Abstract
We investigated the fitness benefits of systemic acquired resistance (SAR) in Arabidopsis thaliana using a mutational and transformational genetic approach. Genetic lines were designed to differ in the genes determining resistance signaling in a common genetic background. Two mutant lines (cpr1 and cpr5) constitutively activate SAR at different points in SAR signaling, and one mutant line (npr1) has impaired SAR. The transgenic line (NPR1-H) has enhanced resistance when SAR is activated, but SAR is still inducible similarly to wild type. The fitness benefits were also investigated under two nutrient levels to test theories that preventing pathogen damage and realized resistance benefits may be affected by nutrient availability. Under low-nutrient conditions and treatment with the pathogenic oomycete, Hyaloperonospora parasitica, wild type had a higher fitness than the mutant that could not activate SAR, demonstrating that normal inducible SAR is beneficial in these conditions; this result, however, was not found under high-nutrient conditions. The mutants with constitutive SAR all failed to show a fitness benefit in comparison to wild type under a H. parasitica pathogen treatment, suggesting that SAR is induced to prevent an excessive fitness cost.
A resistance to a pathogen or herbivore is likely to persist only if the resistance benefits the organism by reducing the fitness loss caused by a pathogen or herbivore. Benefits are assumed in many cases to be balanced by the fitness cost incurred by expressing resistance in the absence of the pathogen/herbivore. While fitness costs of resistance have rarely been unambiguously found in natural populations, less effort has been devoted to finding benefits of resistance.
Benefits have been recorded in a few cases of herbivory resistance for both induced (Agrawal 1999; Heil 2004) and constitutive resistances (Mauricio and Rausher 1997), for herbicide resistance (Baucom and Mauricio 2004), and for pathogen resistance (van Hulten et al. 2006). In other cases, no fitness benefit has been found even though the resistance does reduce damage by herbivores (Simms and Rausher 1987; Elle et al. 1999). In many previous studies of fitness costs, benefits have been presumed (Kakes 1989; Bryant and Julkunen-Tiitto 1995), and it may be a reasonable assumption in most cases that a benefit exists under some environmental conditions, but it is still useful to know under what conditions there is a benefit. Benefits have been shown to depend on resource availability (Elle et al. 1999), a logical result given that costs also depend on resources (Purrington and Bergelson 1997; Heil et al. 2000).
Induced resistance mechanisms are hypothesized to reduce costs and maximize benefits, assuming that such resistance mechanisms have no cost until induction (Rhoades 1979; Karban and Baldwin 1997). Some plants have a mix of inducible and constitutive defenses in different parts of the plant with the constitutive defenses protecting the most valuable reproductive organs while the inducible defenses protect the more expendable roots and leaves (Zangerl and Rutledge 1996). Under an optimal scenario, the level of activation of induced defenses should match the predicted threat. However, induced resistances could also be unbeneficial if a low level of attack on a host stimulates a resistance pathway that is costly, providing excessive protection against a minor threat. The mechanisms of induction and signaling compounds have been extensively studied in multiple plant systems (Farmer et al. 1992; Albrecht et al. 1993; Moura and Ryan 2001; Li et al. 2002); however, this information does not allow easy determination of the costs and benefits as there are many interacting signals leading to a complex response involving many genes (Maleck et al. 2000; Heidel and Baldwin 2004).
Systemic acquired resistance (SAR) is an induced resistance that has been found (with some variation) in all angiosperms tested (Heil 1999). SAR has been shown to have significant fitness costs in a variety of environmental conditions (Heidel et al. 2004). Even though the benefits of SAR against a known pathogen have not been measured beyond the first qualitative observation by Ross (1961), they have been found for the related primed resistance (van Hulten et al. 2006). Pathogen-produced necrotic lesions induce SAR and lead to resistance against a wide spectrum of pathogens, including viruses, bacteria, fungi, and oomycetes (Ryals et al. 1996). The exact mechanism by which SAR is manifested and the range of pathogens that this resistance protects against are not fully understood. However, SAR is believed to be a result of the expression of a variety of defensive proteins and small defensive compounds. Individual components of some of the signaling pathways that lead to resistance to particular pathogens have been successfully identified. A group of proteins upregulated during SAR are known as the pathogenesis-related (PR) proteins and many are known to possess antibacterial or antifungal/oomycete properties (Datta and Muthukrishnan 1999).
SAR is dependent on the production of salicylic acid and the level and activity of NPR1, a protein that interacts with transcription factors that regulate the expression of defense-related genes (Penninckx et al. 1996; Zhang et al. 1999; Fan and Dong 2002; Wang et al. 2005). In Arabidopsis thaliana, SAR can be monitored with the PR-1 gene as a molecular marker. Another inducible resistance mechanism is dependent on production of jasmonic acid and ethylene, involves the marker gene PDF1.2, and is effective against fungal/oomycete pathogens (Penninckx et al. 1996; Clarke et al. 2000). Evidence has accumulated that there are interactions between these pathways whereby induction of one pathway may inactivate the other (Doares et al. 1995a,b; Fidantsef et al. 1999; Thaler et al. 1999).
In A. thaliana, a number of mutants have been isolated in the SAR pathway. The mutant npr1 suppresses SAR against the bacteria Pseudomonas syringae and against the oomycete Hyaloperonospora parasitica (Cao et al. 1994; Bowling et al. 1997), and overexpression of NPR1 in the NPR1-H genotype leads to enhanced resistance to pathogen challenges (Cao et al. 1998). The mutations in cpr1, cpr5, and cpr6 cause SAR to switch from inducible to constitutive resistance (Bowling et al. 1994; Bowling et al. 1997; Clarke et al. 1998, respectively). Analysis of double mutants has determined how these mutations affect the pathways and how the effects are dependent on the signaling compounds salicylic acid and jasmonic acid (Clarke et al. 1998, 2000).
To answer the question of resistance costs and benefits, transgenic and mutant organisms may be useful to discover specific effects of traits that would not be possible otherwise (Tatar 2000). Furthermore, it has been stated that benefits of resistance cannot be determined until genetic manipulation allows inhibiting defense so that plants can be attacked with and without resistance (Stamp 2003). Using the SAR mutants and one transgenic line (Table 1), we were able to perform this experiment. Under a pathogen challenge, we compared the fitness of wild-type plants that have SAR against the fitness of npr1 plants that have suppressed SAR. In addition, we tested the efficacy of induced defense vs. constitutive defense by comparing the fitness of wild-type plants to cpr plants while under pathogen challenge. These benefits are further tested under two nutrient levels to determine the impact of nutrient limitation on benefits.
TABLE 1.
SAR phenotypes
| Genotype | Phenotype |
|---|---|
| Wild type | Normal wild-type SAR |
| npr1 | Cannot induce SAR |
| cpr1 | Constitutive SAR |
| crp5 | Constitutive SAR |
| NPR1-H | SAR is inducible, but stronger than wild type when induced |
MATERIALS AND METHODS
A. thaliana (Brassicaceae) is a short-lived selfing annual plant species found in agricultural fields and waste places throughout the world. The Col-0 ecotype (Landsberg, Germany) was used in this study, since the mutants used were originally isolated in this strain.
Mutant and transgenic genotypes:
The npr1, cpr1, and cpr5 mutants and the NPR1-H transgenic line have been previously described (Heidel et al. 2004), but their SAR phenotypes are summarized in Table 1.
Pathogen:
The oomycete pathogen H. parasitica (formerly Peronospora parasitica) was the pathogen in this study. H. parasitica is a natural pathogen of A. thaliana in Great Britain (Holub et al. 1994) and in the United States where the infection rates have been measured to reach 54% of the plants in a population (Heidel 2002). The isolate Noco2 (Norwich, Norfolk, UK) was used since Col-0 carries no functional resistance gene against this isolate.
Experimental design:
The five genotypes (wild type, cpr1, cpr5, npr1, and NPR1-H) were grown in all combinations of two nutrient treatments and with and without pathogen treatment. Plants (n = 855) were grown in growth chambers at the Duke University Phytotron in 18 flats, each consisting of an array of “cells,” each 4.5 × 5.9 cm and 6.4 cm deep. Every flat was divided as equally as possible among the five genotypes with random placement of genotypes within each flat. Half of the flats received the nutrient supplement described below and half did not. Three or four seeds of one genotype were planted in each cell after 10 days at 4° in water. One week after planting, plants were randomly thinned to one per cell.
An autoclaved 1:1:1 mix of sand, gravel, and turface was used as the growth medium for all plants. In the high-nutrient treatment, each cell received 5 ml of half-strength Hoagland's solution weekly for the first 2 months of the experiment. In the low-nutrient treatment, each cell received 5 ml of distilled water at the same time. All plants were watered as needed at least twice a day during the experiment except during the pathogen treatment when watering was not needed.
Five flats in each nutrient treatment were given the pathogen treatment. For this treatment, 2 μl of a spore suspension of H. parasitica Noco2 was placed on each cotyledon of a seedling. This was done once a day for 3 consecutive days, starting 1 week after planting with spore concentrations of 5.2 × 104/ml, 4.0 × 104/ml, and 12.5 × 104/ml, respectively, on those 3 days. These were the highest concentrations achievable following dilution of the original spore sample gathered that day from stock material. The other four flats in each nutrient treatment had 2 μl of distilled water placed on each cotyledon at the same time at which the other flats were infected. One week after the last inoculation, plants were assayed for disease when the disease phenotype was fully developed. The disease rating was determined by the coverage of conidiospores on the two cotyledons, which were the only leaves present on most plants. The disease rating scale was the following: 0 for no spores, 1 for spores covering <50% of one leaf, 2 for spores covering >50% of one leaf, 3 for spores covering <50% of both leaves, 4 for spores covering <50% of one leaf and >50% of the other leaf, and 5 for spores covering >50% of both leaves. This scale was used for all statistical analyses, but categories 2, 3, and 4 contained few individuals so categories 2, 3, and 4 are joined with category 1 in the figures.
H. parasitica requires high humidity, usually >95% relative humidity, to sporulate and therefore at the start of the experiment the flats had transparent plastic domes over them to make the conditions permissible for the pathogen. Additionally, the domes prevented the spread of conidiospores from infected flats to uninfected flats. The domes raised the humidity level to ∼100%. The temperature inside the domes was kept at ∼16° during the day and 14° during the night by maintaining the temperature in the growth chamber at 9° during the day and 14° during the night. At 2.5 weeks after planting, after the disease rating was measured, the plastic domes were removed and the temperature in the chamber was changed to 22° in the day and 14° in the night with 60% relative humidity to prevent further sporulation and spread of the pathogen. Additionally, all plants were sprayed thoroughly with distilled water when the domes were removed, washing spores off infected plants. No further infections or sporulation was seen once spores were washed off and the humidity lowered. Throughout, the light/dark cycle was 14 hr/10 hr with a light level of 450 mE/sec/m2. Flat locations were randomized within the chamber every week and, starting at 1 month, cells were randomized between flats within nutrient treatments.
At 97 days after planting, all plants had senesced, and seed yield was determined for each plant. The total number of siliques was counted on the main stem and side shoots, respectively. The number of seeds per silique was determined separately for the main and side stems by counting the number of seeds in every fifth intact silique for the high-nutrient treatment and by counting the number of seeds in every intact silique for the low-nutrient treatment. The number of seeds for the main stem and side shoots was determined by multiplying the average seeds per silique by the number of siliques. The seeds in the main stem and side shoots were then added together to give the total number of seeds per plant. Nine plants were not included in seed yield results because they had lost too many seeds from their siliques to make accurate estimations of their seed yield.
RNA analysis:
One leaf was removed from each plant in the experiment at day 45. All leaves of the same genotype within a flat (8–9 plants/genotype/flat if all plants survived) were bulked together for further analysis. RNA was extracted and probed as described by Clarke et al. (1998). Briefly, tissue was frozen in liquid N2 and ground to a powder, and then RNA was extracted with a LiCl buffer. RNA was precipitated with sodium acetate and ethanol and run on a 1.2% agarose gel with formaldehyde. RNA was transferred overnight to a cellulose membrane, crosslinked by UV, and then probed with the appropriate probe containing radiolabeled phosphorous. Probes were used for PR-1, PDF1.2, and UBQ5. UBQ5 (an ubiquitin gene) expression was used as a control since UBQ5 concentration is constant between various treatments and normalization by UBQ5 adjusts for different total amounts of RNA loaded on the gel. Expression of mRNA was quantified using ImageQuant software and sequentially normalized by UBQ5 expression and then by the mean wild-type expression over all treatments. Normalization by wild-type expression reduces the blot effect and results in more accurate estimation of relative expression between genotypes.
Statistical analysis:
The uninfected plants in this study have been previously analyzed separately (Heidel et al. 2004) and are used here only in comparison to the infected plants. Disease rating results were analyzed by logistic regression with genotype and nutrient treatment as fixed effects, and “flat” as a random effect nested within the nutrient treatments and disease rating as an ordinal variable.
The seed yield results were analyzed by analysis of variance (ANOVA), with genotypes, nutrient treatments, and infection treatments as fixed effects (flats nested within nutrient treatments and infection treatments) and flat as a random effect. There were significant differences in mortality between genotypes, so in the measure of fitness, all plants that died before reproduction were given a seed yield of 0. Seed yield data were transformed by various methods (see table legends) to conform to the assumptions of ANOVA. Although the flats are not classical “blocks” since the cells were later randomized between them, the original flat for each cell was found to have a significant effect. Therefore, the original flat for each plant was included in ANOVAs.
PR gene expression results were analyzed by ANOVA with genotype, nutrient treatments, and infection treatments as fixed effects and the blot as a random effect. The blot (particular cellulose membrane used in the RNA hybridization) was treated as an effect because the probe may hybridize and be washed off at different levels between blots. Moreover, different amounts of probe on blots will cause different levels of RNA to be detected between blots even if identical amounts of RNA are on the blots. Expression data were transformed (see table legend) to conform to the assumptions of ANOVA.
In all ANOVAs, all possible interaction terms were tested and if they were not significant, they were removed from the analysis. After ANOVAs, contrast tests between wild type and all other genotypes were performed as orthogonal planned comparisons. All other contrasts were performed as Tukey–Kramer tests. The data used in all figures and the sample sizes for each category are given in supplemental Table 1 at http://www.genetics.org/supplemental/. All statistical analyses were performed using JMP 4 (SAS Institute).
RESULTS
Disease rating of wild-type and SAR mutants:
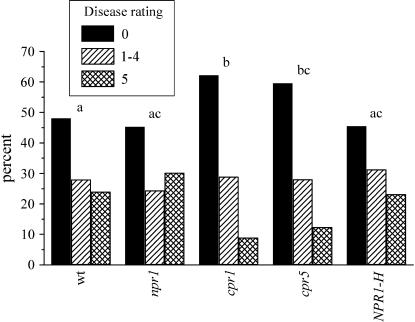
A comparison of the disease ratings demonstrated that both the genotype and the nutrient treatment had significant effects (Table 2 and Figure 1). There was a significantly higher disease rating for plants in the low-nutrient treatment (n = 236) compared to the high-nutrient treatment (n = 228). The absence of a significant genotype by nutrient interaction indicated that a lack of nutrients generally weakened resistance instead of affecting one genotype more than others. The genotypes expressing SAR constitutively (cpr1 and cpr5) had (1) significantly lower disease ratings, (2) a higher percentage of plants with a disease rating of 0, and (3) a lower percentage of plants with a disease rating of 5 than other genotypes. All three points suggest that cpr1 and cpr5 acted as expected and had higher resistance than wild type. However, there was no difference between the disease rating of wild type and the npr1 or NPR1-H genotypes.
TABLE 2.
Effect of genotype and nutrients on disease rating: effect tests
| Source | d.f. | χ2 |
|---|---|---|
| Genotype | 4 | 18.44** |
| Flat (nutrient) | 8 | 14.62 |
| Nutrient | 1 | 3.95* |
*Significant differences at P < 0.05; **significance at P < 0.01.
Figure 1.—
Disease rating across both nutrient treatments. Disease ratings 1–4 were grouped together (bars with diagonals). Letters indicate significant differences in disease rating among genotypes (i.e., “a” is significantly different from “b,” but is not significantly different from “ac”). wt, wild type.
The cpr mutants were initially identified by their expression of resistance in experiments conducted under lower light levels and using a different soil mix than used in this study. Therefore, it was possible that the expression of resistance would be seen only under the original environmental conditions. The lowered disease rating found here for cpr mutants (Table 2; Figure 1) demonstrates that their resistance is expressed in more than one environment.
Seed yield of wild-type and SAR mutants:
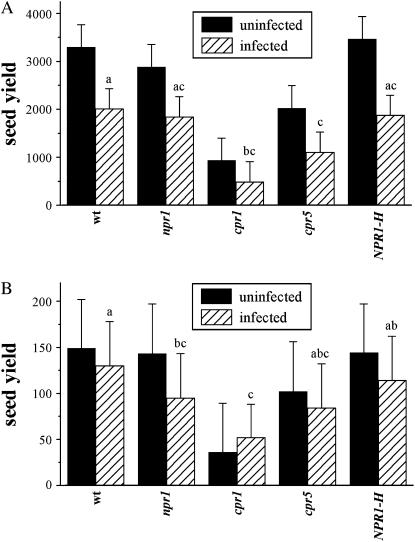
Genotype, flat, nutrient treatment, infection treatment, and genotype × nutrient all had significant effects on seed yield, but the genotype × infection interaction was not significant (Table 3). The infection treatment lowered fitness as expected (Figure 2).
TABLE 3.
Effect of genotype, nutrient, and infection on fitness: effect tests
| Source | d.f. | SS | F-value |
|---|---|---|---|
| Genotype | 4 | 2963 | 35.1*** |
| Nutrient | 1 | 648 | 30.3*** |
| Infection | 1 | 104 | 4.9* |
| Flat (nutrient, infection) | 15 | 2979 | 9.4*** |
| Genotype × nutrient | 4 | 885 | 10.5*** |
ANOVA of effects on fitness with genotype, nutrient, and infection as fixed effects and “flat” as a random effect. Seed-yield data were cube root transformed to conform to assumptions of ANOVA. SS, sum of squares. *Significant F-values at P < 0.05; ***significance at P < 0.001.
Figure 2.—
Fitness of genotypes. Mean seed yield values are shown with ±SE. Letters indicate significantly different values within the infected treatment. (A) Seed yield under high-nutrient treatment. (B) Seed yield under low-nutrient treatment.
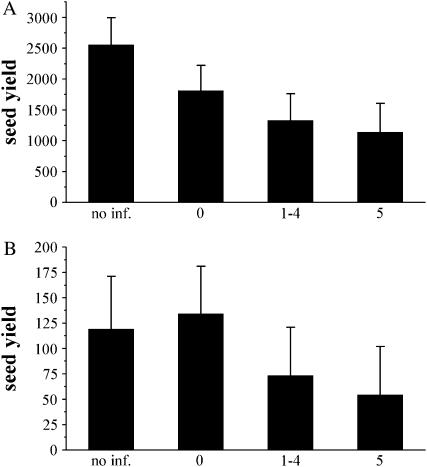
Within infected plants in the high-nutrient treatment, both cpr1 and cpr5 had lower fitness than wild type (Figure 2A; Table 4). Within infected plants in the low-nutrient treatment, cpr1 had lower fitness than both wild type and NPR1-H, and npr1 had lower fitness than wild type (Figure 2B; Table 4). Disease severity as measured by disease rating had a significant effect on fitness (Table 5; Figure 3), indicating the logical negative correlation between increased disease symptoms and lower fitness. In a comparison of fitness of different disease rating categories, plants with a disease rating of 0 had significantly higher seed yield than plants with a disease rating of 3, 4, or 5 by Tukey–Kramer, and there were no other significant differences.
TABLE 4.
Effect of genotype within infected plants: effects tests
| Seed yield in high-nutrient treatment
|
Seed yield in low-nutrient treatment
|
|||||
|---|---|---|---|---|---|---|
| Source | d.f. | SS | F-value | d.f. | SS | F-value |
| Genotype | 4 | 8.17 × 107 | 10.8*** | 4 | 608 | 4.3** |
| Flat | 5 | 1.53 × 108 | 20.3*** | 4 | 4268 | 30.5*** |
Seed-yield data were not transformed in the high-nutrient treatment and were square root transformed in the low-nutrient treatment. SS, sum of squares. **Significant F-values at P < 0.01; ***significance at P < 0.001.
TABLE 5.
Effect of disease rating on fitness within infected treatment
| Source | d.f. | SS | F-value |
|---|---|---|---|
| Genotype | 4 | 1446 | 19.5*** |
| Nutrient | 1 | 162 | 8.8* |
| “Flat” (nutrient) | 8 | 1985 | 13.4*** |
| Disease rating | 5 | 820 | 8.9*** |
| Genotype × nutrient | 4 | 521 | 7.05*** |
Seed-yield data were cube root transformed. Genotype, nutrient, and disease rating (ordinal variable) are fixed effects and “flat” is a random effect. SS, sum of squares. *Significant F-values at P < 0.05; ***significance at P < 0.001.
Figure 3.—
Effect of disease rating on seed yield. Mean seed yield for plants within each disease-rating category (“no inf.” refers to plants in the uninfected treatment) are shown with ±SE. (A) Within high-nutrient treatment category. (B) Within low-nutrient treatment category.
PR-1 and PDF1.2 expression:
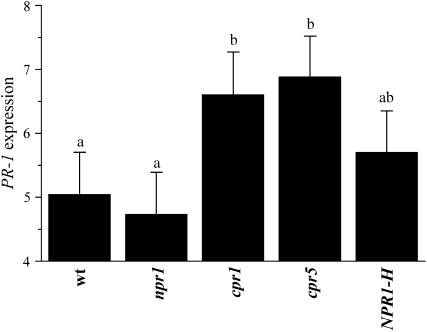
Genotype, nutrient treatment, blot, and genotype × nutrient treatment had significant effects on PR-1 expression with higher expression in the low-nutrient treatment (Table 6; Figure 4). Infection treatment had no significant effect on PR-1 expression. The effect of genotype was largely caused by cpr1 and cpr5. Other than a blot effect (results not shown), there were no significant effects on PDF1.2 expression.
TABLE 6.
Effects on PR-1 expression: effect tests
|
PR-1 expression
|
|||
|---|---|---|---|
| Source | d.f. | SS | F-value |
| Genotype | 4 | 59.5 | 4.70** |
| Nutrient | 1 | 29.8 | 9.41** |
| Infection | 1 | 3.5 | 1.10 |
| Blot | 3 | 42.0 | 4.42** |
Expression data was boxcox transformed. SS, sum of squares. **Significance at P < 0.01.
Figure 4.—
Relative expression of PR-1 across all infection and nutrient treatments. Mean values (±SE) of transformed data are shown. Letters indicate significantly different values.
DISCUSSION
Is SAR beneficial?:
We investigated the benefits of resistance by comparing the response of a mutant that cannot induce SAR, npr1, to wild-type plants. For resistance to be beneficial, resistant plants should have higher fitness than nonresistant plants under a pathogen treatment. The significantly lower fitness of infected npr1 vs. infected wild type under low-nutrient conditions (Figure 2) indicates that normal inducible SAR as it exists in wild-type plants is beneficial under these conditions. There was no benefit found under high-nutrient conditions, but SAR was not found to have a negative effect on fitness. SAR in the wild-type plants could have lowered fitness if the cost of the induced resistance were greater than the benefit of the resistance. These results, in addition to the fact that npr1 plants had lower fitness than wild type in a field trial (Heidel et al. 2004), suggest that normal inducible SAR does have an adaptive purpose.
The NPR1-H genotype allowed us to test whether it is beneficial to have a stronger-than-wild-type induction of SAR. The similar fitness of wild type and NPR1-H under both nutrient treatments demonstrates that stronger SAR is not beneficial, but does not have an extra cost. Perhaps NPR1-H can be beneficial only with a longer and more damaging pathogen attack (Cao et al. 1998).
Why induced resistance?:
We tested whether induced or constitutive defense is more effective against a single pathogen attack by comparing the response of wild-type plants to cpr plants. The constitutive resistance in the cpr plants did not lead to a fitness benefit for the plants in the pathogen treatment (Figure 2), demonstrating that the known fitness cost of constitutive resistance (Heidel et al. 2004) was too high to give a net benefit under the conditions of this experiment. This effect is seen in both nutrient treatments, indicating that this finding is not dependent on nutrient availability. Constitutive resistance might have been beneficial if the resistance had the ability to prevent pathogen damage before the induced resistance was sufficiently induced; however, this was not the case. Similarly to NPR1-H, however, constitutive expression could potentially be beneficial when under a longer pathogen attack, especially when secondary within-plant infections are likely to arise.
These results suggest that SAR is not constitutive because the reduction in pathogen damage from constitutive resistance fails to adequately compensate for the costs despite reducing pathogen damage. These results are partially consistent with the results in a native tobacco species where herbivore resistance induced by methyl jasmonate reduced herbivory and increased fitness in some but not all situations (Baldwin 1998). When resistance is chemically induced with methyl jasmonate, the resistance occurs over a shorter time frame than when resistance is constitutively activated by genetic changes (as in the cpr mutants); therefore, the chemical induction might be less costly, assuming equal intensities of resistances.
Tolerance effects:
Any infection can have deleterious effects due to direct damage by the pathogen and/or as a result of the cost of mounting inducible host defenses. Overall, infection significantly reduced the fitness of all plants (Table 3; Figure 2). Consistent with this, plants with greater disease symptoms had decreased fitness (Table 5; Figure 3). Other than cpr1, the effect of infection on fitness was similar across genotypes as demonstrated by the lack of a significant genotype × infection interaction (Table 4).
However, there is not necessarily a direct linkage between disease severity, as measured here, and pathogen-caused reductions in fitness; two equally strong infections, as determined visually, may have different effects on fitness, depending on undetected levels of damage and tolerance. Tolerance differences have been found in response to herbivory (Tiffin and Rausher 1999) and plant pathogens (Bent et al. 1992; Kover and Schaal 2002; Schurch and Roy 2004), and these tolerance differences can have implications for the study of benefits.
The lack of a linkage between disease severity and reductions in fitness was not an important complication for the cpr mutants since they had lower disease severity than wild type, and disease severity is correlated to fitness in this experiment. Tolerance could not have been the explanation for the fitness differences between the cpr mutants and wild type because similar fitness differences between these genotypes were found without the pathogen (Figure 2). In contrast, tolerance could be important for explaining the data from npr1. Even though the npr1 mutants had a similar disease rating to wild type, they have lower fitness under low-nutrient conditions. This fitness difference could be caused by a less severe infection in wild type due to induced resistance in a manner not captured in our disease rating or by a difference in tolerance (i.e., wild type has a higher tolerance). While it would be useful from a mechanistic view to know whether resistance or tolerance is responsible, it can be argued that, in an evolutionary sense, it is irrelevant because the fitness of npr1 is lower regardless of whether resistance or tolerance is responsible.
Two other studies have measured the fitness effect of H. parasitica infection. Korves and Bergelson (2004) demonstrated that the same strain, Noco2, lowered the seed yield of A. thaliana but paradoxically only in lines carrying the cognate-specific resistance gene and under conditions of intraspecific competition. Lines lacking the R-gene did not have a significant reduction in fitness when infected by the pathogen. In our experiment, all the genotypes lack the R-gene recognizing Noco2, yet there was an overall fitness reduction from infection, as might be typically expected. The difference between the two studies might result from either growth conditions or time of infection. In our study, infection was performed on cotyledons 1 week after germination, while in the Korves and Bergelson (2004) study, infection was carried out on true leaves 2 weeks after germination. However, van Hulten et al. (2006) found that infection by H. parasitica lowers fitness, consistent with our results.
Nutrient effects:
Different theories of how nutrient stress affects resistance have been suggested (Karban and Baldwin 1997). If it were possible to measure the amount of energy and resources devoted to SAR, the effects of nutrient stress might be more easily elucidated. However, determination of the resources and energy used by SAR is difficult, since SAR is a complex resistance mechanism and does not result from the production of a single antimicrobial protein or compound (Maleck et al. 2000). Even if the energy required for biosynthesis of one primary protein can be calculated, the relation of this energy expense to fitness is complex due to further potential interactions with regulatory networks. Without knowing the components of the complete resistance mechanism, it is difficult to test the effects of limited nutrients on the production of SAR resistance mechanisms.
A large number of proteins are produced during SAR (Ward et al. 1991; Maleck et al. 2000), and high concentrations of PR proteins occur in some species following infection (in tobacco, two PR proteins increased to 3% of soluble protein; Vogeli-Lange et al. 1988). It is therefore likely that SAR probably requires a significant amount of nitrogen. Under nitrogen limitation (such as found in the low-nutrient treatment), there are two possible outcomes for PR protein biosynthesis: (1) PR proteins may continue to be synthesized, but the fitness cost of producing such proteins is greater. Or (2) the synthesis of PR proteins can be lowered, decreasing the fitness cost of resistance but also increasing the risk of damage from pathogens. The first option has been found by Purrington and Bergelson (1997) where a plant expressing herbicide resistance had a greater cost under nutrient limitation. In that case, herbicide resistance resulted from changes in the target enzyme that make it less effective in its normal role. In our study, the observation that all genotypes had a higher disease rating under low-nutrient conditions (Figure 2) and that there was no significant genotype × nutrient interaction suggests that resistance was downregulated under low-nutrient conditions independently of the SAR-affecting mutations. An inhibitory mechanism is more likely to exist for SAR than for the herbicide resistance found by Purrington and Bergelson (1997) because herbicide resistance has occurred only very recently, giving little time for regulation to develop. Furthermore, this specific type of herbicide resistance cannot be easily downregulated since the herbicide resistance and the cost directly results from a mutation to an essential enzyme.
Relation of mRNA expression to fitness:
As already noted, connecting the activation of specific molecular pathways to fitness is difficult. Despite this problem, expression of certain mRNAs can be used as markers of costly processes regardless of whether the synthesis of the specific marker mRNA and its protein are costly. In this view, PR-1 is a good marker for costly processes because its expression is higher in cpr1 and cpr5 (which have the lowest fitness relative to wild type), NPR1-H, and npr1 (Figure 4), while PDF1.2 is not a good marker since it has no correlation with resistance costs in this study. This is consistent with previous results in a natural environment, demonstrating that PR-1 is associated with costly resistance while PDF1.2 is not (Heidel et al. 2004). The infection treatment did not have a significant effect on the expression of either PR-1 (Table 6) or PDF1.2. This is probably due to the fact that the leaf samples used for gene expression studies were collected 1 month after disease expression, and by this time differences between infected and uninfected plants may have largely subsided. As demonstrated in this study, induced resistances can be costly if they are left activated for a long time, so it is logical that SAR and its marker genes would be deactivated quickly to conserve resources.
Acknowledgments
We thank Joseph Clarke for generation of the lines used; Janis Antonovics, Amy Lawton-Rauh, Maria Clauss, and two anonymous reviewers for helpful comments on the manuscript; and R. Mosher, M. Kesarwani, and W. Durrant for technical advice. A. Heidel was supported in part by a Cell and Molecular Biology Program fellowship from Duke University and the Duke University Arts and Sciences Research Council. This study was supported by the Duke Biology Department.
References
- Agrawal, A., 1999. Induced responses to herbivory in wild radish: effects on several herbivores and plant fitness. Ecology 80: 1713–1723. [Google Scholar]
- Albrecht, T., A. Kehlen, K. Stahl, H.-D. Knofel, G. Sembdner et al., 1993. Quantification of rapid, transient increases in jasmonic acid in wounded plants using a monoclonal antibody. Planta 191: 86–94. [Google Scholar]
- Baldwin, I. T., 1998. Jasmonate-induced responses are costly but benefit plants under attack in native populations. Proc. Natl. Acad. Sci. USA 95: 8113–8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucom, R., and R. Mauricio, 2004. Fitness costs and benefits of novel herbicide tolerance in a noxious weed. Proc. Natl. Acad. Sci. USA 101: 13386–13390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent, A. F., R. W. Innes, J. R. Ecker and B. J. Staskawicz, 1992. Disease development in ethylene-insensitive Arabidopsis thaliana infected with virulent and avirulent Pseudomonas and Xanthomonas pathogens. Mol. Plant-Microbe Interact. 5: 372–378. [DOI] [PubMed] [Google Scholar]
- Bowling, S. A., A. G. Guo, H. Cao, A. S. Gordon, D. F. Klessig et al., 1994. A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6: 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling, S. A., J. D. Clarke, Y. Liu, D. F. Klessig and X. Dong, 1997. The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9: 1573–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant, J. P., and R. Julkunen-Tiitto, 1995. Ontogenic development of chemical defense by seedling resin birch: energy cost of defense production. J. Chem. Ecol. 21: 883–896. [DOI] [PubMed] [Google Scholar]
- Cao, H., S. A. Bowling, A. S. Gordon and X. Dong, 1994. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., X. Li and X. Dong, 1998. Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc. Natl. Acad. Sci. USA 95: 6531–6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, J. D., Y. Liu, D. F. Klessig and X. Dong, 1998. Uncoupling PR gene expression from NPR1 and bacterial resistance: characterization of the dominant Arabidopsis cpr6–1 mutant. Plant Cell 10: 557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, J. D., S. M. Volko, H. Ledford, F. M. Ausubel and X. Dong, 2000. Roles of salicyclic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 12: 2175–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta, S. K., and S. Muthukrishnan, 1999. Pathogenesis-Related Proteins in Plants. CRC Press, Boca Raton, FL.
- Doares, S. H., T. Syrovets, E. Weiler and C. Ryan, 1995. a Oligogalacturonides and chitosan activate plant defensive genes through the octadecanoid pathway. Proc. Natl. Acad. Sci. USA 92: 4095–4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doares, S. H., J. Narváez-Vásquez, A. Conconi, C. Ryan and C. A. Clarence, 1995. b Salicylic acid inhibits synthesis of proteinase inhibitors in tomato leaves induced by systemin and jasmonic acid. Plant Physiol. 108: 1741–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elle, E., N. M. van Dam and J. D. Hare, 1999. Cost of glandular trichomes, a “resistance” character in Datura wrightii Regel (Solanaceae). Evolution 53: 22–35. [DOI] [PubMed] [Google Scholar]
- Fan, W., and X. Dong, 2002. In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell 14: 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer, E. E., R. R. Johnson and C. A. Ryan, 1992. Regulation of expression of proteinase inhibitor genes by methyl jasmonate and jasmonic acid. Plant Physiol. 98: 995–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidantsef, A. L., M. J. Stout, J. S. Thaler, S. S. Duffey and R. M. Bostock, 1999. Signal interactions in pathogen and insect attack: expression of lipoxygenase, proteinase inhibitor II, and pathogenesis-related protein P4 in the tomato, Lycopersicon esculentum. Physiol. Mol. Plant Pathol. 54: 97–114. [Google Scholar]
- Heidel, A. J., 2002. Fitness costs, benefits and interactions from systemic acquired resistance. Ph.D. Thesis, Duke University, Durham, NC.
- Heidel, A. J., and I. T. Baldwin, 2004. Microarray analysis of SA- and JA-signaling in Nicotiana attenuata's responses to attack by insects from multiple feeding guilds. Plant Cell Environ. 27: 1362–1373. [Google Scholar]
- Heidel, A. J., J. D. Clarke, J. Antonovics and X. Dong, 2004. Fitness costs of mutations affecting the systemic acquired resistance pathway in Arabidopsis thaliana. Genetics 168: 2197–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil, M., 1999. Systemic acquired resistance: available information and open ecological questions. J. Ecol. 87: 341–346. [Google Scholar]
- Heil, M., 2004. Induction of two indirect defences benefits Lima bean (Phaseolus lunatus, Fabaceae) in nature. J. Ecol. 92: 527–536. [Google Scholar]
- Heil, M., A. Hilpert, W. Kaiser and K. E. Linsenmair, 2000. Reduced growth and seed set following chemical induction of pathogen defence: Does systemic acquired resistance (SAR) incur allocation costs? J. Ecol. 88: 645–654. [Google Scholar]
- Holub, E. B., J. L. Beynon and I. R. Crute, 1994. Phenotypic and genotypic characterization of interactions between isolates of Peronospora parasitica and accessions of Arabidopsis thaliana. Mol. Plant-Microbe Interact. 7: 223–239. [Google Scholar]
- Kakes, P., 1989. An analysis of the costs and benefits of the cyanogenic system in Trifolium repens L. Theor. Appl. Genet. 77: 111–118. [DOI] [PubMed] [Google Scholar]
- Karban, R., and I. T. Baldwin, 1997. Induced Responses to Herbivory. University of Chicago Press, Chicago.
- Korves, T., and J. Bergelson, 2004. A novel cost of R gene resistance in the presence of disease. Am. Nat. 163: 489–504. [DOI] [PubMed] [Google Scholar]
- Kover, P. X., and B. A. Schaal, 2002. Genetic variation for disease resistance and tolerance among Arabidopsis thaliana accessions. Proc. Natl. Acad. Sci. USA 99: 11270–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., M. A. Schuler and M. R. Berenbaum, 2002. Jasmonate and salicylate induce expression of herbivore cytochrome P450 genes. Nature 419: 712–715. [DOI] [PubMed] [Google Scholar]
- Maleck, K., A. Levine, T. Eulgem, A. Morgan, J. Schmid et al., 2000. The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26: 403–410. [DOI] [PubMed] [Google Scholar]
- Mauricio, R., and M. D. Rausher, 1997. Experimental manipulation of putative selective agents provides evidence for the role of natural enemies in the evolution of plant defense. Evolution 51: 1435–1444. [DOI] [PubMed] [Google Scholar]
- Moura, D. S., and C. A. Ryan, 2001. Wound-inducible proteinase inhibitors in pepper: differential regulation upon wounding, systemin, and methyl jasmonate. Plant Physiol. 126: 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I. A. M. A., K. Eggermont, F. R. G. Terras, B. P. H. J. Thomma, G. W. De Samlanx et al., 1996. Pathogen-induced systemic acquired activation of a plant defensin gene in Arabidopsis follows a salicyclic acid-independent pathway. Plant Cell 8: 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purrington, C. B., and J. Bergelson, 1997. Fitness consequences of genetically engineered herbicide and antibiotic resistance in Arabidopsis thaliana. Genetics 145: 807–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, D. F., 1979. Evolution of plant chemical defence against herbivores, pp. 1–55 in Herbivores: Their Interaction With Secondary Plant Metabolites, edited by G. A. Rosenthal and D. H. Janzen. Academic Press, New York.
- Ross, F. A., 1961. Systemic acquired resistance induced by localized virus infections in plants. Virology 14: 340–358. [DOI] [PubMed] [Google Scholar]
- Ryals, J. A., U. H. Neuenschwander, M. G. Willits, A. Molina, H. Steiner et al., 1996. Systemic acquired resistance. Plant Cell 8: 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurch, S., and B. A. Roy, 2004. Comparing single- vs. mixed-genotype infections of Mycophaerella graminicola on wheat: effects on pathogen virulence and host tolerance. Evol. Ecol. 18: 1–14. [Google Scholar]
- Simms, E. L., and M. D. Rausher, 1987. Costs and benefits of plant resistance to herbivory. Am. Nat. 130: 570–581. [Google Scholar]
- Stamp, N., 2003. Out of the quagmire of plant defense hypotheses. Q. Rev. Biol. 78: 23–55. [DOI] [PubMed] [Google Scholar]
- Tatar, M., 2000. Transgenic organisms in evolutionary ecology. Trends Ecol. Evol. 15: 207–211. [DOI] [PubMed] [Google Scholar]
- Thaler, J. S., A. L. Fidantsef, S. S. Duffey and R. M. Bostock, 1999. Trade-offs in plant defense against pathogens and herbivores: a field demonstration of chemical elicitors of induced resistance. J. Chem. Ecol. 25: 1597–1609. [Google Scholar]
- Tiffin, P., and M. D. Rausher, 1999. Genetic constraints and selection acting on tolerance to herbivory in the common morning glory Ipomoea purpurea. Am. Nat. 154: 700–716. [DOI] [PubMed] [Google Scholar]
- van Hulten, M., M. Pelser, L. C. van Loon, C. M. J. Pieterse and J. Ton, 2006. Costs and benefits of priming for defense in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 5602–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogeli-Lange, R., A. Hansen-Gehri, T. Boller and F. Meins, Jr., 1988. Induction of the defense-related glucanohydrolases β-1,3-glucanase and chitinase by tobacco mosaic virus infection of tobacco leaves. Plant Sci. 54: 171–176. [Google Scholar]
- Wang, D., N. D. Weaver, M. Kesarwani and X. Dong, 2005. Induction of protein secretory pathway is required for systemic acquired resistance. Science 308: 1036–1040. [DOI] [PubMed] [Google Scholar]
- Ward, E., S. J. Uknes, S. C. Williams, S. S. Dincher, D. L. Wiederhold et al., 1991. Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3: 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangerl, A. R., and C. E. Rutledge, 1996. The probability of attack and patterns of constitutive and induced defense: a test of optimal defense theory. Am. Nat. 147: 599–608. [Google Scholar]
- Zhang, Y., W. Fan, M. Kinkema, X. Li and X. Dong, 1999. Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 96: 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]