Abstract
Mn is an essential component of the oxygen-evolving machinery of photosynthesis and is an essential cofactor of several important enzymes, such as Mn–superoxide dismutase and Mn-catalase. The availability of Mn in the environment varies, and little is known about the mechanisms for maintaining cytoplasmic Mn2+ ion homeostasis. Using a DNA microarray, we screened knockout libraries of His kinases and response regulators of Synechocystis sp PCC 6803 to identify possible participants in this process. We identified a His kinase, ManS, which might sense the extracellular concentration of Mn2+ ions, and a response regulator, ManR, which might regulate the expression of the mntCAB operon for the ABC-type transporter of Mn2+ ions. Furthermore, analysis with the DNA microarray and by reverse transcription PCR suggested that ManS produces a signal that activates ManR, which represses the expression of the mntCAB operon. At low concentrations of Mn2+ ions, ManS does not generate a signal, with resulting inactivation of ManR and subsequent expression of the mntCAB operon.
INTRODUCTION
Mn acts as the catalytic center of the oxygen-evolving photosynthetic machinery (Yachandra et al., 1993), in which four Mn atoms are coordinated within a pocket that is formed by the D1 and D2 proteins (Zouni et al., 2001), and is essential for all organisms, acting as the cofactor of various enzymes such as Mn–superoxide dismutase (Borgstahl et al., 2000), Mn-catalase (Barynin et al., 2001), pyruvate carboxylase, and phosphoenolpyruvate carboxykinase (Frausto da Silva and Williams, 1991; Larson and Pecoraro, 1992). Therefore, the incorporation of Mn by cells is essential, particularly in the case of photosynthetic autotrophs in which oxygenic photosynthesis occurs.
Cyanobacteria are prokaryotes whose machinery for oxygenic photosynthesis is regarded as a useful model for plant chloroplasts. Cyanobacterial cells take up Mn2+ ions via a so-called ABC-type transporter (Bartsevich and Pakrasi, 1995). The mntCAB operon includes mntA, the gene for an ATP binding subunit, mntB, the gene for a hydrophobic subunit, and mntC, the gene for the Mn2+ binding subunit (Bartsevich and Pakrasi, 1995, 1999). However, cyanobacteria are exposed frequently to Mn-limiting conditions, such as in seawater, and expression of the mntCAB operon for the Mn2+ transporter is induced when the external supply of Mn2+ is limited (Bartsevich and Pakrasi, 1996). However, the way in which cyanobacterial cells recognize a deficiency in Mn2+ and the way in which expression of the mntCAB operon is induced subsequently remain to be determined. To address this issue, we postulated that a two-component system might exist in Synechocystis sp PCC 6803 that includes a sensor and a transducer of the signal for Mn2+ deficiency. We generated knockout libraries of His kinases and response regulators and examined gene expression using the DNA microarray technique. We identified a membrane-bound His kinase as a possible sensor and a response regulator of Mn2+ ions that together regulate the expression of the mntCAB operon. Our DNA microarray analysis also suggested that this two-component system acts repressively to regulate gene expression.
RESULTS
Inactivation of ManS Induces the Expression of the mntCAB Operon
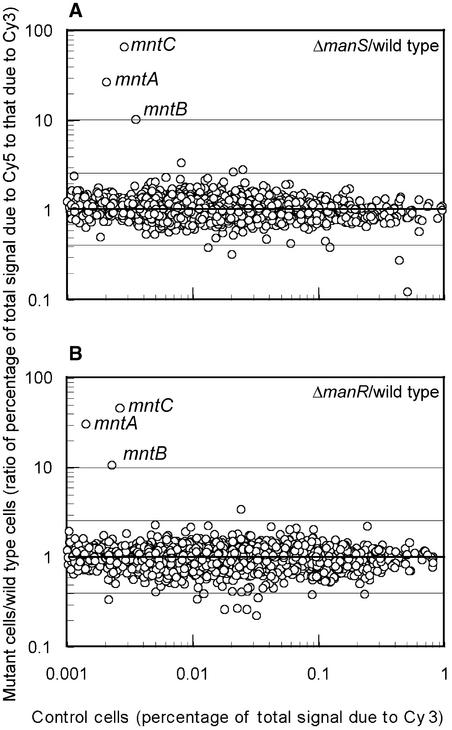
The Synechocystis genome contains 42 putative genes for His kinases. We successfully mutated all of these genes by targeted mutagenesis with a spectinomycin resistance cassette (Suzuki et al., 2000). Using a DNA microarray, we analyzed the effects of each mutation on gene expression in mutant cells that had been grown in BG-11 medium that contained 9 μM Mn2+. No significant alterations in gene expression were evident in most of the mutants. However, one of the mutants, ΔHik27 (according to the terminology described at the CyanoGenes World Wide Web site [http://www.kazusa.or.jp/cyano/Synechocystis/comments/cgi-bin/comshow.cgi?id=slr0640&Kwd=Hik27]), with a mutation in the His kinase encoded by slr0640, was unique. In this mutant, expression was enhanced in only three genes, mntC, mntA, and mntB (Figure 1A), which constitute the mntCAB operon for subunits of the ABC-type Mn2+ transporter (Bartsevich and Pakrasi, 1995). We designated the corresponding His kinase ManS. The results shown in Figure 1A suggest that under normal growth conditions, ManS might transduce a signal that represses the expression of the mntCAB operon and, moreover, that the disappearance of this signal, as a result of the inactivation of ManS, might allow the mntCAB operon to be expressed.
Figure 1.
DNA Microarray Analysis of the Effects of Mutations in the manS and manR Genes on Gene Expression in Synechocystis.
(A) Gene expression in ΔmanS cells compared with wild-type cells.
(B) Gene expression in ΔmanR cells compared with wild-type cells.
Cells were grown in the presence of an adequate concentration of Mn2+ ions (in BG-11 medium that contained 9 μM MnCl2). Both experiments were performed twice, and essentially the same results were obtained in both cases.
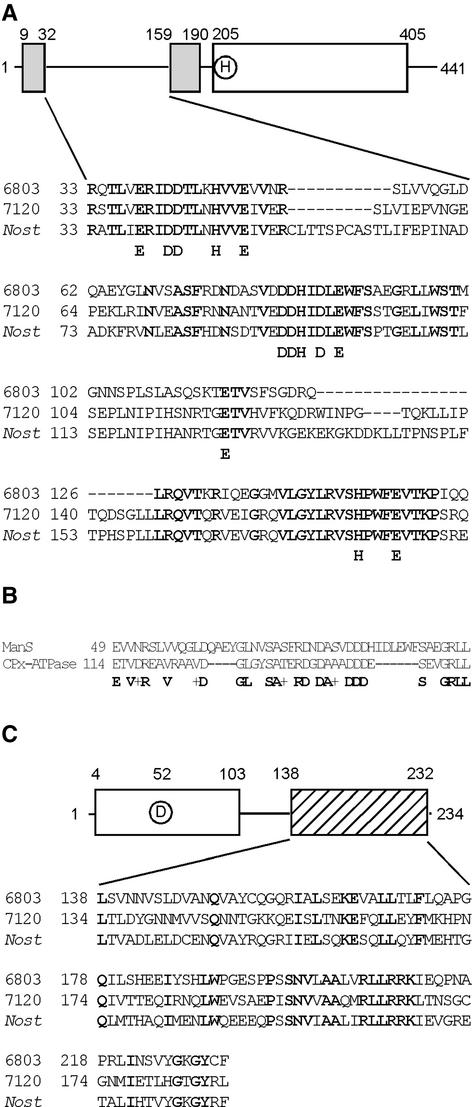
ManS is a typical His kinase with 441 amino acid residues. It has a relative molecular mass of 48 kD (http://www.kazusa.or.jp/cyano/Synechocystis/cgi-bin/geinfo.cgi?type=orf&name=slr0640). The structure of ManS is shown in Figure 2A. There is a His kinase domain in the C-terminal region that includes a phosphorylatable His residue, and there are two membrane-spanning domains in the N-terminal region.
Figure 2.
Structure of ManS and ManR.
(A) Scheme of ManS and alignment of the putative periplasmic domains of ManS and homologous proteins from other cyanobacteria. Boldface letters indicate conserved amino acids. 6803, Synechocystis sp PCC 6803; 7120, Anabaena sp PCC 7120; Nost, Nostoc punctiforme.
(B) Alignment of the putative periplasmic domains of ManS and CPx-type ATPase of Halobacterium. Conserved amino acids are shown on the bottom line. Plus signs indicate the conservation of similar amino acids.
(C) Scheme of ManR and alignment of the putative DNA binding domains of homologous proteins. Boldface letters indicate conserved amino acids.
It is likely that the region between the two membrane-spanning domains is located in the periplasmic space and perceives the extracellular concentration of Mn2+ ions (see Discussion). Figure 2A also shows an alignment of the putative periplasmic domains of ManS and homologous His kinases from Anabaena sp PCC 7120 and Nostoc punctiforme. ManS is 49 and 43% homologous with the His kinases from Anabaena sp PCC 7120 and N. punctiforme, respectively. In these His kinases, the putative periplasmic domain is rich in conserved His, Glu, and Asp residues. These residues might provide ligands to Mn2+, as is the case in Mn–superoxide dismutase from Escherichia coli (Borgstahl et al., 2000), Mn-catalase from Lactobacillus plantarum (Barynin et al., 2001), and the D1 and D2 proteins in the photosystem II complex (Zouni et al., 2001). A homology search revealed similarities between the putative periplasmic domain and the metal binding site of a heavy metal–transporting CPx-type ATPase (Figure 2B) (Solioz and Vulpe, 1996). These results suggest that the region between the two membrane-spanning domains might play a critical role in the binding to Mn2+ ions.
Synechocystis contains 10 to 12 copies of the chromosome per cell (Mann and Carr, 1974). Analysis by PCR with primers specific for the manS gene indicated that this gene had been mutated in all copies of the chromosome in our ΔmanS mutant cells (Figure 3), suggesting that no ManS was present in these cells.
Figure 3.
Positions of Insertions/Replacements in Targeted Mutagenesis of the manS and manR Genes in Synechocystis.
(A) Scheme of the insertion of the spectinomycin resistance gene cassette (Spr) into the manS gene. Two dark gray boxes and a white box represent hydrophobic domains and the His kinase domain, respectively. The encircled H indicates a putative phosphorylatable His residue. Arrows indicate the positions of the forward and reverse primers used for genomic PCR. The numbers represent the positions of amino acids counted from the N terminus of ManS.
(B) Scheme of the insertion of the kanamycin resistance gene cassette (Kmr) into the manR gene. White and hatched boxes represent the receiver region and the DNA binding regions, respectively. The encircled D indicates a putative phosphorylatable Asp residue. Arrows indicate the positions of the forward and reverse primers for genomic PCR. The numbers represent the positions of amino acids counted from the N terminus of ManR.
(C) Determination by genomic PCR of the extent of replacement of wild-type genes by mutated genes. Lanes 1 and 4, DNA from wild-type cells; lanes 2 and 5, DNA from ΔmanS cells; lanes of 3 and 6, DNA from ΔmanR cells; lane M, DNA markers of indicated lengths.
Inactivation of ManR Induces the Expression of the mntCAB Operon
The sequence of the Synechocystis genome suggests the presence of 40 putative genes for response regulators. We successfully mutated most of these genes by targeted mutagenesis with a kanamycin resistance cassette (http://www.kazusa.or.jp/cyano/Synechocystis/comments/cgi-bin/comlist.cgi?sortby=&from=1&to=467). We examined the effects of each mutation on gene expression in mutant cells, which had been grown in BG-11 medium that contained 9 μM Mn2+. One mutation enhanced the expression of only three genes, mntC, mntA, and mntB (Figure 1B). This phenomenon was very similar to the change in gene expression detected in the ΔmanS mutant. We designated the putative response regulator ManR. The results shown in Figure 1B suggest that ManR repressed the expression of the mntCAB operon and that the inactivation of ManR in ΔmanR mutant cells eliminated the repressive effect of ManR, allowing expression of the mntCAB operon.
ManR consists of 234 amino acid residues and has a relative molecular mass of 26 kD (http://www.kazusa.or.jp/cyano/Synechocystis/cgi-bin/geinfo.cgi?type=orf&name=slr1837). As shown in Figure 2C, ManR contains a receiver domain that includes a phosphorylatable Asp residue and a putative DNA binding domain in the C-terminal region. This structure resembles that of OmpR from E. coli, which is a response regulator of osmotic signals (Martínez-Hackert and Stock, 1997). Figure 2C also shows an alignment of the putative DNA binding domain of ManR and homologous response regulators from Anabaena sp PCC 7120 and N. punctiforme. The extent of homology at the amino acid level in this domain is 38 and 54% when ManR from Synechocystis is compared with the homologs from Anabaena sp PCC 7120 and N. punctiforme, respectively. These homologous proteins in Anabaena sp PCC 7120 and N. punctiforme might have functions similar to that of ManR. Analysis by PCR with specific primers for the manR gene indicated that this gene had been mutated in all copies of the chromosome in our ΔmanR mutant cells (Figure 3).
Gene Expression in Wild-Type, ΔmanS, and ΔmanR Cells under Mn2+-Depleted Conditions
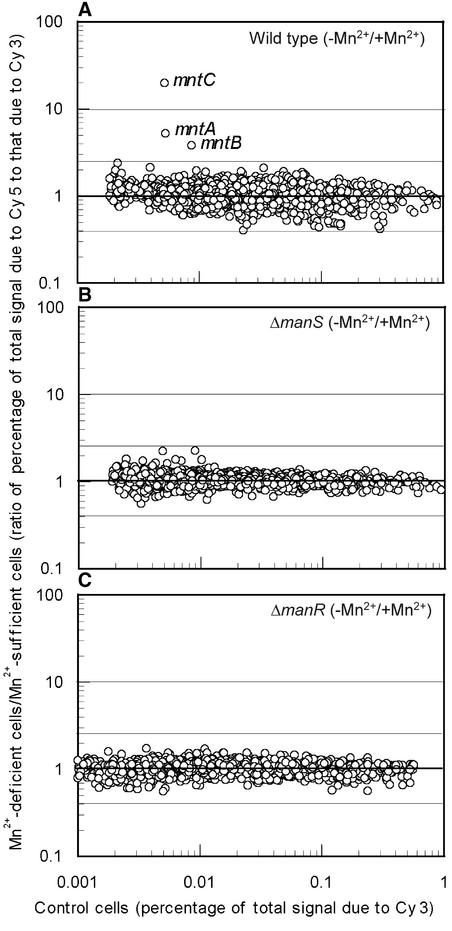
Figure 4 shows the effects of a deficit of Mn2+ ions on gene expression in wild-type, ΔmanS, and ΔmanR cells. Incubation of wild-type cells for 24 h in the absence of Mn2+ ions enhanced the expression of the mntCAB operon but had no obvious effects on other genes (Figure 4A). By contrast, incubation of ΔmanS and ΔmanR cells under the same conditions did not enhance the expression of any genes (Figures 4B and 4C). These observations suggested that ManS and ManR might be involved in the perception and transduction of signals engendered by a depletion of Mn2+ ions in this organism.
Figure 4.
DNA Microarray Analysis of the Effects of Mn2+ Depletion on Gene Expression in Wild-Type, ΔmanS, and ΔmanR Cells.
Gene expression in wild-type (A), ΔmanS (B), and ΔmanR (C) cells that had been incubated for 24 h under Mn2+-depleted conditions (in BG-11 medium to which no MnCl2 had been added) was compared with that in corresponding cells that had been grown in the presence of adequate concentrations of Mn2+ ions (in BG-11 medium that contained 9 μM MnCl2).
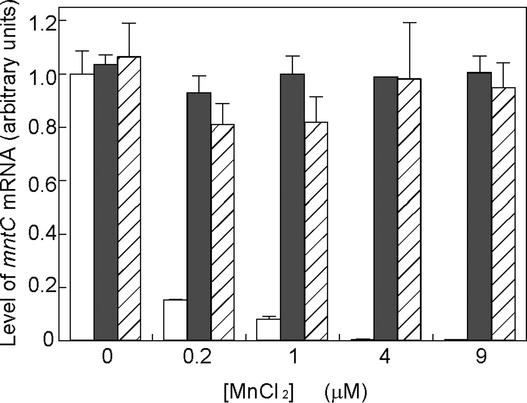
To confirm the changes in levels of expression of the mntCAB operon, we examined, by quantitative reverse transcription (RT) PCR, expression of the mntC gene, which we chose as an indicator of the activity of the promoter of the mntCAB operon (Figure 5). The results demonstrated that, in wild-type cells, the mntC gene was expressed strongly at low concentrations of Mn2+ ions (e.g., 0.2 and 1.0 μM) but not at high concentrations of Mn2+ (e.g., 4 and 9 μM). By contrast, the mntC gene in ΔmanS cells was expressed strongly at concentrations of Mn2+ ions ranging from 0 to 9 μM (Figure 5). These results suggested that, when the concentration of Mn2+ ions is adequate, ManS transduces a signal that leads to repression of the expression of the mntCAB operon that encodes the Mn2+ transporter. Similar results were obtained with ΔmanR mutant cells (Figure 5).
Figure 5.
Effects of Mutations in the manS and manR Genes on the Expression of the mntC Gene, as Determined by Quantitative RT-PCR.
Wild-type, ΔmanS, and ΔmanR cells were cultured with various concentrations of Mn2+ ions for 2 days before analysis. Open, closed, and hatched bars correspond to wild-type, ΔmanS, and ΔmanR cells, respectively.
To study the specificity of the ManS/ManR system for individual cations, we examined the effects of deprivation of Zn2+, Co2+, and Cu2+ ions, compared with Mn2+ ions, on the expression of the mntCAB operon. Table 1 shows that a deficiency only in Mn2+ ions, but in no other metal ions tested, induced the expression of the mntC gene. High concentrations of the tested metal ions, such as 12 μM Zn2+, 3 μM Cu2+, and 5 μM Co2+, had no effects on the expression of the mntC gene in wild-type cells (data not shown). These results indicated that the ManS/ManR system was specific to Mn2+ ions.
Table 1.
Expression of the mntC Gene during Deprivation of Various Metal Ions, as Determined by Quantitative RT-PCR in Wild-Type, ΔmanS, and ΔmanR Cells
| Level of Expression (Arbitrary Units)
|
|||||
|---|---|---|---|---|---|
| Ions Omitted from the Medium
|
|||||
| Cells | Control | Mn2+ | Zn2+ | Co2+ | Cu2+ |
| Wild type | 0.00 ± 0.00 | 1.00 ± 0.18 | 0.01 ± 0.01 | 0.01 ± 0.00 | 0.01 ± 0.00 |
| ΔmanS | 0.94 ± 0.22 | 1.00 ± 0.23 | 1.03 ± 0.18 | 1.12 ± 0.11 | 1.21 ± 0.24 |
| ΔmanR | 1.16 ± 0.34 | 1.00 ± 0.17 | 1.18 ± 0.24 | 1.26 ± 0.47 | 1.05 ± 0.30 |
Cells were incubated for 2 days in BG-11 medium that lacked the indicated metal ions. Quantitative RT-PCR was performed as described in Methods. Levels of expression are given in arbitrary units, relative to the level of expression upon deprivation of Mn2+ ions. The values shown are means of results from four independent experiments with standard deviations.
Complementation of ΔmanS and ΔmanR Mutants
To confirm that the change in the expression of the mntCAB operon was caused by mutation of the manS gene, we transformed ΔmanS mutant cells with the manS gene, which had been inserted in the pVZ321 vector. We found that transformed cells had recovered the ability to express the mntCAB operon when deprived of Mn2+ ions (Figure 6A). These observations indicated that the phenotype observed in the ΔmanS mutant was attributable to the mutation of the manS gene.
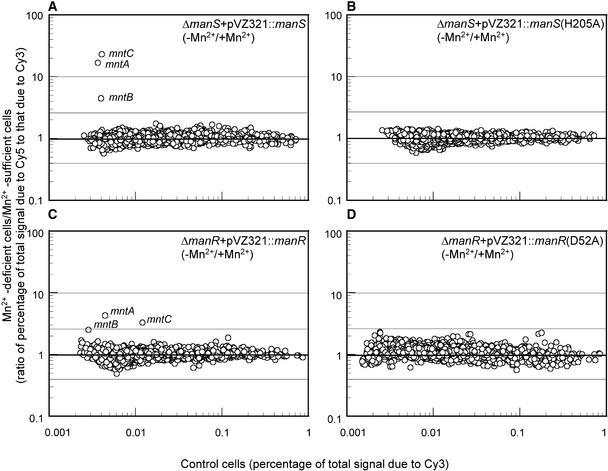
Figure 6.
Complementation of ΔmanS and ΔmanR Mutants Examined by DNA Microarray Analysis of the Mn2+ Deletion–Induced Expression of Genes.
(A) ΔmanS cells transformed with pVZ321-ManS.
(B) ΔmanS cells transformed with pVZ321-ManS(H205A).
(C) ΔmanR cells transformed with pVZ321-ManR.
(D) ΔmanR cells transformed with pVZ321-ManR(D52A).
Gene expression in the transformed cells that had been incubated for 24 h under Mn2+-depleted conditions (in BG-11 medium to which no MnCl2 had been added) was compared with that in corresponding cells that had been grown in BG-11 medium that contained 9 μM MnCl2.
To confirm the role of the phosphorylatable His residue in ManS, we synthesized a mutated manS gene for ManS(H205A), in which the His residue at amino acid 205 (counted from the site of initiation of translation) had been replaced by Ala, and transformed the ΔmanS cells with the pVZ321 vector that ligated the mutated manS gene. As expected, this transformation did not complement the phenotype in terms of enhanced expression of mntCAB genes by depletion of Mn2+ ions (Figure 6B). These observations suggested that the His-205 residue was essential for the transduction of Mn2+ signals.
We performed complementation analysis, as described above for the ΔmanS mutation, of the ΔmanR mutation using the pVZ321 vector that contained the manR gene. We found that transformed cells had recovered the ability to express the mntCAB operon when deprived of Mn2+ ions (Figure 6C). This observation suggested that the phenotype of ΔmanR mutant cells was caused by mutation of the manR gene. To confirm the role of the phosphorylatable Asp residue in ManR, we synthesized a mutated manR gene for ManR(D52A), in which the Asp residue at amino acid 52 (counted from the site of initiation of translation) had been replaced by Ala, and transformed the ΔmanR cells with the pVZ321 vector that ligated the mutated manR gene. As expected, this transformation did not complement the phenotype in terms of enhanced expression of mntCAB genes in response to the depletion of Mn2+ ions in the medium (Figure 6D).
Binding of ManR to the Promoter Region of the mntCAB Operon
To characterize ManS and ManR biochemically, we tried to overexpress ManS and ManR as recombinant proteins in E. coli. We failed to overexpress the full-length ManS protein but were able to overexpress the kinase domain of ManS as a fusion protein with a His tag, and we purified this protein by affinity chromatography. Nevertheless, the fusion protein was not autophosphorylated. By contrast, ManR was expressed successfully as a soluble fusion protein with a His tag at the N terminus. The relative molecular mass of the His-tagged ManR, after purification by affinity chromatography, was estimated by SDS-PAGE to be ∼28.2 kD, which was the same as that deduced from the putative amino acid sequence of the fusion protein.
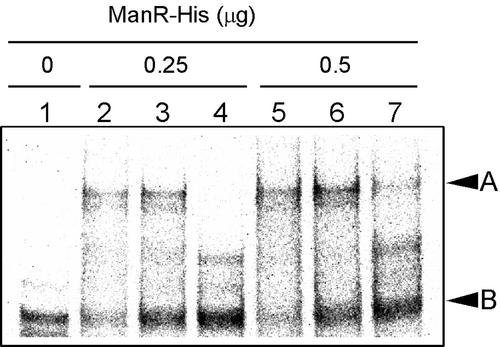
To determine whether ManR is able to interact with the promoter region of the mntCAB operon, we examined the DNA binding activity of His-tagged ManR by a gel mobility shift assay with a 32P-labeled DNA fragment that corresponded to the region from positions −135 to −1 upstream of the codon for the initiation of translation of the mntC gene (Figure 7). We observed a band caused by the complex of His-tagged ManR and the promoter region of the mntCAB operon (Figure 7, lanes 1, 2, and 5, arrowhead A). In the presence of a 10-fold or 100-fold excess of the nonlabeled DNA fragment, the binding of His-tagged ManR to the labeled DNA fragment was reduced significantly (Figure 7, lanes 3, 4, 6, and 7). This result indicated that the His-tagged ManR had bound specifically to the promoter region of the mntCAB operon. We examined the effects of phosphorylation of the Asp residue in ManR on its DNA binding activity by previously incubating ManR with 100 mM acetyl phosphate, as described in Methods. However, this treatment did not enhance the DNA binding activity of ManR (data not shown).
Figure 7.
Gel Mobility Shift Assay of the Binding of ManR to the Promoter of the mntCAB Operon.
His-tagged ManR (ManR-His) and a 32P-labeled DNA fragment of 135 bp that corresponded to the promoter region of the mntCAB operon were synthesized as described in Methods. The labeled DNA fragment (7.7 ng) was incubated with 0.25 or 0.5 μg of ManR-His for 20 min at 25°C in a volume of 20 μL and subjected to gel electrophoresis as described in Methods. Lane 1, no ManR-His; lane 2, 0.25 μg of ManR-His; lane 3, 0.25 μg of ManR-His and 77 ng of nonlabeled DNA fragment; lane 4, 0.25 μg of ManR-His and 770 ng of nonlabeled DNA fragment; lane 5, 0.5 μg of ManR-His; lane 6, 0.5 μg of ManR-His and 77 ng of nonlabeled DNA fragment; lane 7, 0.5 μg of ManR-His and 770 ng of nonlabeled DNA fragment. Arrowheads A and B indicate the positions of the complex of ManR-His with the 135-bp DNA fragment and of the 135-bp DNA fragment, respectively.
Growth of ΔmanS and ΔmanR Cells at Various Concentrations of Mn2+ Ions
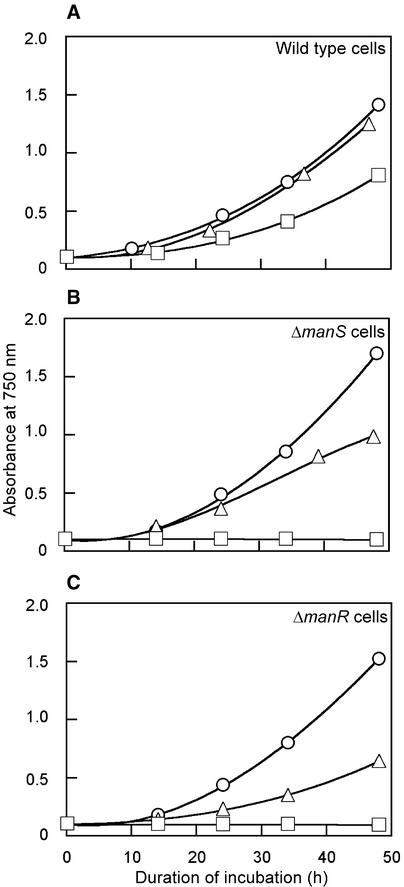
At 9 μM Mn2+, the standard concentration in BG-11 medium, growth rates of wild-type, ΔmanS, and ΔmanR cells were similar. A decrease in the concentration of Mn2+ ions to 0.2 μM did not affect the growth rate of wild-type, ΔmanS, and ΔmanR cells (data not shown). However, at 150 μM Mn2+, the growth rate of ΔmanS and ΔmanR cells decreased by 50%, whereas that of wild-type cells was unaffected. At 1.1 mM Mn2+, neither ΔmanS nor ΔmanR cells grew at all, whereas the growth rate of wild-type cells decreased by only 50% (Figure 8). The sensitivity of ΔmanS and ΔmanR cells to high levels of Mn2+ ions might have been related to the inability of these cells to repress the expression of the mntCAB operon, which might have led to increases in the intracellular concentration of Mn2+ ions that were toxic to cells.
Figure 8.
Effects of Exogenous Mn2+ Ions on the Growth of Wild-Type, ΔmanS, and ΔmanR Cells.
(A) Wild-type cells.
(B) ΔmanS cells.
(C) ΔmanR cells.
Concentrations of Mn2+ ions were 9 μM (circles), 150 μM (triangles), and 1.1 mM (squares). The experiment was performed twice, and essentially the same results were obtained in both cases.
We verified the toxic effects of a high extracellular concentration of Mn2+ ions in ΔmanS and ΔmanR cells by measuring the intracellular concentration of Mn2+ ions. Table 2 shows that elimination of ManS or ManR increased the intracellular level of Mn2+ ions from ∼15 to ∼40 pmol/106 cells when the extracellular concentration of Mn2+ ions was 9 μM and increased it from ∼100 to ∼200 pmol/106 cells when the extracellular concentration of Mn2+ ions was 150 μM. These findings suggested that an increase in the intracellular level of Mn2+ ions might have been toxic and that ΔmanS and ΔmanR cells were unable to adequately control the uptake of Mn2+ ions.
Table 2.
Changes in Levels of Mn2+ Ions in Wild-Type, ΔmanS, and ΔmanR Cells
| Intracellular Level of Mn2+ Ions (pmol/106 cells) |
|||
|---|---|---|---|
| Extracellular (Mn2+) |
Wild Type | ΔmanS | ΔmanR |
| 9 μM | 15 ± 5 | 30 ± 5 | 40 ± 10 |
| 150 μM | 95 ± 20 | 180 ± 30 | 190 ± 50 |
Cells that had been cultivated in BG-11 medium (9 μM MnCl2) were transferred to fresh BG-11 medium or BG-11 medium supplemented with 150 μM MnCl2. Then, levels of Mn2+ ions were measured by atomic absorption spectrometry. The values shown are means of results from three independent experiments with standard deviations.
DISCUSSION
Identification of ManS and ManR by Systematic Genomic Analysis and with a DNA Microarray
The uptake of Mn2+ ions by Synechocystis is enhanced when the extracellular concentration of Mn2+ ions is low (Bartsevich and Pakrasi, 1996). We predicted that a His kinase(s) and a response regulator(s) might be involved in the perception of Mn2+ ions and the transduction of the resulting signal in Synechocystis. The E. coli genome contains 28 putative genes for His kinases and 37 putative genes for response regulators (Mizuno, 1997), whereas the Bacillus subtilis genome contains 36 putative genes for His kinases and 34 putative genes for response regulators (Fabret et al., 1999). Some of these His kinases and response regulators have been characterized, and their functions in the perception and transduction of environmental signals have been clarified (Brown et al., 1995; Jung et al., 2000; Suzuki et al., 2000, 2001; Aguilar et al., 2001; Leonhartsberger et al., 2001). The chromosome of Synechocystis includes 42 genes for His kinases and 40 genes for response regulators (Kaneko et al., 1996; Mizuno et al., 1996). It has been proposed that these proteins act as sensors of environmental stimuli and signal transducers. However, identification of their functions by comparing amino acid sequences with those of proteins encoded by the corresponding genes in E. coli and B. subtilis is difficult because only a few of the His kinases and response regulators in Synechocystis exhibit any obvious homology with proteins from E. coli and B. subtilis. Moreover, unlike the genes in E. coli and B. subtilis, the genes for His kinases and response regulators in Synechocystis are not organized as operons (Kaneko et al., 1996). In addition, the sensor of Mn2+ ions has not been identified in these bacteria.
Que and Helmann (2000) recently identified a transcription factor, MntR, in B. subtilis that regulates, in a repressive manner, the expression of both the mntH gene for the NRAMP-type transporter of Mn2+ ions and the mntABCD operon for the ABC-type transporter of Mn2+ ions. They found that the mntH gene and the mntABCD operon in B. subtilis were expressed constitutively in cells with a mutation in the mntR gene, as was the expression of the mntH gene and the mntABCD operon in wild-type cells when they were subjected to a Mn2+ deficit. A transcription factor homologous with MntR also has been identified in Treponema pallidum (Posey et al., 1999) and in E. coli (Patzer and Hantke, 2001). However, no gene for a transcription factor similar to MntR is present in the genome of Synechocystis.
Our efforts to identify a His kinase and a response regulator that might be involved in the transduction of the Mn2+ signal might be considered a “sledgehammer” approach because it involved the systematic mutation of His kinases and response regulators in combination with screening of the knockout libraries for levels of expression of genes in the mntCAB operon using a DNA microarray. However, using this method, we were able to identify a His kinase and a response regulator as the sensor and the transducer of the Mn2+ signal that regulate the expression of the mntCAB operon.
Kobayashi et al. (2001) used a DNA microarray to analyze the two-component systems in B. subtilis in which individual response regulators were overexpressed when the corresponding His kinase was inactivated. Their analysis identified genes whose expression was regulated by individual two-component systems. However, the sensor of the Mn2+ signal still has not been identified in B. subtilis.
Hypothetical Scheme for the Mn2+ Signal Transduction Pathway in Synechocystis
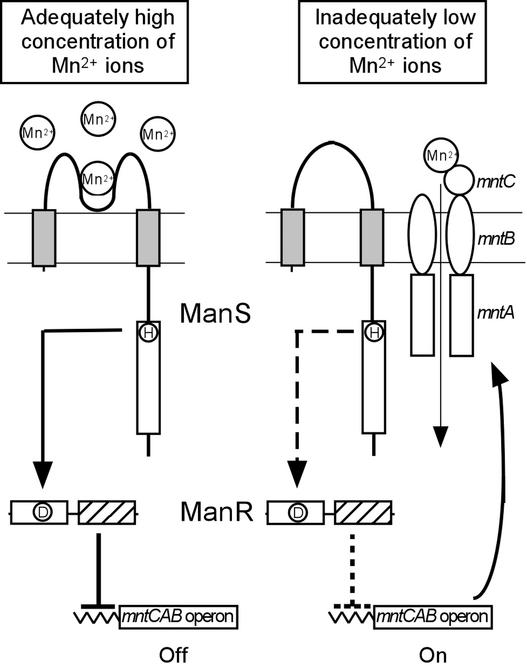
A hypothetical scheme for the regulation of expression of the mntCAB operon by ManS and ManR in Synechocystis is shown in Figure 9. When the extracellular concentration of Mn2+ is adequate (e.g., 9 μM, the standard concentration in BG-11 medium), ManS binds Mn2+ and is activated. Activation might involve phosphorylation of the His residue in the His kinase domain, as observed during the activation of EnvZ in E. coli (Roberts et al., 1994). The activation signal is transmitted to and activates ManR. Activated ManR then represses the transcription of the mntCAB operon. When the extracellular concentration of Mn2+ ions is low, ManS cannot bind Mn2+ ions and remains inactive. Consequently, ManR is not activated, with resulting expression of the mntCAB operon and synthesis of the Mn2+ transporter. In this way, Mn2+ ions are imported to bring the intracellular concentration of Mn2+ ions to an appropriate level.
Figure 9.
Hypothetical Model for the Sensing of Mn2+ Ions, the Transduction of the Mn2+ Signal, and the Regulation of Expression of the mntCAB Operon That Encodes the Mn2+ Transporter.
The presence of two membrane-spanning domains in ManS (closed boxes) was predicted from a hydropathy plot (Kyte and Doolittle, 1982), and the presence of a DNA binding domain in ManR (hatched box) was predicted from the strong similarity of the deduced amino acid sequence to the DNA binding motif of OmpR in E. coli (Martínez-Hackert and Stock, 1997). His and Asp residues that might be involved in a phosphorelay are indicated by the encircled H and D, respectively.
In ΔmanS cells, no activation signal is produced by ManS, irrespective of the extracellular concentration of Mn2+ ions. Thus, ManR remains in an inactive form and the mntCAB operon is expressed constitutively. In ΔmanR cells, ManR cannot repress the mntCAB operon, regardless of the extracellular concentration of Mn2+ ions, and the mntCAB operon is expressed constitutively. This scheme allows us to explain the effects on the expression of the mntCAB operon of the manS and manR mutations (Figure 1) and of a Mn2+ deficit in the medium (Figures 4 and 5). Furthermore, the expression of the mntCAB operon by ManS was activated specifically by Mn2+ ions and not by Zn2+, Co2+, or Cu2+ ions (Table 1). The mutations of the phosphorylatable His residue, His-205, in ManS and of the phosphorylatable Asp residue, Asp-52, in ManR eliminated the transduction of the Mn2+ signal to regulate the expression of the mntCAB operon (Figures 6B and 6D). These observations strongly suggest that a phosphorelay from ManS to ManR plays a critical role in the transduction of the Mn2+ signal. ManR bound to the promoter region of the mntCAB operon in a nonphosphorylated form (Figure 7). The treatment with acetyl phosphate to phosphorylate Asp-52 did not affect the DNA binding ability of ManR in vitro. It seems likely that although nonphosphorylated ManR bound to the promoter region, ManR repressed the transcription in its phosphorylated form. However, it is also a possibility that E. coli might have phosphorylated ManR in vivo and, thus, prephosphorylated ManR bound to the promoter region.
It is unclear why the expression of the mntCAB operon is repressed in Synechocystis cells at high concentrations of Mn2+ ions. It is possible that high concentrations of Mn2+ ions in the cytoplasm are toxic. The results shown in Figure 8 and Table 2 clearly demonstrate the toxic effects of high concentrations of Mn2+ ions. Wild-type Synechocystis cells may cope with the toxic effects of high concentrations of Mn2+ ions by regulating the transport of Mn2+ ions. In both ΔmanS and ΔmanR cells, in which the Mn2+ transporter is induced constitutively, intracellular levels of Mn2+ ions become so high that toxic effects of Mn2+ ions are evident. Thus, the regulatory system that includes ManS and ManR and the Mn2+ transporter is important for the maintenance of the intracellular concentration of Mn2+ ions within a certain range, which is high enough for the formation of the oxygen-evolving machinery but low enough to eliminate any possible toxic effects.
Cation sensors have been reported in several microorganisms. For example, PmrB in Salmonella perceives Fe2+ ions (Wösten et al., 2000), PcoS/CusS in E. coli perceives Cu2+ ions (Brown et al., 1995; Munson et al., 2000), and HydH in E. coli perceives Zn2+ and Pb2+ ions (Leonhartsberger et al., 2001). PhoQ in Salmonella (Véscovi et al., 1996) and KdpD in E. coli (Jung et al., 2000) have been identified as sensors that detect cations, namely, Mg2+ plus Ca2+ and K+, respectively. However, to our knowledge, there have been no previous reports of a sensor of Mn2+ ions.
It has been suggested that the previously identified sensors of metal cations are activated when the concentration of the specific cation exceeds, or falls below, a certain threshold level and that a mutation in such sensors results in repression of the induction of gene expression (Véscovi et al., 1996; Munson et al., 2000; Leonhartsberger et al., 2001). ManS and ManR in Synechocystis, which perceive adequate levels of Mn2+ ions and generate a signal that leads to the repression of gene expression, are examples of a repressor type of two-component system.
The transcriptional repressor MntR in B. subtilis does not include a receiver domain; therefore, it cannot be considered a response regulator (Que and Helmann, 2000). Moreover, the sequence of its DNA binding domain is different from that of the DNA binding domain of ManR from Synechocystis. Indeed, a homology search revealed that the genome of Synechocystis does not contain any gene that is homologous with mntR of B. subtilis. These findings indicate that ManR of Synechocystis and MntR of B. subtilis are very different proteins that, nonetheless, play similar roles in regulating the expression of genes for transporters of Mn2+ ions.
In Arabidopsis, there are 11 genes for His kinases. However, none of these genes is homologous with the manS gene; therefore, none of the His kinases is likely to perceive Mn2+ signals. The mechanism of the perception and transduction of Mn2+ signals in higher plants might differ from that in Synechocystis.
METHODS
Strains and Culture Conditions
Two strains of Synechocystis sp PCC 6803 were used. The Glc-tolerant strain (Williams, 1988) was provided by J.G.K. Williams (DuPont de Nemours Company, Inc., Wilmington, DE), and the Glc-sensitive strain was obtained from the Pasteur Institute (Paris, France). We generated a knockout library of His kinases (Suzuki et al., 2000) using the Glc-tolerant strain and a knockout library of response regulators using the Glc-sensitive strain. The mutant defective in ManS, designated Hik27, is described on the CyanoGenes World Wide Web site at http://www.kazusa.or.jp/cyano/Synechocystis/comments/cgi-bin/comshow.cgi?id=slr0640&Kwd=Hik27, and the mutant defective in ManR, designated Rre16, is described on the CyanoGenes World Wide Web site at http://www.kazusa.or.jp/cyano/Synechocystis/comments/cgi-bin/comshow.cgi?id=slr1837&Kwd=rre16.
Cyanobacterial cells were grown at 34°C in BG-11 medium (Stanier et al., 1971) buffered with 20 mM Hepes-NaOH, pH 7.5, under continuous illumination from incandescent lamps, as described previously (Wada and Murata, 1989). Depletion of Mn2+, Zn2+, Co2+, or Cu2+ ions in the culture medium was achieved by gel filtration as follows. A 4-mL culture of Synechocystis was loaded onto a column that had been filled with 14 mL of Sephadex G-25 (medium gel; Amersham Pharmacia Biotech, Piscataway, NJ) and equilibrated with BG-11 medium prepared without MnCl2, ZnCl2, CoCl2, or CuCl2. Subsequently, 5 mL of BG-11 depleted of MnCl2, ZnCl2, CoCl2, or CuCl2 was loaded on the column. The flow-through fraction that contained Synechocystis cells was collected and used to inoculate BG-11 medium prepared without MnCl2, ZnCl2, CoCl2, or CuCl2, and the culture was incubated for 1 or 2 days under the conditions described above.
Quantitation of Intracellular Mn2+ Ions
Ten-milliliter aliquots of cell cultures were collected, and cells were harvested by centrifugation at 3000g at 30°C. Cells were washed twice in 20 mL of MilliQ-grade water, pelleted by centrifugation as noted above, and resuspended in 5 mL of water. Cells were killed by immersion of the suspension in boiling water for 10 min. The amount of Mn2+ ions in each suspension was determined with an atomic absorption spectrometer (model 207; Hitachi, Tokyo, Japan).
DNA Microarray Analysis
A DNA microarray, CyanoCHIP, was obtained from Takara Shuzo (Kyoto, Japan). This microarray covers 3094 genes (94% of the total genes) of Synechocystis (Kaneko et al., 1996). cDNAs, labeled with fluorescent dyes (Cy3 and Cy5; Amersham Pharmacia Biotech), were prepared from 10 or 20 μg of total RNAs with a CyScribe first-strand cDNA-labeling kit (Amersham Pharmacia Biotech) or an RNA fluorescence labeling core kit (Moloney murine leukemia virus version; Takara Shuzo) according to the manufacturers' instructions. The microarray was incubated first, for prehybridization, at 65°C for 1 h in a solution that contained 4 × SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate), 0.2% SDS, 5 × Denhardt's solution (1× Denhardt's solution is 0.02% Ficoll, 0.02% polyvinylpyrrolidone, and 0.02% BSA), and 100 ng/μL salmon sperm DNA. Then it was rinsed once with 2 × SSC and once with 0.2 × SSC at room temperature. Hybridization of the mixture of Cy3- and Cy5-labeled cDNAs that had been immobilized on the DNA microarray was allowed to proceed at 65°C for 16 h in 20 μL of 4 × SSC, 0.2% SDS, 5 × Denhardt's solution, and 100 ng/μL denatured salmon sperm DNA. After hybridization, the microarray was rinsed with 2 × SSC at room temperature and washed with 2 × SSC at 60°C for 10 min, with 0.2 × SSC plus 0.1% SDS at 60°C for 10 min, and finally with 0.2 × SSC at room temperature. After the final rinse, the microarray was dried with an air spray before scanning with an array scanner (GMS 418; Affymetrix, Woburn, MA). The intensity of each signal was determined with scan-analyzing software (Imagene version 4.1; BioDiscovery, Los Angeles, CA).
Transformation with the manS, manR, Mutated manS, and Mutated manR Genes
DNA fragments that contained the manS and manR genes were amplified by PCR with the genomic DNA of Synechocystis as a template and synthetic oligonucleotides as primers: 5′-CTCGAGCTAACA-TTTCTAAGG-3′ (P1) and 5′-CTCGAGTGTTGTTCAGTGAGC-3′ (P2) for manS and 5′-CTCGAGTTTCAACAACAACCG-3′ (P3) and 5′-CTCGAGGCCTTTCTTCCCAAG-3′ (P4) for manR. The resulting DNA fragments were of 1624 and 1021 bp and included 204- and 200-bp sequences upstream of the site of initiation of translation of the manS and manR genes, respectively. They were cloned into the pCR2.1-TOPO plasmid using a TOPO TA cloning kit (Invitrogen, Tokyo, Japan) to obtain pCRmanS and pCRmanR.
We generated a mutated gene for ManS(H205A) by PCR-based mutagenesis (Barik, 1993). At the first step, two kinds of PCR amplification were conducted with pCRmanS as a template and the following sets of primers: primer P1 and 5′-CGCAATGGGGTTTCT-AAGTTCGGCGGAGGC-3′, which contained the site of H205A in which the codon CAC for His had been replaced by GCC for Ala, and 5′-TTTTCGCGATAATGACGC-3′ and primer P2. Then, the second step of PCR amplification was performed with the resulting products of the first step as templates and P1 and P2 as primers. We also generated a mutated gene for ManR(D52A) by PCR-based mutagenesis. At the first step, two kinds of PCR amplification were conducted with pCRmanR as a template and the following sets of primers: primer P3 and 5′-GGGCAACATCCAAGCAAGGATTAACAAGTC-3′, which contained the site of D52A in which the codon CAT for His had been replaced by GCT for Ala; and 5′-GACTTGTTAATCCTTGCTTGGATG-TTGCCC-3′, which contained the same mutation, and primer P4. Then, the second step of PCR amplification was performed with the resulting products of the first step as templates and P3 and P4 as primers. These products of PCR were cloned into the pCR2.1-TOPO plasmid to obtain pCRmanR-mutated and pCRmanR-mutated.
manS, manR, mutated manS, and mutated manR were excised from pCRmanS, pCRmanR, pCRmanS-mutated, and pCRmanR-mutated, respectively, with XhoI and inserted into a cyanobacterial autonomous replication plasmid, pVZ321 (Zinchenko et al., 1999), which had been digested with XhoI. The resulting plasmids were introduced into ΔmanS and ΔmanR cells by triparental gene transfer (Zinchenko et al., 1999).
Genomic PCR for Examination of the Segregation of Wild-Type Genes in Mutant Cells
Chromosomal DNA was extracted from wild-type, ΔmanS, and ΔmanR cells and purified with ISOPLANT II (Nippon Gene, Osaka, Japan) according to the manufacturer's instructions. The primers used for PCR for amplification of the gene for ManS were 5′-CAC-TCATGATTCAGGCCACCCGT-3′ and 5′-CTACTCGAGCTATCCAAG-AGGAGCGGC-3′, and those for ManR were 5′-TTCCTCGAGTTT-CAACAACAACCG-3′ and 5′-ATACTCGAGGCCTTTCTTCCCAAG-3′.
Quantitative Reverse Transcription PCR
Quantitative reverse transcription PCR was performed with the GeneAmp 5700 system (Perkin-Elmer Biosystems, Foster City, CA) and a SYBR Green Assay kit (Perkin-Elmer Biosystems) according to the instructions from the manufacturer. Reverse transcription reactions were performed in a 100-μL reaction mixture that contained 1 μg of total RNA, 50 mM Tris-HCl, pH 8.3, 10 mM KCl, 4 mM DTT, 10 mM MgCl2, 0.5 mM dATP, 0.5 mM dTTP, 0.5 mM dCTP, 0.5 mM dGTP, 300 pmol of random hexamers, 100 units of ribonuclease inhibitor, and 120 units of reverse transcriptase XL from adenovirus (Takara Shuzo). After incubation at 25°C for 10 min, the reverse transcription reaction was allowed to proceed at 48°C for 30 min. The reaction was stopped by heating at 95°C for 5 min. The primers used for quantitative reverse transcription PCR were 5′-GCAGTCTACGCGAGCAGTTAAAG-3′ and 5′-GCTCCTTCGCAACTCACCAA-3′ and were based on the sequence of the mntC gene (Bartsevich and Pakrasi, 1995).
Overproduction in Escherichia coli and Purification of His-Tagged ManR
The nucleotide sequence of the manR gene was obtained from the CyanoBase World Wide Web site at http://www.kazusa.or.jp/cyano/Synechocystis/cgi-bin/geinfo.cgi?type=orf&name=slr1837. The chromosomal DNA of Synechocystis was extracted from wild-type cells, purified with ISOPLANT II, and used as a template for amplification by PCR. A 0.77-kb fragment of DNA that included the manR gene was amplified by PCR with the primers 5′-CATATGGCC-AATATCCTCTTAGTGGACG-3′ and 5′-GGATCCCTAATTGGCCTC-AAAACAGTACC-3′. The first primer was designed to introduce a NdeI site with an ATG codon for the initiation of translation, and the second primer was designed to introduce a BamHI site with a TAG codon for the termination of translation. These newly introduced sites are underlined above. The amplified DNA fragment was ligated into the TA cloning vector pT7Blue-T (Novagen, Madison, WI). The resulting plasmid was used for transformation of E. coli JM109. Both strands of the cloned fragment were sequenced with an automated DNA sequencer (ABI310; Perkin-Elmer Biosystems) to confirm the sequence of the PCR product. The plasmid was digested with NdeI and BamHI, and the resulting 770-bp DNA fragment was cloned into the vector pET16b (Novagen) for subsequent expression of a fusion protein with a His tag at its N terminus, after pET16b had been digested with NdeI and BamHI. The product was designated pETmanR and used for the transformation of E. coli BL21(DE3)pLysS. Transformed cells were grown at 37°C in 400 mL of Terrific broth (1.2% tryptone, 2.4% yeast extract, 0.4% glycerol, 72 mM K2HPO4, and 17 mM KH2PO4) supplemented with 200 μg/mL carbenicillin and 60 μg/mL chloramphenicol. When the absorbance at 600 nm of the culture reached 0.6, isopropyl β-d-thiogalactopyranoside was added to a final concentration of 0.4 mM, and the culture was incubated for an additional 6 h at 20°C.
All procedures for purification of the fusion protein were performed at 0 to 4°C. The transformed E. coli cells were disrupted by sonication (Sonifier 250; Branson, Danbury, CT) for 5 min, with six pauses of 1 min each, on ice in 20 mL of 20 mM Tris-HCl buffer, pH 7.9, that contained 5 mM imidazole, 0.5 M NaCl, and 1 mM phenylmethylsulfonyl fluoride. The lysate was centrifuged at 11,000g for 15 min, and then the supernatant was centrifuged at 16,000g for 30 min. The resulting supernatant was loaded onto a 2-mL His•Bind Resin column (Novagen), and affinity chromatography was performed according to the manufacturer's instructions (His•Bind Kits; Novagen). Fractions that contained the fusion protein were desalted by passing them through a prepacked Sephadex G-25M column (PD-10; Amersham Pharmacia Biotech), which had been equilibrated with 50 mM Tris-HCl buffer, pH 8.0, that contained 5 mM EDTA (disodium salt), 50 mM KCl, and 10% (v/v) glycerol. The eluate was frozen in liquid N2 and stored at −80°C before use.
Concentrations of protein were determined as described by Bradford (1976) with BSA as the standard. The purity of the fusion protein was examined by SDS-PAGE (Laemmli, 1970).
Gel Mobility Shift Assay
A 135-bp DNA fragment that corresponded to positions −135 to −1 upstream of the site of the initiation of translation of the mntC gene was synthesized by PCR amplification with primers 5′-GGGGCT-CGAGCAGAAAGTTTTAGTGGTCAG-3′ and 5′-TTTTAAGCTTAGTGA-TAATGGTCTTTCAT-3′. A XhoI site in the first primer and a HindIII site in the second primer are underlined. The resulting fragment of DNA was digested with HindIII and XhoI and then was end labeled with α-32P-dCTP (6000 Ci/mmol; Amersham Pharmacia Biotech) using the Klenow fragment of DNA polymerase (Takara Shuzo). Unincorporated radioisotope was removed by gel-filtration chromatography on a Centri-Sep spin column (Perkin-Elmer Biosystems). Approximately 7.7 ng (0.08 pmol) of the end-labeled DNA fragment (∼2000 cpm) was mixed with 0.25 μg (9.1 pmol) or 0.5 μg (18.2 pmol) of His-tagged ManR, which had been incubated previously in the presence of 100 mM acetyl phosphate or in its absence in 20 μL of 50 mM Tris-HCl buffer, pH 8.0, that contained 1 mM EDTA, 150 mM KCl, 1 mM DTT, 1 μg/mL poly(dI-dC) (Amersham Pharmacia Biotech), and various amounts of nonlabeled DNA fragment. Each reaction mixture was incubated at 25°C for 20 min and then loaded onto a 6% nondenaturing polyacrylamide gel. Electrophoresis was performed at 4°C in TAE buffer (40 mM Tris-acetate and 1 mM EDTA). Then the gel was dried and analyzed with a Bio-Image analyzer (BAS 2000; Fuji Photo Film, Tokyo, Japan).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purpose. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Acknowledgments
This work was supported, in part, by Grants-in-Aid for Scientific Research (Grant 13854002 to N.M.), for Scientific Research on Priority Areas (Genome Biology; Grant 13206081 to I.S.), and for the Genome Frontier Project (to M.K.) from the Ministry of Education, Science, Sports, and Culture of Japan, and by grants from the Russian Foundation for Basic Research (Grants 01-04-48081 to V.Z. and 00-04-48421a to D.A.L.).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.006262.
References
- Aguilar, P.S., Hernandez-Arriaga, A.M., Cybulski, L.E., Erazo, A.C., and de Mendoza, D. (2001). Molecular basis of thermosensing: A two-component signal transduction thermometer in Bacillus subtilis. EMBO J. 20, 1681–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barik, S. (1993). Site-directed mutagenesis by double polymerase chain reaction: Megaprimer method. In Methods in Molecular Biology, Vol. 15, B.A. White, ed (Clifton, NJ: Humana Press), pp. 277–286. [DOI] [PubMed]
- Bartsevich, V.V., and Pakrasi, H.B. (1995). Molecular identification of an ABC transporter complex for manganese: Analysis of a cyanobacterial mutant strain impaired in the photosynthetic oxygen evolution process. EMBO J. 14, 1845–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsevich, V.V., and Pakrasi, H.B. (1996). Manganese transport in the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 271, 26057–26061. [DOI] [PubMed] [Google Scholar]
- Bartsevich, V.V., and Pakrasi, H.B. (1999). Membrane topology of MntB, the transmembrane protein component of an ABC transporter system for manganese in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 181, 3591–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barynin, V.V., Whittaker, M.M., Antonyuk, S.V., Lamzin, V.S., Harrison, P.M., Artymiuk, P.J., and Whittaker, J.W. (2001). Crystal structure of manganese catalase from Lactobacillus plantarum. Structure 9, 725–738. [DOI] [PubMed] [Google Scholar]
- Borgstahl, G.E.O., Pokross, M., Chehab, R., Sekher, A., and Snell, E.H. (2000). Cryo-trapping the six-coordinate, distorted-octahedral active site of manganese superoxide dismutase. J. Mol. Biol. 296, 951–959. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Brown, N.L., Barrett, S.R., Camakaris, J., Lee, B.T., and Rouch, D.A. (1995). Molecular genetics and transport analysis of the copper-resistance determinant (pco) from Escherichia coli plasmid pRJ1004. Mol. Microbiol. 17, 1153–1166. [DOI] [PubMed] [Google Scholar]
- Fabret, C., Feher, V.A., and Hoch, J.A. (1999). Two-component signal transduction in Bacillus subtilis: How one organism sees its world. J. Bacteriol. 181, 1975–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frausto da Silva, J.J.R., and Williams, R.J.P. (1991). The Biological Chemistry of the Elements: The Inorganic Chemistry of Life. (New York: Oxford University Press).
- Jung, K., Veen, M., and Altendorf, K. (2000). K+ and ionic strength directly influence the autophosphorylation activity of the putative turgor sensor KdpD of Escherichia coli. J. Biol. Chem. 275, 40142–40147. [DOI] [PubMed] [Google Scholar]
- Kaneko, T., et al. (1996). Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3, 109–136. [DOI] [PubMed] [Google Scholar]
- Kobayashi, K., Ogura, M., Yamaguchi, H., Yoshida, K., Ogasawara, N., Tanaka, T., and Fujita, Y. (2001). Comprehensive DNA microarray analysis of Bacillus subtilis two-component regulatory systems. J. Bacteriol. 183, 7365–7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte, J., and Doolittle, R.F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Larson, E.J., and Pecoraro, V.L. (1992). Manganese Redox Enzymes. (New York: VCH Publishers).
- Leonhartsberger, S., Huber, A., Lottspeich, F., and Böck, A. (2001). The hydH/G genes from Escherichia coli code for a zinc and lead responsive two-component regulatory system. J. Mol. Biol. 307, 93–105. [DOI] [PubMed] [Google Scholar]
- Mann, N., and Carr, N.G. (1974). Control of macromolecular composition and cell division in the blue-green alga Anacystis nidulans. J. Gen. Microbiol. 83, 399–405. [DOI] [PubMed] [Google Scholar]
- Martínez-Hackert, E., and Stock, A.M. (1997). The DNA-binding domain of OmpR: Crystal structures of a winged helix transcription factor. Structure 15, 109–124. [DOI] [PubMed] [Google Scholar]
- Mizuno, T. (1997). Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res. 4, 161–168. [DOI] [PubMed] [Google Scholar]
- Mizuno, T., Kaneko, T., and Tabata, S. (1996). Compilation of all genes encoding bacterial two-component signal transducers in the genome of the cyanobacterium, Synechocystis sp. strain PCC 6803. DNA Res. 3, 407–414. [DOI] [PubMed] [Google Scholar]
- Munson, G.P., Lam, D.L., Outten, F.W., and O'Halloran, T.V. (2000). Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J. Bacteriol. 182, 5864–5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzer, S.I., and Hantke, K. (2001). Dual repression by Fe2+-Fur and Mn2+-MntR of the mntH gene, encoding an NRAMP-like Mn2+ transporter in Escherichia coli. J. Bacteriol. 183, 4806–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posey, J.E., Hardham, J.M., Norris, S.J., and Gherardini, F.C. (1999). Characterization of a manganese-dependent regulatory protein, TroR, from Treponema pallidum. Proc. Natl. Acad. Sci. USA 96, 10887–10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que, Q., and Helmann, J.D. (2000). Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol. Microbiol. 35, 1454–1468. [DOI] [PubMed] [Google Scholar]
- Roberts, D.L., Bennett, D.W., and Forst, S.A. (1994). Identification of the site of phosphorylation on the osmosensor, EnvZ, of Escherichia coli. J. Biol. Chem. 269, 8728–8733. [PubMed] [Google Scholar]
- Solioz, M., and Vulpe, C. (1996). CPx-type ATPases: A class of P-type ATPases that pump heavy metals. Trends Biochem. Sci. 21, 237–241. [PubMed] [Google Scholar]
- Stanier, R.Y., Kunisawa, R., Mandel, M., and Cohen-Bazire, G. (1971). Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 35, 171–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, I., Kanesaki, Y., Mikami, K., Kanehisa, M., and Murata, N. (2001). Cold-regulated genes under control of the cold sensor Hik33 in Synechocystis. Mol. Microbiol. 40, 235–244. [DOI] [PubMed] [Google Scholar]
- Suzuki, I., Los, D.A., Kanesaki, Y., Mikami, K., and Murata, N. (2000). The pathway for perception and transduction of low-temperature signals in Synechocystis. EMBO J. 19, 1327–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Véscovi, E.G., Soncini, F.C., and Groisman, E.A. (1996). Mg2+ as an extracellular signal: Environmental regulation of Salmonella virulence. Cell 84, 165–174. [DOI] [PubMed] [Google Scholar]
- Wada, H., and Murata, N. (1989). Synechocystis PCC6803 mutants defective in desaturation of fatty acid. Plant Cell Physiol. 30, 971–978. [Google Scholar]
- Williams, J.G.K. (1988). Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol. 167, 766–778. [Google Scholar]
- Wösten, M.M., Kox, L.F., Chamnongpol, S., Soncini, F.C., and Groisman, E.A. (2000). A signal transduction system that responds to extracellular iron. Cell 103, 113–125. [DOI] [PubMed] [Google Scholar]
- Yachandra, V.K., DeRose, V.J., Latimer, M.J., Mukerji, I., Sauer, K., and Klein, M.P. (1993). Where plants make oxygen: A structural model for the photosynthetic oxygen-evolving manganese cluster. Science 260, 675–679. [DOI] [PubMed] [Google Scholar]
- Zinchenko, V.V., Piven, I.V., Melnik, V.A., and Shestakov, S.V. (1999). Vectors for the complementation analysis of cyanobacterial mutants. Russian J. Genet. 35, 228–232. [Google Scholar]
- Zouni, A., Witt, H.T., Kern, J., Fromme, P., Krauss, N., Saenger, W., and Orth, P. (2001). Crystal structure of photosystem II from Synechococcus elongatus at 3.8 Å resolution. Nature 409, 739–743. [DOI] [PubMed] [Google Scholar]