Abstract
Comparisons of gene orders between species permit estimation of the rate of chromosomal evolution since their divergence from a common ancestor. We have compared gene orders on three chromosomes of Drosophila pseudoobscura with its close relative, D. miranda, and the distant outgroup species, D. melanogaster, by using the public genome sequences of D. pseudoobscura and D. melanogaster and ∼50 in situ hybridizations of gene probes in D. miranda. We find no evidence for extensive transfer of genes among chromosomes in D. miranda. The rates of chromosomal rearrangements between D. miranda and D. pseudoobscura are far higher than those found before in Drosophila and approach those for nematodes, the fastest rates among higher eukaryotes. In addition, we find that the D. pseudoobscura chromosome with the highest level of inversion polymorphism (Muller's element C) does not show an unusually fast rate of evolution with respect to chromosome structure, suggesting that this classic case of inversion polymorphism reflects selection rather than mutational processes. On the basis of our results, we propose possible ancestral arrangements for the D. pseudoobscura C chromosome, which are different from those in the current literature. We also describe a new method for correcting for rearrangements that are not detected with a limited set of markers.
HOMOLOGOUS chromosome arms in the genus Drosophila are referred to as Muller's elements A–F (Muller 1940; Sturtevant and Novitsky 1941), with element A being the X chromosome of Drosophila melanogaster, elements B and C the two arms of its chromosome 2, D and E the two arms of chromosome 3, and element F being chromosome 4. Comparisons of the localizations of genes have allowed the establishment of homologies among chromosome arms over a wide range of species (see Ashburner 1989, p. 23, Table 5.2). The ancestral state is one with five major acrocentric chromosomes (elements A–E) and a dot chromosome (element F).
The chromosome arms in Drosophila have a high level of conservation of gene content, despite much rearrangement by inversions within arms (Ranz et al. 2001; González et al. 2002), and a lower rate of evolution of new karyotypes generated by fusions of pairs of elements (Ashburner 1989; Powell 1997). One such type of derived chromosomes is represented by the so-called neo-sex chromosomes, which involve the fusion of an autosome with a sex chromosome (Ashburner 1989). Several species of Drosophila have neo-sex chromosomes, which provide good models for studying the evolution of genes that were originally located on autosomes, but are now found on a new secondary sex chromosome pair (Steinemann and Steinemann 1998; McAllister and Charlesworth 1999). An especially useful model species of this kind is D. miranda, a close relative of D. pseudoobscura, whose element C (chromosome arm 2R of D. melanogaster, equivalent to chromosome 3 of D. pseudoobscura) has been fused with the Y chromosome, creating a neo-Y chromosome (Dobzhansky 1935; Macknight 1939; Steinemann and Steinemann 1998). Its free homolog is called the neo-X or X2 chromosome, to distinguish it from the true X chromosome of D. miranda, which is homologous to that of D. pseudoobscura.
The nonrecombining neo-Y chromosome of D. miranda shows an intermediate stage of degeneration, reflecting the effects of the reduction of the efficacy of selection on nonrecombining chromosomes (Charlesworth and Charlesworth 2000), with partial loss of gene function, accumulation of transposable elements, and very low DNA sequence variability (Steinemann and Steinemann 1992; Yi and Charlesworth 2000a; Bachtrog and Charlesworth 2002; Bachtrog 2005). This progressive inactivation of the neo-Y chromosome favors the evolution of mechanisms of dosage compensation on its homolog, the neo-X chromosome (Charlesworth 1996; Steinemann and Steinemann 1998).
An apparent exception in D. miranda to the principle of chromosomal homology, involving the transposition of a gene from the neo-X chromosome to the ancestral X chromosome, was suggested to provide a novel means of dosage compensation, evolved in response to the degeneration of its paralog on the neo-Y chromosome (Yi and Charlesworth 2000b). This led us to investigate the prevalence of the transposition or duplication of genes from the neo-X chromosome onto other chromosomes. In addition, we have explored the frequency of these events on other chromosomes, to determine if this phenomenon was confined to genes derived from element C, as this model of dosage compensation would predict (Yi and Charlesworth 2000b). We have also studied the extent of chromosomal variation for inversions within D. miranda, which complements the first systematic survey performed by Yi et al. (2003), and have examined the extent of evolution of inversions between D. miranda and two other Drosophila species (D. melanogaster and D. pseudoobscura).
Overall, our results show little evidence for an unusually high rate of duplications or transpositions of genes from the neo-X chromosome. They reveal the existence of two new inversion polymorphisms within D. miranda, as well as an unusually high rate of rearrangements involving inversions in the lineage leading to D. miranda and D. pseudoobscura. They also shed light on the probable ancestral arrangement of chromosome 3 of D. pseudoobscura, which harbors most of the inversion polymorphisms in this species (Powell 1992).
MATERIALS AND METHODS
Strains:
Eighteen D. miranda lines derived from single wild-caught females were investigated in our survey of inversion polymorphism. Twelve of them were described previously (Yi and Charlesworth 2000a): 0101.3, 0101.4, 0101.5, and 0101.7 (Port Coquitlam, BC, Canada); 0101.9, MA28, and MA32 (Mather, CA); SP138, SP235, and SP295 (Spray, OR); and MSH22 and MSH38 (Mt. St. Helena, CA). The remaining six lines (MA03.1, MA03.2, MA03.3, MA03.4, MA03.5, and MA03.6) were collected in July 2003 by B. Charlesworth and Deborah Charlesworth at Mather, California. The identity of these new strains as D. miranda was established by Doris Bachtrog, using species-specific PCR primers (D. Bachtrog, personal communication). Stocks were maintained in bottle culture on banana medium at 18°.
Probes for in situ hybridization:
DNA was extracted from single D. miranda male flies by means of Puregene (Gentra Systems, Minneapolis). Standard PCR procedures were used (Expand High-Fidelity PCR System; Roche Diagnostics, Lewes, UK). Primers were designed for regions conserved between D. melanogaster and D. pseudoobscura, using the publicly available genome sequences (see below). Details of the primers and PCR conditions are available on request. The products were gel purified with Qiaquick (QIAGEN, Crawley, UK) and cloned with TOPO-TA (Invitrogen, San Diego). DNA sequencing was performed, to identify each cloned fragment, on an ABI3730 sequencer using Dyenamic (Amersham Biosciences, Little Chalfont, UK). For preparing the probes used in the experiments described, new products were obtained from the selected clones by PCR amplification with M13 primers; these were gel purified and labeled with the relevant kits (see below).
In situ hybridization:
The chromosomal location of the genes was characterized by in situ hybridization of salivary chromosome squashes from third-instar larvae from various D. miranda strains, following the protocol described by Maside et al. (2001). Markers were initially selected from D. pseudoobscura sequences available in GenBank; however, the release of its whole genome sequence allowed us to randomly choose new loci along the different chromosomes. The probes were labeled with Biotin High Prime (Roche Diagnostics) and subsequently detected by staining with diaminobenzidine and peroxidase (Vector Laboratories, Peterborough, UK).
The localization of gene probes in D. miranda was established using the polytene chromosome maps of Das et al. (1982). The positions of their orthologs in D. melanogaster were obtained from FlyBase (http://flybase.bio.indiana.edu/), and the relative locations of these loci on D. pseudoobscura elements C and E were determined using the annotation of the D. pseudoobscura genome release 1.0 (http://flybase.net/annot/dpse_release1.04.txt), where these chromosome arms are represented by a single ultrascaffold each. The locations of markers on elements A and D of D. pseudoobscura were obtained by combining the genome annotation (which for these chromosomes comprises 4 and 5 ultrascaffolds, respectively) with in situ data from the literature (Segarra and Aguadé 1992; Segarra et al. 1996; Machado et al. 2002), since the relative location and orientation of the multiple ultrascaffolds on these arms are unknown. In addition, the localization of Gapdh2 in this species was determined by B. Charlesworth and S. Assimacopoulos (unpublished data), using in situ hybridization and the polytene map of Kastritsis and Crumpacker (1966). Details of the positions of the markers, and the estimated proportions of the chromosomes in each segment, are given in the online supplemental material (supplemental tables at http://www.genetics.org/supplemental/).
Southern blotting:
Genomic DNA was extracted separately from 20–30 D. miranda males and females (line MSH22), using Puregene (Gentra Systems). Three to five enzymes, not cutting within the probe sequence, were selected for restriction digestion purposes. Five micrograms of DNA were digested in each reaction (37° for 5 hr), sodium acetate precipitated, and resuspended in H2O.
Electrophoresis was performed in a 0.8% agarose gel at 80 V for 16 hr. DNA was subsequently denatured, transferred, and UV crosslinked (Sambrook et al. 1989) onto a positively charged membrane (Roche Diagnostics). DNA hybridization and detection were carried out using the Renaissance Random Primer fluorescein labeling kit with antifluorescein-AP (PerkinElmer Life Sciences, Boston).
Inversion analyses:
Line MSH22 was selected as a tester strain for screening the newly collected strains of D. miranda because of its apparent lack of inversions (Yi et al. 2003). Crosses between males of this tester line and females of six newly collected D. miranda strains (see above) were made to investigate the presence of inversion polymorphisms. Eight larvae per line were scored after chromosomal staining with orcein following the protocol of Yi et al. (2003). The breakpoints of the inversions were determined using the maps of Das et al. (1982).
Chromosomal rearrangements distinguishing species:
We used four different methods to calculate the number of inversions between species:
GRIMM (Tesler 2002): This computes the minimum possible number of rearrangement steps between a pair of species and determines a possible sequence of breaks for this number of steps (http://www-cse.ucsd.edu/groups/bioinformatics/GRIMM/grimm_instr.html).
Nadeau and Taylor (1984): This method estimates the true number of chromosomal rearrangements between two species from the lengths of conserved segments separating successive markers, assuming that the distribution of markers across the chromosomes is random and that breaks are thrown down randomly among these markers. The aim is to correct for inversions that have been overlooked because their breakpoints fall within a segment, as do the following two methods.
Ranz et al. (1997): This method estimates the number of inversions by a maximum-likelihood method that makes use of the fraction of segments between markers that are conserved. This assumes that the number of potential breakpoints in a given segment follows a Poisson distribution and that the mean of this distribution is proportional to the length of the segment. The likelihood of the observed configuration is calculated for an assumed number, n, of inversions on the chromosome in question.
This article: This is a new method, which relaxes the assumption of a Poisson distribution made by Ranz et al. (1997), which is likely to be true only if n is large. The chromosome is divided into a set of segments defined by the markers of a focal species, for which the lengths of the intervals between markers are known, e.g., those in D. pseudoobscura. As in the previous two methods, a segment is classified as conserved if the same pair of markers bound the segment in the species to which the focal species is being compared (the telomeres and proximal end of the euchromatin can be used as conserved markers). The length of the ith segment is li. The probability that a break occurs in this segment for the focal species is obviously
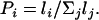 |
We assume that 2n breaks are to be assigned to the chromosome, where n is constrained to be greater than the minimum number given by the GRIMM algorithm. Two breaks are assigned to their respective intervals by sampling two independent uniform random numbers, using the cumulative distribution generated by the Pi to decide where to place the breaks. If both breaks fall in the same segment, the segment is left as conserved, but the occurrence of an inversion is recorded. If they occur in different segments, each is scored as unconserved. This procedure is repeated until 2n breaks have been assigned.
A hit is scored if the number of conserved segments matches the observed number for the species pair, and the procedure is repeated a large number of times, so that the fraction of hits for a given value of n can be estimated reasonably well; i.e., there are at least 30 hits for the maximum-likelihood value, but in most cases these numbered in the hundreds or thousands (this sometimes required as many 109 runs). This fraction gives the likelihood of n, L(n). By repeating this for successive values of n, a likelihood profile for the number of inversions is obtained, yielding 2-unit support limits from the differences in log-likelihoods. The advantage of this method is that no assumptions other than random and independent break positions need be made; the drawback is that it becomes very demanding of computing time if n is large. In this case, however, the method of Ranz et al. (1997) can probably be safely used. A FORTRAN program to conduct this analysis is available on request from B. Charlesworth, and the code is provided in the online supplemental material at http://www.genetics.org/supplemental/.
The last three methods require knowledge of the physical lengths between consecutive markers in the reference species. This information was obtained from FlyBase for the melanogaster–miranda and melanogaster–pseudoobscura comparisons and from the annotation of the D. pseudoobscura genome for the miranda–pseudoobscura comparisons involving elements C and E. Given that the annotation process is not yet complete for element A in this species, we measured the distance between markers in the drawn D. miranda map (Das et al. 1982) as an approximation to the physical map. To calibrate it, it we used the estimate of the chromosome length of this element in D. pseudoobscura (Richards et al. 2005; supplemental Table S3 at http://www.genetics.org/supplemental/).
To estimate what fraction of A was included in the pericentric inversions involving element D (see results) we assumed, as a crude approximation, that ∼1.1 Mb (the equivalent of one melanogaster chromosomal division) of element A has been moved to element D in D. pseudoobscura.
RESULTS
Duplications and/or transposition:
An unusual rate of gene duplications and/or transpositions from the neo-X chromosome onto other chromosomal locations was proposed as a possible mechanism for evolving dosage compensation for genes that are degenerating on the neo-Y chromosome of D. miranda (Yi and Charlesworth 2000b). This hypothesis predicts that such events should preferentially involve genes derived from Muller's element C, which constitutes the neo-X and neo-Y chromosome pair in this species. To test this, we performed in situ hybridizations using a large number of DNA markers. Together with previously published results (Segarra and Aguadé 1992; Segarra et al. 1996; Steinemann and Steinemann 1998, 1999; Bachtrog 2003; Yi et al. 2003), we have data for 31 genes on the neo-X chromosome, 12 on chromosome 2 (element E), 10 on chromosome 4 (element B), and 13 and 4 on the left and right arms of the X chromosome, respectively (elements A and D).
There are a total of 39 loci from both arms of the X chromosome and the two autosomes, whose locations have been determined by means of in situ hybridization, which provide evidence concerning any possible transpositions or duplications (Table 1). These include some previously published localizations, as noted in the Table 1 footnotes. The single case of duplication/transposition that we found in this part of the study involves the smoothened gene (smo), whose expected location, according to the expected chromosomal homology (Ashburner 1989), is on chromosome 4 (element B). We detected two copies of smo on this chromosome (at positions 64D and 68E) plus a third one on chromosome 2 (element E) (47A), although we do not know which of the two locations on chromosome 4 is the ancestral one. Interestingly, this gene is also duplicated in the D. pseudoobscura genome release 1.0 (http://flybase.bio.indiana.edu/cgi-bin/gbrowse_fb/dpse), where it shows two copies, one on chromosome 4 (GA11066) and another on chromosome 2 that is not yet annotated (between GA21431 and GA15018, whose orthologs in D. melanogaster are Actn3 and CG18622, respectively). This second copy exhibits numerous stop codons along its sequence and is probably not functional.
TABLE 1.
In situ hybridization results (standard arrangement)
| Gene | Chromosome D. miranda | Location D. miranda |
|---|---|---|
| Gapdh2 | XL | 2D |
| sisAa | XL | 2F |
| scutea | XL | 3C |
| Annx | XL | 4D |
| sesBa | XL | 5B |
| cyp1a | XL | 5F |
| runta | XL | 6D |
| whiteb | XL | 7C |
| exu2c | XL | 7D |
| pera | XL | 8A |
| swallowa | XL | 8A |
| elava | XL | 8B |
| su-fb | XL | 8F |
| Est-5B | XR | 17F |
| Hsp83 (Hsp82) | XR | 19A |
| Pgdb | XR | 21F |
| zb | XR | 21F |
| Amyreld | neo-X | 22B |
| Alk | neo-X | 22B |
| evee | neo-X | 23A |
| vlc | neo-X | 23B |
| stan | neo-X | 25A |
| grau | neo-X | 25C |
| Amyd | neo-X | 27A |
| nompA | neo-X | 27C |
| Toll-7 | neo-X | 28A |
| Rya | neo-X | 28A |
| CG30152 | neo-X | 28C–D |
| exu1 | neo-X | 28C–D |
| CG13437 | neo-X | 28C–D |
| CG11136 | neo-X | 28C–D |
| CG11159 | neo-X | 28C–D |
| CG16799 | neo-X | 28C–D |
| Updo | neo-X | 28E |
| UbaI | neo-X | 28E |
| jeb | neo-X | 29E |
| gek | neo-X | 29F |
| Pcl | neo-X | 30C |
| Asx | neo-X | 31B |
| ene | neo-X | 31C |
| cnk | neo-X | 31D |
| CycBe | neo-X | 32A |
| tud | neo-X | 33A |
| dpn | neo-X | 33C |
| roboe | neo-X | 33C |
| sax | neo-X | 33C |
| Lcpf | neo-X | 33C |
| cos | neo-X | 34A |
| sry-aa | 2 | 35C |
| RpL32 (rp49) | 2 | 35F–36A |
| T1 | 2 | 37F–38A |
| Bruce | 2 | 40A |
| Nop56 | 2 | 43A |
| rosy (Xdh) | 2 | 44C |
| hyd | 2 | 47B–C |
| bcd | 2 | 48A |
| ftz | 2 | 48D |
| ninaE (rh1) | 2 | 52C |
| hb | 2 | 53B |
| Gld | 2 | 53C |
| smo | 4 | 47A, 64D, 68E |
| Uro | 4 | 57B |
| Eno | 4 | 61C |
| dpp | 4 | 63C |
| Gpdh | 4 | 66C |
| Adha | 4 | 68C |
| Adhr | 4 | 68C |
| ade3 (Gart) | 4 | 68E |
| amd | 4 | 68F |
| Ddc | 4 | 68F |
Names in parentheses represent the names used in previous studies. Gene order between CG30152 and CG16799 is described by Bachtrog (2003). The localizations of dpn and exu1 are slightly different from those given by Bachtrog (2005).
Localization by Yi et al. (2003).
Localization by Segarra and Aguadé (1992).
Localization by Bachtrog and Charlesworth (2003).
Localization by Steinemann and Steinemann (1999).
Localization by Bachtrog and Charlesworth (2002).
Localization by Steinemann and Steinemann (1998).
The existence of duplications of this gene in D. miranda was further checked by Southern blotting using three different restriction enzymes (XbaI, PstI, and ApaI). The exact number of copies was difficult to determine, although we detected at least three bands, in agreement with what we found using in situ hybridization. The digestion with PstI exhibited a fourth band; we cannot, however, rule out the possibility of this being caused by a new restriction site in one of the extra copies.
As shown in Table 1, none of the element C loci exhibit signs of transposition; all the genes under study hybridized only to their putative homologs (the neo-sex chromosomes), contrasting with the pattern previously described for these chromosomes (Yi and Charlesworth 2000b). It seems, however, that these apparent transpositions/duplications are artifacts of the in situ hybridization results. The first of them, the apparent transposition of the exuperantia 1 gene (exu1) from the neo-X chromosomes (element C) onto the ancestral X (element A) (Yi and Charlesworth 2000b), was subsequently explained by the existence of a misleading hybridization signal on element A. The true location of this locus is on element C, as expected from the conservation of chromosome element content (Bachtrog and Charlesworth 2003).
A second case, where deadpan (dpn) apparently showed two different signals (Yi and Charlesworth 2000b), one corresponding to its ancestral location on element C and a new one on element E (representing a recent transposition), appears to be an error caused by the proximity of this locus to the centromere, which makes the hybridization signal difficult to interpret. We have reinvestigated its location by in situ hybridization in 12 of the D. miranda lines and detected a single site on the neo-X chromosome (see Table 1), but none on chromosome 2.
These data thus provide no support for the idea that there is frequent transposition of genes from the neo-X chromosome of D. miranda. However, when investigating the DNA sequence variation on the neo-sex chromosomes (C. Bartolomé and B. Charlesworth, unpublished data) we identified a possible duplication of the genghis khan gene (gek). Its localization by in situ hybridization did not reveal transpositions onto other chromosomes, and the only staining clearly visible was that corresponding to the neo-X copy at 29F. Blasts against the D. pseudoobscura genome (http://flybase.net/blast/) showed that gek is present as a single copy in this species.
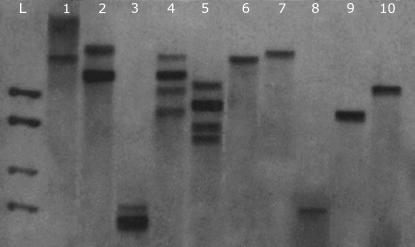
We performed Southern blotting with five different restriction digestion reactions (ApaI, BamHI, EcoRI, HindIII, and Nde) to examine whether there were differences in the banding patterns of D. miranda males and females and found that males indeed exhibited a greater number of copies than females, indicating that this locus is duplicated on the neo-Y chromosome but not on the neo-X chromosome (see Figure 1). The possibility of a transposon insertion on the neo-Y copy being responsible for this pattern cannot be ruled out. However, this seems unlikely because the presence of a transposable element would generate two bands instead of one (and not three, as can clearly be seen in Figure 1, lanes 4 and 5). In addition, it would be odd to find restriction sites in such a short fragment for each of the five restriction enzymes used.
Figure 1.
Southern blotting results for determining the banding patterns of gek in D. miranda males and females. Lane L, kilobase ladder; lanes 1–5, digests of male genomic DNA. The enzymes used were ApaI (1), BamHI (2), EcoRI (3), HindIII (4), and Nde (5). Lanes 6–10, digests of female genomic DNA using the same set.
Intraspecific inversion polymorphisms:
Another aspect of chromosomal variation that can be studied in this species is the extent of inversion polymorphism, which has been very poorly documented in D. miranda (Yi et al. 2003). We have analyzed six lines recently collected from a natural population (see materials and methods), in which we have detected two polymorphic inversions (Table 2) in addition to the four described by Yi et al. (2003). One of these inversions, X2-2, is the second inversion found on the neo-X chromosome. It seems to be fixed in strain MSH22, since it is present in all the test crosses with these lines (all the female larvae examined carried the inversion). Reexamination of our records shows that only one female from this strain was tested in a cross to the standard used in the previous study (MSH38), so that the presence of the inversion was presumably overlooked, possibly because of nonvirginity of the MSH38 females used in the cross. The banding pattern in a squash of a male of line MSH22, used for the in situ localization of Uro, was reexamined and showed the presence of this inversion when compared with the standard arrangement shown for a male in Steinemann and Steinemann (1998, Figure 8).
TABLE 2.
Inversion polymorphisms in six new lines of D. miranda
| Chromosome | Inversion | Breakpoints | Lines showing inversion in crosses with MSH22 |
|---|---|---|---|
| XR | XR-1a | 18D–20Ca | MA03.2, MA03.6 |
| X2 | X2-2 | 29F–34A | MA03.1, MA03.2, MA03.3, MA03.4, MA03.5, MA03.6 |
| 2 | 2-1a | 52A–54Da | MA03.3 |
| 4 | 4-1 | 55D–58C | MA03.5 |
Inversions are described by Yi et al. (2003).
The second inversion newly identified in these strains, 4-1, segregates exclusively in line MA03.5, in four of eight larvae, suggesting that it is a low-frequency variant. We also detected two other inversions at low frequencies, previously described by Yi et al. (2003). XR-1 was observed in two lines, MA03.2 and MA03.6, in five and four (of eight) females, respectively. The remaining inversion polymorphism, 2-1, was found in a single larva of line MA03.3.
The localizations of grau and stan in Table 1 were affected by the presence of the X2-1 inversion (Yi et al. 2003), since they are both situated within this inversion and were hybridized to lines that carried it. Taking account of its presence, we assign grau to 25C and stan to 25A. These are the only localizations that might have been affected by the presence of an inversion relative to the banding pattern in the map of Das et al. (1982), which we have used as our standard.
These findings suggest that the scale of the chromosomal rearrangements within D. miranda is relatively small, in contrast with what we observe when we compare the location of D. miranda genes with their orthologs in D. pseudoobscura and D. melanogaster (see below).
Interspecific chromosomal rearrangements:
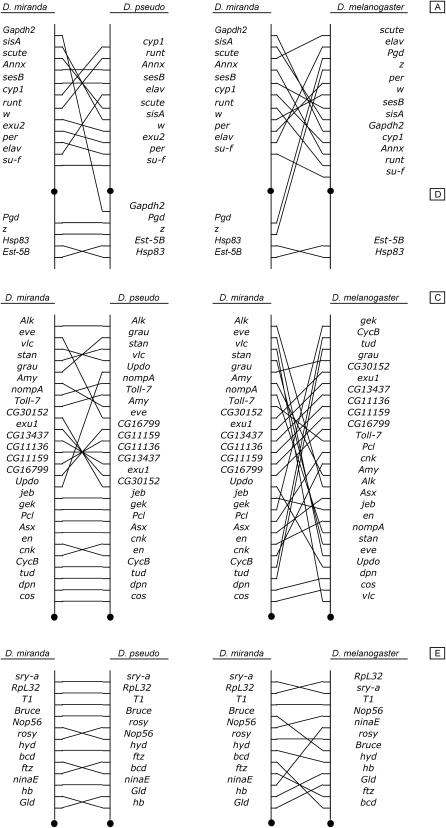
Comparisons of the in situ localizations of genes in D. miranda with the order of genes revealed by the D. pseudoobscura genome sequence and published in situ localizations in D. pseudoobscura (see materials and methods) reveal extensive reshuffling, involving numerous inversion events (Table 1 and Figure 2). Although the locations of the genes are well preserved among chromosomal arms, suggesting that most of these inversions are paracentric, there are a few exceptions with respect to elements A and D, which form the X chromosome of these two species.
Figure 2.
Schematic of the relative order of markers and chromosomal rearrangements between D. miranda and two other species (D. pseudoobscura and D. melanogaster). The solid circle represents the centromere; the position of the markers shows only their relative order along the chromosomes. The locations of the genes and the sizes of Muller elements are not scaled. The arrangements depicted correspond to the Standard–D. miranda and the Arrowhead–D. pseudoobscura arrangements.
A pericentric inversion caused Pgd and z to be moved to XR from XL, which is in common to D. miranda and D. pseudoobscura (Segarra and Aguadé 1992). We also have evidence for a second pericentric inversion that occurred after the split between D. pseudoobscura and D. miranda (Figure 2). In this case, one of the markers located in the distal region of element A in D. miranda (Gapdh2) hybridizes to the proximal end of element D in D. pseudoobscura. Examination of the polytene chromosome map of D. pseudoobscura (Kastritsis and Crumpacker 1966) shows the presence of a proximal group of about four bands in division 18 that is not present at the base of XL in D. miranda and that resembles the group of bands including the site of Gapdh2 and distal to it in D. miranda (2C–D). These are distally located, implying that at least one other inversion has occurred in this region of the genome. Gapdh2 is located on A in D. melanogaster and this suggests that the inversion occurred in the D. pseudoobscura lineage subsequent to the divergence from D. miranda. Since there is no evidence for any polymorphism for an inversion in this region, it must be fixed in D. pseudoobscura.
Muller's element E seems to be the most conserved element among the three species, particularly in its distal portion (sections 35–40 of D. miranda). We found what appears to be the small inversion distinguishing D. miranda and D. pseudoobscura described by Dobzhansky and Tan (1936); this explains the inverted positions of Nop56 and rosy (Segarra et al. 1996) in these species (sections 43A–44C of D. miranda).
As expected from their phylogeny (Powell 1997), D. miranda and D. pseudoobscura show more similarities in their chromosomal organization than do D. miranda and D. melanogaster. This is particularly clear for the proximal region of element C (sections 29–34 of D. miranda). However, the high level of inversion polymorphism on this chromosome in D. pseudoobscura (Powell 1992; Schaeffer and Anderson 2005) means that we have to be cautious about which arrangement in D. pseudoobscura is ancestral, a classic subject of debate. Given that the recent consensus is that Tree Line (TL) or Santa Cruz (SC) is the ancestral arrangement in D. pseudoobscura (Popadic and Anderson 1994; Schaeffer and Anderson 2005), and that the Arrowhead (AR) arrangement used for the D. pseudoobscura genome sequence (Richards et al. 2005) differs from this by four inversions, this high degree of synteny may seem somewhat surprising, but it reflects mainly the fact that the breakpoints involved in the path connecting TL to AR are medial to distal in their location (see Figure 1 of Schaeffer and Anderson 2005). The identity of the common ancestor of the D. pseudoobscura arrangements is examined further in the discussion.
We also found conservation of some short syntenic blocks between D. melanogaster and D. miranda, which exhibit the opposite orientation to that of D. pseudoobscura (such as CG30152–CG16799), indicating that these inversions are probably the result of the numerous rearrangements undergone by this chromosome in D. pseudoobscura. In addition, we see some reorganization of the D. miranda chromosomes, involving small segments, which are not shared by the other two species. For example, there is a small proximal inversion on element D, involving the Est-5B-Hsp83 region, also described by Segarra et al. (1996). This appears to be distinct from the proximal XR-1 inversion found to be polymorphic in D. miranda by Yi et al. (2003), since Est-5B is located distal to this inversion.
Rates of chromosomal evolution:
Table 3 summarizes the numbers of between-species inversions and rates of rearrangement in elements A, C, and E obtained by the four different algorithms described in materials and methods, using the localizations shown in Figure 2. (For pairs of markers that are so close that their relative location in D. miranda could not be determined with confidence, we used only one member of the pairs for comparison with the other species.) Given the relatively small number of markers on element B, and the incomplete assembly of this chromosome in the D. pseudoobscura genome sequence, we did not attempt to estimate the rate of rearrangement of this chromosome.
TABLE 3.
Number of inversions and rate of chromosomal evolution
| GRIMMa
|
Nadeau–Taylor
|
Ranz et al.b
|
This studyc
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | C | E | A | C | E | A | C | E | A | C | E | |
| No. of inversions | ||||||||||||
| D. miranda–D. pseudoobscura | 7 | 5 | 3 | 11 | 15 | 10 | 7 ± 3 | 6 ± 2 | 5 ± 2 | 7 (7–13) | 6 (5–9) | 7 (3–11) |
| D. miranda–D. melanogaster | 7 | 13 | 6 | 7 | 29 | 27 | 14 ± 5 | 27 ± 8 | 20 ± 9 | 17 (8–27) | — | 21 (8–49) |
| D. pseudoobscura–D. melanogaster | 7 | 14 | 6 | 42 | 29 | 13 | 33 ± 15 | 27 ± 8 | 12 ± 5 | 34 (13–74) | — | 11 (6–24) |
| Breaks/Mb/MY | ||||||||||||
| D. miranda–D. pseudoobscura | 0.130 | 0.126 | 0.047 | 0.204 | 0.379 | 0.158 | 0.130 ± 0.056 | 0.152 ± 0.051 | 0.079 ± 0.032 | 0.130 (0.130–0.242) | 0.152 (0.126–0.227) | 0.110 (0.047–0.174) |
| D. miranda–D. melanogaster | 0.011 | 0.021 | 0.007 | 0.011 | 0.048 | 0.032 | 0.021 ± 0.008 | 0.044 ± 0.013 | 0.024 ± 0.011 | 0.026 (0.012–0.041) | — | 0.025 (0.010–0.059) |
| D. pseudoobscura–D. melanogaster | 0.011 | 0.023 | 0.007 | 0.064 | 0.048 | 0.016 | 0.050 ± 0.023 | 0.044 ± 0.013 | 0.014 ± 0.060 | 0.052 (0.020–0.113) | — | 0.013 (0.007–0.029) |
Minimum number of inversions.
Plus or minus standard deviation (±SD).
Lower and upper 2-unit support limits are indicated in parentheses.
To obtain comparable estimates of rates of evolution for the different elements, we divided the numbers of breaks by the sizes of each chromosome in megabases (Richards et al. 2005; supplemental Table S3 at http://www.genetics.org/supplemental/), assuming a divergence time of 2 million years (MY) between D. miranda and D. pseudoobscura (Barrio et al. 1992) and ∼30 MY between D. melanogaster and these species (Throckmorton 1975). Although the results show differences according to the approach used for inferring the number of inversions (see discussion), all the miranda–pseudoobscura comparisons indicate an extremely high rate of evolution since the split of these species. This high rate of chromosomal rearrangement becomes even more dramatic if we assume a greater divergence time (54.9 MY) between D. melanogaster and the obscura group (Tamura et al. 2004).
DISCUSSION
Conservation of chromosomal elements:
In agreement with other studies of various groups of Drosophila species (Steinemann 1982; Vieira et al. 1997a; Ranz et al. 2001; Richards et al. 2005), we find that the majority of genes of D. miranda have remained on the Muller elements expected from their locations in D. pseudoobscura and D. melanogaster, although these elements are subject to internal rearrangements via paracentric inversions. There is no evidence for translocations of material among chromosomes of the type proposed by Dobzhansky and Tan (1936), in agreement with the results of Steinemann (1982) for 13 probes.
We have found only one new case (smo) of cross-element duplication, for which the ancestral copy is located on chromosome 4. We did not confirm the transposition of dpn from the neo-X proposed by Yi and Charlesworth (2000b). There does seem to be a duplication involving zipper, for which in situ hybridization data suggest an additional location at the distal end of XL in D. miranda, as well as at the probable ancestral location at the base of X2 (D. Bachtrog and B. Charlesworth, unpublished data). However, there is no statistical support for an excess of duplications/transpositions from X2 to other chromosomes, as predicted by the proposal of Yi and Charlesworth (2000b) for a method of compensating for the loss of gene activity of degenerating neo-Y-linked genes. Of course, our results do not falsify this hypothesis, but imply that such events are insufficiently frequent to be detected at the level of resolution provided by our current data set.
Our data do, however, provide evidence that D. pseudoobscura has undergone a pericentric inversion, which brought a small number of polytene bands that include Gapdh2 onto the proximal region of the XR chromosome from XL. This is in addition to the larger pericentric inversion described by Segarra and Aguadé (1992), which includes Pgd and z (Figure 2) and is shared by both D. miranda and D. pseudoobcura. We can only speculate as to the reason for the fixation of these inversions, but it is worth pointing out that there is evidence that pericentric inversions may have such small effects on fitness when heterozygous that they can persist in populations (Aulard et al. 2004). One possibility is that movement of part of XL onto the newly evolving XR, resulting from the fusion of element D to A in the ancestor of D. miranda and D. pseudoobcura, might have facilitated dosage compensation of genes on the fused D, whose neo-Y counterparts were degenerating, by allowing spreading of the dosage compensation machinery (Marin et al. 2000) from A to D. This does not, however, explain the subsequent transfer of A-derived material to the proximal portion of XR in D. pseudoobscura.
Ancestral states of chromosomes with respect to inversion polymorphisms:
The majority of chromosomal differences between the three species that we have analyzed involve paracentric inversions. These are by far the most frequent type of arrangement in Drosophila, at both the intra- and the interspecific levels (Powell 1992, 1997). To obtain accurate estimates of evolutionary rates, it is useful to know the ancestral states of each species used for comparisons, in cases where there are inversion polymorphisms within species. Before discussing the estimates of rates of evolution, we therefore consider first the question of the ancestral states of each species used here.
Our results on inversion polymorphisms within D. miranda, and those of Yi et al. (2003), suggest that these are relatively few and are present on all chromosomal arms except XL, mostly at low frequencies. The one exception is the polymorphism for the X2-1 inversion on element C, which is present at an intermediate frequency. The pattern of polymorphism at the eve locus, which is slightly distal to this inversion, suggested that X2-1 inversion may in fact be the ancestral arrangement (Yi et al. 2003). However, this pattern is not seen in data on DNA sequence polymorphism at grau and stan, which are located between the breakpoints of this inversion (C. Bartolomé and B. Charlesworth, unpublished data). The question of whether or not the standard arrangement is ancestral is thus currently unresolved. It seems reasonable, however, to assume that the high-frequency arrangements for the other inversions, corresponding to the standard arrangement in the map of Das et al. (1982), are the ancestral states.
This pattern contrasts with the pattern of inversion polymorphisms found in the close relative D. pseudoobscura, where only the third chromosome (element C) is significantly polymorphic, with the exception of the inversion complex associated with the sex-ratio distorter chromosome SR of element D (Powell 1992). More than 30 gene arrangements have been described for element C, 10 of which are abundant and widely distributed (Powell 1992).
This raises the question of the identity of the ancestral arrangement of element C in D. pseudoobscura. Schaeffer and Anderson (2005) describe the relations between the known locations of genes and chromosomal breakpoints of the major inversions of element C in D. pseudoobscura. Their Figure 1 for the markers in common with our study (en, exu1, Amy, and eve) shows that the order of these in D. miranda is similar to that in TL, Standard (ST), and Hypothetical (HY), but not AR or SC. Thus, any one of TL, HY, and ST is more parsimoniously inferred to be closer to the ancestral state than the other rearrangements.
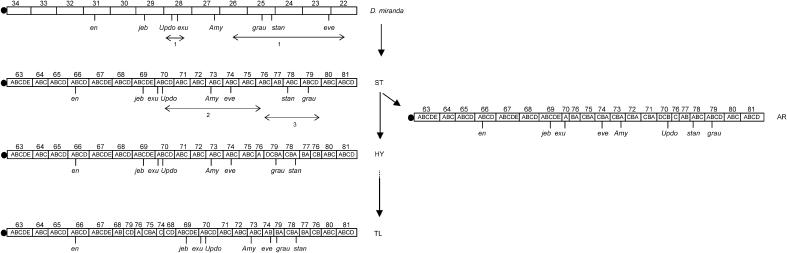
To examine this question further, we used the locations in D. miranda of some additional markers (jeb, Updo, grau, and stan), which were not used by Schaeffer and Anderson (2005) but whose locations in D. pseudoobscura are known from the genome sequence. Updo is very close to vg, used by Schaeffer and Anderson (2005), which they placed in 70B of the D. pseudoobscura map. We located it to division 28 of the D. miranda map, adjacent to division 29, which contains jeb. In D. pseudoobscura, jeb is in division 69, which is adjacent to the whole of division 70 in ST and HY (see our Figure 3 and Figure 1 of Schaeffer and Anderson 2005). But 70B is located distally in AR, as a result of the inversion that generated it from ST. Together with the other evidence mentioned above, this rules out AR as the ancestral arrangement.
Figure 3.
This shows the most parsimonious interpretation of the relations between the major rearrangements in D. pseudoobscura and the D. miranda–Standard arrangement. Double arrows indicate putative inversions between arrangements: 1, inversions D. miranda–ST; 2, inversion ST–AR; 3, inversion ST–HY. The exact location of the genes in D. miranda is shown in Table 1; the locations of the genes in the ST, HY, and TL arrangements were inferred from their approximate position in the D. pseudoobscura Genome Project (AR).
By measuring the distance between en and the closest of these markers (jeb) on the drawn D. miranda map (Das et al. 1982), we calculated that this segment represents ∼14% of the length of element C. A very similar result was obtained for AR in D. pseudoobscura from the genome sequence (16% of element C). Given that HY, ST, and AR are all identical in arrangement in this region, and that AR has already been eliminated as a potential ancestor, this similarity is inconsistent with either SC or TL being the ancestral state, since they have a large fraction of element C (parts of divisions 74–76 and part of 79) inserted into this region (Schaeffer and Anderson 2005). Our localization of grau and stan shows that their orientation is reversed between the standard D. miranda arrangement and ST, but not HY, of D. pseudoobscura (Figure 3). The relative orientation of eve with respect to grau and stan in ST and AR can be explained by an inversion including all three genes that moved eve proximally, if the D. miranda standard arrangement is the ancestral arrangement (Figure 3), so that ST is the most parsimonious ancestor for the D. pseudoobscura arrangements (HY would require a further inversion to change the order of grau and stan back again; see Figure 1 of Schaeffer and Anderson 2005). If X2-1 was ancestral in D. miranda, HY would be the most parsimonious ancestor. Parsimony thus cannot distinguish between ST and HY as the most likely ancestor of the D. pseudoobscura arrangements without further evidence on the nature of the ancestral arrangement in D. miranda.
Popadic and Anderson (1994) based their conclusion that either TL or SC was likely to be the ancestral arrangement on phylogenetic reconstructions, using sequence data on the Amy1 gene that is located within the inversions they considered and the restriction mapping results for the Amy1 region from Aquadro et al. (1991). We have reexamined this approach using two sets of published data, using MEGA (Kumar et al. 2001) to construct neighbor-joining trees on whole sequences. We first used Amy1 sequences from the six different arrangements used by Popadic and Anderson (1994; GenBank accession nos. U20330, U20331, U20333–U20335, and U203370). We also included two more sequences from this region: AR (D. pseudoobscura Genome Project) and D. miranda (Steinemann and Steinemann 1999; accession no. Y15603).
The results for a maximum sequence length of 955 sites show very similar rates of divergence among D. miranda and the sequences from D. pseudoobscura, using the Kimura two-parameter approach and the number of differences (not shown here); the values range between 0.022 (ST–D. miranda) and 0.033 (TL–D. miranda). If we remove the ST sequence from the study as being shorter than the rest, we obtain analogous results (2602 sites, range 0.023–0.027 for AR–D. miranda and TL–D. miranda, respectively). There is no support from the tree for placing TL or SC as the closest to the split from D. miranda.
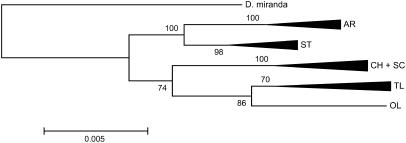
The second data set includes numerous sequences from seven genes and an intergenic region (F6) contained in the rearrangements (Schaeffer et al. 2003; accession nos. AF476112–AF476921). For each gene, we randomly selected six samples from each arrangement [except for SC and Olympic (OL), for which there was only one available]; we then concatenated all genes and F6 sequences from each sample, giving a total of as many as 2857 sites in each. The results of the analyses were nearly identical to those of the previous one (range 0.026–0.032 for ST–D. miranda and SC–D. miranda, respectively). Although ST seems to exhibit a somewhat lower divergence from D. miranda than other arrangements, there is no power to discriminate which arrangement could be ancestral within D. pseudoobscura. In fact, the similarity among divergence values suggests that the D. pseudoobscura inversions radiated out from a common core, well after the divergence from D. miranda (Figure 4), but with an early separation into two clades, composed of ST with its derived inversions and SC and its derived inversions (see Figure 1 of Schaeffer and Anderson 2005). The conclusion from the location data that either ST or HY is closest to the ancestral state is not challenged by the sequence data.
Figure 4.
Neighbor-joining tree obtained by concatenating six random samples of each arrangement from the Schaeffer et al. (2003) data set. It includes seven genes (en, exu1, vg, Amy1, eve, mef2, and EcR) and an intergenic region (F6). AR, Arrowhead; ST, Standard; CH, Chiricaua; SC, Santa Cruz; TL, Tree Line; OL, Olympic.
D. melanogaster is also highly polymorphic for paracentric inversions, although in this case they affect all chromosomal arms, with an average of five to seven different arrangements per population in Africa (Lemeunier and Aulard 1992; Aulard et al. 2002). Given the high degree of synteny between D. simulans and D. melanogaster (Ashburner 1989), it seems reasonable to assume that the standard arrangements in D. melanogaster are ancestral.
Rates of chromosomal rearrangement by inversions:
Although the number of loci we studied is insufficient to determine the exact number of inversions that have occurred, especially in the comparisons with D. melanogaster, the locations of genetic markers in the different genomes allow us to examine the approximate extent of chromosomal reorganization.
Use of the GRIMM algorithm (Tesler 2002) for direct counts of the minimum numbers of breakpoints among species pairs suggests that Muller's element E has the most conserved gene order among the three species used for comparison (Table 3). This is in agreement with the results obtained by Segarra et al. (1996), who detected a high degree of conservation for this element among the obscura species group. In addition, this chromosome exhibits the largest number of syntenic blocks (>20) between D. melanogaster and D. pseudoobscura (Richards et al. 2005; supplemental Figure S2 at http://www.genetics.org/supplemental/), which is also consistent with our data. However, this apparent conservation of element E implies that there is no genuswide pattern of chromosomal evolution, suggested by González et al. (2002) and Vieira et al. (1997a,b), who found that this autosome shows the highest rate of rearrangements between D. melanogaster and D. repleta and among three species from the D. virilis group, respectively.
In contrast, element A appears to show more drastic changes in its chromosomal organization, not only within the obscura group but also between the former and D. melanogaster, as also suggested by Segarra et al. (Segarra and Aguade 1992; Segarra et al. 1995). This observation is supported by the finding that this element shows the shortest syntenic blocks between D. pseudoobscura and D. melanogaster (Richards et al. 2005; supplemental Figure S2 at http://www.genetics.org/supplemental/). These findings might be taken as evidence of the faster evolution of X-linked rearrangements (element A) compared with that for autosomal ones, as proposed by Charlesworth et al. (1987).
However, these conclusions should be qualified in light of the fact that the various methods for correcting for undetected arrangements all suggest that the elements analyzed here have comparable rates of evolution to the other chromosomes, especially since the confidence intervals provided by the relatively assumption-free method we have developed are very wide (Table 3).
The great similarity of the proximal regions of element C in D. pseudoobscura and D. miranda (Figure 2), in contrast to the extensive reshuffling that has taken place between these species and D. melanogaster (see also Richards et al. 2005), seems to suggest that a relevant fraction of internal rearrangements between the obscura group and D. melanogaster took place either in the D. melanogaster lineage or in the ancestor of the pseudoobscura subgroup before the split of D. miranda and D. pseudoobscura.
As far as the more distal part of this chromosome is concerned, which has undergone far more rearrangements between D. miranda and D. pseudoobscura, we need to take into account the fact that either ST or HY is likely to be the ancestral state for D. pseudoobscura, as discussed above. Given that these differ by one or two rearrangements from AR, the standard of comparison, our estimate of the minimum numbers of changes between these species for element C in Table 3 should be reduced by one or two, depending on whether or not X2-1 is the ancestral state in D. miranda (see above). Thus, the minimum number of differences between element C in D. miranda and D. pseudoobscura is probably not more than four and possibly only three.
This would make the level of difference quite comparable to that for E. This comparatively low rate of divergence for element C is very interesting, in view of the fact that this is the most highly polymorphic chromosome in D. pseudoobscura and its sibling species, D. persimilis (Powell 1992), and is of course the most intensively studied of all examples of inversion polymorphisms. It strongly suggests that natural selection is the reason for the abundance of these inversion polymorphisms on this chromosome, not any internal genetic factors, consistent with the large body of evidence for selection acting on this system (Powell 1992; Schaeffer et al. 2003).
It is also apparent, however, that our results suggest an unusually high rate of evolution due to inversion breakpoints in the D. miranda/D. pseudoobscura lineage. There are some differences between the results generated by the different methods of estimation. The GRIMM algorithm yields simply the minimum number of inversions needed to account for the difference in order of the markers between a pair of species. This is likely to be much below the true value for distant comparisons. The method of Nadeau and Taylor (1984) assumes a random distribution of markers, which is inappropriate for most of our data, so that either the Ranz et al. (1997) method or our own method is to be preferred. The latter is more robust, but cannot be applied when there are a large number of rearrangements, in which case the Ranz et al. method should work well. Fortunately, the two methods yield very similar results, except for a wider upper confidence interval for our method.
Even after making the correction for the ancestral states of element C, this element has an estimated rate of evolution of 0.076 breaks/Mb/MY between D. miranda and D. pseudoobscura by the GRIMM method and 0.101 by the Ranz method and our method. Elements A and E yield comparable or higher rates. In fact, the rates are of the same order of magnitude as the lower range of estimates for Caenorhabditis given by (Coghlan and Wolfe 2002) (0.4–1.0 breaks/Mb/MY). This high rate in nematodes may be explained partly by the fact that Caenorhabditis elegans and C. briggsae are both highly self-fertilizing, which is expected to be more permissive than outcrossing for the evolution of chromosome rearrangements with mildly deleterious heterozygous fitness effects (Charlesworth 1992). However, recent mapping studies of the highly selfing plant Arabidopsis thaliana and its self-incompatible close relative A. lyrata show a maximum rate of evolution of inversions in the A. thaliana lineage of only 0.015 breaks/Mb/MY (Koch and Kiefer 2005; D. Charlesworth, personal communication). Inbreeding per se does not, therefore, appear to cause a high rate of accumulation of inversion breakpoints.
The estimated rates for the divergence of these two species from D. melanogaster are, in contrast, much lower and similar to estimates from other species groups using comparable methods (Ranz et al. 1997, 2001). This strongly suggests that the high rate of evolution is associated with the obscura group and probably has a relatively recent origin. It was noted by Richards et al. (2005) that there may be a causal relation between a type of repeated sequence distributed over the D. pseudoobscura genome and interruptions of synteny. This suggests that, in contrast to the conclusion concerning inversion polymorphism in D. pseudoobscura, the elevation of the rate of chromosomal evolution may be mutational in origin.
Acknowledgments
We are grateful to Xulio Maside, Alfredo Ruiz, and Doris Bachtrog for helpful comments and discussions and to Jorge Amigo Lechuga for writing a Perl script to run the Ranz et al. (1997) maximum-likelihood method. We are particularly indebted to Xulio Maside for checking the in situ localizations of the markers. We also thank two anonymous reviewers for suggestions that improved the original manuscript. This work was funded by grants to B.C. from the Royal Society and the Biotechnology and Biological Sciences Research Council (United Kingdom).
References
- Aquadro, C. F., A. L. Weaver, S. W. Schaeffer and W. W. Anderson, 1991. Molecular evolution of inversions in Drosophila pseudoobscura: the amylase gene region. Proc. Natl. Acad. Sci. USA 88: 305–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M., 1989. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Aulard, S., J. R. David and F. Lemeunier, 2002. Chromosomal inversion polymorphism in Afrotropical populations of Drosophila melanogaster. Genet. Res. 79: 49–63. [DOI] [PubMed] [Google Scholar]
- Aulard, S., P. Vaudin, V. Ladeveze, N. Chaminade, G. Periquet et al., 2004. Maintenance of a large pericentric inversion generated by the hobo transposable element in a transgenic line of Drosophila melanogaster. Heredity 92: 151–155. [DOI] [PubMed] [Google Scholar]
- Bachtrog, D., 2003. Adaptation shapes patterns of genome evolution on sexual and asexual chromosomes in Drosophila. Nat. Genet. 34: 215–219. [DOI] [PubMed] [Google Scholar]
- Bachtrog, D., 2005. Sex chromosome evolution: molecular aspects of Y-chromosome degeneration in Drosophila. Genome Res. 15: 1393–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog, D., and B. Charlesworth, 2002. Reduced adaptation of a non-recombining neo-Y chromosome. Nature 416: 323–326. [DOI] [PubMed] [Google Scholar]
- Bachtrog, D., and B. Charlesworth, 2003. On the genomic location of the exuperantia1 gene in Drosophila miranda: the limits of in situ hybridization experiments. Genetics 164: 1237–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrio, E., A. Latorre, A. Moya and F. J. Ayala, 1992. Phylogenetic reconstruction of the Drosophila obscura group, on the basis of mitochondrial DNA. Mol. Biol. Evol. 9: 621–635. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., 1992. Evolutionary rates in partially self-fertilizing species. Am. Nat. 140: 126–148. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., 1996. The evolution of chromosomal sex determination and dosage compensation. Curr. Biol. 6: 149–162. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., and D. Charlesworth, 2000. The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B 355: 1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., J. A. Coyne and N. Barton, 1987. The relative rates of evolution of sex chromosomes and autosomes. Am. Nat. 130: 113–146. [Google Scholar]
- Coghlan, A., and K. H. Wolfe, 2002. Fourfold faster rate of genome rearrangement in nematodes than in Drosophila. Genome Res. 12: 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, M., D. Mutsudi, A. K. Duttagupta and A. S. Mukherjee, 1982. Segmental heterogeneity in replication and transcription of the X2 chromosome of Drosophila miranda and conservativeness in the evolution of dosage compensation. Chromosoma 87: 373–388. [Google Scholar]
- Dobzhansky, T., 1935. Drosophila miranda, a new species. Genetics 20: 377–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky, T., and C. C. Tan, 1936. Studies on hybrid sterility. III. A comparison of the gene arrangement in two species, D. pseudoobscura and D. miranda. Z. Indukt. Abstammungs. Vererbungsl. 72: 88–114. [Google Scholar]
- González, J., J. M. Ranz and A. Ruiz, 2002. Chromosomal elements evolve at different rates in the Drosophila genome. Genetics 161: 1137–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastritsis, C. D., and D. W. Crumpacker, 1966. Gene arrangements in the third chromosome of Drosophila pseudoobscura. J. Hered. 75: 151–158. [DOI] [PubMed] [Google Scholar]
- Koch, M. A., and M. Kiefer, 2005. Genome evolution among cruciferous plants: a lecture from the comparison of the genetic maps of three diploid species: Capsella rubella, Arabidopsis lyrata subsp. petraea, and A. thaliana. Am. J. Bot. 92: 761–767. [DOI] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura, I. B. Jakobsen and M. Nei, 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17: 1244–1245. [DOI] [PubMed] [Google Scholar]
- Lemeunier, F., and S. Aulard, 1992. Inversion polymorphisms in Drosophila melanogaster, pp. 339–405 in Drosophila inversion polymorphisms, edited by C. B. Krimbas and J. R. Powell. CRC Press, Boca Raton, FL.
- Machado, C. A., R. M. Kliman, J. A. Markert and J. Hey, 2002. Inferring the history of speciation from multilocus DNA sequence data: the case of Drosophila pseudoobscura and close relatives. Mol. Biol. Evol. 19: 472–488. [DOI] [PubMed] [Google Scholar]
- Macknight, R. H., 1939. The sex-determining mechanism of Drosophila miranda. Genetics 24: 180–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin, I., M. L. Siegal and B. S. Baker, 2000. The evolution of dosage-compensation mechanisms. BioEssays 22: 1106–1114. [DOI] [PubMed] [Google Scholar]
- Maside, X., C. Bartolomé, S. Assimacopoulos and B. Charlesworth, 2001. Rates of movement and distribution of transposable elements in Drosophila melanogaster: in situ hybridization vs. Southern blotting data. Genet. Res. 78: 121–136. [DOI] [PubMed] [Google Scholar]
- McAllister, B. F., and B. Charlesworth, 1999. Reduced sequence variability on the neo-Y chromosome of Drosophila americana americana. Genetics 153: 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, J. H., 1940. Bearings of the Drosophila work on systematics, pp. 185–268 in New Systematics, edited by J. Huxley. Clarendon Press, Oxford.
- Nadeau, J. H., and B. A. Taylor, 1984. Lengths of chromosomal segments conserved since divergence of man and mouse. Proc. Natl. Acad. Sci. USA 81: 814–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popadic, A., and W. W. Anderson, 1994. The history of a genetic system. Proc. Natl. Acad. Sci. USA 91: 6819–6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, J. R., 1992. Inversion polymorphisms in Drosophila pseudoobscura and Drosophila persimilis, pp. 73–126 in Drosophila Inversion Polymorphisms, edited by C. B. Krimbas and J. R. Powell. CRC Press, Boca Raton, FL.
- Powell, J. R., 1997. Progress and Prospects in Evolutionary Biology: The Drosophila Model. Oxford University Press, Oxford.
- Ranz, J. M., C. Segarra and A. Ruiz, 1997. Chromosomal homology and molecular organization of Muller's elements D and E in the Drosophila repleta species group. Genetics 145: 281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz, J. M., F. Casals and A. Ruiz, 2001. How malleable is the eukaryotic genome? Extreme rate of chromosomal rearrangement in the genus Drosophila. Genome Res. 11: 230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, S., Y. Liu, B. R. Bettencourt, P. Hradecky, S. Letovsky et al., 2005. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res. 15: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schaeffer, S. W., and W. W. Anderson, 2005. Mechanisms of genetic exchange within the chromosomal inversions of Drosophila pseudoobscura. Genetics 171: 1729–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer, S. W., M. P. Goetting-Minesky, M. Kovacevic, J. R. Peoples, J. L. Graybill et al., 2003. Evolutionary genomics of inversions in Drosophila pseudoobscura: evidence for epistasis. Proc. Natl. Acad. Sci. USA 100: 8319–8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segarra, C., and M. Aguadé, 1992. Molecular organization of the X chromosome in different species of the obscura group of Drosophila. Genetics 130: 513–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segarra, C., E. R. Lozovskaya, G. Ribo, M. Aguadé and D. L. Hartl, 1995. P1 clones from Drosophila melanogaster as markers to study the chromosomal evolution of Muller's A element in two species of the obscura group of Drosophila. Chromosoma 104: 129–136. [DOI] [PubMed] [Google Scholar]
- Segarra, C., G. Ribo and M. Aguadé, 1996. Differentiation of Muller's chromosomal elements D and E in the obscura group of Drosophila. Genetics 144: 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinemann, M., 1982. Analysis of chromosomal homologies between two species of the subgenus Sophophora: D. miranda and D. melanogaster. Chromosoma 87: 77–88. [DOI] [PubMed] [Google Scholar]
- Steinemann, M., and S. Steinemann, 1992. Degenerating Y chromosome of Drosophila miranda: a trap for retrotransposons. Proc. Natl. Acad. Sci. USA 89: 7591–7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinemann, M., and S. Steinemann, 1998. Enigma of Y chromosome degeneration: neo-Y and neo-X chromosomes of Drosophila miranda a model for sex chromosome evolution. Genetica 102–103: 409–420. [PubMed] [Google Scholar]
- Steinemann, S., and M. Steinemann, 1999. The Amylase gene cluster on the evolving sex chromosomes of Drosophila miranda. Genetics 151: 151–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant, A. H., and E. Novitsky, 1941. The homologies of the chromosome elements in the genus Drosophila. Genetics 26: 517–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., S. Subramanian and S. Kumar, 2004. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol. Biol. Evol. 21: 36–44. [DOI] [PubMed] [Google Scholar]
- Tesler, G., 2002. GRIMM: genome rearrangements web server. Bioinformatics 18: 492–493. [DOI] [PubMed] [Google Scholar]
- Throckmorton, L. H., 1975. The phylogeny, ecology, and geography of Drosophila, pp. 421–469 in Handbook of Genetics, edited by R. C. King. Plenum, New York.
- Vieira, J., C. P. Vieira, D. L. Hartl and E. R. Lozovskaya, 1997. a Discordant rates of chromosome evolution in the Drosophila virilis species group. Genetics 147: 223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira, J., C. P. Vieira, D. L. Hartl and E. R. Lozovskaya, 1997. b A framework physical map of Drosophila virilis based on P1 clones: applications in genome evolution. Chromosoma 106: 99–107. [DOI] [PubMed] [Google Scholar]
- Yi, S., and B. Charlesworth, 2000. a Contrasting patterns of molecular evolution of the genes on the new and old sex chromosomes of Drosophila miranda. Mol. Biol. Evol. 17: 703–717. [DOI] [PubMed] [Google Scholar]
- Yi, S., and B. Charlesworth, 2000. b A selective sweep associated with a recent gene transposition in Drosophila miranda. Genetics 156: 1753–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, S., D. Bachtrog and B. Charlesworth, 2003. A survey of chromosomal and nucleotide sequence variation in Drosophila miranda. Genetics 164: 1369–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]