Abstract
Rice (Oryza sativa) was cultivated by Asian Neolithic farmers >11,000 years ago, and different cultures have selected for divergent starch qualities in the rice grain during and after the domestication process. An intron 1 splice donor site mutation of the Waxy gene is responsible for the absence of amylose in glutinous rice varieties. This mutation appears to have also played an important role in the origin of low amylose, nonglutinous temperate japonica rice varieties, which form a primary component of Northeast Asian cuisines. Waxy DNA sequence analyses indicate that the splice donor mutation is prevalent in temperate japonica rice varieties, but rare or absent in tropical japonica, indica, aus, and aromatic varieties. Sequence analysis across a 500-kb genomic region centered on Waxy reveals patterns consistent with a selective sweep in the temperate japonicas associated with the mutation. The size of the selective sweep (>250 kb) indicates very strong selection in this region, with an inferred selection coefficient that is higher than similar estimates from maize domestication genes or wild species. These findings demonstrate that selection pressures associated with crop domestication regimes can exceed by one to two orders of magnitude those observed for genes under even strong selection in natural systems.
BEGINNING with Charles Darwin (Darwin 1859, 1897), there has been intense interest in the study of crop species for understanding the process of evolution (Vavilov 1992). Crops can provide unique insights not only into the evolution of “domestication traits” favored by early farming cultures (e.g., loss of seed dispersal mechanisms, increased yield), but also into the subsequent diversification of traits arising from diverse human cultural preferences (e.g., grain color, taste) (Simmonds 1976). The study of genes directly responsible for phenotypic variation within a crop species offers opportunities to infer the origin and dispersal of specific traits selected upon during and after domestication. Moreover, such studies can provide a means to directly assess the genomic impact of human-imposed selection, including the physical boundaries of genomic regions affected by selection under a domestication regime (Wang et al. 1999).
Rice (Oryza sativa L.) is the major staple food for over one-third of the world's population (Khush 1997). It is also among the oldest domesticated crops, with archeological evidence indicating that it was cultivated >11,000 years ago by Asian Neolithic farmers (Mannion 1999). Traditionally, three major “variety groups” or subspecies have been recognized within O. sativa: indica varieties, typical of the Indian subcontinent; tropical japonica (javanica) varieties, most common in Southeast Asia and South China; and temperate japonica varieties, which predominate across northeastern Asia (Glazmann 1987; Khush 1997; Garris et al. 2005). A number of additional genetically distinct variety groups are also recognized, including aromatic varieties of the Indian subcontinent (e.g., basmati) and the aus varieties of Bangladesh and West Bengal (Khush 1997). Rice was domesticated from the wild species O. rufipogon, and there is growing evidence that indica and japonica rice varieties arose through two separate domestication events (Khush 1997; Garris et al. 2005). Temperate japonica varieties are believed to have arisen subsequently from tropical japonicas, as rice cultivation spread northward across Asia following domestication in South Asia (Khush 1997; Garris et al. 2005).
Starch is one of the key quality components of cereal grains, and it has been a target of selection during both domestication and subsequent crop diversification (Whitt et al. 2002; Wilson et al. 2004). In rice, varieties vary widely in the relative proportions of two types of endosperm starch: the unbranched starch amylose (0–30%) and the branched starch amylopectin (70–100%) (Juliano 1985). Rice varieties with high amylose levels (∼20–30%) tend to form discrete, noncohesive grains when cooked, whereas varieties with lower amylose levels form cohesive cooked grains. Higher amylose levels are associated with many South and Southeast Asian rices classified as indica and tropical japonica variety groups; high amylose also occurs in the crop's wild progenitor, O. rufipogon (Morishima et al. 1992). Lower amylose levels (∼10–20%) are more common in Northeast Asia, where a more cohesive cooked grain is often preferred; they are characteristic of the temperate japonica variety groups that predominate in this region (Juliano and Villareal 1993). Varieties possessing only trace amounts of amylose in the endosperm (<1%) are known as glutinous (sticky) rice; these varieties are favored by the upland peoples of Laos and northern Thailand, and they are widely used in festival foods and desserts throughout Asia (Golomb 1976; Juliano and Villareal 1993).
The glutinous phenotype has been shown to result from drastically reduced synthesis of amylose arising from a mutation in the Waxy (Wx) gene, which encodes a granule-bound starch synthase (Sano 1984). Glutinous rice contains a G-to-T mutation at the 5′ splice site of Wx intron 1, which leads to incomplete post-transcriptional processing of the pre-mRNA (Wang et al. 1995; Bligh et al. 1998; Cai et al. 1998; Hirano et al. 1998; Isshiki et al. 1998). Glutinous rice varieties do not have detectable levels of spliced mRNA as a result of this mutation (Wang et al. 1995; Bligh et al. 1998). Interestingly, some degree of amylose synthesis is restored in cultivars that carry the mutation but display cryptic splice-site activation, which results in alternative Wx pre-mRNA splicing patterns (Wang et al. 1995; Bligh et al. 1998; Cai et al. 1998). Thus, the occurrence of the splice donor mutation is characteristic of both glutinous varieties, as well as of some nonglutinous, low-amylose varieties.
In a previous article we described a population genetic and phylogeographic analysis of the O. sativa Wx gene and documented selection for the intron 1 splice donor mutation associated with the origin of glutinous rice (Olsen and Purugganan 2002). Here, we demonstrate that this Wx splice-site mutation has also been under selection in most low-amylose rice landraces that form the temperate japonica varieties cultivated widely in Northeast Asia. Selection for the splice donor mutation in this variety group is associated with a selective sweep that spans ∼250 kb in the Wx genomic region at the top of chromosome 6; the sweep appears to have affected genetic diversity across at least 39 genes. The estimated selection coefficient at the Wx gene, as well as the selection coefficients observed in other domestication genes in maize, indicate that the selection regimes associated with crop domestication and diversification are significantly greater than those observed for even strong cases of natural selection.
MATERIALS AND METHODS
Sampling and classification:
A set of 96 diverse Oryza accessions was selected to represent a broad range of the genetic diversity found in O. sativa and its wild ancestor, O. rufipogon. One sample of O. nivara was included for comparison, and O. barthii and O. meridionalis were included as outgroups. O. sativa samples represent a subset of those analyzed by Garris et al. (2005) and include representatives of the five major subpopulations identified in that study, including 21 indica, 18 tropical japonica, 21 temperate japonica, six aus, and six aromatic landraces from diverse geographical locations in Asia (supplemental Table S1 at http://www.genetics.org/supplemental/). The 21 samples of O. rufipogon represented 8 samples collected in Nepal; 8 from China; and 1 each from Malaysia, India, Laos, and Papua New Guinea. Seed was obtained from the International Rice Germplasm Collection (IRGC) in the Philippines; from the National Small Grains Collection in Aberdeen, Idaho; from the Rice Genetic Resources Unit in Japan; or from Hee Jong Koh in Seoul, Korea (supplemental Table S1 at http://www.genetics.org/supplemental/). O. sativa accessions were purified by single-seed descent for two generations prior to the initiation of the study, while the wild species were grown from seeds obtained directly from the germplasm repositories or collected in the field. Plants were grown in the Guterman greenhouse at Cornell University, and DNA was extracted from single plants using the protocol described in McCouch et al. (1988) with minor modifications.
PCR and DNA sequencing:
Accessions were sequenced at Waxy and portions of 12 flanking loci that were located at ∼40-kb intervals upstream and downstream of Waxy. All primers were designed from the Nipponbare (temperate japonica) genomic sequence available on the Gramene website (http://www.gramene.org), using Primer3 (Rozen and Skaletsky 2000). For Wx, primers were designed to amplify 21 partially overlapping portions of the gene (∼50–100 bp overlap between portions), together spanning ∼1 kb of 5′ promoter sequence, the entire transcriptional unit, and ∼500 bp of 3′ sequence. For flanking loci, primers were designed to amplify ∼500-bp portions of genes with putative or known function; most primer pairs were anchored on exons and spanned one or more intronic regions. All PCR primers were blasted against the rice genomic sequence in Gramene to ensure that only the targeted genomic region would be amplified.
DNA samples (4 ng/reaction) were amplified using M13F- and M13R-tailed PCR primers in 10-μl reactions with titanium DNA polymerase (CLONTECH, Mountain View, CA). PCR products were purified using Acroprep glass fiber filter plates (Pall, East Hills, NY). The purified PCR products were sequenced using M13F and M13R primers and Big Dye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA); sequencing products were separated on a 3700 or 3730xl DNA analyzer (Applied Biosystems). All PCR and DNA sequencing was performed by Genaissance Pharmaceuticals (New Haven, CT). GenBank accession numbers for sequenced regions are DQ280684–DQ281821.
Sequence editing and alignment was performed using the Phred and Phrap programs (Codon Code, Dedham, MA) and BioLign Version 2.09.1 (Tom Hall, North Carolina State University). Heterozygous sites were identified using Polyphred (Deborah Nickerson, University of Washington) and by visual inspection of chromatograms for double peaks. O. sativa is predominantly selfing and is therefore highly homozygous. For all heterozygous O. sativa sequences, phase could be determined through simple haplotype subtraction, and one of the two haplotype sequences was arbitrarily selected to represent the accession in subsequent genetic analyses. In contrast to cultivated rice, wild Oryza species show substantial heterozygosity, and 12 of the 21 sequenced O. rufipogon accessions showed evidence of allele-specific priming at one or more amplified portions across Waxy; these accessions with potentially incomplete sequence data were excluded from all analyses, leaving 9 homozygous O. rufipogon accessions in the analyzed data sets.
Genetic diversity analysis:
Population genetic analyses were conducted for Wx and for each of the 12 flanking genes using DnaSP 4.1 (Rozas and Rozas 1999). All analyses included the published Nipponbare (temperate japonica) genomic sequence. Levels of nucleotide diversity per silent site were estimated as π for each of the variety groups of cultivated rice and for O. rufipogon. Silent-site nucleotide diversity between accessions with and without the Wx intron 1 splice donor mutation was also compared. Patterns of nucleotide diversity were further assessed using Tajima's (1989) D statistic. To examine the genomic impact of selection for the splice donor mutation, extended haplotype homozygosity (EHH) analysis (Sabeti et al. 2002) was used to compare the physical boundaries of haplotypes with and without the splice donor mutation. If favored by positive directional selection, haplotypes carrying the splice donor mutation are expected to manifest an extended block of linkage disequilibrium around the mutation. Selection for the splice donor mutation was also quantified by estimating the selection coefficient on the basis of the observed size of the selective sweep (Kaplan et al. 1989).
Amylose content analysis:
Apparent amylose content was determined using rice flour samples according to the method of Webb (1972) with modifications as described by Perez and Juliano (1975). Digested samples were evaluated using an AutoAnalyzer3 (Seal Analytical, Mequon, WI).
RESULTS AND DISCUSSION
Nucleotide variation at the Waxy gene in rice:
We examined genetic variation in a 5.3-kb region encompassing the entire Wx gene in domesticated Asian rice, O. sativa, and in its wild ancestor, O. rufipogon. The rice samples include the major variety groups within O. sativa (Garris et al. 2005), including 21 accessions of indica, 18 accessions of tropical japonica, 22 accessions of temperate japonica (including the published Nipponbare sequence), and 6 accessions each of aus and aromatic rice varieties (supplemental Table S1 at http://www.genetics.org/supplemental/).
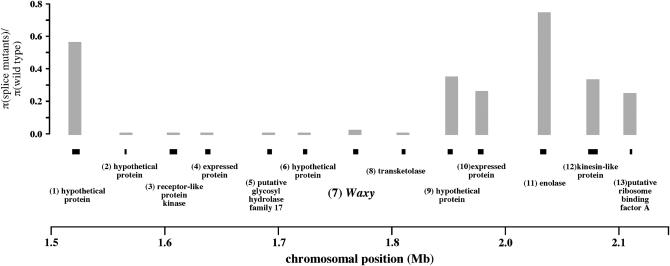
We observed 101 polymorphisms in the O. sativa Wx gene, and the silent-site nucleotide diversity, π, is 0.006 (Table 1). Estimates of π for two of the three major rice variety groups—tropical japonicas and temperate japonicas—are low (π = 0.0002 and 0.0011, respectively). Most of the diversity in the temperate japonicas is due to one accession carrying a haplotype found predominantly in the aus variety group; without this one allele, the levels of silent-site diversity are substantially lower (π = 0.00006). The other major variety group, indica, is at least fivefold more diverse than the japonica variety groups (π = 0.0059). In the nine O. rufipogon accessions included in these analyses, there are 165 single-nucleotide polymorphisms, with silent-site π equal to 0.0213. We also estimated genetic variation in ∼500-bp portions of 12 genes upstream and downstream of Wx in the O. sativa and O. rufipogon genomes (Figure 1); these fragments are spaced ∼40 kb apart. Estimated values of silent-site π in O. sativa for these flanking gene fragments range from 0 to 0.009 (Table 2).
TABLE 1.
Nucleotide variation at the Waxy gene
| Group | Sample size (n) | Nucleotide diversity (π)a | Haplotype diversity | Tajima's D |
|---|---|---|---|---|
| O. sativa | 73 | 0.0056 | 0.845 | +0.4991 |
| variety indica | 21 | 0.0059 | 0.952 | +0.3726 |
| variety tropical japonica | 18 | 0.0002 | 0.739 | −1.5037 |
| variety temperate japonica | 22 | 0.00109 | 0.476 | −2.6149 |
| variety aus | 6 | 0.0028 | 0.733 | +2.0874 |
| variety aromatic | 6 | 0.0037 | 0.800 | −1.5374 |
| O. rufipogon | 9 | 0.0213 | 0.972 | +1.1864 |
Based on silent sites.
Figure 1.
Nucleotide variation across the O. sativa Wx genomic region on chromosome 6. Approximately 500-bp portions of 12 flanking genes were sequenced, along with the entire Wx gene. The approximate genomic locations of the genes are indicated by solid bars, with gene identities as follows: (1) hypothetical protein, (2) hypothetical protein, (3) receptor-like protein kinase, (4) expressed protein, (5) putative glycosyl hydrolase family 17, (6) hypothetical protein, (7) Waxy, (8) transketolase, (9) hypothetical protein, (10) expressed protein, (11) enolase, (12) kinesin-like protein, and (13) putative ribosome-binding factor A. Shaded bars indicate the ratios of silent-site nucleotide diversities for those individuals with and without the Wx splice donor mutation. The smallest bars are equivalent to a ratio of 0.
TABLE 2.
Nucleotide diversities at Waxy and 12 flanking genes for O. sativa and O. rufipogon
| Silent-site nucleotide diversity (π)
|
||||||||
|---|---|---|---|---|---|---|---|---|
|
O. sativa
|
O. rufipogon: | |||||||
| Genea | Genomic position (chromosome 6) (Mb) | indica (N = 21) | temperate japonica (N = 22) | tropical japonica (N = 18) | aus (N = 6) | aromatic (N = 6) | All sativa (N = 73)b | (N = 9) |
| 1. Hypothetical protein | 1.522 | 0.00236 | 0.00060 | 0.00205 | 0.00147 | 0.00082 | 0.00247 | 0.00259 |
| 2. Hypothetical protein | 1.565 | 0.00242 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00071 | 0.00281 |
| 3. Receptor-like protein kinase | 1.606 | 0.00141 | 0.00048 | 0.00141 | 0.00093 | 0.00149 | 0.00127 | 0.00093 |
| 4. Expressed protein | 1.636 | 0.00200 | 0.00000 | 0.00446 | 0.00000 | 0.00387 | 0.00238 | 0.00473 |
| 5. Putative glycosyl hydrolase | 1.688 | 0.00235 | 0.00080 | 0.00313 | 0.00133 | 0.00000 | 0.00351 | 0.00393 |
| 6. Hypothetical protein | 1.720 | 0.00000 | 0.00034 | 0.00161 | 0.00000 | 0.00328 | 0.00112 | 0.00077 |
| 7. Waxy | 1.763–1.768 | 0.00594 | 0.00109 | 0.00015 | 0.00279 | 0.00372 | 0.00559 | 0.02127 |
| 8. Transketolase | 1.810 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00089 |
| 9. Hypothetical protein | 1.850 | 0.00564 | 0.00155 | 0.00262 | 0.00000 | 0.00570 | 0.00864 | 0.00891 |
| 10. Expressed protein | 1.878 | 0.00371 | 0.00030 | 0.00037 | 0.00466 | 0.00333 | 0.00435 | 0.00589 |
| 11. Enolase | 1.933 | 0.00431 | 0.00473 | 0.00755 | 0.00134 | 0.00000 | 0.00905 | 0.00281 |
| 12. Kinesin-like protein | 1.980 | 0.00395 | 0.00182 | 0.00085 | 0.00407 | 0.00000 | 0.00474 | 0.00245 |
| 13. Putative ribosome-binding factor | 2.020 | 0.00150 | 0.00019 | 0.00022 | 0.00219 | 0.00000 | 0.00203 | 0.00194 |
Numbers refer to nomenclature in Figure 1.
Includes O. sativa variety groups indica, temperate japonica, tropical japonica, aus, and aromatic.
Selective sweep at the Wx genomic region:
Previous studies have shown that a splice donor mutation in intron 1 of the Wx gene is associated with the absence of amylose-characterizing glutinous rice varieties (Wang et al. 1995; Bligh et al. 1998; Cai et al. 1998; Hirano et al. 1998; Isshiki et al. 1998). We find that this splice donor mutation is observed with a frequency of 29% in our O. sativa sample overall and that there is reduced variation in the Wx gene for O. sativa accessions that harbor this mutation. The level of silent-site nucleotide diversity at Wx for accessions carrying the splice donor mutation is π = 0.0002, while the estimate of π for wild-type accessions is 0.0064. This represents a 97% reduction in molecular variation at the Wx gene among individuals that carry the splice donor mutation. This pattern is consistent with positive directional selection for the splice donor mutation.
While this reduced nucleotide variation is consistent with a selective sweep at Wx, it could also potentially arise through demographic effects, such as a population bottleneck occurring among the samples that harbor the Wx splice donor mutation. Any such demographic factors would affect genetic diversity across the entire rice genome. To test this possibility, we measured the ratio of silent-site nucleotide diversities between individuals with and without the Wx splice donor polymorphism for 111 gene fragments distributed across the O. sativa genome (A. L. Caicedo, unpublished results), in comparison to Wx. While the ratio at Wx is 0.03, the genomic-average ratio is >10 times greater, at 0.46, indicating that the pattern observed at Wx is not typical of the genome overall.
A selective sweep is expected to affect genetic diversity not only at the specific locus containing a favored mutation but also at surrounding loci, to the extent that they are in linkage disequilibrium with the target of selection. To assess the physical boundaries of the selective sweep around Wx, we compared the ratio of silent-site nucleotide diversity for individuals with and without the splice donor mutation in 12 genes spanning a 500-kb region around Wx. There is a distinct valley of reduced nucleotide variation, spanning ∼250 kb, among individuals carrying the splice donor mutation (Figure 1). Analysis of variation within the japonica groups shows a similar pattern of variation across the Wx genomic region (supplemental Table S2 at http://www.genetics.org/supplemental/).
To confirm that this reduced nucleotide variation across the Wx genomic region is not an artifact of demographic history, we compared mutant-to-wild-type polymorphism ratios for the sequenced loci in the 500-kb Wx genomic region to 111 gene fragments across the O. sativa genome (A. L. Caicedo, unpublished results). The average ratio is 0.22 for the 500-kb Wx genomic region, compared to 0.46 in the genome-wide assessment; this difference is statistically significant (Kruskal–Wallis nonparametric rank test, H = 5.01608, d.f. = 1, P < 0.05). Thus, the reduced nucleotide diversity in the Wx genomic region associated with the splice donor mutation is not due to any demographic factor that affects the entire genome.
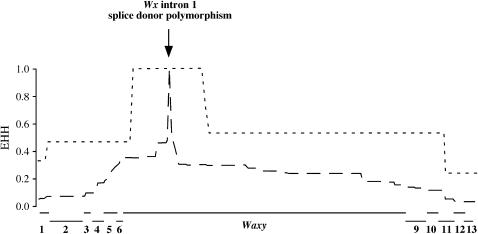
In addition to reducing levels of genetic diversity, positive directional selection is also expected to lead to elevated linkage disequilibrium (LD) around a recently arisen, favored mutation; this increased LD is manifested as a region of EHH associated with the favored mutation (Sabeti et al. 2002). We calculated EHH across the Wx genomic region for accessions with and without the splice donor mutation; the calculation was centered on the putative selected site. The EHH analysis reveals a wide block of LD around the splice donor site for those accessions containing the splice donor mutation, whereas LD decays rapidly on either side of the mutation in wild-type accessions (Figure 2). This pattern strongly suggests positive directional selection specifically for this mutation.
Figure 2.
EHH across the Wx genomic region. Short-dashed and long-dashed lines indicate EHH values for individuals with and without the Wx intron 1 splice donor mutation, respectively. The relative position of the Wx splice donor mutation is indicated. Numbers along x-axis indicate flanking genes (see labels in Figure 1); horizontal lines indicate which polymorphisms correspond to a particular gene.
To assess the degree to which this dramatic difference in LD decay is unique to the splice donor site, we repeated the EHH analysis for all polymorphic sites across the Wx gene that were represented by two alternative nucleotide sites with both sites present at ≥10% frequency. We compared the areas under the curves for the portions of the plots defined by the largest span of polymorphisms where EHH = 1. For the splice donor site mutation, relative EHH = 1 extends for a span of 31 polymorphic sites, and within this region, the difference in area between the splice donor and wild-type polymorphisms is 18.73. This value is significantly greater than the mean value across Wx (9.97 ± 1.18, N = 75), indicating that the extended LD associated with the splice donor mutation is not typical of a Wx nucleotide polymorphism.
Selection for the Wx splice donor mutation in temperate japonica varieties:
The Wx splice donor mutation is not randomly distributed with respect to the major O. sativa variety groups. Over 90% of indica and tropical japonica varieties in the sample carry wild-type Wx alleles (Table 3), as do all aus and aromatic accessions examined. In contrast, the majority of temperate japonicas contain the Wx splice donor mutation; individuals with the mutation represent 85% of the landraces in temperate japonicas (Table 3). This nonrandom distribution of the splice donor polymorphism across variety groups is statistically significant (χ2 = 33.721, d.f. = 4, P < 8.5 × 10−7), and it suggests that there has been selection for the mutation in the temperate japonica variety group.
TABLE 3.
Distribution of Wx intron 1 splice donor mutant and wild-type alleles among rice variety groups
| Group | Mutant | Wild type | Unknown |
|---|---|---|---|
| indica | 2 | 18 | 1 |
| tropical japonica | 1 | 16 | 1 |
| temperate japonica | 17 | 3 | 2 |
| aus | 0 | 6 | 0 |
| aromatic | 0 | 6 | 0 |
The distribution of mutant vs. wild-type Wx alleles is significant in a chi-square test (χ2 = 33.721, d.f. = 4, P < 8.5 × 10−7) for the three major variety groups (indica, tropical japonica, temperate japonica); aus and aromatic varieties were excluded from the statistical test because of small sample size.
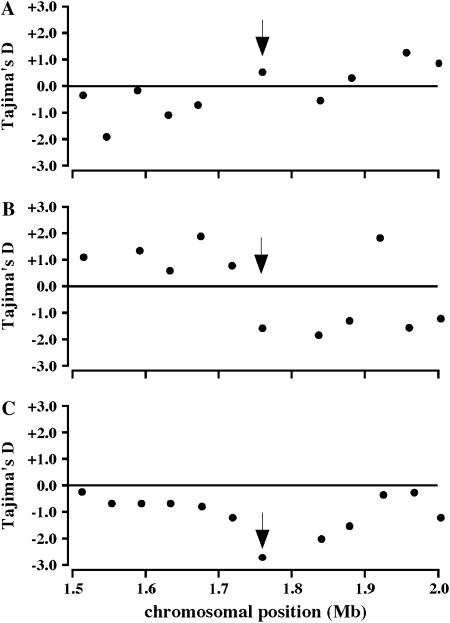
A statistical analysis of nucleotide diversity across the Wx genomic region provides further evidence that there has been selection for the Wx splice donor mutation in the temperate japonica variety group. Tajima's (1989) D statistic describes the frequency distribution of single nucleotide polymorphisms. In temperate japonica accessions, the Wx gene shows a negative deviation in Tajima's D (D = −2.61492) (Table 1, Figure 3), a pattern consistent with recent directional selection on Wx. Moreover, there is a negative trough of Tajima's D across the entire Wx genomic region in temperate japonicas, with Wx located at the minimum (Figure 3). In contrast, no such pattern is observed for either indicas or tropical japonicas, although Tajima's D is also negative for the Wx gene in the latter variety group (Table 1, Figure 3). These patterns are consistent with a recent selective sweep at the Wx gene associated specifically with temperate japonica varieties.
Figure 3.
Levels of Tajima's D in O. sativa major variety groups. Levels of Tajima's D for (A) indica, (B) tropical japonica, and (C) temperate japonica variety groups. The arrow indicates the position of the Wx gene.
If there were selection for the Wx splice-site mutation in temperate japonicas, one would expect a lower amylose content in grains from this variety group compared to indicas and tropical japonicas. Consistent with this prediction, the levels of apparent amylose content (AC) among a random selection of individuals in our sample set is significantly lower for temperate japonica (mean AC = 10.5 ± 5.2, n = 13) than for indica (mean AC = 23.4 ± 1.5, n = 14) and tropical japonica (mean AC = 19.5 ± 1.4, n = 7) variety groups. It should be noted that that four of the temperate japonica samples tested had trace amylose levels and are likely glutinous rice varieties; nonetheless, if we exclude these four samples, the remaining temperate japonicas still have significantly lower levels of apparent amylose content (mean AC = 13.5 ± 1.39, n = 9). This analysis also indicates that most temperate japonicas possessing the Wx intron 1 splice donor mutation have low but detectable levels of amylose, as previously reported (Olsen and Purugganan 2002).
Strength of selection at the O. sativa Wx genomic region:
There are several methods to estimate the strength of selection on a gene, s, from molecular population genetic data. One such approach utilizes the reduced variation surrounding a target of directional selection that arises from genetic hitchhiking of neutral sites linked to the selected site. The distance between a selected and hitchhiking neutral site (d) is dependent on the strength of selection (either as the selection coefficient s or as the population selection parameter α = 2Nes) and on the recombination rate (c or as the population recombination parameter C = 2Nec) in this genomic region. The relationship among these variables is d = 0.01α/C = 0.01s/c (Kaplan et al. 1989), and this relationship has been used to calculate the selection coefficient associated with selective sweeps (Wang et al. 1999). The selection coefficient is equivalent to the fractional increase in progeny carrying the allele in the next generation due to selection; an s of 1 indicates a doubling in allele frequency. It should be noted that inferred values are likely to be imprecise; they do, however, provide an indication of the relative strengths of selection in various systems.
We can calculate the strength of selection using either direct estimates of recombination or the population recombination parameter. Using crossover data between wx mutants, a direct estimate of the intragenic recombination frequency for Wx has been estimated previously as c = 3.67 × 10−7/bp (Inukai et al. 2000). If we assume the span of the affected genomic region to be 250 kb, then the mean distance between the selected site and the farthest hitchhiking site would be 125 kb, and the inferred selection coefficient would be ∼4.59.
We can also use estimated population parameters to calculate s, which has the advantage of taking into account demographic features of the system such as selfing and population history by incorporating Ne into the estimated parameters. The estimated population recombination rate of the Wx gene among those O. sativa individuals that have the splice donor mutation is R = 2C = 0.0103/bp. This provides an estimate of α = 2Nes = 64,375. For O. sativa temperate japonica, Ne is estimated to be ∼7600. This estimate is based on data from 111 sequenced gene fragments and on a coalescent framework that models a population bottleneck in this variety group from an ancestral tropical japonica gene pool, conditional on the domestication event in rice occurring 10,000–12,000 years ago (S. Williamson and A. L. Caicedo, unpublished data). This approach leads to an inferred selection coefficient of 4.24, which is similar to the value estimated from the crossover frequency at Wx.
As a predominantly selfing species, rice would be expected to show lower effective recombination than an outcrossing species (resulting from high homozygosity), leading to a larger selective sweep for a given s value. However, the smaller Ne associated with inbreeding also weakens the effect of selection as a result of drift, and these two forces (lower recombination vs. weaker selection) appear to counterbalance each other.
Table 4 presents a comparison of these inferred selection coefficients with values calculated from selective sweeps reported in wild species (Wootton et al. 2002; Nair et al. 2003; Saez et al. 2003; Schlenke and Begun 2004), as well as for two domestication-related genes in maize (Wang et al. 1999; Clark et al. 2004; Palaisa et al. 2004). All of these estimates use the size of the swept region to estimate s and so are broadly comparable. The inferred selection estimate for the Wx splice donor mutation is higher than the highest selection coefficients observed in wild systems—selection for drug resistance in the malarial parasite Plasmodium falciparum (Table 4). Like rice, P. falciparum is a selfing species, and so the comparison between these two taxa is particularly appropriate. Selection coefficients for the pfcrt drug resistance gene in this species are in the range of 0.1 to 0.7 (Wootton et al. 2002), which is an order of magnitude lower than selection coefficient estimates for the rice Waxy gene.
TABLE 4.
Inferred selection coefficients for domesticated and natural systems
| System | s | Sweep size(kb) | Reference |
|---|---|---|---|
| Domesticated systems | |||
| O. sativa Wxa | 4.59 | ∼250 | This study |
| O. sativa Wxb | 4.24 | ∼250 | This study |
| Z. mays Y1 | 1.2 | >600 | Palaisa et al. (2004) |
| Z. mays tb1 | 0.05 | ∼60–90 | Wang et al. (1999); Clark et al. (2004) |
| Natural systems | |||
| Drosphila melanogaster Sod | 0.02–0.103 | ∼41–54 | Saez et al. (2003) |
| D. simulans Cyp6g1 | 0.022 | ∼100 | Schlenke and Begun (2004) |
| P. falciparum pfcrt | 0.1–0.7 | >200 | Wootton et al. (2002) |
| P. falciparum dhfr | 0.1 | ∼100 | Nair et al. (2003) |
Estimate from wx crossover frequency.
Estimate from population recombination parameter (C = 2Nec).
We can also compare the selection coefficients at the rice Wx gene to examples in maize, another cereal crop species. For the maize tb1 gene, the selected site appears to be in the promoter region, and we use the genome-wide recombination estimate for maize (Fu et al. 2002), which may be a more appropriate value for the intergenic recombination rate estimates (J. Doebley, personal communication). In the maize Y1 genomic region the mean recombination rate across a 1.2-Mb region containing the gene was calculated as 4 × 10−8, using observed recombination in a mapping population (Palaisa et al. 2004). Selection coefficients for both rice Wx and maize Y1, both crop diversification genes, are of the same order of magnitude and higher than that for the domestication gene tb1 (see Table 4).
Rice domestication, the impact of selection, and the evolution of Wx:
Human culture was irrevocably altered by the domestication of cereal crops, whose cultivation directly led to the development of sedentary agriculturalist societies from hunter–gatherer groups (Harlan 1975). There has been a recent concerted effort to scan genomes for loci that may underlie the domestication process (Wright et al. 2005) and the associated phenotypes that they confer. The levels and composition of starch were major targets of selection, both during the process of domestication and with the subsequent diversification of crops to fill agroecological and cultural niches (Whitt et al. 2002; Wilson et al. 2004). Selection on cereal crop starches has been documented in two clear examples. In maize, multiple genes in the starch biosynthetic pathway, including ae1, bt2, and su1, appear to have been under directional selection (Whitt et al. 2002; Wilson et al. 2004), possibly associated with specific culinary use (tortilla production) by the early Mexican domesticators of this crop. In rice, the origin of the glutinous phenotype (<1% amylose), favored by upland peoples of Southeast Asia, arose through selection for the Wx intron 1 splice donor mutation, which disrupts the synthesis of the amylose biosynthetic enzyme granule-bound starch synthase (Olsen and Purugganan 2002).
Much of the rice in Northeast Asia belongs to the temperate japonica rice variety group (Khush 1997), and this major rice variety group has reduced levels of amylose compared to individuals in the indica and tropical japonica variety groups (Juliano and Villareal 1993). Previous molecular genetic data demonstrate that the low-to-moderate amylose content arises from the partial suppression of the Wx splice donor mutation (Wang et al. 1995; Bligh et al. 1998; Cai et al. 1998), which is controlled in part by modifier genes, including the Dull locus (Dung et al. 2000; Isshiki et al. 2000). Thus, the Wx splice donor mutation appears to have played a key role in the evolution of the nonglutinous temperate japonica variety group as well as of many glutinous rice varieties.
The temperate japonicas are not fixed for the Wx splice donor mutant alleles. In our sample, three identified alleles do not carry the splice donor mutation. One of these is characteristic of the aus variety group, and its occurrence in a temperate japonica variety may reflect introgression from cross-breeding between variety groups. The other two alleles are equivalent to a sequence previously identified as the “progenitor” haplotype from which the splice donor mutation arose (Olsen and Purugganan 2002). This mutation-progenitor haplotype predominates in the tropical japonica variety group, and its very high frequency accounts for the low genetic diversity of that variety group. The majority of the Wx alleles in tropical japonicas correspond to this or to closely related derivative haplotypes. The reduction in variation at Wx in the tropical japonicas may suggest that this progenitor haplotype was also selected for in this variety group, but the evidence for a recent selective sweep is not compelling.
Selection for the Wx splice donor mutation has had a profound impact on the Wx genomic region, resulting in (i) a broad ∼250-kb region of reduced nucleotide variation (Figure 1), (ii) extended linkage disequilibrium associated with the favored mutation (Figure 2), and (iii) a trough of excess low-frequency polymorphisms in the genomic region surrounding the Wx locus (Figure 3). The value of selection coefficient in the Wx region appears to be very high and may be associated with the high cultural and culinary value of highly cohesive rice grains in Northeast Asian cultures. Studies have shown that Northeast Asians, where temperate japonicas are predominantly cultivated, have clear preferences for low-amylose rice (Juliano and Hicks 1996). Moreover, lower amylose levels result in more cohesive or stickier rice grains when cooked, making it easier to manipulate rice grains during eating. The reduced amylose levels as a result of selection on Wx in temperate japonica may thus be associated with the cultural use of chopsticks by Northeast Asians in rice consumption.
The size of the selective sweep in rice Wx is similar to those observed in the outcrossing crop species Zea mays for tb1 and Y1 (Table 4), despite the fact that rice is a selfing species and might be expected to show greater linkage disequilibrium associated with lower effective recombination rates. For the maize tb1 locus, the selective sweep affected variation in an ∼60- to 90-kb genomic region upstream of the gene (Wang et al. 1999; Clark et al. 2004); for the Y1 locus, genomic diversity is reduced across an ∼600-kb region, primarily downstream of the target gene (Palaisa et al. 2004). As with both of these maize loci, the selective sweep around Wx in O. sativa is asymmetrical, extending only ∼40 kb downstream but ∼200 kb upstream of Wx. Such asymmetries are commonly seen in theoretical simulations of genetic hitchhiking arising from positive selection, particularly in cases of low effective population sizes (Kim and Stephan 2002).
While the size of domestication-related selective sweeps may be comparable between rice and maize, these two species may in fact show very different genomic responses to selection as a result of differences in genome structure. In maize, the low density of genes across the genome limits the hitchhiking impact of a selective sweep to relatively few linked genes. For example, no other protein-coding genes (outside of those found in inserted transposon sequences) are affected by the selection on tb1 (Wang et al. 1999; Clark et al. 2004). In contrast, reduced nucleotide diversity in the Wx genomic region spans ∼39 genes as a result of genetic hitchhiking. Thus, the higher gene density of rice results in selective sweeps affecting gene diversity levels among a relatively large number of loci. This finding suggests that “selective interference” (Hill and Robertson 1966)—in which the evolutionary response to selection is slowed by countervailing selection pressures at linked genes—may prove to be more pronounced in rice than in other domesticated cereals.
Taken together, our results illustrate the utility of using domesticated systems in studying the evolutionary dynamics of strong selection regimes and their impacts on genome structure and diversity. They also provide glimpses into the co-evolution of the human species and other organisms, evolutionary interactions that together lay the foundation for human civilizations.
Acknowledgments
We thank members of the Purugganan and McCouch laboratories and Brandon Gaut for a critical reading of the manuscript. We also thank Vince Schulz of Genaissance Pharmaceuticals for managing the high-throughput sequencing component of the project. This research was funded in part by a grant from the National Science Foundation Plant Genome Research Program to M.D.P, S.R.M, Carlos Bustamante, and Rasmus Nielsen.
References
- Bligh, H. F. J., P. D. Larkin, P. S. Roach, C. A. Jones, H. Fu et al., 1998. Use of alternate splice sites in granule-bound starch synthase mRNA from low-amylose rice varieties. Plant Mol. Biol. 38: 407–415. [DOI] [PubMed] [Google Scholar]
- Cai, X. L., Z. Y. Wang, Y. Y. Xing, J. L. Zhang and M. M. Hong, 1998. Aberrant splicing of intron 1 leads to the heterogeneous 5′ UTR and decreased expression of waxy gene in rice cultivars of intermediate amylose content. Plant J. 14: 459–465. [DOI] [PubMed] [Google Scholar]
- Clark, R. M., E. Linton, J. Messing and J. F. Doebley, 2004. Pattern of diversity in the genomic region near the maize domestication gene tb1. Proc. Natl. Acad. Sci. USA 101: 700–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin, C., 1859. On the Origin of Species (1988 reprint). Harvard University Press, Cambridge, MA.
- Darwin, C., 1897. The Variation of Animals and Plants Under Domestication. D. Appleton, New York.
- Dung, L. V., I. Mikami, E. Amano and Y. Sano, 2000. Study on the response of dull endosperm 2–2, du2–2, to two wx alleles in rice. Breeding Sci. 50: 215–219. [Google Scholar]
- Fu, H., Z. Zheng and H. Dooner, 2002. Recombination rates between adjacent genic and retrotransposon regions in maize vary by 2 orders of magnitude. Proc. Natl. Acad. Sci. USA 99: 1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garris, A. J., T. H. Tai, J. Coburn, S. Kresovich and S. R. McCouch, 2005. Genetic structure and diversity in Oryza sativa L. Genetics 169: 1631–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazmann, J. C., 1987. Isozymes and classification of Asian rice varieties. Theor. Appl. Genet. 74: 21–30. [DOI] [PubMed] [Google Scholar]
- Golomb, L., 1976. The origin, spread and persistence of glutinous rice as a staple crop in mainland Southeast Asia. J. Southeast Asian Stud. 7: 1–15. [Google Scholar]
- Harlan, J. R., 1975. Crops and Man. American Society of Agronomy, Madison, WI.
- Hill, W. G, and A. Robertson, 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8: 269–294. [PubMed] [Google Scholar]
- Hirano, H. Y., M. Eiguchi and Y. Sano, 1998. A single base change altered the regulation of the waxy gene at the posttranscriptional level during the domestication of rice. Mol. Biol. Evol. 15: 978–987. [DOI] [PubMed] [Google Scholar]
- Inukai, T., A. Sako, H. Y. Hirano and Y. Sano, 2000. Analysis of intragenic recombination at wx in rice: correlation between the molecular and genetic maps within the locus. Genome 43: 589–596. [DOI] [PubMed] [Google Scholar]
- Isshiki, M., K. Morino, M. Nakajima, R. J. Okagaki, S. R. Wessler et al., 1998. A naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5′ splice site of the first intron. Plant J. 15: 133–138. [DOI] [PubMed] [Google Scholar]
- Isshiki, M., M. Nakajima, H. Satoh and K. Shimamoto, 2000. dull: rice mutants with tissue-specific effects on the splicing of the waxy pre-mRNA. Plant J. 23: 451–460. [DOI] [PubMed] [Google Scholar]
- Juliano, B. O., 1985 Rice: Chemistry and Technology. American Association of Cereal Chemists, St. Paul.
- Juliano, B. O., and P. A. Hicks, 1996. Rice functional properties and food products. Food Rev. Int. 12: 71–103. [Google Scholar]
- Juliano, B. O., and C. P. Villareal, 1993. Grain quality evaluation of world rices. IRRI, Manila, Philippines.
- Kaplan, N. L., R. R. Hudson and C. H. Langley, 1989. The hitchhiking effect revisited. Genetics 123: 887–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khush, G. S., 1997. Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 35: 25–34. [PubMed] [Google Scholar]
- Kim, Y., and W. Stephan, 2002. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics 160: 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannion, A. M., 1999. Domestication and the origins of agriculture: an appraisal. Prog. Phys. Geogr. 23: 37–56. [Google Scholar]
- McCouch, S. R., G. Kochert, Z. Yu, Z. Wang, G. S. Khush et al., 1988. Molecular mapping of rice chromosomes. Theor. Appl. Genet. 76: 815–829. [DOI] [PubMed] [Google Scholar]
- Morishima, H., Y. Sano and H. I. Oka, 1992. Evolutionary studies in cultivated rice and its wild relatives. Oxf. Surv. Evol. Biol. 8: 135–184. [Google Scholar]
- Nair, S., J. T. Williams, A. Brockman, L. Paiphun, M. Mayxay et al., 2003. A selective sweep driven by pyrimethamine treatment in southeast Asian malaria parasites. Mol. Biol. Evol. 20: 1526–1536. [DOI] [PubMed] [Google Scholar]
- Olsen, K. M., and M. D. Purugganan, 2002. Molecular evidence on the origin and evolution of glutinous rice. Genetics 162: 941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaisa, K., M. Morgante, S. Tingey and A. Rafalski, 2004. Long-range patterns of diversity and linkage disequilibrium surrounding the maize Y1 gene are indicative of an asymmetric selective sweep. Proc. Natl. Acad. Sci. USA 101: 9885–9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, C. M. and B. O. Juliano, 1975 Modification of the simplified amylose test for milled rice. Starch/Starke 30: 424–426.
- Rozas, J., and R. Rozas, 1999. DnaSP: an integrated program for molecular population genetics and molecular evolution analysis, version 3. Bioinformatics 15: 174–175. [DOI] [PubMed] [Google Scholar]
- Rozen, S., and H. J. Skaletsky, 2000. Primer3 on the WWW for general users and for biologist programmers, pp. 365–386 in Bioinformatics Methods and Protocols: Methods in Molecular Biology, edited by S. Krawetz and S. Misener. Humana Press, Totowa, NJ. [DOI] [PubMed]
- Sabeti, P. C., D. E. Reich, J. M. Higgins, H. Z. P. Levine, D. J. Richter et al., 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 419: 832–837. [DOI] [PubMed] [Google Scholar]
- Saez, A. G., A. Tatarenkov, E. Barrio, N. H. Becerra and F. J. Ayala, 2003. Patterns of DNA sequence polymorphism at Sod vicinities in Drosophila melanogaster: unraveling the footprint of a recent selective sweep. Proc. Natl. Acad. Sci. USA 100: 1793–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano, Y., 1984. Differential regulation of waxy gene expression in rice endosperm. Theor. Appl. Genet. 68: 467–473. [DOI] [PubMed] [Google Scholar]
- Schlenke, T. A., and D. J. Begun, 2004. Strong selective sweep associated with a transposon insertion in Drosophila simulans. Proc. Natl. Acad. Sci. USA 101: 1626–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds, N. W., 1976. Evolution of Crop Plants. Longman, New York.
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavilov, N. I., 1992. Proiskhozhdenie i Geografiia Kulturnykh Rastenii (Origin and Geography of Cultivated Plants), translated by Doris Löve. Cambridge University Press, New York.
- Wang, R. L., A. Stec, J. Hey, L. Lukens and J. Doebley, 1999. The limits of selection during maize domestication. Nature 398: 236–239. [DOI] [PubMed] [Google Scholar]
- Wang, Z. Y., F. Q. Zheng, G. Z. Shen, J. P. Gao, D. P. Snustad et al., 1995. The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J. 7: 613–622. [DOI] [PubMed] [Google Scholar]
- Webb, B. D., 1972. Automated system of amylose analysis in whole-kernel rice. Cereal Sci. Today 17: 284. [Google Scholar]
- Whitt, S. R., L. M. Wilson, M. I. Tenaillon, B. S. Gaut and E. S. Buckler, 2002. Genetic diversity and selection in the maize starch pathway. Proc. Natl. Acad. Sci. USA 99: 12959–12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, L. M., S. R. Whitt, A. M. Ibanez, T. R. Rocheford, M. M. Goodman et al., 2004. Dissection of maize kernel composition and starch production by candidate gene association. Plant Cell 16: 2719–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton, J. C., X. Feng, M. T. Ferdig, R. A. Cooper, M. Jianbing et al., 2002. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 418: 320–323. [DOI] [PubMed] [Google Scholar]
- Wright, S., I. V. Bi, S. Schroeder, M. Yamasaki, J. F. Doebley et al., 2005. The effects of artificial selection of the maize genome. Science 308: 1310–1314. [DOI] [PubMed] [Google Scholar]