Abstract
Scp160p is a multiple KH-domain RNA-binding protein in yeast that has been demonstrated previously to associate with both soluble and membrane-bound polyribosomes as an mRNP component. One key question that has remained unanswered, however, is whether the mRNAs in these mRNP complexes are random or specific. We have addressed this question using microarray analyses of RNAs released from affinity isolated Scp160p-containing complexes, compared with total RNA controls from the same lysates. Our results, confirmed by quantitative RT–PCR analysis, clearly demonstrate that Scp160p associates with specific rather than with random messages, and that among the enriched targets are DHH1, YOR338W, BIK1, YOL155C and NAM8. Furthermore, loss of Scp160p resulted in a significant change in both the abundance and distribution between soluble and membrane-associated fractions for at least one of these messages (YOR338W), and in a subtle yet significant shift from soluble polyribosomes to soluble mRNPs for at least two of these target messages (DHH1 and YOR338W). Together, these data not only identify specific mRNA targets associated with Scp160p in vivo, they demonstrate that the association of Scp160p with these messages is biologically relevant.
INTRODUCTION
The gene SCP160 encodes a 160 kDa protein (Scp160p) originally postulated to function in the maintenance of ploidy in yeast (1). More recently, however, a variety of experimental approaches have all demonstrated that Scp160p associates with polyribosomes as a component of large cytoplasmic complexes, believed to be mRNPs (2–5). In addition to Scp160p, these complexes also contain the polyA binding protein Pab1p, and Bfr1p (4). As would be expected of mRNPs, these complexes are resistant to EDTA, but sensitive to both RNase and high salt (3,4). Together, these data support the hypothesis that Scp160p functions at some level of cytoplasmic mRNA metabolism, and that the scp160 null phenotype, which includes abnormal cell size and shape, increased DNA content, and missegregation of genetic markers through meiosis, may reflect the indirect result of aberrant target gene regulation, rather than a direct loss of Scp160p function from many different biological processes.
Subcellular fractionation studies have demonstrated that Scp160p partitions between the soluble and membrane-bound compartments (2,4,5). Similarly, fluorescence microscopy studies using both anti-Scp160p antibodies and GFP-tagged Scp160p, have demonstrated that while some diffuse signal is evident in the cytosol, a significant enrichment of signal is seen around the nuclear envelope (1,4,5), which is the site of the endoplasmic reticulum in yeast. Finally, localization of Scp160p to the endoplasmic reticulum has been demonstrated to be both RNA-dependent (4), and microtubule-dependent (5). Together, these data support the conclusion that Scp160p associates with both soluble and rough endoplasmic reticulum-bound polyribosomes in vivo.
Although little is currently known about the structure of the Scp160p protein, sequence alignment studies have revealed the presence of 14 tandem copies of the hnRNP K homology (KH) domain (2,6), a highly conserved motif found in many RNA-binding proteins (7). Indeed, Scp160p demonstrates significant amino acid sequence homology to a large and extended family of multiple KH-domain proteins, collectively known as vigilins (3,8–12). Although all vigilin proteins studied to date have been reported to bind nucleic acid, in most cases both the type of nucleic acid bound, and the functional significance of the interaction, remain unclear. One notable exception is Xenopus vigilin, which was demonstrated recently not only to bind specifically to a defined sequence in the 3′ untranslated region of the vitellogenin message, but also to inhibit cleavage of that sequence by the mRNA endonuclease polysomal ribonuclease 1 (13). In vitro studies have previously demonstrated that Scp160p can bind directly to ribohomopolymers, as well as to yeast ribosomal RNA, but not to tRNA (2). Although both we and others have hypothesized previously that Scp160p associates with mRNAs in vivo (2–5), whether those mRNAs are random or specific, and whether these associations are biologically significant, has remained unclear.
We report here the results of experiments that directly address both of these questions. In brief, we have asked (i) Do Scp160p-associated mRNPs contain random or specific subsets of yeast messages, and, if specific, what are they? and (ii) Is there any detectable impact of scp160 loss on its target messages? To address the first question, we utilized affinity isolation of Scp160p-associated mRNPs, followed by microarray and quantitative RT–PCR analyses of the mRNAs released from these complexes. We found not only that yeast mRNA sequences are present in these samples, but also that the sequences present are specific, not random. To address the second question, we used quantitative RT–PCR analyses of the RNAs from cell lysates as well as from defined sucrose gradient fractions representing both wild-type and scp160-null cells to demonstrate a significant shift from the membrane fraction to the soluble fraction for one Scp160p-associated mRNA, and a subtle yet significant shift in the polyribosome association profiles of at least two Scp160p-associated mRNAs relative to a non-target control. Together, these results not only confirm that Scp160p associates with specific mRNAs in yeast, but also that these interactions are biologically meaningful.
MATERIALS AND METHODS
Yeast strains and culture conditions
The yeast strains used in this study included JJ52 (MATα gal7Δ102 ura3-52 trp1-289 ade1 lys1 leu2-3,112, a generous gift from Drs Mark Parthun and Judith Jaehning, University of Colorado Health Sciences Center) and JFy1511, which was derived from JJ52 by substitution of an N-terminally FLAG-tagged allele of SCP160 in place of the wild-type allele (3). All studies comparing wild-type versus scp160-null cells were performed using diploid strains of W303-derived cells homozygous for a genomic scp160 deletion, that either did (JFy4100), or did not, carry a plasmid borne copy of wild-type SCP160 (JF3116, URA3), respectively. Due to concerns over potential and progressive aneuploidy in the scp160-null strains, these strains were always generated fresh from JFy4100 just prior to use by plasmid curing on medium containing 5-fluororotic acid (5FOA) (14).
Affinity isolation of Scp160p-containing complexes
Scp160p-containing complexes were isolated from the yeast strain JFy1511 by virtue of a FLAG-tag engineered onto the N-terminus of Scp160p, as described previously (3). In brief, cells were grown to early log phase (OD600 ∼1) in rich medium, harvested by centrifugation, and lysed in T75 buffer (25 mM Tris pH 7.5, 75 mM NaCl) containing 30 mM EDTA to disrupt the polyribosomes. After centrifugation, the supernatant was passed over an S-300 gel-filtration column, and the void material (>1300 kDa) was pooled and loaded onto a 1 ml M2 α-FLAG column (Sigma Aldrich). Finally, after washing the column with 100 ml of T75 buffer, the FLAG-Scp160p-containing complexes were eluted using 184 mg/ml FLAG peptide (asp-tyr-lys-asp-asp-asp-asp-lys, purchased from the Emory Microchemical Facility) in T75 buffer.
32P-labeled first-strand cDNA synthesis
Total RNA derived from Scp160p-containing complexes by the method of Schmitt et al. (15) was used as template in a first-strand cDNA synthesis reaction using Maxiscript™ reverse transcriptase (Ambion) and [α-32P]dCTP, as recommended by the manufacturer. After a 10 min incubation at 37°C, each reaction was subjected to centrifugation through a microspin™ G-50 column (Amersham Pharmacia Biotech) to remove unincorporated nucleotides. Finally, each sample was mixed with loading buffer (300 mM NaOH, 6 mM EDTA, 18% Ficoll, 0.25% xylene cyanol, 0.15% bromocresol green), electrophoresed through a 1.2% agarose gel under alkaline denaturing conditions (50 mM NaOH, 1 mM EDTA), dried, and visualized by autoradiography.
Microarray analysis of Scp160p-associated RNAs
RNA was prepared as described previously (15) from both affinity-isolated Scp160p-containing complexes, and from the corresponding whole cell lysates, and used as template for the production of double-stranded cDNA, using the Superscript Choice™ system (Invitrogen), as recommended by the manufacturer. Following isolation, this cDNA was used as template for the production of biotin-labeled cRNA, using the RNA Transcript Labeling kit (ENZO), as recommended by the manufacturer. Finally, the labeled cRNA was purified using RNeasy spin columns (Qiagen), fragmented (using 40 mM Tris–acetate, pH 8.1, 100 mM KOAc, 30 mM MgOAc), and hybridized with Affymetrix Yeast Genome S98 chips, as recommended by the manufacturer. After washing, staining, and scanning the probed arrays, database profiles representing the Scp160p-associated versus corresponding total RNA controls were compared electronically, as recommended by the manufacturer.
Quantitative RT–PCR
As an independent approach to test the relative abundance of candidate target messages in different RNA pools (Scp160p-associated versus total, or specific sucrose gradient or subcellular fractions), we performed quantitative RT–PCR using a LightCycler (Roche), as recommended by the manufacturer. In brief, RNA was isolated from the desired samples as described previously (15), and used as template for the generation of single-stranded cDNA using oligo-dT as a primer and Superscript™II RNase H– reverse transcriptase (Invitrogen). Next, equivalent amounts of total cDNA were used as template in PCR reactions with different primer sets, each designed to amplify a 210–361 bp gene-specific fragment in the LightCycler, in parallel with a dilution series designed to serve as a standard curve for quantitation in each experiment. The correlation coefficient for each standard curve was ≥0.99. The primers used were: for YGR023W (YCR023WF1, 5′-ATTGTCACAGCTTATCGTAGTA-3′ and YGR023WR1, 5′-ATCTATGCCTCTGTTATGGAAG-3′), for YOR338W (YOR338F1, 5′-ATAATGTCCCCAAGTATATTCC-3′ and YOR338R1, 5′-AAAGATCACATGGAAGTCTCAG-3′), for YOL155C (YOL155CF1, 5′-TCAAGATAATCACCTCTCAAATA-3′ and YOL155CR1, 5′-AAGTATTGGCATTCAAACCTGC-3′), for YDR247W (YDR247WF1, 5′-GTCTCCAAAATCCTACGTGTA-3′ and YDR247WR1, 5′-GTTGGAAATCTTAGAACATTCGT-3′), for YBR150C (YBR150CF1, 5′-TCTTGCAATTTTCAAGCATTCC-3′ and YBR150CR1, 5′-TATTGTTACTGTTACTGTAGTTAT-3′), for YHR086W (NAM8, YHR086WF1, 5′-GAGTTGACATTGTAGGAGATAT-3′ and YHR086 WR1, 5′-GACACAGCTTCGTCAGGTCT-3′), for YDL 160C (DHH1, DHH1F2, 5′-ATAAGTCATTGTATGTGGCTGA-3′ and DHH1R2, 5′-AATGTTCCTGCTGTGGAGGA-3′), for YOL059W (YOL059WF1, 5′-TACTATCAAGAATCCGCTGGT-3′ and YOL059WR1, 5′-GTAGACTATCTGGTAGACTGC-3′), for YCL029C (BIK1, YCL029CF1, 5′-AACGATTCGCTCAGTAAAGAAT-3′ and YCL029CR1, 5′-ATCGCAATGCTCACACCACTG-3′), for YKL203C (TOR2, TOR2F1 5′-CGCTCGTTACGCATCTCATTT-3′ and TOR2R1, 5′-CCAATGCCACCATTACCACA-3′), for YBL109W (YBL109WF1, 5′-CCTAACACT ACCCTAACCCTA-3′ and YBL109WR1, 5′-CAGATGGTGGATGGTAGAGT-3′), for YGR110W (YGR110WF1, 5′-TGGACAGCAGCTATTCGAT-3′ and YGR110WR1, 5′-GTTATCCAGGTACAAGTGATG-3′), and for YHR174W (ENO2, YHR174WT3F4, 5′-GCGCGGAATTAACCCTCACTAAAGGGAACGAGCTCGAAGGCTGCTGACGCTTTG-3′ and YHR174WT7R4, 5′-GCGCGGTAATACGACTCACTATAGGGAGGGGATCCTTACAACTTGTCACCGTGGTG-3′).
Calculations to determine fold enrichment of specific sequences in Scp160p complexes
The enrichment values for candidate Scp160p-associated messages listed in Table 1 were calculated as follows. First, ratios were generated using as numerator the data values from microarrays hybridized with probes representing RNAs from FLAG-Scp160p affinity isolated complexes, and as denominator the corresponding values from microarrays hybridized with probes representing total RNAs from the same whole cell lysates (prior to affinity isolation). Next, ratios were generated from data sets representing mock affinity isolated complexes versus total RNAs derived from cells expressing native rather than FLAG-tagged Scp160p. The values listed in Table 1 represent the fold enrichment calculated for each candidate sequence using the FLAG-tagged cell preparations, corrected for any background detected using the untagged cell preparations.
Table 1. Yeast mRNA and ORF sequences enriched ≥2.5-fold in Scp160p-containing complexes relative to total mRNA (data from duplicate microarray analyses).
| Sequence identifier | Gene name | Fold increase | Probe set | Comments |
|---|---|---|---|---|
| SCIIYLEFT_6 | – | 10.5, 4.5 | 6949_f_at | Saccharomyces cerevisiae chromosome II, complete chromosome sequence. Found forward in NC_001134 between 6001 and 6215 |
| NGR056C | – | 8.8, 3.2 | 4653_at | SAGE ORF found reverse in NC_001139 between 708217 and 708372 (16) |
| *YGR023W | MTL1 | 6.8, 4.4 | 4996_at | Acts in concert with Mid2p to transduce cell wall stress signals |
| YOR338W | ORF | 6.2, 4.5 | 8228_at | Similarity to YAL034c |
| YML062C | MFT1 | 5.9, 3.2 | 9692_at | Protein involved in mitochondrial import of fusion proteins |
| gJL04_0 | – | 5.6, 2.9 | 3128_at | Saccharomyces cerevisiae chromosome X, complete chromosome sequence. Found forward in NC_001142 between 196825 and 197824 |
| YER188C | – | 5.4, 3.9 | 3325_f_at | Strong similarity to subtelomeric encoded proteins |
| YOL155C | ORF | 5.4, 3.4 | 8724_at | Similarity to glucan 1,4-glucosidase Sta1p and YAR066W |
| *YDR247W | ORF | 5.3, 4.6 | 6212-at | Strong similarity to Sks1p |
| *YBR150C | TBS1 | 5.1, 4.1 | 7196_at | Probable Zn-finger protein |
| YOL054W | PSH1 | 5.0, 3.4 | 8597_at | Weak similarity to transcription factors |
| NGR053C | – | 4.8, 3.1 | 4652_at | SAGE ORF found reverse in NC_001139 between 707915 and 708058 (16) |
| YJR151C | DAN4 | 4.8, 2.9 | 10853_at | Similarity to mucin proteins, YKL224c, Sta1p |
| YPR065W | ROX1 | 4.7, 2.5 | 7685_at | Site-specific DNA binding protein, repressor |
| YJL116C | NCA3 | 4.6, 2.8 | 11133_at | With NCA2, regulates proper expression of subunits 6 (Atp6p) and 8 (Atp8p) of the Fo-F1 ATP synthase |
| NGR052C | – | 4.5, 2.9 | 4651_at | Non-annotated SAGE ORF found reverse in NC_001139 between 707583 and 707720 (16) |
| YGL125W | MET13 | 4.4, 4.3 | 5117_at | Putative methylenetetrahydrofolate reductase |
| YDR173C | ARG82 | 4.2, 3.3 | 6319-at | Regulator of arginine-responsive genes with ARG80 and ARG81 |
| YGL128C | CWC23 | 4.2, 2.6 | 5160_at | Weak similarity to rat cysteine string protein |
| YDR162C | NBP2 | 4.2, 2.5 | 6308_at | Nap1p-binding protein |
| YGL139W | ORF | 4.0, 2.6 | 5150_at | Strong similarity to hypothetical protein YPL221w |
| YBL081W | ORF | 4.0, 3.7 | 7461_at | Hypothetical protein |
| YLR170C | APS1 | 3.9, 3.3 | 0150_at | Clathrin associated protein complex small subunit |
| TY1B | – | 3.8, 4.5 | 3407_at | Saccharomyces cerevisiae chromosome IV, complete chromosome sequence. Found forward in NC_001136 between 804494 and 805681 |
| YCR062W | ORF | 3.8, 3.4 | 6803-at | Similarity to Ytp1p protein |
| YIL135C | ORF | 3.7, 3.0 | 4221_at | Similarity to Ymk1p |
| YDR543C | ORF | 3.7, 3.4 | 3356_f_at | Strong similarity to subtelomeric encoded proteins |
| YML015C | TAF11 | 3.7, 2.6 | 9649_at | TFIID subunit |
| YPR084W | ORF | 3.7, 3.0 | 7660_at | Hypothetical protein |
| NKL008W | – | 3.6, 4.7 | 10439_at | SAGE ORF found forward in NC_001143 between 308848 and 309084 (16) |
| NOR010W | – | 3.6, 4.4 | 8040_at | SAGE ORF found forward in NC_001147 between 524856 and 525080 (16) |
| YMR201C | RAD14 | 3.6, 3.1 | 9428_at | Human xeroderma pigmentosum group A DNA repair gene homolog |
| YCR037C | PHO87 | 3.6, 2.9 | 6827_at | May collaborate with Pho86p and Pho84p in inorganic phosphate uptake, protein contains 12 predicted transmembrane domains |
| YIL130W | GIN1 | 3.6, 2.7 | 4226_at | Similarity to Put3p and to hypothetical protein YJL206c |
| NGL021W | – | 3.5, 6.3 | 4621_at | SAGE ORF found forward in NC_001139 between 274428 and 274583 (16) |
| YOR223W | ORF | 3.5, 3.0 | 8335_at | Protein of unknown function |
| YFL046W | ORF | 3.5, 2.6 | 5434_at | Weak similarity to middle part of Caenorhabditis elegans myosin heavy chain A |
| YPL039W | ORF | 3.4, 4.8 | 7804_at | Hypothetical protein |
| YGL164C | ORF | 3.4, 2.7 | 5171_at | Similarity to Schizosaccharomyces pombe hypothetical protein SPAC31A2.10 |
| YOL071W | EMI5 | 3.3, 3.6 | 8626_at | Similarity to hypothetical Schizosaccharomyces pombe protein |
| YDR201W | SPC19 | 3.2, 2.8 | 6258_at | Component of spindle pole |
| YHR086W | NAM8 | 3.1, 3.4 | 4468_at | Required in meiosis specific splicing of MER2 and MER3 |
| YBR212W | NGR1 | 3.1, 2.9 | 7168_at | RNA/single-stranded DNA binding |
| YPR081C | GRS2 | 3.0, 2.6 | 7657_at | Strong similarity to glycyl-tRNA synthetases |
| YDR205W | MSC2 | 3.0, 2.6 | 6262_at | Cation:cation antiporter |
| YLR403W | SFP1 | 3.0, 2.7 | 9938_at | Transcription factor |
| YBR040W | FIG1 | 3.0, 3.0 | 7308_at | Integral membrane protein |
| YOR256C | ORF | 2.9, 2.6 | 8277_at | Strong similarity to secretory protein Ssp134p |
| YOR011W | AUS1 | 2.9, 2.5 | 8528_at | Strong similarity to ATP-dependent permeases |
| YMR181C | ORF | 2.9, 2.6 | 9451_at | Similarity to YPL229w |
| YLR459W | CDC91 | 2.8, 2.5 | 9863_at | Cell division control protein |
| YOR342C | ORF | 2.8, 2.8 | 8186_at | Weak similarity to YAl037w |
| YDL160C | DHH1 | 2.8, 5.7 | 6660_at | Putative RNA helicase of DEAD box family, required for Rap1p localization to telomeres |
| YFR053C | HXK1 | 2.8, 3.2 | 5307_at | Hexokinase I (PI) also called hexokinase A |
| YNL330C | RPD3 | 2.8, 2.5 | 9200_at | Histone deacetylase |
| YDR441C | APT2 | 2.8, 2.7 | 6048_at | Molecular function unknown |
| YOR196C | LIP5 | 2.7, 2.8 | 8353_at | Involved in lipoic acid metabolism |
| YHR061C | GIC1 | 2.7, 3.1 | 4489_at | GTPase-interacting component 1 |
| YOL090W | MSH2 | 2.7, 2.5 | 8653_at | MutS homolog encoding major mismatch repair activity in mitosis and meiosis |
| YIL161W | ORF | 2.6, 2.5 | 4240_at | Hypothetical protein |
| YGR275W | RTT102 | 2.6, 2.6 | 4705_at | Hypothetical protein |
| YBR293W | ORF | 2.6, 3.2 | 7066_at | Probable multidrug resistance protein |
| *YOL059W | GPD2 | 2.5, 3.6 | 8592_at | Glycerol-3-P dehydrogenase (NAD+) |
| YNL288W | CAF40 | 2.5, 2.8 | 9151_at | Similarity to Caenorhabditis elegans hypothetical protein |
| YLL020C | ORF | 2.5, 2.6 | 10368_s_at | Questionable ORF |
| NBR046W | – | 2.5, 3.0 | 7032_at | Non-annotated SAGE ORF found forward in NC_001134 between 649944 and 650114 (16) |
| YCL038C | AUT4 | 2.5, 3.7 | 6894_at | Molecular function unknown |
| YGR166W | KRE11 | 2.5, 2.6 | 4821_at | Molecular function unknown |
| YNR009W | ORF | 2.5, 2.5 | 8857_at | Hypothetical protein |
Comments were derived from the Affymetrix web site (https://www.affymetrix.com/site/login/login.affx) and/or from the Saccharomyces Genome Database (SGD) web site (http://genome-www.stanford.edu/Saccharomyces/). Gene names, where available, were taken from SGD. SGD entries with no gene name assigned are listed as ORF. Sequences confirmed as enriched by quantitative RT–PCR reactions (Table 2) are indicated here in bold type. Sequences for which quantitative RT–PCR reactions failed to confirm ≥2.5-fold enrichment are indicated by an asterisk (*). Sequences that are designated by unique identifiers in the Affymetrix database but not in the SGD are listed as (–) under gene name.
The enrichment values for candidate Scp160p-associated messages listed in Table 2 were calculated as follows. First, quantitative RT–PCR reactions were performed using the primers listed above with cDNA samples representing both affinity-isolated FLAG-Scp160p complexes, and total RNAs from the same whole cell lysates. In each case, the appropriate total RNA sample was used to generate the standard curve. Next, the amount of each candidate sequence detected in a given cDNA sample was normalized to the amount of a control sequence, YHR174W (ENO2, enolase), detected in the same sample. Finally, to calculate fold enrichment, ratios were generated representing the normalized values for each candidate sequence in the FLAG-Scp160p affinity-isolated sample versus in the corresponding total cell RNA sample. Values presented in Table 2 represent these ratios, corrected for any background detected by parallel analyses of samples derived from cells expressing native, untagged Scp160p.
Table 2. Yeast mRNA and ORF sequences confirmed by quantitative RT–PCR as specifically enriched in Scp160p-containing complexes.
| Gene name(s) | Fold specific enrichment | N | Comments |
|---|---|---|---|
| YDL160C (DHH1) | 17.92 ± 5.59 | 6 | Putative RNA helicase of DEAD box family, required for Rap1p localization to telomeres |
| YCL029C (BIK1) | 12.04 ± 9.82 | 5 | Microtubule-associated protein required for microtubule function during mating and mitosis |
| YOR338W (ORF) | 9.58 ± 4.78 | 6 | Hypothetical ORF |
| YOL155C (ORF) | 4.15 ± 1.78 | 6 | Hypothetical ORF |
| YHR086W (NAM8) | 2.85 ± 0.94 | 6 | Required in meiosis-specific splicing of MER2 and MER3, double strand breaks, synaptonemal complexes |
Comments were derived from the SGD web site (http://genome-www.stanford.edu/Saccharomyces/).
Fractionation of soluble and membrane-associated populations of Scp160p
Cells were lysed by vortex agitation with an equal volume of acid-washed glass beads in 1× polyribosome lysis buffer (25 mM Tris–HCl pH 7.2, 50 mM KCl, 30 mM MgCl2, 5 mM β-mercaptoethanol, 100 µg/ml cyclohexamide). Each lysate was transferred to a clean microfuge tube and centrifuged in an Eppendorf microfuge, Brinkmann, model 54150, with a 24 place rotor, at 3000 r.p.m. for 5 min at 4°C to remove glass beads and cell debris. Each supernatant was then transferred to a fresh tube and centrifuged at 12 000 r.p.m. in the same microfuge for 10 min at 4°C to separate the soluble and membrane-associated fractions. The supernatant resulting from this high speed spin contained the soluble pool of Scp160p. The pellet contained the membrane-associated pool, which was solubilized by resuspension with a 10 min incubation (on ice) in lysis buffer containing 1.5% Triton X-100, followed by a 10 min, 4°C spin at 14 000 r.p.m. in the Eppendorf microfuge to remove residual insolubles.
Sucrose gradient fractionation of yeast lysates
Sucrose gradient fractionation of soluble cell lysates was performed as described previously (3). In brief, a 100 ml culture of yeast was grown in rich (YPD) medium to early log phase (OD600 ∼ 1.5), at which time cyclohexamide (Sigma) was added directly to the culture to a final concentration of 100 µg/ml. The culture was incubated on ice for 25 min, and cells were harvested by centrifugation in a Sorvall RC5B centrifuge using an SLA 1500 rotor (4000 r.p.m., 1541 g, for 10 min). Following two washes in 10 ml of water containing 100 µg/ml cyclohexamide, cells were lysed as described above in 1× polyribosome lysis buffer. Each lysate was transferred to a clean microfuge tube and centrifuged in an Eppendorf microfuge (same as above) at 6500 r.p.m. for 10 min at 4°C, followed by a second spin of the supernatant at 14 000 r.p.m., also for 10 min at 4°C. Finally, 100 µl of supernatant were loaded onto a 15–45% sucrose gradient (11 ml) made using a Gradient Master automatic system. Each gradient was centrifuged using a SW41ti rotor in a Beckman Optima LE-80K ultracentrifuge at 39 000 r.p.m. (197 000 g) for 2.5 h at 4°C, after which fractions were harvested using an Isco gradient fractionator. Gradient RNA profiles were monitored by following absorbance at 254 nm.
Sucrose gradient fractionation of membrane-associated Scp160p complexes was achieved using a high speed (12 000 r.p.m.) spin, in the Eppendorf microfuge described above, to separate membrane pellets, as described above. Prior to loading onto the sucrose gradient, these pellets were resuspended and incubated for 10 min (on ice) in lysis buffer containing 1.5% Triton X, then centrifuged a second time (10 min, 4°C) at 14 000 r.p.m. in the Eppendorf microfuge to remove insolubles.
RESULTS
Polyadenylated RNA is present in Scp160p-containing complexes
To address directly the question of whether Scp160p-containing complexes include mRNA, we exploited the presence of a FLAG epitope tag engineered onto the N-terminus of Scp160p (see Materials and Methods) to affinity isolate these complexes, essentially as described previously (3). FLAG-Scp160p has been demonstrated previously to function in vivo indistinguishably from the untagged native protein (3). As a control for specificity, cells expressing native, untagged Scp160p also were subjected to the affinity isolation procedure. Total RNA was then released from both isolates and subjected to reverse transcription using an oligo-dT primer in the presence of [α-32P]dCTP. As illustrated in Figure 1, a strong smear, centered at approximately 1500 bases in size, was observed in the lane representing FLAG-Scp160p, but not in the control lane, although a larger fraction of the control reaction sample was loaded. Parallel reverse transcription reactions using total RNA isolated from whole cell soluble lysates of both strains resulted in indistinguishable strong smears (data not shown).
Figure 1.
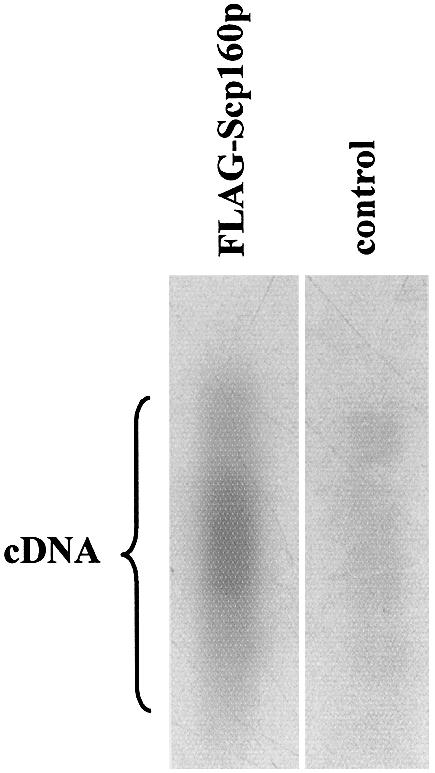
First-strand cDNA synthesized from Scp160p complex-derived RNA. RNAs, derived either from anti-FLAG affinity isolated FLAG-Scp160p-containing mRNP complexes (FLAG-Scp160p lane), or from control samples mock isolated from cells expressing untagged Scp160p (control lane), were used as template in first-strand cDNA synthesis reactions with oligo-dT and 32P. The center of the visible smear represents ∼1500 nt.
Identification and confirmation of specific mRNA sequences associated with Scp160p
To address the question of sequence specificity, RNA samples derived from FLAG-Scp160p-containing complexes versus total RNA from the same cell lysates were used as templates to generate probes for hybridization to Affymetrix YG-S98 yeast gene chips (see Materials and Methods). As a control, corresponding pools of RNA derived from cells expressing native, rather than FLAG-tagged Scp160p, also were prepared. The results, determined by comparing the hybridization results of each Scp160p complex-derived sample against its corresponding total RNA control (see Materials and Methods), were striking. First, although many strong hybridization spots were detected in both test and control samples, the patterns were different, indicating that the Scp160p complex-derived samples did not simply contain a random subset of total cellular mRNAs. Furthermore, those sequences most abundant in the mock-isolated samples were completely distinct from those most abundant in the FLAG-Scp160p complex-derived samples (data not shown), demonstrating specificity of the isolation procedure. In sum, of the >6000 putative yeast gene sequences interrogated on the microarrays in duplicate experiments, only ∼1% (69 sequences) appeared >2.5-fold enriched in the FLAG-Scp160p complex-derived samples in both experiments (Table 1).
To test a subset of these candidates with an independent technology, we performed quantitative RT–PCR using a Roche LightCycler with primers designed to amplify small fragments from the 3′ ends of each of 12 candidate enriched messages, eight of which are listed in Table 1 (YGR023W, YOR338W, YOL155C, YDR247W, YBR150C, YHR086W, YDL160C, YOL059W), and four of which (YCL029C, YKL203C, YBL109W, YGR110W) are not listed because they appeared enriched in only one of the two microarray experiments performed. Templates analyzed by quantitative RT–PCR were mRNAs derived from three or more independent Scp160p complex isolation procedures, each compared against its corresponding total mRNA control. Those five sequences that were confirmed as enriched by quantitative RT–PCR are presented in Table 2. To ensure specificity of these values, mock enrichment procedures also were performed using cells expressing native rather than FLAG-tagged Scp160p, and apparent fold enrichment of each candidate message in those ‘mock’ samples was calculated and subtracted as background from the values presented in Table 2. As indicated by asterisks in Table 1, four of the eight messages tested from that group did not confirm as enriched by >2.5-fold when measured by quantitative RT–PCR. An additional three of the four ‘single microarray candidates’ tested also did not confirm as enriched by quantitative RT–PCR.
Impact of scp160-loss on DHH1 and YOR338W
To address the question of biological significance of Scp160p association with its target messages, we first checked both message abundance and message distribution between the soluble and membrane-associated pools for two target sequences, DHH1 and YOR338W, comparing wild-type versus scp160-null yeast. As illustrated in Table 3, although no significant change was seen for DHH1, YOR338W demonstrated a significant increase in abundance in scp160-null cells. Furthermore, the distribution of that signal was shifted significantly away from the membrane pellets, and toward the soluble fraction.
Table 3. Relative abundance and distribution of DHH1 and YOL338W messages in SCP160+ and scp160-null cells.
| Sample | SCP160+ | scp160-null |
|---|---|---|
| DHH1 total | 1.00 ± 0.12 | 1.30 ± 0.30 |
| DHH1 soluble | 24.67 ± 3.32 | 19.37 ± 1.53 |
| DHH1 pellet | 75.33 ± 3.32 | 80.63 ± 1.53 |
| YOL338W total* | 1.00 ± 0.23 | 2.53 ± 0.23 |
| YOL338W soluble* | 11.73 ± 3.41 | 23.97 ± 3.07 |
| YOL338W pellet | 88.27 ± 3.41 | 76.03 ± 3.07 |
Values presented represent the averages and standard deviations from analyses of three independent samples in each category, each normalized to an enolase control, as described in Materials and Methods. Values representing total RNA samples were further scaled to set each wild-type value to 1.0, and the corresponding soluble and pellet values for each set sere scaled to represent percentages totaling 100. Samples demonstrating statistically significant differences (two-sided t-test) between SCP160+ and scp160-null cells are indicated with an asterisk.
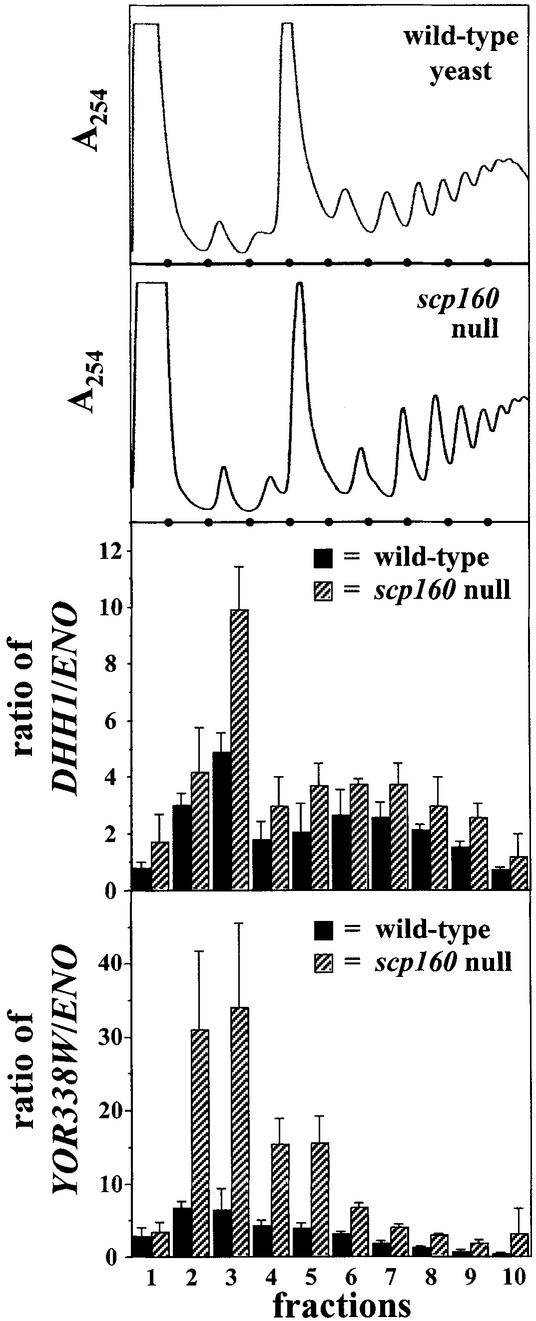
Next, we utilized sucrose-gradient fractionation to explore the subcellular distribution of DHH1 and YOR338W in both wild-type and scp160-null yeast. In brief, both strains were grown to early log phase (OD ∼ 1), lysed as described previously (3,4), and the soluble portions subjected to sucrose gradient fractionation, as described previously (3,4). Total RNA was isolated from each fraction, and subjected to quantitative RT–PCR using a Roche LightCycler with the appropriate primers (Fig. 2; see Materials and Methods). For each fraction, the target sequence signal detected was normalized to the corresponding signal from a non-target control sequence (enolase, ENO2), so that the data points presented represent ratios. As illustrated (Fig. 2, bottom two panels), cells devoid of Scp160p (striped bars) demonstrated a marked enrichment of both DHH1 and YOR338W in the lighter gradient fractions (representing mRNPs), as compared with their wild-type counterparts (solid bars).
Figure 2.
Impact of scp160-loss on subcellular distribution of the DHH1 and YOR338W messages. The relative abundance of DHH1 and YOR338W messages, normalized to ENO2, was followed in sucrose gradient fractionated lysates representing both wild-type (solid bars) and scp160-null (striped bars) cells using quantitative RT–PCR. All values plotted represent averages (with standard deviation) of three independent analyses. The top two panels illustrate representative OD254nm profiles from wild-type and scp160-null yeast, respectively. Solid circles are positioned at the bottom of each of these panels to indicate division points between different fractions.
Finally, we performed parallel sucrose gradient fractionation experiments on samples derived from the membrane-associated compartments of both wild-type versus scp160-null cells. No reproducible differences in the distribution of DHH1 or YOR338W signals were observed in these experiments (data not shown).
DISCUSSION
The results reported here demonstrate two main points. First, Scp160p associates with specific rather than random mRNAs in yeast. Second, loss of Scp160p results in a detectable change in the abundance and membrane association of at least one of its target messages (YOR338W), and in the soluble polyribosome association profiles of at least two of its target messages (DHH1 and YOR338W), relative to a non-target control (ENO2). Each of these findings represents an important step forward in our effort to understand the biological function of Scp160p.
The first point, that Scp160p associates with only ∼1% of yeast mRNAs, is important because it rules out the possibility that Scp160p is a general translation factor in yeast. This point is made even stronger considering that only 5 of the 12 candidate targets tested confirmed by quantitative RT–PCR, so that close to half of the other potential target messages currently indicated by microarray analysis alone might also fail to confirm. The actual percentage of messages in yeast that associate with Scp160p may therefore be <1%. Furthermore, considering the disparity between the microarray data obtained and quantitative RT–PCR results, it is reasonable to assume that genuine target messages may also have been missed by the microarray experiments.
Perhaps more important, although the set of Scp160p-associated messages we have presented may not be comprehensive, our data provide a ready list of potential targets for further study—targets that will likely offer additional insights into the mechanism and impact of Scp160p function in vivo. For example, among the enriched Scp160p-associated messages we have identified and confirmed are DHH1, a putative RNA helicase with close homologs in mammals, including human; BIK1, a putative microtubule binding protein required for microtubule function in mitosis and mating; and NAM8, an RNA-binding protein required for the meiosis-specific splicing of MER2 and MER3. Others (YOR338W and YOL155C) remain hypothetical open reading frames (ORFs); through studies of their interplay with Scp160p, we may also gain insight into their functions, which are currently unknown. Clearly many more interesting potential targets also remain to be studied.
The second point, that loss of Scp160p results in a change in abundance and membrane association for at least one target message (YOR338W), as well as a subtle but significant change in the soluble polyribosome association profile of at least two target messages (DHH1 and YOR338W) relative to a non-target control (ENO2), is equally important, because it demonstrates that the interaction of these messages with Scp160p is biologically meaningful. These data are consistent with the conclusion that Scp160p-loss results in a shift of at least some of its target messages from membrane associated polyribosomes to free mRNPs in the soluble pool. At minimum, these data strengthen the argument that Scp160p functions in some aspect of cytoplasmic mRNA metabolism, perhaps including translation. Whether the observed shift in polyribosome association reflects altered translational efficiency, stability, or some other property of the target messages, remains to be determined. Furthermore, future studies will be required to determine whether other target messages (e.g. BIK1, NAM8, YOL155C, and others as yet unconfirmed) will demonstrate similar or distinct responses to the loss of Scp160p.
Although the results presented here represent a significant step forward, much work remains to be completed if we are to understand the biological role(s) of Scp160p in yeast, and of its counterparts in other species. For example, although we have demonstrated clear association of specific mRNA sequences with Scp160p-containing complexes, we do not yet know whether these interactions are direct or indirect. Preliminary in vitro RNA-binding studies between recombinant Scp160p and labeled transcript suggest that direct binding can occur with target sequences, although the specificity of that binding is unclear. Furthermore, what features these confirmed target messages exhibit, and perhaps share in common, that enable each to associate with Scp160p, remain to be defined. It is also possible, if not probable, considering the large size and significant number of non-Scp160p proteins apparent in Scp160p-containing mRNP complexes (3,4), that some determinants of specificity may derive from other components of these complexes, not only from Scp160p and transcript. What these other components are, and how they may contribute to the specificity of message association, remains to be defined. In addition, we have, to date, selected target messages for study based only upon their degree of apparent enrichment in Scp160p-containing complexes (e.g. ≥2.5-fold); however, these may not be the most biologically important messages impacted by Scp160p. Alternative approaches will be required to define pools of messages that not only associate with Scp160p, but that are also specifically impacted in any given way by Scp160p-loss.
Finally, in the microarray studies reported here, by lysing cells in the presence of EDTA, which disrupts polyribosomes, we have intentionally mixed the soluble and membrane-associated Scp160p pools prior to Scp160p complex isolation. This strategy of isolation was designed to give a ‘whole cell’ representation of Scp160p, and to minimize the number of microarrays required to perform the experiments. However, it is entirely possible that the membrane-associated and soluble populations of Scp160p may interact with different subsets of mRNA. Future experiments will focus on exploring separately the mRNA and protein components of soluble versus membrane-associated Scp160p-containing complexes, in order to compare and contrast these two populations.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Drs S. Warren, Peng Jin and Yue Feng for numerous helpful discussions and for allowing us access to their sucrose gradient analysis systems, and to Drs S. Warren and D. Wallace for allowing us access to their Roche LightCycler systems. This work was supported in part by funds from the Emory University Research Committee, and in part by an award from the National Science Foundation (MCB-0112911.01), both to J.L.F.-K.
REFERENCES
- 1.Wintersberger U., Kuhne,C. and Karwan,A. (1995) Scp160p, a new yeast protein associated with the nuclear membrane and the endoplasmic reticulum, is necessary for maintenance of exact ploidy. Yeast, 11, 929–944. [DOI] [PubMed] [Google Scholar]
- 2.Weber V., Wernitznig,A., Hager,G., Harata,M., Frank,P. and Wintersberger,U. (1997) Purification and nucleic-acid-binding properties of a Saccharomyces cerevisiae protein involved in the control of ploidy. Eur. J. Biochem., 249, 309–317. [DOI] [PubMed] [Google Scholar]
- 3.Lang B.D. and Fridovich-Keil,J.L. (2000) Scp160p, a multiple KH-domain protein, is a component of mRNP complexes in yeast. Nucleic Acids Res., 28, 1576–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lang B.D., Li,A.-M., Black-Brewster,H.D. and Fridovich-Keil,J.L. (2001) The brefeldin A resistance protein Bfr1p is a component of polyribosome-associated mRNP complexes in yeast. Nucleic Acids Res., 29, 2567–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frey S., Pool,M. and Seedorf,M. (2001) Scp160p, an RNA-binding, polysome-associated protein, localizes to the endoplasmic reticulum of Saccharomyces cerevisiae in a microtubule-dependent manner. J. Biol. Chem., 276, 15905–15912. [DOI] [PubMed] [Google Scholar]
- 6.Currie J.R. and Brown,W.T. (1999) KH domain-containing proteins of yeast: absence of a fragile X gene homologue. Am. J. Med. Genet., 84, 272–276. [PubMed] [Google Scholar]
- 7.Siomi H., Matunis,M.J., Michael,W.M. and Dreyfuss,G. (1993) The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res., 21, 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt C., Henkel,B., Poschl,E., Zorbas,H., Purschke,W.G., Gloe,T.R. and Muller,P.K. (1992) Complete cDNA sequence of chicken vigilin, a novel protein with amplified and evolutionary conserved domains. Eur. J. Biochem., 206, 625–634. [DOI] [PubMed] [Google Scholar]
- 9.Plenz G., Kugler,S., Schnittger,S., Rieder,H., Fonatsch,C. and Muller,P.K. (1994) The human vigilin gene: identification, chromosomal localization and expression pattern. Hum. Genet., 93, 575–582. [DOI] [PubMed] [Google Scholar]
- 10.Dodson R.E. and Shapiro,D.J. (1997) Vigilin, a ubiquitous protein with 14 K homology domains, is the estrogen-inducible vitellogenin mRNA 3′-untranslated region-binding protein. J. Biol. Chem., 272, 12249–12252. [DOI] [PubMed] [Google Scholar]
- 11.Cortes A., Huertas,D., Fanti,L., Pimpinelli,S., Marsellach,F.X., Pina,B. and Azorin,F. (1999) DDP1, a single-stranded nucleic acid-binding protein of Drosophila, associates with pericentric heterochromatin and is functionally homologous to the yeast Scp160p, which is involved in the control of cell ploidy. EMBO J., 18, 3820–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cortes A. and Azorin,F. (2000) DDP1, a heterochromatin-associated multi-KH-domain protein of Drosophila melanogaster, interacts specifically with centromeric satellite DNA sequences. Mol. Cell. Biol., 20, 3860–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cunningham K.S., Dodson,R.E., Nagel,M.A., Shapiro,D.J. and Schoenberg,D.R. (2000) Vigilin binding selectively inhibits cleavage of the vitellogenin mRNA 3′ untranslated region by the mRNA endonuclease polysomal ribonuclease 1. Proc. Natl Acad. Sci. USA, 97, 12498–12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guthrie C. and Fink,G.R. (1991) Guide to yeast genetics and molecular biology. Methods Enzymol., 194, 302–318. [PubMed] [Google Scholar]
- 15.Schmitt M.E., Brown,T.A. and Trumpower,B.L. (1990) A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res., 18, 3091–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Velculescu V.E., Zhang,L., Zhou,W., Vogelstein,J., Basrai,M.A., Bassett,D.E.J., Hieter,P., Vogelstein,B. and Kinzler,K.W. (1997) Characterization of the yeast transcriptome. Cell, 8, 243–251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.