmic incompatibility types in Drosophila simulans. Genetics 129: 399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama, E. N., and J. F. Powell, 1996. Intraspecific nuclear DNA variation in Drosophila. Mol. Biol. Evol. 13: 261–277. [DOI] [PubMed] [Google Scholar]
- Morton, R. A., M. Choudhary, M. L. Cariou and R. S. Singh, 2004. A reanalysis of protein polymorphism in Drosophila melanogaster, D. simulans, D. sechellia and D. mauritiana: effects of population size and selection. Genetica 120: 101–114. [DOI] [PubMed] [Google Scholar]
- Mousset, S., N. Derome and M. Veuille, 2004. A neutrality test based on the mismatch distribution. Mol. Biol. Evol. 21: 724–731. [DOI] [PubMed] [Google Scholar]
- Quesada, H., U. E. Ramirez, J. Rozas and M. Aguade, 2003. Large-scale adaptive hitchhiking upon high recombination in Drosophila simulans. Genetics 165: 895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, R. A., and H. Harpending, 1992. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 9: 552–569. [DOI] [PubMed] [Google Scholar]
- Rouault, J., P. Capy and J.-M. Jallon, 2000. Variations of male cuticular hydrocarbons with geoclimatic variables: an adaptive mechanism in Drosophila melanogaster? Genetica 110: 117–130. [DOI] [PubMed] [Google Scholar]
- Rouault, J. D., C. Marican, C. Wicker-Thomas and J. M. Jallon, 2004. Relations between cuticular hydrocarbon (HC) polymorphism, resistance against desiccation and breeding temperature; a model for HC evolution in D. melanogaster and D. simulans. Genetica 120: 195–212. [DOI] [PubMed] [Google Scholar]
- Rozas, J., M. Gullaud, G. Blandin and M. Aguade, 2001. DNA variation at the rp49 gene region of Drosophila simulans: evolutionary inferences from an unusual haplotype structure. Genetics 158: 1147–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., J. C. Sanchez-Delbarrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- Schlenke, T. A, and D. J. Begun, 2004. Strong selective sweep associated with a transposon insertion in Drosophila simulans. Proc. Natl. Acad. Sci. USA 101: 1626–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, S., and L. Excoffier, 1999. Estimation of past demographic parameters from the distribution of pairwise differences when mutation rates vary among sites: application to human mitochondrial DNA. Genetics 152: 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöfl, G., and C. Schlötterer, 2004. Patterns of microsatellite variability among X chromosomes and autosomes indicate a high frequency of beneficial mutations in non-African D. simulans. Mol. Biol. Evol. 21: 1384–1390. [DOI] [PubMed] [Google Scholar]
- Slatkin, M., and R. H. Hudson, 1991. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129: 555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solignac, M., 2004. Mitochondrial DNA in the Drosophila melanogaster complex. Genetica 120: 41–50. [DOI] [PubMed] [Google Scholar]
- Tajima, F., 1989. a Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1989. b The effect of change in population size on DNA polymorphism. Genetics 123: 597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teissier, G., 1957. Discriminative biometrical characters in French and Japanese Drosophila melanogaster. Cytologia (Suppl.): 502–505.
- True, J. R., J. M. Mercer and C. C. Laurie, 1996. Differences in crossover frequency and distribution among three sibling species of Drosophila. Genetics 142: 507–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veuille, M., V. Bénassi, S. Aulard and F. Depaulis, 1998. Allele-specific population structure of Drosophila melanogaster Alcohol dehydrogenase at the molecular level. Genetics 149: 971–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall, J. D., 1999. Recombination and the power of statistical tests of neutrality. Genet. Res. 74: 65–79. [Google Scholar]
- Wall, J. D., P. Andolfatto and M. Przeworski, 2002. Testing models of selection and demography in Drosophila simulans. Genetics 162: 203–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson, G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]