Abstract
Transcriptional silencing involves the formation of specialized repressive chromatin structures. Previous studies have shown that the histone H3–H4 chaperone known as chromatin assembly factor 1 (CAF-1) contributes to transcriptional silencing in yeast, although the molecular basis for this was unknown. In this work we have identified mutations in the nonconserved C terminus of antisilencing function 1 (Asf1) that result in enhanced silencing of HMR and telomere-proximal reporters, overcoming the requirement for CAF-1 in transcriptional silencing. We show that CAF-1 mutants have a drastic reduction in DNA-bound histone H3 levels, resulting in reduced recruitment of Sir2 and Sir4 to the silent loci. C-terminal mutants of another histone H3–H4 chaperone Asf1 restore the H3 levels and Sir protein recruitment to the silent loci in CAF-1 mutants, probably as a consequence of the weakened interaction between these Asf1 mutants and histone H3. As such, these studies have identified the nature of the molecular defect in the silent chromatin structure that results from inactivation of the histone chaperone CAF-1.
THE eukaryotic genome is packaged into chromatin, the foundation of which is a regular array of repeating units termed nucleosomes. The nucleosome is composed of ∼147 bp of DNA wound almost twice around an octamer of histone proteins with two molecules each of histones H2A, H2B, H3, and H4 (Luger et al. 1997). The packaging of the DNA into chromatin has a profound influence on transcriptional regulation (Peterson and Laniel 2004; Cairns 2005). A clear example of this is provided by heterochromatin or silenced chromatin, where additional proteins bind to the nucleosomal array to generate a specialized chromatin structure that is transcriptionally “silent.”
The formation of silent chromatin structures has been extensively studied in budding yeast (Rusche et al. 2003). The budding-yeast genome has three regions that are transcriptionally silenced: the mating-type loci HML and HMR, the rDNA, and the telomere-proximal regions. Here we focus on the mating-type loci and telomere-proximal regions. Silencing at the mating-type loci and telomere-proximal regions is established by the recruitment of the Silent information regulator (Sir) proteins to the silencers, which are DNA sequences that dictate the region to be silenced. Sir4 preexists in a soluble complex with Sir2, while Sir3 is thought to be recruited to DNA via Sir4 (Moazed et al. 1997; Ghidelli et al. 2001; Hoppe et al. 2002). Once bound to the silencer, the Sir proteins spread throughout the silent locus, with Sir3 and Sir4 binding to the unacetylated N-terminal tails of histones H3 and H4 (Hecht et al. 1995; Carmen et al. 2002). The enzymatic activity of Sir2 as a NAD-dependent histone deacetylase promotes spreading of the silent chromatin structure at the telomere-proximal regions and mating-type loci by deacetylating the histones to create high-affinity binding sites for Sir3 and Sir4 (Smith et al. 1998; Imai et al. 2000; Landry et al. 2000). The extent of spreading of the Sir proteins is limited by boundaries between silenced and expressed chromatin. One mechanism by which the boundaries of silent chromatin structure appear to be set is the localized recruitment of histone acetyltransferases, resulting in acetylated histones that are refractory to binding of Sir proteins (Donze and Kamakaka 2001). Consistent with this idea, Sir3 spreads further from a telomere when the gene encoding the histone acetyltransferase Sas2 is deleted (Kimura et al. 2002; Suka et al. 2002). Similarly, the binding of bromodomain factor 1 (Bdf1) to acetylated chromatin prevents deacetylation by Sir2 and therefore prevents the spreading of the Sir proteins (Ladurner et al. 2003).
The silent chromatin structure is maintained and inherited through cell division. Every time the DNA replicates, the histones are reassembled onto the newly replicated DNA (Cairns 2005). This process is mediated in part by histone chaperones. The histone chaperone chromatin assembly factor 1 (CAF-1) was discovered by its biochemical ability to deposit histones H3 and H4 onto newly replicated DNA (Smith and Stillman 1989). CAF-1 is also likely to assemble chromatin following DNA replication in vivo, as it localizes to sites of DNA replication (Marheineke and Krude 1998) and is found in a complex with histones that are specifically assembled following DNA replication (Tagami et al. 2004). Furthermore, bulk chromatin from yeast lacking CAF-1 is more accessible to digestion by micrococcal nuclease and DNAseI, and the endogenous 2μ plasmid is less supercoiled, consistent with a role for CAF-1 in global chromatin assembly in vivo (Hoek and Stillman 2003; Adkins and Tyler 2004; Nabatiyan and Krude 2004). In vitro, CAF-1-mediated chromatin assembly following DNA replication is dependent on another histone H3–H4 chaperone termed antisilencing function 1 (Asf1) (Tyler et al. 1999). Like CAF-1, Asf1 also localizes to DNA replication forks in vivo (Schulz and Tyler 2006). The binding partners and phenotypes of yeast lacking ASF1 have implicated Asf1 in many processes. These binding partners include the DNA damage checkpoint protein Rad53, the bromodomain factor Bdf1, the histone information regulator Hir1, and the histone acetyl transferase something about silencing (SAS-I) complex in addition to CAF-1 and histones H3 and H4 (Tyler et al. 1999, 2001; Meijsing and Ehrenhofer-Murray 2001; Osada et al. 2001; Sharp et al. 2001; Chimura et al. 2002; Mello et al. 2002). Yeast lacking ASF1 are sensitive to DNA damaging agents and replicational stress and show transcriptional defects (Le et al. 1997; Singer et al. 1998; Tyler et al. 1999; Sutton et al. 2001; Chimura et al. 2002; Adkins et al. 2004; Ramey et al. 2004; Zabaronick and Tyler 2005). These phenotypes presumably reflect the role of Asf1 in mediating chromatin assembly and/or disassembly during replication, DNA repair, and transcriptional regulation.
Both Asf1 and CAF-1 contribute to transcriptional silencing, but the molecular basis for this is unknown. Deletion of any of the three genes encoding the CAF-1 complex, CAC1, CAC2, or MSI1/CAC3, results in a loss of transcriptional silencing (Kaufman et al. 1997). The silencing defect in CAF-1 mutants appears to be due to the transient loss of silencing (Enomoto and Berman 1998) and an increased frequency of switching the expression state of telomeric reporter genes (Monson et al. 1997). As such, silencing is established in the absence of CAF-1, but CAF-1 is important for the maintenance of silencing through the cell cycle and the inheritance of silencing through DNA replication. Overexpression of Asf1 weakens silencing of reporters at the HMR and telomere-proximal loci (Le et al. 1997; Singer et al. 1998). Deletion of ASF1 causes a nominal defect in silencing (Le et al. 1997; Singer et al. 1998), while deletion of ASF1 in addition to inactivation of CAF-1 or mutation of the silencing enhancer HMR-E leads to a further defect in silencing (Tyler et al. 1999; Meijsing and Ehrenhofer-Murray 2001).
To investigate the molecular basis for the contribution of CAF-1 and Asf1 to transcriptional silencing, we screened for insertion mutations in yeast ASF1 that alter its silencing abilities. The biochemical and genetic characterization of Asf1 mutants that bypass the requirement for CAF-1 in silencing has allowed us to discern the molecular contribution of the CAF-1 and Asf1 histone chaperones to the silent chromatin structure. Specifically, CAF-1 is required to deposit a foundation of nucleosomes for recruitment of the Sir proteins; in the absence of CAF-1 there is a striking reduction in histone H3 and Sir protein occupancy at the silent loci. This role for CAF-1 in transcriptional silencing can be bypassed by dominant Asf1 mutants that result in increased chromatin assembly and recruitment of Sir proteins in CAF-1 mutants.
MATERIALS AND METHODS
Transposon insertion mutagenesis:
The ASF1 ORF and promoter inserted into pRS314 were subjected to insertion mutagenesis using the GPS-LS linker-scanning system (New England Biolabs, Beverly, MA). One-third of the resulting 15-bp insertions introduced an in-frame stop codon. The mutagenized plasmids were then transformed into asf1Δcac1Δ yeast, plated onto low adenine plates, and visually screened for color changes.
Silencing assays:
Yeast strains (see Table 1) were grown to log phase and adjusted to an OD600nm of 1.0. Strains were then spotted on plates in 10-fold serial dilutions on rich media, media lacking TRP (both labeled “control” in Figures 1, 2, and 4) or 5′ fluoroorotic acid (5′FOA), or low adenine (low ade). Yeast were grown for 2–4 days at 30° and then placed at 4° for 7 days to allow for the color to develop. To assay the degree of sectoring, cells were streaked onto −TRP/low-ade plates, grown for 3–5 days until the colony size was large enough to visualize a sector, and then placed at 4° to develop the colony color.
TABLE 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Genotype |
|---|---|
| ACN026 | W303 MATα ade2-1 leu2-3,112, lys5, ura3-52, ade3∷Gal10∷HO ΔHO∷Ade1, Δhml∷ADE1 Δhmr∷ade1 ASF1-13myc:KAN |
| BAT003 | W303 MATaade2-1 can1-100 his3-11 leu2-3,112 trp1-1 ura3-1 Asf1-185T-13myc∷KAN |
| BAT004 | W303 MATα ade2-1 leu2-3,112 his3-11 trp1-1 ura3-1 cac1∷LEU2 TELVIIL∷URA3 HMRa∷ADE2 Asf1-185T-13myc∷KAN |
| BAT005 | W303 MATaade2-1 can1-100 his3-11 leu2-3,112 trp1-1 ura3-1 Asf1-152T-13myc∷KAN |
| BAT006 | W303 MATα ade2-1 leu2-3,112 his3-11 trp1-1 ura3-1 cac1∷LEU2 TEVIIL∷URA3 459bpT-13myc∷KAN |
| BAT0033 | W303 MATα ade2-1 leu2-3,112 his3-11 trp1-1 ura3-1 asf1∷his5+ bdf1∷KAN TEVIIL∷URA HMRa∷ADE2 |
| BAT0034 | W303 MATα ade2-1 leu2-3,112 his3-11 trp1-1 ura3-1 asf1∷his5+ cac1∷LEU2 hir1∷KAN TEVIIL∷URA HMRa∷ADE2 |
| BAT0043 | W303 MATα ade2-1 leu2-3,112 his3-11 trp1-1 ura3-1 cac1∷LEU2 TEVIIL∷URA HMRa∷ADE2 558bpT-13myc∷KAN SIR2-3HA∷TRP |
| BAT0044 | W303 MATα ade2-1 leu2-3,112 his3-11 trp1-1 ura3-1 cac1∷LEU2 TEVIIL∷URA HMRa∷ADE2 459bpT-13myc∷KAN SIR2-3HA∷TRP |
| BAT0045 | W303 MATα ade2-1 leu2-3,112 his3-11 trp1-1 ura3-1 cac1∷LEU2 TEVIIL∷URA HMRa∷ADE2 SIR2-3HA∷TRP |
| BAT0046 | W303 MATα ade2-1 LYS2 leu2-3,12 his3-11 trp1-1 ura3-1 TELVIIL∷URA3 HMRa∷ADE2 can1-100 SIR2-3HA∷TRP |
| BAT0052 | W303 MATα ade2-1 leu2-3,12 his3-11 trp1-1 ura3-1 asf1∷his5+ bdf1∷KAN cac1∷LEU2 TELVIIL∷URA3 HMRa∷ADE2 |
| BAT0053 | W303 MATaade2-1 leu2-3,12 his3-11 trp1-1 ura3-1 asf1∷his5+ sir2∷KAN cac1∷LEU2 TELVIIL∷URA3 HMRa∷ADE2 |
| BAT055 | W303 MATα ade2-1 leu2-3,12 his3-11 trp1-1 ura3-1 ASF1-152T∷KAN cac1∷LEU2 TELVIIL∷URA3 HMRa∷ADE2 can1-100 |
| BAT056 | W303 MATα ade2-1 leu2-3,12 his3-11 trp1-1 ura3-1 ASF1-185T∷KAN cac1∷LEU2 TELVIIL∷URA3 HMRa∷ADE2 can1-100 |
| JKT0049 | W303 MATα ade2-1 leu2-3,112 his3-11 trp1-1 ura3-1 cac1∷LEU2 TELVIIL∷URA HMRa∷ADE2 (Tamburini et al. 2005) |
| ROY1169 | W303 MATα ade2-1 leu2-3,12 his3-11 trp1-1 ura3-1 cac1∷LEU2 asf1∷his5+ TELVIIL∷URA3 HMRa∷ADE2 can1-100 (Tyler et al. 1999) |
| ROY1171 | W303 MATα ade2-1 LYS2 leu2-3,112 his3-11 trp1-1 ura3-1 cac1∷LEU2 TELVIIL∷URA3 HMRa∷ADE2 can1-100 (Tyler et al. 1999) |
| ROY1172 | W303 MATα ade2-1 LYS2 leu2-3,112 his3-11 trp1-1 ura3-1 TELVIIL∷URA3 HMRa∷ADE2 can1-100 (Tyler et al. 1999) |
DNA damage sensitivity:
Yeast strains were grown to log phase and adjusted to an OD600nm of 1.0. Strains were then spotted on plates in 10-fold serial dilutions on media lacking TRP (labeled control in Figure 2) and with 0.005 and 0.01% methyl methanesulfonate (MMS). Yeast were grown for 2–4 days at 30°.
Phosphatase assay:
Phosphatase activity was measured exactly as described previously (Adkins et al. 2004).
Flow cytometry analysis:
Approximately 5 × 106 cells per sample were stained with propidium iodide (Stone and Pillus 1996). Ten thousand cells per sample were scanned using a Beckman–Coulter XL-MCL machine.
Immunoprecipitation:
Immunoprecipitation analyses were performed exactly as described previously (Tamburini et al. 2005).
Chromatin immunoprecipitation:
Chromatin immunoprecipitation (ChIP) was performed as described previously (Kuo and Allis 1999) with the following alterations. Yeast cells were grown overnight in rich media, diluted, and grown 2–3 hr at 30° until cells reached an OD600nm of 1.0. Each ChIP reaction was performed in duplicate with 1 × 108 cells for H3 and Sir2-HA immunoprecipitations and with 9 × 108 for Sir4 immunoprecipitations, with a crosslinking time of 15 min. To immunoprecipitate Sir2-HA, Sir4, and H3 we used 4 μl of anti-mouse HA antibody (Covance), 1 μl of antiserum to Sir4 (Hoppe et al. 2002), and 2 μl of antiserum to the C terminus of H3 (Abcam), respectively. Samples were analyzed using a 1:625–1:2500 dilution of the immunoprecipitated samples and a 1:240,000 dilution of the input samples. The linear range of PCR amplification was determined, using twofold dilutions in initial experiments, to be 25 cycles. The primer pairs were used as described previously (Hoppe et al. 2002). Agarose gels (4.5–5%) were used to analyze small PCR products using a 1:1 mixture of GenePure LE Agarose (ISC Bioexpress) and the low melting temperature NuSieve GTG Agarose (Cambrex) certified for the recovery of nucleic acids <1 kb. The DNA was resolved on the agarose gels with 1.5% eithidium bromide and quantitated from an unsaturated image using Labworks (UVB Bioimaging Systems). To calculate the percentage of total chromatin bound, immunoprecipitated chromatin (IP) and total chromatin (input) were multiplied by the respective dilution factors, and then the ratio of immunoprecipitated chromatin to total chromatin was calculated and multiplied by 100 to get a percentage of total chromatin value. In Figure 5, error bars represent the standard deviation from the average of three independent experiments and asterisks represent statistical significance, where one asterisk represents a P-value of <0.1, two asterisks represent a P-value of <0.05, and three asterisks represents a P-value of <0.001.
Figure 5.
Reduced histone H3 and Sir levels at the silent loci in CAF-1 mutants are restored by the 152T and 185T mutations of Asf1. (A) Chromatin immunoprecipitation (ChIP) analysis of Sir2 levels. Quantitation of Sir2-HA ChIP analysis from strains ROY1172 (No tag), BAT046 (WT), BAT043 (cac1Δ Asf1-185T), BAT044 (cac1Δ Asf1-152T), and BAT045 (cac1Δ) is shown at the indicated regions of the genome; primer pairs used were as described previously (Hoppe et al. 2002), where TEL0.35 is 0.35 kb away from the end of TELVIR, etc., and the HMR-E primer pairs are located within the HMRa locus flanking the E silencers. Percentage of total chromatin was calculated as the percentage of total chromatin precipitated. Error bars represent the standard deviation of at least three independent experiments. (B) Chromatin immunoprecipitation (ChIP) analysis of Sir4 levels: quantitation of Sir4 antibody ChIP from the same strains used in A. (C) Chromatin immunoprecipitation (ChIP) analysis of H3 levels: quantitation of H3 C-terminal Antibody ChIP from the same strains used in A. The ratio of immunoprecipitated DNA (IP) to input was calculated as a percentage of the total chromatin, and the value for the wild-type samples was normalized to 1. Error bars represent the standard deviation of multiple independent experiments. (D) Western blot analysis of Sir2-HA. Soluble protein extracts from the yeast strains used in A were Western blotted with either an anti-HA antibody or a GAPDH antibody (as a loading control) to visualize Sir2 or GAPDH levels, respectively.
RESULTS
Isolation of Asf1 mutants with enhanced silencing capabilities:
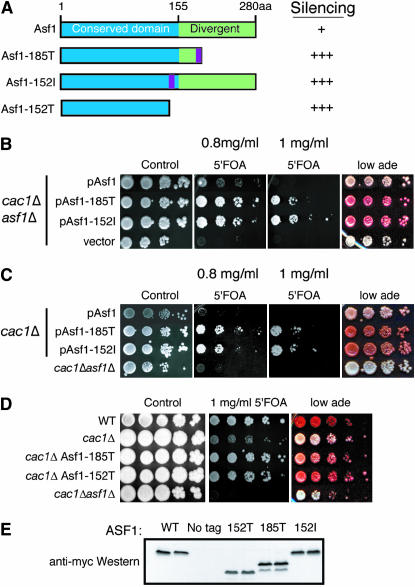
To better understand the molecular role of Asf1 in transcriptional silencing, we used transposon-insertion mutagenesis of the ASF1 open reading frame to identify mutations that alter Asf1-mediated silencing. We generated a library of insertion mutations into a CEN/ARS plasmid carrying the ASF1 open reading frame driven by the ASF1 promoter. This ASF1 plasmid library was introduced into a strain deleted for ASF1 and CAC1; CAF-1, via deletion of CAC1, uncovers the role of Asf1 in transcriptional silencing (Tyler et al. 1999). To assay silencing, the strain carried a URA3 gene inserted proximal to the telomere and an ADE2 gene inserted into the HMR locus. A defect in silencing of TELVIIL∷URA3 leads to death on 5′FOA, while a defect in silencing of HMR∷ADE2 leads to a change in colony color from pink to white. We visually screened 2500 colonies derived from the ASF1 plasmid library for a change from the light pink colonies of cac1 mutants on low-adenine plates.
We identified many isolates from the ASF1 plasmid library with a defect in transcriptional silencing that were also sensitive to DNA damaging agents. Sequencing of the plasmids indicated that many of the insertions introduced a 5′ in-frame stop codon generating extremely short Asf1 truncations and these were not pursued further. We isolated two Asf1 mutants with an enhanced ability to silence as compared to the wild-type Asf1 protein expressed from the same CEN vector. One of these, isolated five independent times, carried an insertion mutation, termed “185T,” resulting in an in-frame stop codon and truncation of Asf1 after amino acid 185 (Figure 1A). Another Asf1 mutant that had enhanced silencing, termed “152I,” had a 15-bp insertion encoding amino acids V F L H Y between amino acids 152 and 153 of Asf1 (Figure 1A). The 185T and 152I mutants had an improved ability to grow on 5′FOA and had a darker pink colony color as compared to cac1 mutants carrying the pAsf1 plasmid (Figure 1B). As such, the 152I and 185T Asf1 mutants had increased abilities to mediate transcriptional silencing, as compared to the wild-type Asf1 protein, at the telomere-proximal and HMR loci. Interestingly, the Asf1 mutants with enhanced silencing abilities are dominant, as they mediated enhanced silencing even when the endogenous Asf1 protein was also present (Figure 1C). The increase in silencing caused by the 152I and 185T Asf1 mutants was not simply due to increased amounts of Asf1, as an extra plasmid-borne copy of the wild-type ASF1 gene (pAsf1) had no effect on silencing in our assays (Figure 1C).
Figure 1.
Mutations in Asf1 alter transcriptional silencing. (A) Schematic of Asf1 mutants and their observed phenotype for transcriptional silencing. The purple bars represent positions of the insertion mutations. (B) Asf1 mutants with enhanced silencing abilities of TELVIIL∷URA3 and HMR∷ADE2 cac1Δasf1Δ yeast (ROY1169) carrying the wild-type pAsf1 plasmid, mutant pAsf1 plasmid (as indicated), or the empty vector (vector) were plated in 10-fold serial dilutions onto rich media (control), media with only 25% of the normal amount of adenine (low ade), or 5′FOA. (C) The 152I and 185T Asf1 mutants are dominant. cac1Δ yeast (JKT0049) carrying the wild-type pAsf1 plasmid, mutant pAsf1 plasmid (as indicated), or cac1Δasf1Δ yeast (ROY1169) were plated in 10-fold serial dilutions onto rich media (control) and plates with the indicated concentration of 5′FOA. (D) The 185T and 152T Asf1 mutants bypass the requirement for CAF-1 in telomeric and HMR silencing. Yeast strains ROY1172 (WT), ROY1171 (cac1Δ), BAT004 (cac1Δ Asf1-185T) or BAT055 (cac1Δ Asf1-185T), BAT006 (cac1Δ Asf1-152T) or BAT056 (cac1Δ Asf1-152T), and ROY1169 (cac1Δasf1Δ) were plated in 10-fold serial dilutions onto rich media (control) or media with 5′FOA. (E) Anti-myc Western blot of yeast strains ACN026 (WT), JKT0049 (no tag), BAT006 (152T), BAT004 (185T), and ROY1169 transformed with the pAsf1152I vector (152I). A total of 7.5 mg/ml of total protein was loaded for each sample.
To verify the phenotypes of these mutations, we integrated the insertion mutations into the endogenous copy of the ASF1 gene. We also mutated the endogenous ASF1 gene to generate Asf1 truncated at 185 (185T) and 152 (152T) amino acids (Figure 1D). To determine the degree to which silencing is enhanced by the 185T and 152I/T Asf1 mutants in cac1Δ cells, we compared their ability to silence transcription to that of wild-type cells. We found that cac1Δ Asf1-185T, cac1Δ Asf1-152T, and cac1Δ Asf1-152I strains were able to grow as well as wild-type strains on 5′FOA and were nearly as pink as wild-type strains on low adenine (Figure 1D). This indicates that the 185T and 152I/T Asf1 mutants compensate for the lack of CAF-1 for silencing. 13Myc epitopes were introduced onto the C terminus of all the Asf1 mutants to verify equal expression levels to endogenous Asf1. Although the plasmid-borne 152I mutation expressed similar levels of Asf1 protein as compared to the wild-type Asf1 protein, we were able to detect only small amounts of expression of the integrated 152I mutants (Figure 1E, data not shown). Therefore, all further studies of the 152I Asf1 mutant used the plasmid-borne copy of this mutation. The plasmid-borne 152I and integrated 152T and 185T Asf1 mutants expressed the same protein levels as wild-type Asf1 (Figure 1E). Wherever it was experimentally possible, further studies used the integrated ASF1 mutants 152T and 185T. Since the Asf1 152T and 152I mutants were phenotypically identical we used 152T and 185T to assess the function of the Asf1 C-terminal region. Because the integrated ASF1 mutations also result in enhanced silencing we conclude that the altered silencing was due to the mutations in ASF1, not due to secondary mutations elsewhere in the genome or altered expression levels of ASF1.
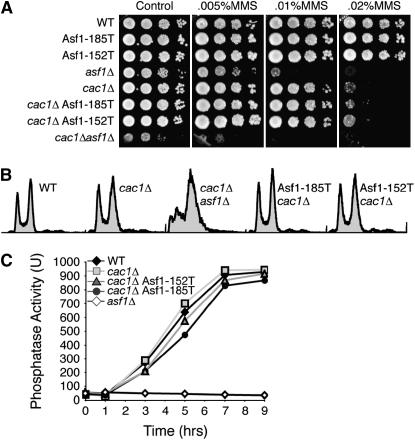
To determine whether the effects of the Asf1 mutations were specific to transcriptional silencing, we examined whether the mutants retained other known properties of wild-type Asf1. We verified that like wild-type Asf1 (Sutton et al. 2001), the mutant Asf1 proteins localized to the nucleus, using immunofluorescence analysis (data not shown). Furthermore, the Asf1 mutants, by contrast to yeast deleted for ASF1 (Tyler et al. 1999), had no obvious cell cycle defect or sensitivity to DNA damaging agents (Figure 2, A and B). Furthermore, the 185T and 152T Asf1 mutants do not compensate for the lack of CAF-1 for resistance to DNA damaging agents (Figure 2A).
Figure 2.
Mutations in Asf1 have no defect in DNA damage resistance, cell cycle progression, or nucleosome disassembly. (A) Tenfold serial dilution analysis of yeast strains ROY1172 (WT), BAT003 (Asf1-185T), BAT005 (Asf1-152T), ROY1170 (asf1Δ), JKT0049 (cac1Δ), BAT004 (cac1Δ Asf1-185T), BAT006 (cac1Δ Asf1-152T), and ROY1169 (cac1Δasf1Δ) were plated on the indicated amounts of methyl methanesulfonate. (B) Flow cytometry profiles of asynchronous log phase cultures of yeast strains ROY1172 (WT), ROY1171 (cac1Δ), ROY1169 (asf1Δ cac1Δ), BAT004 (Asf1-185T cac1Δ), and BAT006 (Asf1-152T cac1Δ). (C) Phosphatase activity of the gene product of PHO5 was measured using a colorimetric assay at increasing times after phosphate depletion from the media. Yeast strains measured were ROY1172 (WT), ROY1171 (cac1Δ), BAT006 (cac1Δ Asf1-152T), and BAT004 (cac1Δ Asf1-185T).
Asf1 mediates the disassembly of chromatin at the PHO5 promoter to allow production of the acid phosphatase encoded by PHO5 (Adkins et al. 2004). We found that both the Asf1-152T and the Asf1-185T mutants activate the PHO5 promoter as effectively as wild-type Asf1, as measured by phosphatase activity (Figure 2C). These data indicate that the I52I, 152T, and 185T mutations are not globally disrupting all the functions of Asf1, but instead are specifically affecting transcriptional silencing mediated by Asf1.
The 185T and 152I/T Asf1 mutants bypass the requirement for CAF-1 for the maintenance of transcriptional silencing:
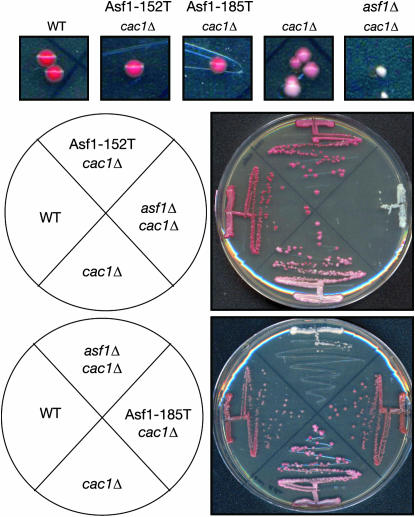
To understand how the Asf1 mutants are influencing transcriptional silencing, we examined the colony sectoring due to expression of HMR∷ADE2 in more detail. CAF-1 has been shown to be important for the maintenance/inheritance, but not the establishment, of silencing (Monson et al. 1997; Enomoto and Berman 1998). This is apparent from the white colonies with red sectors seen in the cac1 mutant carrying the HMR∷ADE2 reporter (Figure 3), as was previously observed for cac1 mutants carrying the TELVR∷ADE2 reporter (Monson et al. 1997). Colonies of the cac1Δ Asf1-185T and cac1Δ Asf1-152T strains were mostly red (Figure 3), indicating that they were able to maintain their silent chromatin structure better than a cac1 mutant. When we observed the Asf1-152I mutation on a plasmid in this same assay we saw identical results, further demonstrating that the Asf1-152I and Asf1-152T alleles are comparable (data not shown). This result indicates that the 185T and 152I/T Asf1 mutants can almost entirely rescue the defect in maintenance/inheritance of silencing caused by lack of CAF-1.
Figure 3.
Asf1 mutants bypass the defect in maintenance of transcriptional silencing due to CAF-1 inactivation. Magnification of colonies is shown for analysis of color and sectoring due to HMR∷ADE2. Images of colonies of yeast strains ROY1172 (WT), ROY1171 (cac1Δ), ROY1169 (cac1Δasf1Δ), BAT055 (cac1Δ ASF1-152T), and BAT056 (cac1Δ ASF1-185T) are shown. Comparison of colony color due to silencing of HMR∷ADE2 is shown. Streaks of strains described above are shown, according to the schematic, with each plate including the indicated Asf1 mutant.
Enhanced silencing by the 185T and 152I Asf1 mutants requires Sir2:
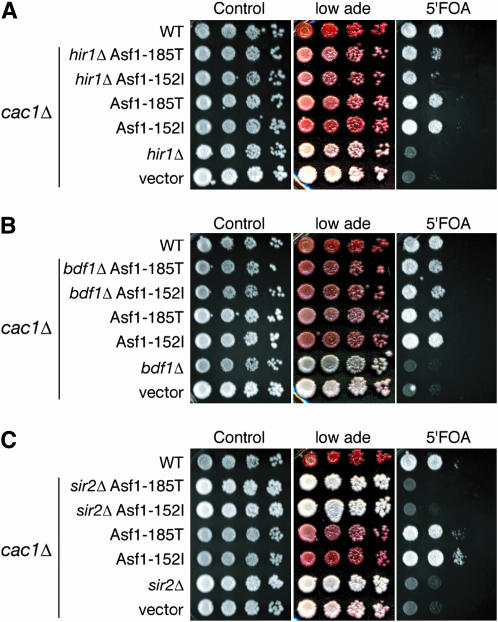
To gain insight into the mechanism whereby the 185T and 152I/T Asf1 mutants lead to increased transcriptional silencing we sought to identify other gene products required for the increased transcriptional silencing. To do this, we deleted several factors that have been shown to interact with Asf1 and/or have a known role in transcriptional silencing and tested their effect on silencing in combination with the Asf1 mutants. We reasoned that if the mechanism of increased silencing mediated by the Asf1 mutants required these other factors, then their deletion should abrogate the increased silencing. Hir1 binds to the N terminus of Asf1 and it has been proposed that Hir1 and Asf1 are in the same pathway for silencing since deletion of both HIR1 and ASF1 together caused no additional defect in mating efficiency over each single deletion (in combination with a CAC1 deletion) (Sharp et al. 2001). We found that deletion of HIR1 only slightly reduced the enhanced transcriptional silencing mediated by the 152I or 185T Asf1 mutants (Figure 4A). This result indicates that Hir1, or the interaction between Asf1 and Hir1, may be partially required for the enhanced transcriptional silencing due to the Asf1 mutations.
Figure 4.
Sir2, but not Hir1 or Bdf1, is required for the enhanced silencing due to Asf1 mutants. (A) Hir1 is only partially required for enhanced silencing by the 185T and 152I Asf1 mutants. All strains except WT have CAC1 deleted. Yeast strains ROY1172 with the pRS314 empty vector (WT), BAT034 with pAsf1-185T (hir1Δ Asf1-185T), BAT034 with pAsf1-152I (hir1Δ Asf1-152I), ROY1169 with pAsf1-185T (Asf1-185T), ROY1169 with pAsf1-152I (Asf1-152I), BAT034 with pAsf1 (hir1Δ), and ROY1171 with empty vector (vector) were plated in 10-fold serial dilutions onto media with 75% less adenine (low ade) or 1.0 mg/ml of 5′FOA. (B) Bdf1 is not required for enhanced silencing by the 185T and 152I Asf1 mutants. All strains except WT have CAC1 deleted. Yeast strains ROY1172 with the pRS314 empty vector (WT), BAT052 with pAsf1-185T (bdf1Δ Asf1-185T), BAT052 with pAsf1-152I (bdf1Δ Asf1-152I), ROY1169 with pAsf1-185T (Asf1-185T), ROY1169 with pAsf1-152I (Asf1-152I), BAT052 with pAsf1 (bdf1Δ), and ROY1171 with empty vector (vector) were plated in 10-fold serial dilutions onto media with 75% less adenine (low ade) or 1.0 mg/ml of 5′FOA. (C) Sir2 is required for enhanced silencing by the 185T and 152I Asf1 mutants. All strains except WT have CAC1 deleted. Yeast strains ROY1172 with the pRS314 empty vector (WT), BAT053 with pAsf1-185T (sir2Δ Asf1-185T), BAT053 with pAsf1-152I (sir2Δ Asf1-152I), ROY1169 with pAsf1-185T (Asf1-185T), ROY1169 with pAsf1-152I (Asf1-152I), BAT053 with pAsf1 (sir2Δ), and ROY1171 with empty vector (vector) were plated in 10-fold serial dilutions onto media with low adenine or 1.0 mg/ml 5′FOA.
We performed a similar epistasis analysis with BDF1 encoding one of the missing bromodomains of TAF1, the yeast counterpart of TAFII250. Asf1 interacts with Bdf1 via its bromodomain and Bdf1 contributes to transcriptional silencing by defining the boundary between heterochromatin and euchromatin (Chimura et al. 2002; Ladurner et al. 2003). Contrary to a previous report, we found that the asf1Δbdf1Δ strain is viable (Chimura et al. 2002), presumably due to differences within the strain background. Deletion of BDF1 did not reduce the enhanced transcriptional silencing mediated by the 185T and 152I Asf1 mutants (Figure 4B), indicating that neither Bdf1 nor the interaction between Bdf1 and Asf1 is required for the enhanced transcriptional silencing due to the Asf1 mutations.
When we performed the same type of epistasis analysis with the central silencing protein Sir2, we found that deletion of SIR2 abolishes the enhanced transcriptional silencing that is due to the 185T and 152I Asf1 mutants (Figure 4C). This result indicates that Sir2 is required for the mechanism whereby the 185T and 152I Asf1 mutants increase transcriptional silencing at the telomere-proximal region and the HMR locus. Upon finding this result we concluded our search and focused on the requirement of Sir2 in bypassing a cac1 deletion for transcriptional silencing.
The 185T and 152I/T Asf1 mutants overcome a defect in histone deposition and Sir2 recruitment in yeast lacking CAF-1:
To investigate how Sir2 contributes to the increased silencing by the Asf1 mutants, we measured Sir2 recruitment to the silent loci. We used ChIP analysis against Sir2 C-terminally tagged with three HA epitopes, using PCR to amplify regions at increasing distances from the telomere and at the silencer HMR-E. We found that yeast deleted for CAC1 have a significantly reduced Sir2 occupancy at the telomere-proximal and HMR loci, as compared to wild-type cells (Figure 5A). Furthermore, when we looked at Sir2 occupancy at increasing distances from the telomere, we did not observe any significant shift in the location of the boundary between the regions with Sir2 and without Sir2 (Figure 5A) in the Asf1 mutants, as previously observed in other mutants that affect silencing (Kimura et al. 2002; Ladurner et al. 2003). Because Sir2 is absolutely required for silencing, this reduced Sir2 occupancy in cac1 mutant yeast is likely to be the molecular reason for the defect in the maintenance/inheritance of silencing in CAF-1 mutants. Strikingly, the cac1Δ Asf1-185T and cac1Δ Asf1-152T strains had a Sir2 occupancy that was not significantly different from that of wild type (Figure 5A). This result indicates that the 185T and 152T Asf1 mutants can restore Sir2 occupancy to wild-type levels in the cac1 mutant.
To investigate whether the defect in Sir protein recruitment in the cac1 mutant was specific to Sir2, we examined recruitment of Sir4. To do this we performed ChIP analysis using an antibody specific to Sir4. We found a significant reduction in Sir4 recruitment to the HMR-E and telomere-proximal region in the absence of CAF-1 (Figure 5B). As was the case with Sir2, we found that Sir4 recruitment was restored to wild-type levels by the additional mutation of Asf1-152T or Asf1-185T in a cac1 mutant (Figure 5B). The restored Sir2 and Sir4 occupancy resulting from the 185T and 152T Asf1 mutants correlates well with the restored transcriptional silencing (Figures 1 and 3A) and increased maintenance of silencing due to these Asf1 mutants (Figure 3). Consequently, the enhanced transcriptional silencing due to the 185T and 152T Asf1 mutants in the absence of CAF-1 is likely to result from increased Sir2 and Sir4 occupancy at the silent regions.
Next, we wanted to determine the mechanism whereby the 185T and 152I/T Asf1 mutants restored Sir protein occupancy at silent regions in CAF-1 mutants. We compared global levels of Sir2 proteins in WT, cac1Δ, cac1Δ Asf1-185T, and cac1Δ Asf1-152T strains and found no significant difference (Figure 5D). Furthermore, transcript levels of the SIR genes are not significantly altered by deletion of CAC2 or ASF1 (Zabaronick and Tyler 2005). Sir2 is recruited to silent regions via its interaction with Sir4 (Ghidelli et al. 2001; Hoppe et al. 2002), which in turn is recruited via its interaction with histones H3 and H4 on chromatin (Hecht et al. 1995). To investigate whether reduced Sir2 and Sir4 recruitment in the cac1 mutant (Figure 5, A and B) is due to reduced histone occupancy we measured histone occupancy by ChIP. Our ChIP analyses used an antibody to the C terminus of histone H3 that has been used extensively to measure histone occupancy on DNA (Reinke and Horz 2003; Adkins et al. 2004; Bernstein et al. 2004; Korber et al. 2004; Lee et al. 2004; Schwabish and Struhl 2004; Radonjic et al. 2005; Zhang et al. 2005). We found that histone H3 occupancy is greatly reduced in the cac1 mutant at all regions that we examined, including the telomere-proximal and HMR loci and an open reading frame not found within a silenced region, ALD6 (Figure 5C). Given that histone H3 is always tightly associated with histone H4, and given their central location in the nucleosome, the reduced histone H3 occupancy indicates a reduced nucleosome density in cac1 mutants. Our results strongly suggest that the reduced Sir2 and Sir4 occupancy at silent regions and the resulting defect in transcriptional silencing in CAF-1 mutants are a consequence of reduced nucleosome density. When we examined histone H3 occupancy in the cac1Δ Asf1-185T and cac1Δ Asf1-152T strains, we found no significant difference from wild type (Figure 5C). This result indicates that the 185T and 152T Asf1 mutants restore histone occupancy in a cac1Δ strain, which in turn restores Sir2 and Sir4 occupancy and silencing.
The Asf1-histone H3 interaction is disrupted in Asf1-152T and Asf1185T mutants:
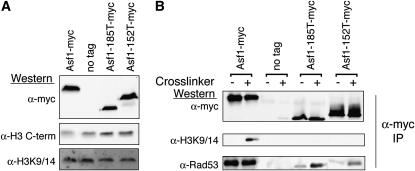
To investigate how the 185T and 152I/T Asf1 mutants result in increased histone occupancy, we first verified that the amount of total Asf1 protein and total soluble H3 present was equivalent between samples. This allowed us to rule out the possibility that the Asf1 185T and 152T mutants were altering expression levels of histones (Figure 6A). Since the Asf1 and histone levels were comparable between samples we tested whether the interaction between Asf1 and histone H3 was affected by these mutations. To do this, we measured the level of histone H3 that coimmunoprecipitated with the endogenous wild-type and mutant Asf1 proteins. We had previously found that the stringency of our immunoprecipitation conditions required the addition of an in vivo protein–protein crosslinker, dithiobis(succinimidyl propionate) (DSP), to detect the Asf1–histone H3 interaction (Tamburini et al. 2005) (Figure 6B). However, we found that even with the crosslinker, we were unable to detect co-immunoprecipitating histone H3 with the Asf1-152T or Asf1-185T proteins (Figure 6B). It is unlikely that the Asf1 mutants are globally misfolded, because they were still able to co-immunoprecipitate Rad53 (Emili et al. 2001; Hu et al. 2001) (Figure 6B). Interestingly, the interaction between Rad53 and Asf1-152T or Asf1-185T was weaker than between wild-type Asf1 and Rad53, suggesting that the unstructured C-terminal tail of Asf1 may act to stabilize protein–protein interactions. We propose that the interaction between Asf1 and histone H3 still exists, but that it is less stable, allowing Asf1 to place histones onto the DNA more readily. These results indicate that the 152T and 185T mutations reduce the stability of the histone–Asf1 interaction.
Figure 6.
The Asf1–histone H3 interaction is reduced in Asf1-185T and 152T mutants. The amount of soluble histones is invariable between the Asf1 mutant strains used. (A) Western blot analysis of total Asf1 and histone proteins using an H3 C-terminal antibody and an antibody that recognizes H3 acetylated on lysine residues 9 and 14 was performed with soluble protein extracts from yeast strains ACN026 (WT), ROY1172 (no tag), BAT004 (Asf1-185T), and BAT006 (Asf1-152T). The amount of total protein loaded from each sample was 7.5 mg/ml. (B) Co-immunoprecipitation analysis of histones with Asf1. Yeast strains used in A were immunoprecipitated with an anti-myc antibody and Western blot analysis was used to visualize H3 and Rad53. “Crosslinker” indicates the use of a protein–protein crosslinker, DSP. Newly synthesized soluble histones are acetylated on lysine 9 of histone H3; therefore, the anti-H3 acetyl lysine 9/14 antibody was used to detect all soluble histones bound to Asf1, as used previously (Tamburini et al. 2005).
DISCUSSION
This work discerns the molecular basis for the mechanism whereby chromatin assembly factors mediate transcriptional silencing. Our data indicate that CAF-1 promotes silencing by assembling chromatin with a nucleosome density sufficient to recruit enough Sir proteins to mediate the formation of a stable silent chromatin structure. Mutations within the C terminus of the chromatin assembly factor Asf1 suppress the requirement for CAF-1 in silencing, presumably via their weakened interaction with histones that results in a restored nucleosome density and increased Sir protein recruitment.
CAF-1 promotes transcriptional silencing by depositing histones that serve as the binding sites for Sir proteins:
Although the defect in the maintenance/inheritance of transcriptional silencing in CAF-1 mutants has been well documented, the molecular mechanism was unclear (Monson et al. 1997; Enomoto and Berman 1998). The Sir2–Sir4 complex and Sir3 are required for the maintenance of silencing and are recruited to DNA via interactions with histones H3 and H4 (Hecht et al. 1995). In the absence of CAF-1, nucleosome occupancy is reduced and the resulting Sir recruitment is reduced (Figure 5, A–C). It was interesting to note that the level of Sir proteins on the silent chromatin in CAF-1 mutants was not reduced to the same extent as the level of histone proteins (Figure 5, A–C). Sir proteins appear to be limiting for silencing in wild-type cells (see, for example, Renauld et al. 1993; Fritze et al. 1997; Smith et al. 1998). As such it is unlikely that the maximal Sir:histone ratio is reached at the silent chromatin in wild-type cells. In CAF-1 mutants, histone deposition is reduced compared to that in wild type (Figure 5C). Total available pools of Sir protein in the CAF-1 mutant cell are the same as that in a wild-type cell (Figure 5D). Consequently, CAF-1 mutants have a higher Sir:histone ratio at the silent chromatin than wild type. Even though the Sir:histone ratio at the silent chromatin is higher in the CAF-1 mutants, the absolute levels of Sir proteins on the chromatin are still less than that of wild type (because there are less histones for Sir to bind to) and we propose that this is the reason for the silencing defect in CAF-1 mutants. The Sir:histone ratio at the silent chromatin in the CAF-1 mutants, although higher than that in wild type, still presumably does not reach the maximal Sir:histone ratio possible, because overexpression of Sir proteins in CAF-1 mutants restores the silencing defect (Monson et al. 1997; Enomoto and Berman 1998; Smith et al. 1999).
Given the reduced histone occupancy and Sir2 and Sir4 occupancy at the silent regions in CAF-1 mutants (Figure 5, A–C), we propose that CAF-1-mediated chromatin assembly following DNA replication generates a nucleosome density sufficient for recruitment of enough Sir proteins to mediate the maintenance of the silent chromatin structure. Because silencing can be established in CAF-1 mutants, it appears that a silent chromatin structure with suboptimal stability can be generated in the absence of CAF-1, but is not stably maintained. This is likely to be a consequence of the reduced levels of Sir2 and Sir4 recruitment in CAF-1 mutants because reduced levels of Sir2 or Sir3 lead to a similar defect in the maintenance of silencing (Holmes and Broach 1996; Enomoto and Berman 1998). In accordance with this idea, overexpression of the SIR3 or SIR2 gene is able to suppress the silencing defect seen in CAF-1 mutants (Monson et al. 1997; Enomoto and Berman 1998; Smith et al. 1999) (data not shown). Overexpression of SIR3 may force Sir3 to the silent regions, increasing the occupancy of Sir3 protein to a level sufficient for maintenance of the silent state in CAF-1 mutants. Furthermore, it has been shown previously that overexpression of histones H3 and H4 can partially rescue the silencing defect in CAF-1 mutants (Kaufman et al. 1998), perhaps resulting in an increased histone occupancy on the DNA, which would facilitate recruitment of the Sir proteins for silencing. Similarly, deletion of one copy of the genes expressing histone H3 and H4 exacerbates the silencing defect of CAF-1 mutants (Kaufman et al. 1998), presumably resulting in an even lower histone occupancy on the DNA and further reduced Sir protein recruitment.
Our data provide the first direct indication of a defect in histone deposition in CAF-1 mutants, supporting a role for CAF-1 in chromatin assembly in vivo. The reduced occupancy of histone H3 on chromatin in CAF-1 mutants (Figure 5C) is consistent with (i) the biochemical function of CAF-1 as a chromatin assembly factor that deposits histones H3 and H4 (Smith and Stillman 1989, 1991), (ii) the increased accessibility of chromatin upon CAF-1 inactivation to micrococcal nuclease and DNAse I and reduced supercoiling of the endogenous 2μ plasmid (Hoek and Stillman 2003; Adkins and Tyler 2004; Nabatiyan and Krude 2004), and (iii) the increased psoralen accessibility observed at the rDNA loci in CAF-1 mutants (Smith et al. 1999). CAF-1 is recruited to sites of DNA replication via its interaction with proliferating cell nuclear antigen (PCNA) (Shibahara and Stillman 1999). Consistent with the idea that silencing defects in CAF-1 mutants are the consequence of improper chromatin assembly following DNA replication is the fact that PCNA alleles also have silencing defects that are in the same genetic pathway as CAF-1 mutants (Zhang et al. 2000).
Mutations that reduce global histone methylation or acetylation indirectly influence silencing (Lacoste et al. 2002; Ng et al. 2002; van Leeuwen et al. 2002). Sir proteins bind preferentially to unmodified histones, and consequently the loss of histone modifications causes promiscuous binding of the Sir proteins throughout the genome. Because the pools of silencing proteins appear to be limiting (Buck and Shore 1995; Strahl-Bolsinger et al. 1997; Smith et al. 1998), the result is a reduced effective concentration of Sir proteins and weakened silencing. Although histone methylation has not been examined in CAF-1 mutants, there is no detectable defect in histone acetylation in CAF-1 mutants (Adkins and Tyler 2004). This argues against the possibility that reduced Sir2 occupancy in the silent regions of cac1 mutants may be due to increased histone acetylation at the silent loci preventing Sir binding or reduced histone acetylation elsewhere in the genome sequestering the Sir proteins. However, it is possible that there is increased histone acetylation restricted to the silent regions that was undetectable by the previous analysis, causing the decrease in Sir2 recruitment. Silencing defects can also be due to transcriptional misregulation of gene products required for silencing. It is noteworthy that we did not observe altered amounts of Sir2 protein upon deletion of CAC1 (Figure 5D), nor did we observe altered transcript levels of any of the known silencing proteins by microarray analyses in cac2 or asf1 mutants (Zabaronick and Tyler 2005). The strong correlations between the reduced histone occupancy, Sir protein occupancy, and transcriptional silencing in the CAF-1 mutants and their restoration by the Asf1-152T and 185T mutants strongly support our proposal that reduced histone deposition is the molecular basis for the silencing defect in CAF-1 mutants.
The C terminus of Asf1 contributes to the histone interaction and transcriptional silencing:
Our studies provide the first evidence that the C terminus of Asf1 is functionally relevant. While other studies have found that Asf1 is functional without the C terminus of the protein (Umehara et al. 2002; Daganzo et al. 2003; Mousson et al. 2005), we have uncovered a role in transcriptional silencing where mutations in the Asf1 C terminus are able to compensate for the defect in silencing caused by deletion of CAC1. We observed an apparent reduced affinity interaction between the Asf1 mutants and histone H3, suggesting that the C terminus of Asf1 may contribute to the stability of the interaction with histone H3. There are at least two possible scenarios for how a reduced interaction between histone H3 and Asf1 could lead to higher histone levels on the DNA when CAF-1 is inactive and the resulting increased Sir2 and Sir4 recruitment and restored transcriptional silencing that we observed. First, the mutant Asf1 proteins may be less efficient at removing the histones from the DNA during chromatin disassembly, resulting in increased histone occupancy on the DNA. Second, the weakened interaction between histones and mutant Asf1 may promote histone deposition onto the DNA. However, only an increased assembly activity of the mutants could account for the dominant nature of the mutant Asf1 proteins over the wild-type Asf1 protein (Figure 1C). Furthermore, a decreased disassembly activity would yield a silencing phenotype more like the ASF1 deletion, not the enhanced silencing that is observed with the 185T and 152I/T Asf1 mutants. Consequently, we propose that the C-terminal mutations result in Asf1 proteins with increased chromatin assembly activity. The 185T and 152I/T Asf1 mutants that bypass the requirement for CAF-1 for silencing are reminiscent of specific mutations within the N-terminal tail and core of histones H3 and H4 that also bypass the requirement for CAF-1 for telomeric silencing (Smith et al. 2002). It will be interesting to determine whether those particular histone mutants also lead to increased histone deposition onto the silent chromatin in CAF-1 mutants, perhaps via reducing their affinity for Asf1.
While the crystal structure and NMR structure of the conserved N-terminal domain of yeast Asf1 and human Asf1a, respectively, have been solved, little is known about the regions of Asf1 that mediate protein interactions important for its function (Daganzo et al. 2003; Mousson et al. 2005). Mutational analysis directed against unstructured loop regions within the Asf1 conserved domain has revealed the binding site for human HirA on Asf1a to be in the cleft of the protein adjacent to the hydrophobic groove and near the N- and C-terminal regions of the conserved domain (Daganzo et al. 2003). In addition, the N-terminal conserved region of Asf1 is important for histone H3–H4 binding, while the nonconserved acidic C terminus of Asf1 contributes to binding of histones H3, H4, H2A, and H2B in vitro (Umehara et al. 2002). Recent data also suggest that the hydrophobic groove within Asf1 is important for histone binding, specifically valine 94, aspartic acid 54, and arginine 108 (Mousson et al. 2005). Since the 152T and 185T Asf1 mutants were not able to detectably bind to histone H3 in our assay, it is possible that the C-terminal tail of Asf1 stabilizes the interaction of histone H3 in the “groove” of Asf1 (Daganzo et al. 2003). Moreover, histone binding to Asf1 truncated at amino acid 155 is easily disrupted in high-salt conditions, suggesting that the highly acidic C terminus is involved in stabilizing electrostatic contacts (Daganzo et al. 2003). Interestingly, the interaction between Rad53 and Asf1-152T or Asf1-185T was weaker than between wild-type Asf1 and Rad53 (Figure 6B), possibly because the C-terminal tail of Asf1 is also involved in stabilizing this interaction.
Here we demonstrate that the C-terminal region of the Asf1 protein has distinct silencing function in vivo. The Asf1 mutants that had enhanced silencing activity and weakened histone interaction were insertions between amino acids 152 and 153, truncation at 152, and truncation at 185. Amino acid 152 is found at the C terminus of the solved protein structure and is part of an unstructured region (Daganzo et al. 2003). Because insertion and truncation at amino acid 152 provided indistinguishable defects in silencing, it would appear that the sequences around amino acid 152 are important for silencing. Furthermore, the silencing defect in the truncation at amino acid 185 indicates that sequences C-terminal to amino acid 185 are also important for silencing, perhaps contributing to the stability of histone binding. One possibility is that the region of Asf1 around amino acid 152 and the C terminus of Asf1 are close together in three-dimensional space, where they both contribute to the same aspect of silencing. However, such a prediction awaits the structure of the full-length Asf1 protein.
In summary, CAF-1 contributes to the recruitment of Sir proteins to the silent loci via the interaction with histone proteins to mediate transcriptional silencing. Furthermore, mutations that weaken the interaction between Asf1 and the histones result in restored histone levels, Sir2 recruitment, and transcriptional silencing at the silent loci in CAF-1 mutants. These studies provide molecular insight into the interplay between the formation of the underlying nucleosome structure and specialized chromatin structures.
Acknowledgments
We thank Christine English, Melissa Adkins, and Josh Ramey for critical reading of this manuscript. We are grateful to Danesh Moazed for the Sir4 antibody. We thank Judit Kiss for technical assistance with screening the mutants and Melissa Adkins for assistance with the phosphatase assay. This work was supported by a grant from the National Institutes of Health (GM64475) to J.K.T. J.K.T. is a Leukemia and Lymphoma Society Scholar.
References
- Adkins, M. W., and J. K. Tyler, 2004. The histone chaperone Asf1p mediates global chromatin disassembly in vivo. J. Biol. Chem. 279: 52069–52074. [DOI] [PubMed] [Google Scholar]
- Adkins, M. W., S. R. Howar and J. K. Tyler, 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14: 657–666. [DOI] [PubMed] [Google Scholar]
- Bernstein, B. E., C. L. Liu, E. L. Humphrey, E. O. Perlstein and S. L. Schreiber, 2004. Global nucleosome occupancy in yeast. Genome Biol. 5: R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck, S. W., and D. Shore, 1995. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 9: 370–384. [DOI] [PubMed] [Google Scholar]
- Cairns, B. R., 2005. Chromatin remodeling complexes: strength in diversity, precision through specialization. Curr. Opin. Genet. Dev. 15: 185–190. [DOI] [PubMed] [Google Scholar]
- Carmen, A. A., L. Milne and M. Grunstein, 2002. Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J. Biol. Chem. 277: 4778–4781. [DOI] [PubMed] [Google Scholar]
- Chimura, T., T. Kuzuhara and M. Horikoshi, 2002. Identification and characterization of CIA/ASF1 as an interactor of bromodomains associated with TFIID. Proc. Natl. Acad. Sci. USA 99: 9334–9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daganzo, S. M., J. P. Erzberger, W. M. Lam, E. Skordalakes, R. Zhang et al., 2003. Structure and function of the conserved core of histone deposition protein Asf1. Curr. Biol. 13: 2148–2158. [DOI] [PubMed] [Google Scholar]
- Donze, D., and R. T. Kamakaka, 2001. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 20: 520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emili, A., D. M. Schieltz, J. R. Yates, 3rd and L. H. Hartwell, 2001. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol. Cell 7: 13–20. [DOI] [PubMed] [Google Scholar]
- Enomoto, S., and J. Berman, 1998. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 12: 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritze, C. E., K. Verschueren, R. Strich and R. Easton Esposito, 1997. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 16: 6495–6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghidelli, S., D. Donze, N. Dhillon and R. T. Kamakaka, 2001. Sir2p exists in two nucleosome-binding complexes with distinct deacetylase activities. EMBO J. 20: 4522–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht, A., T. Laroche, S. Strahl-Bolsinger, S. M. Gasser and M. Grunstein, 1995. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80: 583–592. [DOI] [PubMed] [Google Scholar]
- Hoek, M., and B. Stillman, 2003. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc. Natl. Acad. Sci. USA 100: 12183–12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, S. G., and J. R. Broach, 1996. Silencers are required for inheritance of the repressed state in yeast. Genes Dev. 10: 1021–1032. [DOI] [PubMed] [Google Scholar]
- Hoppe, G. J., J. C. Tanny, A. D. Rudner, S. A. Gerber, S. Danaie et al., 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 22: 4167–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, F., A. A. Alcasabas and S. J. Elledge, 2001. Asf1 links Rad53 to control of chromatin assembly. Genes Dev. 15: 1061–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai, S., C. M. Armstrong, M. Kaeberlein and L. Guarente, 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800. [DOI] [PubMed] [Google Scholar]
- Kaufman, P. D., R. Kobayashi and B. Stillman, 1997. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 11: 345–357. [DOI] [PubMed] [Google Scholar]
- Kaufman, P. D., J. L. Cohen and M. A. Osley, 1998. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol. Cell. Biol. 18: 4793–4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, A., T. Umehara and M. Horikoshi, 2002. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 32: 370–377. [DOI] [PubMed] [Google Scholar]
- Korber, P., T. Luckenbach, D. Blaschke and W. Horz, 2004. Evidence for histone eviction in trans upon induction of the yeast PHO5 promoter. Mol. Cell. Biol. 24: 10965–10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, M. H., and C. D. Allis, 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19: 425–433. [DOI] [PubMed] [Google Scholar]
- Lacoste, N., R. T. Utley, J. M. Hunter, G. G. Poirier and J. Cote, 2002. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J. Biol. Chem. 277: 30421–30424. [DOI] [PubMed] [Google Scholar]
- Ladurner, A. G., C. Inouye, R. Jain and R. Tjian, 2003. Bromodomains mediate an acetyl-histone encoded antisilencing function at heterochromatin boundaries. Mol. Cell 11: 365–376. [DOI] [PubMed] [Google Scholar]
- Landry, J., J. T. Slama and R. Sternglanz, 2000. Role of NAD(+) in the deacetylase activity of the SIR2-like proteins. Biochem. Biophys. Res. Commun. 278: 685–690. [DOI] [PubMed] [Google Scholar]
- Le, S., C. Davis, J. B. Konopka and R. Sternglanz, 1997. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast 13: 1029–1042. [DOI] [PubMed] [Google Scholar]
- Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl and J. D. Lieb, 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36: 900–905. [DOI] [PubMed] [Google Scholar]
- Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent and T. J. Richmond, 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260. [DOI] [PubMed] [Google Scholar]
- Marheineke, K., and T. Krude, 1998. Nucleosome assembly activity and intracellular localization of human CAF-1 changes during the cell division cycle. J. Biol. Chem. 273: 15279–15286. [DOI] [PubMed] [Google Scholar]
- Meijsing, S. H., and A. E. Ehrenhofer-Murray, 2001. The silencing complex SAS-I links histone acetylation to the assembly of repressed chromatin by CAF-I and Asf1 in Saccharomyces cerevisiae. Genes Dev. 15: 3169–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello, J. A., H. H. Sillje, D. M. Roche, D. B. Kirschner, E. A. Nigg et al., 2002. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 3: 329–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed, D., A. Kistler, A. Axelrod, J. Rine and A. D. Johnson, 1997. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc. Natl. Acad. Sci. USA 94: 2186–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monson, E. K., D. de Bruin and V. A. Zakian, 1997. The yeast Cac1 protein is required for the stable inheritance of transcriptionally repressed chromatin at telomeres. Proc. Natl. Acad. Sci. USA 94: 13081–13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousson, F., A. Lautrette, J. Y. Thuret, M. Agez, R. Courbeyrette et al., 2005. Structural basis for the interaction of Asf1 with histone H3 and its functional implications. Proc. Natl. Acad. Sci. USA 102: 5975–5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabatiyan, A., and T. Krude, 2004. Silencing of chromatin assembly factor 1 in human cells leads to cell death and loss of chromatin assembly during DNA synthesis. Mol. Cell. Biol. 24: 2853–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, H. H., Q. Feng, H. Wang, H. Erdjument-Bromage, P. Tempst et al., 2002. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 16: 1518–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada, S., A. Sutton, N. Muster, C. E. Brown, J. R. Yates, 3rd et al., 2001. The yeast SAS (something about silencing) protein complex contains a MYST-type putative acetyltransferase and functions with chromatin assembly factor ASF1. Genes Dev. 15: 3155–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, C. L., and M. A. Laniel, 2004. Histones and histone modifications. Curr. Biol. 14: R546–R551. [DOI] [PubMed] [Google Scholar]
- Radonjic, M., J. C. Andrau, P. Lijnzaad, P. Kemmeren, T. T. Kockelkorn et al., 2005. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol. Cell 18: 171–183. [DOI] [PubMed] [Google Scholar]
- Ramey, C. J., S. Howar, M. Adkins, J. Linger, J. Spicer et al., 2004. Activation of the DNA damage checkpoint in yeast lacking the histone chaperone anti-silencing function 1. Mol. Cell. Biol. 24: 10313–10327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke, H., and W. Horz, 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11: 1599–1607. [DOI] [PubMed] [Google Scholar]
- Renauld, H., O. M. Aparicio, P. D. Zierath, B. L. Billington, S. K. Chhablani et al., 1993. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 7: 1133–1145. [DOI] [PubMed] [Google Scholar]
- Rusche, L. N., A. L. Kirchmaier and J. Rine, 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72: 481–516. [DOI] [PubMed] [Google Scholar]
- Schulz, L. L., and J. K. Tyler, 2006. The histone chaperone ASF1 localizes to active DNA replication forks to mediate efficient DNA replication. FASEB J. 20: 488–490. [DOI] [PubMed] [Google Scholar]
- Schwabish, M. A., and K. Struhl, 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 24: 10111–10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, J. A., E. T. Fouts, D. C. Krawitz and P. D. Kaufman, 2001. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr. Biol. 11: 463–473. [DOI] [PubMed] [Google Scholar]
- Shibahara, K., and B. Stillman, 1999. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96: 575–585. [DOI] [PubMed] [Google Scholar]
- Singer, M. S., A. Kahana, A. J. Wolf, L. L. Meisinger, S. E. Peterson et al., 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150: 613–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C. M., Z. W. Haimberger, C. O. Johnson, A. J. Wolf, P. R. Gafken et al., 2002. Heritable chromatin structure: mapping “memory” in histones H3 and H4. Proc. Natl. Acad. Sci. USA 99(Suppl. 4): 16454–16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J. S., C. B. Brachmann, L. Pillus and J. D. Boeke, 1998. Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics 149: 1205–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J. S., E. Caputo and J. D. Boeke, 1999. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol. 19: 3184–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S., and B. Stillman, 1989. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 58: 15–25. [DOI] [PubMed] [Google Scholar]
- Smith, S., and B. Stillman, 1991. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 10: 971–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, E. M., and L. Pillus, 1996. Activation of an MAP kinase cascade leads to Sir3p hyperphosphorylation and strengthens transcriptional silencing. J. Cell Biol. 135: 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger, S., A. Hecht, K. Luo and M. Grunstein, 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11: 83–93. [DOI] [PubMed] [Google Scholar]
- Suka, N., K. Luo and M. Grunstein, 2002. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 32: 378–383. [DOI] [PubMed] [Google Scholar]
- Sutton, A., J. Bucaria, M. A. Osley and R. Sternglanz, 2001. Yeast asf1 protein is required for cell cycle regulation of histone gene transcription. Genetics 158: 587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami, H., D. Ray-Gallet, G. Almouzni and Y. Nakatani, 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116: 51–61. [DOI] [PubMed] [Google Scholar]
- Tamburini, B. A., J. J. Carson, M. W. Adkins and J. K. Tyler, 2005. Functional conservation and specialization among eukaryotic anti-silencing function 1 histone chaperones. Eukaryot. Cell 4: 1583–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler, J. K., C. R. Adams, S. R. Chen, R. Kobayashi, R. T. Kamakaka et al., 1999. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402: 555–560. [DOI] [PubMed] [Google Scholar]
- Tyler, J. K., K. A. Collins, J. Prasad-Sinha, E. Amiott, M. Bulger et al., 2001. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol. Biol. Cell 21: 6574–6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara, T., T. Chimura, N. Ichikawa and M. Horikoshi, 2002. Polyanionic stretch-deleted histone chaperone cia1/Asf1p is functional both in vivo and in vitro. Genes Cells 7: 59–73. [DOI] [PubMed] [Google Scholar]
- van Leeuwen, F., P. R. Gafken and D. E. Gottschling, 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109: 745–756. [DOI] [PubMed] [Google Scholar]
- Zabaronick, S. R., and J. K. Tyler, 2005. The histone chaperone anti-silencing function 1 is a global regulator of transcription independent of passage through S phase. Mol. Cell. Biol. 25: 652–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., S. Schroeder, N. Fong and D. L. Bentley, 2005. Altered nucleosome occupancy and histone H3K4 methylation in response to ‘transcriptional stress’. EMBO J. 24: 2379–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., K. Shibahara and B. Stillman, 2000. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature 408: 221–225. [DOI] [PubMed] [Google Scholar]