Abstract
Historical hybridization events between the two subspecies of cattle, Bos taurus and B. indicus, have occurred in several regions of the world, while other populations have remained nonadmixed. We typed closely linked X chromosome microsatellites in cattle populations with differing histories of admixture from Africa, Europe, the Near East, and India. Haplotype breakdown will occur as admixed populations age, and longer ancestral haplotypes will remain intact in more recently admixed populations compared to older ones. We genotyped male animals from these populations, obtaining unambiguous haplotypes, and measured levels of linkage disequilibrium (LD) and ancestral mosaicism. Extensive LD, likely to be the result of ongoing admixture, was discovered in hybrid cattle populations from the perimeter of the tsetse zone in West Africa. A Bayesian method to assign microsatellite allele ancestry was used to designate the likely origin of each chromosomal segment and assess the relative ages of admixture in the populations. A gradient of the age of admixture in the African continent emerged, where older admixture has produced more fragmented haplotypes in the south, and longer intact haplotypes, indicating more recent hybridization, feature in the northwest.
CATTLE have been domesticated at least twice from two distinct wild aurochs populations (Loftus et al. 1994, 1999; Bradley et al. 1996). The two extant taxa are Bos taurus (taurine) and B. indicus. The former were domesticated in the Middle East, Anatolia, and probably Africa ∼10,000 years ago and are now found throughout northern Eurasia and certain regions of Africa. B. indicus or zebu cattle were domesticated from a different variant of wild progenitor on the Indian subcontinent.
Substantial genetic exchange has taken place between the two types. Zebu cattle were brought from southern Asia to the Middle East and the breeds that are found in this area today are affected by hybridization that took place perhaps >3000 years ago (Loftus et al. 1999). Nuclear genetic markers including Y chromosome polymorphisms (Bradley et al. 1994; Hanotte et al. 2000) and autosomal microsatellites (MacHugh et al. 1997) have been used to show that there has been a widespread introgression of zebu cattle into the African continent, which would have initially been inhabited only by African B. taurus, pure examples of which remain only in certain parts of North and West Africa.
The genetic exchange between taurine and zebu cattle represents an appropriate model to study the effects of postadmixture haplotypic decay through recombination, mutation, and genetic drift. Four of the domestic cattle hybrid zones, the Middle East plus East, West, and southern Africa, have breeds with roughly equal amounts, but different ages of admixture (Loftus et al. 1999; Hanotte et al. 2002). The first-generation population created after an admixture event will contain intact chromosomes from each of the inputting parental populations. However, as mating occurs within the hybrid population these intact chromosomes are mixed by recombination and gradually become more mosaic with each generation. Therefore, the extent of genomic blocks undisturbed by hybrid recombination will contain information about the history of admixture.
The examination of closely linked markers can be used to estimate how parental haplotypes have been broken down over time in the hybrid population. Microsatellite allele frequency spectra have been shown to differ sharply between B. indicus and B. taurus, facilitating the assignment of ancestry at specific chromosome positions. Moreover, extended haplotypes may be unambiguously assigned to X chromosomes in males.
Here we investigate differences in haplotypic mosaicism among 346 X chromosomes typed in male cattle from regions of differing phylogeographic history. We estimate microsatellite allele ancestry at 10 loci within a 10-cM region to examine the breakdown of genomic blocks and assess levels of linkage disequilibrium (LD) in populations with differing histories of admixture. We relate the different levels of haplotypic mosaicism and LD to the different time depths and manners of introgression. For simplicity, we use the term LD to describe both traditional linkage disequilibrium and pairwise disequilibrium between nonlinked markers.
MATERIALS AND METHODS
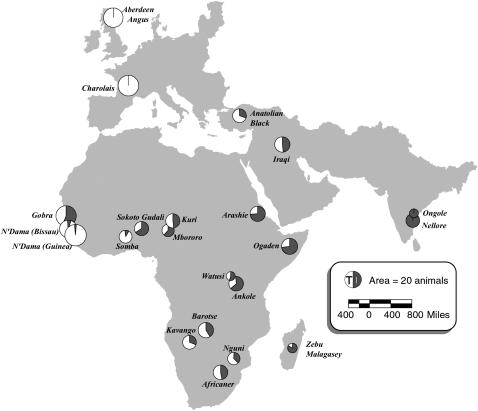
Three hundred forty-six male cattle DNA samples from 22 breeds were obtained as described previously (Table 1) (MacHugh et al. 1997; Loftus et al. 1999; Ritz et al. 2000; Hanotte et al. 2002). The approximate locations of the sample sites and previously calculated admixture estimates (mR) are shown in Figure 1 (MacHugh et al. 1997; Loftus et al. 1999; Hanotte et al. 2002; Freeman et al. 2004). For LD analysis, the novel data obtained here were combined with microsatellite data from previously published studies (MacHugh et al. 1997; Loftus et al. 1999; Hanotte et al. 2002; Freeman et al. 2004). Of the 22 breeds, 2 were unadmixed European B. taurus, 3 were primarily unadmixed West African B. taurus, and two were primarily unadmixed B. indicus. The remaining 15 breeds were known to have been formed by admixture between B. taurus and B. indicus. Figure 1 shows the geographic location of the breeds.
TABLE 1.
Cattle breeds included in this study
| Breed name | N | E(τ) | SD(τ) | Regional designation |
|---|---|---|---|---|
| Aberdeen Angus | 29 | NA | NA | European Bos taurus |
| Charolais | 33 | NA | NA | European B. taurus |
| N'Dama (Guinea) | 36 | NA | NA | West African B. taurus |
| N'Dama (Guinea Bissau) | 22 | NA | NA | West African B. taurus |
| Somba | 10 | NA | NA | West African B. taurus |
| Gobra | 32 | 6.67 | 2.70 | West African hybrids |
| Sokoto Gudali | 13 | 9.90 | 4.62 | West African hybrids |
| Mbororo | 9 | 4.27 | 3.24 | West African hybrids |
| Kuri | 13 | 13.00 | 5.01 | West African hybrids |
| Arashie | 16 | 10.71 | 4.79 | East African hybrids |
| Ogaden | 18 | 10.23 | 4.76 | East African hybrids |
| Ankole | 14 | 7.99 | 4.92 | East African hybrids |
| Watusi | 3 | 15.45 | 7.23 | East African hybrids |
| Barotse | 16 | 18.90 | 5.04 | Southern African hybrids |
| Kavango | 12 | 21.30 | 4.78 | Southern African hybrids |
| Nguni | 9 | 15.27 | 5.87 | Southern African hybrids |
| Afrikaner | 14 | 8.87 | 4.28 | Southern African hybrids |
| Zebu Malagasy | 4 | 17.92 | 6.65 | Madagascar |
| Anatolian Black | 12 | 13.13 | 5.05 | Near Eastern hybrids |
| Iraqi | 16 | 17.76 | 4.98 | Near Eastern hybrids |
| Nellore | 12 | NA | NA | B. indicus, of Indian descent |
| Ongole | 3 | NA | NA | Indian B. indicus |
N is the number of individuals in each sample, E(τ) and SD(τ) are the posterior mean and standard deviation of τ, respectively, and τ is a measure of the age of admixture. Breed names in italics are those breeds that were genotyped using radioactively labeled PCR products.
Figure 1.—
Map showing approximate sampling sites for cattle populations included in this study. The areas of the pie graphs are proportional to the number of animals in the population. The pie graphs represent a previously calculated mR value for each breed, which is a measure of Bos taurus/B. indicus admixture that has been calculated using European B. taurus and Indian B. indicus breeds as parental populations. The Nellore breed was sampled in Brazil, but is composed of Ongole individuals that were brought to South America (Felius 1995; Ritz et al. 2000).
Ten X chromosome microsatellites were selected from the Meat Animal Research Center (MARC) database (http://sol.marc.usda.gov/genome/genome.html) (Bishop et al. 1994; Sun et al. 1994; Kappes et al. 1997). All of the microsatellites selected have been linkage mapped to a 10-cM region on the X chromosome (Kappes et al. 1997; Sønstegard et al. 1997, 2001; Ihara et al. 2004). Six of the markers have also been included in radiation hybrid maps of the bovine X chromosome (BtaX) (Amaral et al. 2002; Williams et al. 2002), and all but one of the markers can be found in the Ensembl bovine genome assembly (http://www.ensembl.org/index.html). Approximate distances between markers and marker order were taken from the genome assembly after observing agreement between this and the linkage and radiation hybrid maps for most markers. To control for potential misorientation of contigs in the preliminary assembly, a comparison with the syntenic chromosomal region in humans was examined. Again, order seemed to be mostly conserved. Thus, for the purposes of this article, the marker order and positions listed in Table 2 were used. Two methods of genotyping were used, with common standards enabling combination of the data. Populations in italics in Table 1 were genotyped using a radioactive PCR method described in MacHugh et al. (1997), while the remaining populations were genotyped using an ABI 377 system following the protocol of Hanotte et al. (2002).
TABLE 2.
Primer sequences and optimal PCR conditions for 10 X chromosome microsatellites
| Locus name | Approximate chromosome position (Mb) | Accession no. | UniSTS | Primers (5′–3′) | Amplicon size | PCR [MgCl2] | TA PCR |
|---|---|---|---|---|---|---|---|
| BMS1616 | 7 | G18677 | 27189 | CAGTGTGTATAGCATGATTCCGAGTTGGTCTGCATTCATACATTAA | 92–116 | 1.25 | 54°–55° |
| BL1045 | 9.2 | 251338 | TGCCAGAACAAGTCACAAGCCTCCAAGGGTGTCTCTATCCC | 92–100 | 1.5 | 59° | |
| UWCA19 | 9.4 | U01794 | 251002 | CTGTGATAACCTATATGGGAAAGGAGAGGACAGCAAAGTGATTCAGT | 97–105 | 1.5 | 58° |
| BL1098 | 10 | 251324 | CCACAACTTCCAGAAGCCTCCAGAAACCACCCAAACTAACC | 203–235 | 1.25 | 56° | |
| BMS960 | 11 | G18760 | 74452 | TGTAAATAGCTCCCCTCCTGCCTAGCAATTTGTTGTTCATGGC | 106–128 | 1.75 | 57° |
| XBM701 | 11.7 | 251432 | TTCCCCTTGAAATCATCTGGTCCAAGTTACCAAAATTGACCC | 148–168 | 2.0 | 58° | |
| BMS2713 | 12.2 | G18462 | 69721 | TGAATACCTGTTTCCAGCCCCTCCTAAGTCCAGGAAGCCC | 143–155 | 1.5 | 56° |
| XBM7 | 251010 | CTGTATTAGAGTTCCCTGGAGAAAGCCAACATGCCCTGTAGAAT | 180–202 | 1.5 | 57° | ||
| XBM361 | 14.8 | 251341 | ACACACTGAGAATTAAACTACAAAAACCACACAACTGAAGCCACTTTAAGC | 150–162 | 1.25 | 58°–59° | |
| BMS649 | 16.3 | G18728 | 37262 | CTCGGACTCATAACGCACATCGAGCAACAAGAGTGAAGGT | 116–134 | 1.75 | 59° |
Magnesium chloride concentration is shown by [MgCl2] and annealing temperature is denoted by TA. The UniSTS column gives accession numbers for the database of sequence tagged sites (http://www.ncbi.nlm.nih.gov/genome/sts).
Pairwise linkage disequilibrium between locus pairs was carried out using an extension of Fisher's exact test implemented by the GENEPOP program (v3.2a) (Raymond and Rousset 1995). Populations were defined according to the regional groups in Table 1. For a given pair of loci within one population, the possible genotypes are represented by GENEPOP in a contingency table. For each locus pair within each population, the unbiased estimate of the P-value is calculated using a Markov chain as the sum of the probabilities of all tables (with the same marginal values as the observed one) with a lower or equal probability than the observed table.
Statistical inference on admixture and the ancestry crossover rate in the admixed cattle populations was made in the computer program ADMIXMAP by simulating posterior samples of the model parameters using Markov chain Monte Carlo simulation (Hoggart et al. 2003, 2004). ADMIXMAP implements a full Bayesian hierarchical model for admixture at the population, individual, and locus levels and also models the allele frequencies in the unadmixed subpopulations that have contributed to the admixed population under study. The stochastic variation of ancestry between loci on each chromosome is modeled by independent Poisson arrival processes, one for each unadmixed population contributing to the admixed population under study, in our case B. taurus and B. indicus. Thus inference is made on parameters representing population and individual admixture, indicator parameters for locus ancestry for all loci and all individuals, parameters of multinomial distributions representing allele and haplotype frequencies in the two populations, and τ, the sum-of-intensities of the two Poisson arrival processes. While admixture was modeled separately for each individual in a population (independent and identically distributed from the population level admixture distribution), τ was assumed to be the same for all individuals in the population.
For an individual with admixture proportion θ from B. taurus the intensities per morgan of the arrival processes for B. taurus and B. indicus are 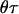 and
and 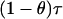 , respectively. The ancestry crossover rate ρ (the rate per morgan of transitions between ancestry of one subpopulation to ancestry of the other) is given by
, respectively. The ancestry crossover rate ρ (the rate per morgan of transitions between ancestry of one subpopulation to ancestry of the other) is given by 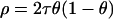 ; thus the larger τ is the more frequent the crossovers. Since the X chromosome undergoes recombination only in female gametes, a different τ will be applicable to the X chromosome than to the autosomes. However, in our data sets the autosomal markers are unlinked and thus τ was estimated from the linked markers typed in the 10-cM region of the X chromosome alone. In contrast, individual admixture proportions (θ) were estimated from both the autosomal and X chromosome markers. When introgression occurs in a single pulse the expectation of τ on the autosomes is equal to the number of generations since admixture (Falush et al. 2003). However, admixture in the cattle populations is not believed to have occurred as a single event but rather through continuous gene flow of B. indicus into B. taurus populations. The effect of continuous gene flow on τ is unclear and thus it cannot be interpreted as the number of generations since admixture but as a relative measure of the age of admixture among the populations studied. Given the lack of interpretation of τ it was assigned an uninformative prior, log τ-uniform. Prior distributions for population-specific allele frequencies for B. taurus and B. indicus were set from allele counts in the five B. taurus populations (both European and West African) and two B. indicus populations listed in Table 1. While it is known that the West African B. taurus populations contain a degree of indicus admixture, this is very small (mR = 3, 7, and 10% in Guinean N'Dama, Guinea Bissau N'Dama, and Somba, respectively) and thus will have minimal effect on the allele counts. Furthermore, specifying prior distributions from the allele counts rather than fixing them at their mean values allows for error in the frequencies to be accounted for. Uninformative uniform priors were assigned for the population level admixture.
; thus the larger τ is the more frequent the crossovers. Since the X chromosome undergoes recombination only in female gametes, a different τ will be applicable to the X chromosome than to the autosomes. However, in our data sets the autosomal markers are unlinked and thus τ was estimated from the linked markers typed in the 10-cM region of the X chromosome alone. In contrast, individual admixture proportions (θ) were estimated from both the autosomal and X chromosome markers. When introgression occurs in a single pulse the expectation of τ on the autosomes is equal to the number of generations since admixture (Falush et al. 2003). However, admixture in the cattle populations is not believed to have occurred as a single event but rather through continuous gene flow of B. indicus into B. taurus populations. The effect of continuous gene flow on τ is unclear and thus it cannot be interpreted as the number of generations since admixture but as a relative measure of the age of admixture among the populations studied. Given the lack of interpretation of τ it was assigned an uninformative prior, log τ-uniform. Prior distributions for population-specific allele frequencies for B. taurus and B. indicus were set from allele counts in the five B. taurus populations (both European and West African) and two B. indicus populations listed in Table 1. While it is known that the West African B. taurus populations contain a degree of indicus admixture, this is very small (mR = 3, 7, and 10% in Guinean N'Dama, Guinea Bissau N'Dama, and Somba, respectively) and thus will have minimal effect on the allele counts. Furthermore, specifying prior distributions from the allele counts rather than fixing them at their mean values allows for error in the frequencies to be accounted for. Uninformative uniform priors were assigned for the population level admixture.
The ADMIXMAP analyses assume that all LD between markers can be attributed to admixture LD and LD due to ancestral mosaicism but ignore background LD. The X chromosome markers used in our analyses are between 0.1 and 2.5 Mb apart (Table 2), and at such distances it is a reasonable approximation to ignore background LD.
Statistical testing was carried out using SPSS (v. 12.0) and values of τ were plotted with sampling location using the ArcView GIS software package. Equal interval contours join areas with similar values of τ (Environmental Systems Research Institute, Redlands, CA). A synthetic map was finalized using the Adobe Illustrator (v. 8.0) package.
RESULTS
A set of 10 linked X chromosome microsatellites that have been mapped within a 10-cM region of BtaX were genotyped in 346 male cattle samples from geographical locations shown in Figure 1 and detailed in Table 1. The data were combined with unlinked autosomal microsatellite genotypic data from previous publications.
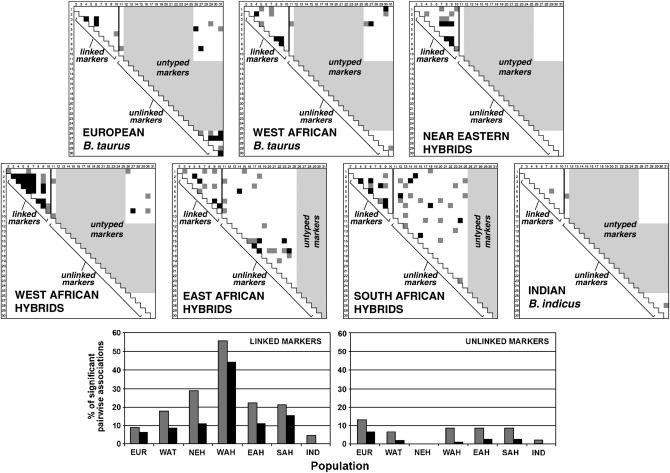
The strength of pairwise LD between all typed markers was estimated and the results were divided into two groups according to the significance of association between pairs of markers: P < 0.05 and P < 0.01. Figure 2 shows matrices of LD significance levels for all possible pairwise comparisons of the 10 X chromosome loci in their chromosomal order and for the unlinked markers. The samples were divided into regional groupings according to Table 1, excluding the Zebu Malagasy breed, which does not fall easily into an appropriate category and is represented only by four individuals. The unlinked autosomal microsatellite markers provide a measure of the background levels of LD in each regional group. LD measures for linked markers are shown on the left-hand side of the thick solid line in Figure 2, in the order that they are located on the X chromosome.
Figure 2.—
Linkage disequilibrium plots showing LD assessed by the significance of allelic associations at pairs of loci. The areas with lightest shading represent untyped markers. Shaded and solid squares mark locus pairs with alleles associating with P-values of <0.01 and <0.05, respectively. The matrix plots facilitate comparison of LD levels among the different regional groups. Additionally, the differences between LD among the sets of linked markers and unlinked markers are also evident. To further clarify the differences both between regional groups and between sets of linked and unlinked markers, the percentages of significant associations (at the two designated levels) are also shown in two histograms. The cattle populations are grouped according to Table 1.
In general, and as expected, physically linked markers tend to have higher levels of association than markers from different chromosomes. This trend is evident in all groups except European B. taurus. This group exhibits slightly lower levels of LD among linked markers than unlinked markers, but the difference is small. The trend is apparent in all of the hybrid populations (Near Eastern, West African, East African, and South African), but most especially in the West African hybrid population, which exhibits especially high levels of LD among the X chromosome microsatellites. Nearly 45% of all possible pairwise comparisons between linked markers are in LD at a significance level of 0.01 in this group. This is over three times as many as is seen in any of the other population groups.
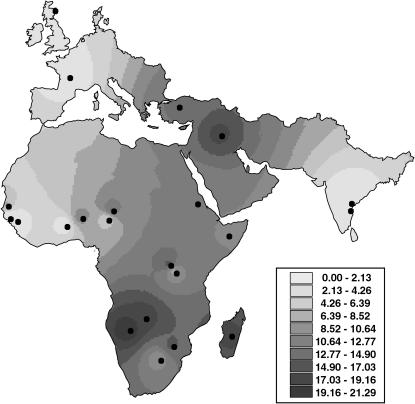
Estimates of τ obtained from ADMIXMAP enable comparison of time since admixture among the different sampled populations, but cannot be taken as absolute values, as a single event is unlikely to be an accurate model of past hybridization processes. Values of τ are shown on a synthetic contour map (Figure 3). Despite the relatively small number of hybrid breeds (15), some geographical congruency can be observed for values of τ.
Figure 3.—
Map showing values of τ. τ is the number of generations since a single admixture event. In these cattle populations admixture probably occurred in a continuous manner; therefore, τ is interpreted as a relative measure of the age of admixture among the populations studied (see materials and methods). Higher values of τ have darker shading and suggest older admixture in those areas.
The posterior means and standard deviations of the sum-of-intensities parameter τ for the admixed breeds are shown in Table 1. We see that within the African continent there are two main trends among the hybrid populations. First, values of τ tend to be higher in the south of the continent, and second, there is a decreasing cline in values of τ from east to west in the northern part of the continent. To test for significant difference between hybrid breeds in the north and south we pooled the estimates of τ using the inverse variance method (Deeks et al. 2001). The pooled estimate of τ for hybrid breeds in the northern part of the continent (Mbororo, Gobra, Sokoto Gudali, Arashie, and Kuri) is 7.6 (standard deviation 1.66), and the pooled estimate for hybrids in the south (Ogaden, Ankole, Afrikaner, Nguni, Watusi, Zebu Malagasey, Barotse, and Kavango) is 13.9 (standard deviation 1.85). The test for difference between the means of the groups is significant, P = 0.012. The pooled estimate for τ in the two Near Eastern hybrids is 15.4 (standard deviation 3.54), with the highest value being observed in the easternmost Iraqi breed (τ = 17.8).
DISCUSSION
Cattle populations represent an ideal model for the study of admixture-generated linkage disequilibrium and haplotype decay, as it is well established that they were domesticated from divergent wild progenitors leading to two main interfertile lineages: B. taurus and B. indicus. Since domestication, the two types have been combined in many different hybrid breeds but there are also pure extant taurine and zebu populations that provide good proxies for the original ancestral populations (MacHugh et al. 1997).
The amount of admixture in the breeds used in this study had previously been estimated using autosomal microsatellites (MacHugh et al. 1997; Loftus et al. 1999; Hanotte et al. 2002; Freeman et al. 2004) (Figure 1). Here we set out to examine the pattern of haplotype decay in the bovine genome after admixture by genotyping closely linked microsatellite markers. The level of divergence between the contributing parental populations will determine whether it is possible to assign definite ancestry to an allele in a hybrid individual. In a previous study, we found that >80% of microsatellite alleles surveyed at 20 loci in European, Near Eastern, and Indian cattle populations showed >50% frequency differential (scaled by the absolute allele frequency) between B. indicus and B. taurus (Kumar et al. 2003). All of the markers used here are located within a 10-cM region of the X chromosome and were typed only in males. This allowed the unambiguous assignment of a haplotype to an individual without pedigree information.
We found LD in this region of the X chromosome in all of the cattle populations and between linked and unlinked markers in all of the populations except for the Near Eastern hybrids. These results concur with those of Farnir et al. (2000), who surveyed 284 autosomal markers and found that intrachromosomal LD extends over several tens of centimorgans and is common between nonsyntenic loci in dairy cattle populations. In that case, the high levels of overall LD may have been a result of a small effective population size (Lonjou et al. 1999), which could be as low as 50 for 1.2 million Holstein–Friesian females, or admixture (Farnir et al. 2000).
Recurrent introgression has been cited previously as the cause of higher than expected levels of LD in African–American human populations (Lautenberger et al. 2000). Here, the very high level in the West African hybrid population may partially be explained by recent and continuous gene flow to this region (Pfaff et al. 2001). Pfaff and colleagues argue that maintenance of LD over relatively large (∼10 cM) chromosomal segments is a not typical of a single admixture event, but is a characteristic of a continuous gene flow pattern of admixture. While the initial introgression of B. indicus chromosomes into Africa is likely ancient, zebu–taurine admixture in West Africa probably began only recently in many populations where it had previously been constrained by the tsetse/trypanosomiasis infestation zone, a barrier to introgression. Understanding the patterns of LD in the hybrid populations from the surround of the West African tsetse zone will prove useful if these populations are used to map genes that influence disease traits (Laan and Paabo 1997).
In the absence of mutation, every portion of a chromosome in a hybrid individual should be directly traceable to one of the individual's ancestors. When two distinct gene pools meet, a mosaic genome is produced (Chapman and Thompson 2002), containing intact segments from each contributing founder population (Nordborg and Tavare 2002). Genomes that have undergone recent hybridization will have largely intact ancestral haplotypes, while recombination will act to disrupt these haplotypes in subsequent generations (Wiehe et al. 2000). If admixture is ancient, reciprocal recombination, genetic drift, and mutation will have eroded the association between markers from parental populations and a greater density of markers will be required to detect significant associations.
Here we employed a Bayesian method to assign probable ancestry to each allele in every X chromosome haplotype. The amount of fragmentation in haplotypes was assessed by the posterior distribution of τ, and this was used as a measure for the age of admixture. The mean values of τ (Table 1) for each of the cattle breeds are between 4.27 and 21.3, which is substantially less than the number of generations since admixture is believed to have first occurred. However, as discussed earlier, in the case of cattle a single admixture event may not accurately model the historical reality and τ has expectation only equal to the number of generations since admixture in the case of a single admixture event. Furthermore, the prior has the effect of favoring small values of τ.
The values obtained appear to show some geographical consistency, whereby most hybrid populations from southern Africa and the Near East appear to have experienced older admixture than those in the more northern part of the African continent. Furthermore there is a decline in age of admixture from east to west in the latter. The differences between the east and the west of the continent support a scenario of earlier introgression of B. indicus material into the former populations, as the degree of recombination would be expected to accumulate with time. This finding is supported by high levels of LD in West African hybrids and by previous work that has shown that the major B. indicus introductions occurred in the east, probably in the region of the Horn (Hanotte et al. 2002).
The difference in τ-values between the south and the north of Africa may be a signal of different histories of cattle in the two regions. However, it is well established that the earliest domesticated cattle of Africa were found in the north of the continent and were B. taurus in nature and that introgression of B. indicus into the continent via the Horn of Africa was secondary. Our results suggest that these earliest hybrids may have been the animals that later gave rise to southern populations. This scenario implies that B. indicus introgression occurred before pastoralism spread extensively southward and indeed, despite extensive sampling, there is no evidence for the presence of pure African B. taurus in the south of the continent (Hanotte et al. 2002).
On the basis of archaeological evidence the origins of B. indicus introgression in Africa are controversial. Some authors have suggested that they were found as early as 3500–2000 years before the present (YBP) (Hassan 2000; Marshall 2000; Paris 2000), but these studies are inconclusive, and the earliest verified archaeological date for their presence is in the Horn of Africa in the second century a.d. (Marshall 2000). The archaeological record shows that B. taurus cattle were present in northern Africa and the Sahara from ∼6000 YBP (Hassan 2000) and spread as far south as Kenya by 3000 YBP (Hassan 2000; Mitchell 2005). Further southward movement may have been hampered by ecological factors such as the many sub-Saharan disease challenges to which domestic cattle would be maladapted (Gifford-Gonzalez 2000; Mitchell 2005), and relatively few cattle were present in the south until 1000 a.d. (Smith 2000). It is therefore possible that nomadic pastoralists in eastern Africa were herding animals composed of the two cattle lineages before the expansion of domestic cattle to the south.
Our sample from Madagascar suggests that admixture here was older but this sample is very small, n = 4, and should not be overinterpreted. There is limited archaeological evidence for early arrival of B. indicus cattle to Madagascar and zebu cattle occupy a place of prominence in Malagasy ritual and culture, which is compatible with an extensive history on the island (Hemmer 1990; van der Zwan and Evers 1998).
The employment of haplotypic mosaicism as an indicator of the antiquity of admixture receives some confirmation from the Near Eastern results. The region between the primary centers of B. indicus and B. taurus domestication is an area of ancient interaction between the two lineages. As such, the sample from Iraq yields one of the highest values of τ within the haplotypes.
Acknowledgments
We thank David Lynn, Colm O'hUigin, Paul McKeigue, and Mark Shriver for helpful discussions and Claude Gaillard for provision of samples. This material is based on works supported by Science Foundation Ireland under grant no. 02-IN.1-B256. A. R. Freeman was funded by a Wellcome studentship in biodiversity, no. 054275/Z/98/Z.
References
- Amaral, M. E., S. R. Kata and J. E. Womack, 2002. A radiation hybrid map of bovine X chromosome (BTAX). Mamm. Genome 13: 268–271. [DOI] [PubMed] [Google Scholar]
- Bishop, M. D., S. M. Kappes, J. W. Keele, R. T. Stone, S. L. Sunden et al., 1994. A genetic linkage map for cattle. Genetics 136: 619–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley, D. G., D. E. MacHugh, R. T. Loftus, R. S. Sow, C. H. Hoste et al., 1994. Zebu-taurine variation in Y chromosomal DNA: a sensitive assay for genetic introgression in west African trypanotolerant cattle populations. Anim. Genet. 25: 7–12. [PubMed] [Google Scholar]
- Bradley, D. G., D. E. MacHugh, P. Cunningham and R. T. Loftus, 1996. Mitochondrial diversity and the origins of African and European cattle. Proc. Natl. Acad. Sci. USA 93: 5131–5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, N. H., and E. A. Thompson, 2002. The effect of population history on the lengths of ancestral chromosome segments. Genetics 162: 449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks, J., D. Altman and M. Bradburn, 2001. Statistical methods for examining heterogeneity and combining results from several studies in meta-analysis, pp. 285–372 in Systematic Reviews in Health Care. Meta-Analysis in Context, edited by M. Egger, G. Davey Smith and D. Altman. BMJ Books, London.
- Falush, D., M. Stephens and J. K. Pritchard, 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnir, F., W. Coppieters, J. J. Arranz, P. Berzi, N. Cambisano et al., 2000. Extensive genome-wide linkage disequilibrium in cattle. Genome Res. 10: 220–227. [DOI] [PubMed] [Google Scholar]
- Felius, M., 1995. Cattle Breeds—An Encyclopedia. Misset, Doetinchem, The Netherlands.
- Freeman, A. R., C. M. Meghan, D. E. MacHugh, R. T. Loftus, M. D. Achukwi et al., 2004. Admixture and diversity in West African cattle populations. Mol. Ecol. 13: 3477. [DOI] [PubMed] [Google Scholar]
- Gifford-Gonzalez, D., 2000. Animal disease challenges to the emergence of pastoralism in sub-Saharan Africa. Afr. Archaeol. Rev. 17: 95–139. [Google Scholar]
- Hanotte, O., C. L. Tawah, D. G. Bradley, M. Okomo, Y. Verjee et al., 2000. Geographic distribution and frequency of a taurine Bos taurus and an indicine Bos indicus Y specific allele amongst sub-Saharan African cattle breeds. Mol. Ecol. 9: 387–396. [DOI] [PubMed] [Google Scholar]
- Hanotte, O., D. G. Bradley, J. W. Ochieng, Y. Verjee, E. W. Hill et al., 2002. African pastoralism: genetic imprints of origins and migrations. Science 296: 336–339. [DOI] [PubMed] [Google Scholar]
- Hassan, F. A., 2000. Climate and cattle in North Africa: a first approximation, pp. 61–86 in The Origins and Development of African Livestock: Archaeology, Genetics, Linguistics and Ethnography, edited by R. M. Blench and K. C. MacDonald. UCL Press, London.
- Hemmer, H. (Editor), 1990. Domestication: The Decline of Environmental Appreciation, p. 11. Cambridge University Press, Cambridge/London/New York.
- Hoggart, C. J., E. J. Parra, M. D. Shriver, C. Bonilla, R. A. Kittles et al., 2003. Control of confounding of genetic associations in stratified populations. Am. J. Hum. Genet. 72: 1492–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggart, C. J., M. D. Shriver, R. A. Kittles, D. G. Clayton and P. M. McKeigue, 2004. Design and analysis of admixture mapping studies. Am. J. Hum. Genet. 74: 965–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara, N., A. Takasuga, K. Mizoshita, H. Takeda, M. Sugimoto et al., 2004. A comprehensive genetic map of the cattle genome based on 3802 microsatellites. Genome Res. 14: 1987–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes, S. M., J. W. Keele, R. T. Stone, R. A. McGraw, T. S. Sonstegard et al., 1997. A second-generation linkage map of the bovine genome. Genome Res. 7: 235–249. [DOI] [PubMed] [Google Scholar]
- Kumar, P., A. R. Freeman, R. T. Loftus, C. Gaillard, D. Q. Fuller et al., 2003. Admixture analysis of South Asian cattle. Heredity 91: 43–50. [DOI] [PubMed] [Google Scholar]
- Laan, M., and S. Paabo, 1997. Demographic history and linkage disequilibrium in human populations. Nat. Genet. 17: 435–438. [DOI] [PubMed] [Google Scholar]
- Lautenberger, J. A., J. C. Stephens, S. J. O'Brien and M. W. Smith, 2000. Significant admixture linkage disequilibrium across 30 cM around the FY locus in African Americans. Am. J. Hum. Genet. 66: 969–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus, R. T., D. E. MacHugh, D. G. Bradley, P. M. Sharp and P. Cunningham, 1994. Evidence for two independent domestications of cattle. Proc. Natl. Acad. Sci. USA 91: 2757–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus, R. T., O. Ertugrul, A. H. Harba, M. A. El-Barody, D. E. MacHugh et al., 1999. A microsatellite survey of cattle from a centre of origin: the Near East. Mol. Ecol. 8: 2015–2022. [DOI] [PubMed] [Google Scholar]
- Lonjou, C., A. Collins and N. E. Morton, 1999. Allelic association between marker loci. Proc. Natl. Acad. Sci. USA 96: 1621–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacHugh, D. E., M. D. Shriver, R. T. Loftus, P. Cunningham and D. G. Bradley, 1997. Microsatellite DNA variation and the evolution, domestication and phylogeography of taurine and zebu cattle (Bos taurus and Bos indicus). Genetics 146: 1071–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, F., 2000. The origins and spread of domestic animals in East Africa, pp. 191–221 in The Origins and Development of African Livestock: Archaeology, Genetics, Linguistics and Ethnography, edited by R. M. Blench and K. C. MacDonald. UCL Press, London.
- Mitchell, P., 2005. African Connections, pp. 33–63. AltaMira, Walnut Creek, CA.
- Nordborg, M., and S. Tavare, 2002. Linkage disequilibrium: what history has to tell us. Trends Genet. 18: 83–90. [DOI] [PubMed] [Google Scholar]
- Paris, F., 2000. African livestock remains from Saharan mortuary context, pp. 111–126 in The Origins and Development of African Livestock: Archaeology, Genetics, Linguistics and Ethnography, edited by R. M. Blench and K. C. MacDonald. UCL Press, London.
- Pfaff, C. L., E. J. Parra, C. Bonilla, K. Hiester, P. M. McKeigue et al., 2001. Population structure in admixed populations: effect of admixture dynamics on the pattern of linkage disequilibrium. Am. J. Hum. Genet. 68: 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond, M., and F. Rousset, 1995. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J. Hered. 86: 248–249. [Google Scholar]
- Ritz, L. R., M. L. Glowatzki-Mullis, D. E. MacHugh and C. Gaillard, 2000. Phylogenetic analysis of the tribe Bovini using microsatellites. Anim. Genet. 31: 178–185. [DOI] [PubMed] [Google Scholar]
- Smith, A. B., 2000. The origins of the domesticated animals of southern Africa, pp. 222–238 in The Origins and Development of African Livestock: Archaeology, Genetics, Linguistics and Ethnography, edited by R. M. Blench and K. C. MacDonald. UCL Press, London.
- Sønstegard, T. S., N. L. Lopez-Corrales, S. M. Kappes, R. T. Stone, S. Ambady et al., 1997. An integrated genetic and physical map of the bovine X chromosome. Mamm. Genome 8: 16–20. [DOI] [PubMed] [Google Scholar]
- Sønstegard, T. S., W. Barendse, G. L. Bennett, G. A. Brockmann, S. Davis et al., 2001. Consensus and comprehensive linkage maps of the bovine sex chromosomes. Anim. Genet. 32: 115–117. [DOI] [PubMed] [Google Scholar]
- Sun, H. S., M. R. Dentine, W. Barendse and B. W. Kirkpatrick, 1994. UWCA19 and UWCA20: polymorphic bovine microsatellites. Anim. Genet. 25: 121. [DOI] [PubMed] [Google Scholar]
- van der Zwan, N., and S. Evers, 1998. Madagascar: The Zebu as Guide Through Past and Present. Berg en Dal, The Netherlands.
- Wiehe, T., J. Mountain, P. Parham and M. Slatkin, 2000. Distinguishing recombination and intragenic gene conversion by linkage disequilibrium patterns. Genet. Res. 75: 61–73. [DOI] [PubMed] [Google Scholar]
- Williams, J. L., A. Eggen, L. Ferretti, C. J. Farr, M. Gautier et al., 2002. A bovine whole-genome radiation hybrid panel and outline map. Mamm. Genome 13: 469–474. [DOI] [PubMed] [Google Scholar]