Abstract
Small interfering RNAs (siRNAs) are powerful RNA interference (RNAi) reagents for directed post- transcriptional gene silencing. Exogenous siRNA is frequently used in RNAi studies. However, due to profound differences in the activity of siRNAs targeted to different regions of a gene, several reagents may have to be screened for optimal activity. This approach is expensive due to the cost of chemical synthesis of RNAs. We report a technically simple, quick and cost-effective method for the production of siRNAs that makes use of in vitro transcription and deoxyribozyme digestion of the transcripts to produce the desired sequence and length. The method allows for several siRNAs to be produced in parallel at much reduced costs. The siRNAs produced with this method were tested in MDA-MB-231 human breast cancer cells for efficacy against the type 1 insulin-like growth factor receptor (IGF1R) mRNA and they caused dose-dependent inhibition of IGF1R expression comparable to that induced by chemically synthesised siRNAs of the same sequence. This method is also useful for producing long RNA fragments of defined length and sequence that may be difficult to synthesise chemically, and also for producing large quantities of RNAs for applications including structural studies and the study of interactions between RNA and other molecules, such as proteins, other nucleic acids and drugs.
INTRODUCTION
Small interfering RNAs (siRNAs) have emerged as powerful RNA interference (RNAi) reagents for directed post- transcriptional gene silencing (1–5) and inhibition of viral propagation (6–10). Exogenous siRNAs are frequently used in RNAi studies. Using chemically synthesised RNA oligonucleotides, Elbashir et al. (1) described a systematic analysis of the optimal Drosophila siRNA duplexes. Exogenous siRNAs typically consist of a double-stranded region of 19 bp and 2 nt 3′ overhangs: TT-3′/UU-3′ overhangs are generally preferred over other sequences (1). Due to profound differences in the activities of siRNAs targeted to different regions of a gene (11–14), several reagents may have to be screened for optimal activity. This approach is expensive due to the cost of chemical synthesis of RNAs. Intracellular expression of siRNA (15–17) is laborious and the efficacy of the cloned siRNA is difficult to predict. Furthermore, intracellular expression is largely suitable where stable expression of the reagents is needed. Therefore, it is highly desirable to develop methods for cost-effective production of siRNAs.
We report a technically simple and cost-effective method for the production of siRNAs that makes use of in vitro transcription and deoxyribozyme digestion of the transcripts to produce the desired sequence and length. The method allows for several siRNAs to be produced in parallel at much reduced cost and produces high yields of effective siRNAs. This method is also useful for the production of long RNA fragments of defined length and sequence that may be difficult to obtain by chemical synthesis, and also for the production of large quantities of RNAs for applications including structural studies and the study of interactions between RNA and other molecules, such as proteins, other nucleic acids and drugs.
MATERIALS AND METHODS
Oligonucleotides and purification
Oligodeoxyribonucleotides were synthesised on an ABI394 DNA/RNA synthesiser using standard nucleotide CE- phosphoramidites (Cruachem) and were desalted through NAP-5 columns (Amersham) at least twice. The purity and integrity of the oligonucleotides was evaluated on a 12% (w/v) denaturing polyacrylamide gel after end-labelling with 32P in the presence of [γ-32P]ATP (>5000 Ci/mmol; Amersham) and T4 polynucleotide kinase (Roche Diagnostics). Oligonucleotide stocks were maintained at 100 pmol/µl concentration in water at –20°C. See Table 1 for a list of oligonucleotides and deoxyribozymes. Chemically synthesised, HPLC-purified RNA/DNA chimeric oligonucleotides were obtained from Cruachem and were HPLC purified. The sequence was the same as that for enzymatically prepared RNAs, except that each strand had 19 bases of RNA with two 3′ deoxythymidine nucleotides. To produce siRNAs, complementary strands were annealed in 100 mM potassium acetate, 30 mM HEPES–KOH pH 7.4, 2 mM magnesium acetate by incubating at 85°C for 1 min followed by 37°C for 1 h. Duplex formation was confirmed by electrophoresis through 5% (w/v) low melting temperature agarose (NuSieve GTG; FMC BioProducts).
Table 1. Template oligonucleotide and deoxyribozyme sequences.
Target sequences are in bold and the mutant nucleotides are underlined.
In vitro transcriptions
These were carried out at 37°C for 4–5 h or overnight using MegaScript or MegaShortScript (Ambion) kits mainly as instructed by the supplier. Two complementary oligonucleotides were annealed to produce template for transcription reactions as follows: IGF-S1 and IGF-S2 to produce sense strand (S); IGF-AS1 and IGF-AS2 to produce antisense strand (AS); IGF-S1M and IGF-S2M to produce mutant sense strand (SM); IGF-AS1M to produce mutant antisense strand. An aliquot of 100 pmol of the annealed template was used in a 20 µl reaction containing a 11.25 mM concentration of each NTP and 3–4 µl of the T7 enzyme mix supplied with the Ambion kits. In labelling reactions, 1 µl of either [α-32P]UTP (3000 Ci/mmol; Amersham) or [α-33P]UTP (2500 Ci/mmol; Amersham) was added as tracer to assess the quality of transcripts. After transcription, 1 U of RNase-free DNase (Ambion) was added to digest the template (37°C for 15 min) that was found to interfere with the subsequent deoxyribozyme digestion (not shown). The reaction was diluted to 40 µl with water and purified through a Sephadex G25 Microspin column (Amersham). Typical RNA yields at this stage were ∼200 µM.
Deoxyribozyme reactions
During in vitro transcription reactions using T7/T3 or SP6 promoters/RNA polymerases, a 5′ leader sequence is added to the transcripts. This needs to be removed to obtain single-stranded RNA templates for siRNA of desired sequence and length. We used deoxyribozymes to remove unwanted nucleotides from transcripts as follows: IGF-817S was used to cleave the S and SM strand; IGF-817AS was used to cleave the AS and ASM strand. Typically, a deoxyribozyme reaction was carried out in a 20 µl volume containing ∼100–120 µM transcripts and 100 µM deoxyribozyme in 100 mM NaCl, 30 mM MgCl2, 20 mM Tris–HCl pH 7.2 and 20 U of RNasin (Promega). The reaction was incubated overnight at room temperature or in a thermal cycler (MJ Research) (cycling at 50°C for 1 min, 20°C for 2 min and 30°C 10 min). Finally, 1 U of DNase I (Ambion) was added to digest deoxyribozyme that could interfere with the annealing of the siRNA strands by competing with complementary sequences. The product was purified through a Sephadex G25 Microspin column (Amersham). Sense and antisense strands were prepared in separate reactions and were annealed at 70°C for 2 min and cooled to room temperature to obtain siRNA duplexes. siRNA duplexes were stored at ∼50 µM concentration in 0.5× deoxyribozyme reaction buffer at –20°C. Overall, starting with a 20 µl transcription reaction for each strand, ∼160 µl of ∼50 µM siRNA duplexes were obtained.
Cell culture and transfection
Human estrogen receptor negative breast cancer cell line MDA-MB-231 (Cancer Research UK) was used to test the efficacy of siRNA duplexes. The cells were cultured in RPMI 1640 medium with 10% (v/v) foetal calf serum and were negative on testing for mycoplasma infection. Cells were transfected with siRNA using Oligofectamine™ (Invitrogen) according to the manufacturer’s instructions. After 48 h the cells were lysed and analysed as previously described (18) by immunoblotting for the type 1 insulin-like growth factor receptor (IGF1R) (Santa-Cruz) and β-tubulin (Sigma).
RESULTS AND DISCUSSION
We report an approach to producing siRNA of desired sequence and length using in vitro transcription and deoxyribozyme digestion of the transcript. The method is cost-effective and produces high yields of effective siRNAs. The use of oligonucleotides to produce short transcripts (19) was recently used to produce siRNAs (17,20). This useful approach is limited by specific sequence requirements: all siRNAs made with this method start with a 5′-G residue and require a C-3′ residue at position 19 (i.e. 5′-G-N17-C-3′) to allow annealing with the complementary RNA which also has to start with a 5′-G residue. Therefore, if the most commonly used TT-3′/UU-3′ overhang is employed in a siRNA, the mRNA sequence must be 5′-AAG-N17-C-3′). Furthermore, the efficacy of siRNAs targeted to different regions of a gene varies dramatically (11–14), limiting the number of sequences that can be targeted using siRNAs generated by this method, substantially reducing the chances of identifying optimally effective siRNAs.
We used deoxyribozymes to overcome this limitation and enable the synthesis of any sequence by a simple enzymatic process. Deoxyribozymes consist of a catalytic domain flanked by two substrate-specific 7–8 nt binding arms. The target RNA is cleaved between two specific nucleotides (21,22). The requirement for this dinucleotide sequence is different for different deoxyribozyme groups, making them flexible tools for digesting a variety of sequences. For example, 10-23 type deoxyribozymes can digest an RNA strand containing the sequences 5′-AT, 5′-AC, 5′-GC and 5′-GT, 8-17 type deoxyribozymes cleave at 5′-AG and Bipartite II deoxyribozymes optimally cleave at 5′-AA. The underlined nucleotides are unpaired and the second nucleotides pair with a complementary nucleotide in the deoxyribozyme. In optimally active Bipartite II deoxyribozymes, five unpaired nucleotides separate the binding arms including those at the catalytic site (22). Therefore, in principle siRNA sequences starting and ending with any nucleotide can be chosen.
We have investigated this approach by targeting the IGF1R gene with siRNAs produced using T7 RNA polymerase-generated transcripts and 8-17 type deoxyribozymes.
The accessibility profile of a region of the IGF1R mRNA was obtained by hybridisation of the mRNA to an array of anti-IGF1R antisense oligonucleotides (23). A structurally accessible region was selected for targeting with siRNAs (data not shown). The template oligonucleotides for producing RNA transcripts were designed such that the resultant siRNA contained a 19 bp double-stranded region and 3′-UU overhang at both ends (1).
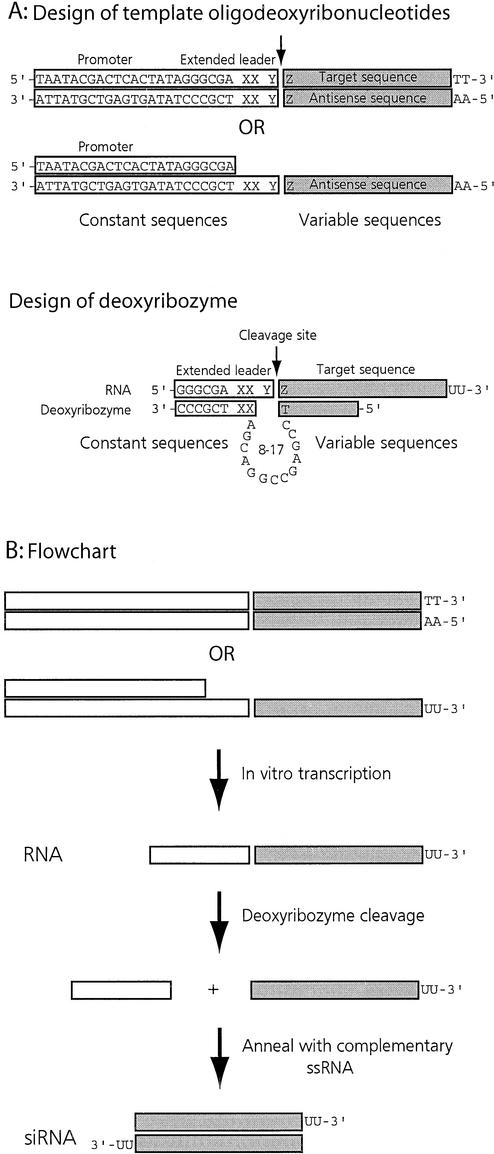
The two strands of the siRNA were produced in separate in vitro transcription reactions. For each strand, two complementary oligodeoxyribonucleotides were designed to provide template for RNA polymerase. The oligonucleotides incorporated the sequence for binding T7 RNA polymerase and the sequence of the igf1r gene. The commonly used hexa-nucleotide leader sequence of the T7 promoter, 5′-GGGAGA or 5′-GGGCGA, appears in the transcripts. The 3′-arm of the deoxyribozyme was designed to bind to this promoter sequence in RNA and the 5′-arm was complementary to the target gene sequence cassette. However, 8-17 deoxyribozymes have been shown to work optimally with 7–8 nt binding arms (21) and therefore we extended the T7 leader sequence by 2 nt to provide a longer annealing site for the 3′-arm of the deoxyribozyme. We chose 5′-AT to extend the promoter sequence since catalytic sites surrounded by A or U appear to be a better substrate (21); 5′-T (of the 5′-AT extension) also helped to trace the 5′ cleavage product with the use of either [α-32P]UTP or [α-33P]UTP in in vitro transcription reactions. A third nucleotide, 5′-A, was added that, with the first nucleotide of the target sequence (5′-G in this case), made the catalytic site for the deoxyribozyme (5′-AG). Therefore, the sequence of the 3′-binding arm of the deoxyribozyme and that of the catalytic domain remains constant for one type of deoxyribozyme and the only variable is the sequence of the 5′-binding arm that changes with the target sequence. 5′-TT/3′-AA was added after the target gene sequence to provide a 5′-UU overhang in the siRNA. Two 21mer complementary RNA transcripts were annealed to produce siRNA with a 19 bp region and 3′-UU overhangs. A simple guide to designing siRNAs with this method is given in Figure 1.
Figure 1.
(A) Design of the template oligonucleotides and the deoxyribozymes. The 5′-GGGCGA leader sequence of the T7 promoter is added to the transcripts. This sequence varies with the type of promoter used [e.g. T3 or SP6 promoter (27)]. The leader sequence was extended by 3 nt (5′-XXY). 5′-XX nucleotides (we used 5′-AT) are added to increase the length of the sequence to provide appropriate binding sequence for the deoxyribozyme. The nucleotide 5′-Y makes the unpaired base that with the first base of the target sequence, 5′-Z, makes the catalytic site for the deoxyribozyme. The catalytic domain shown is for 8-17 deoxyribozymes. 5′-TT is to give the 2 nt 3′ overhang in the siRNA. (B) Flowchart. Two oligonucleotides are annealed to produce the template for RNA polymerase. Either two fully complementary oligonucleotides or those complementary over the promoter/leader sequence can be used as templates for in vitro transcription. Each strand of the siRNA is prepared in separate in vitro transcription reactions. The transcripts are digested with an appropriate deoxyribozyme to remove unwanted ribonucleotides. The two complementary RNA strands are annealed to obtain siRNA.
The annealed RNAs (siRNAs) were tested for their ability to inhibit IGF1R production in MDA-MB-231 breast cancer cells (Fig. 2) and their activity was compared with chemically synthesised siRNAs. The cells were transfected with siRNA duplexes, lysed after 48 h and IGF1R levels were measured by immunoblotting with anti-IGF1Rβ antibody. The bands were quantified by densitometry and IGF1R levels were corrected for loading differences. We compared the effect of the fully matched duplex S/AS with chemically synthesised duplex R2 (sequence as S/AS, except that it had -TT-3′ overhangs instead of -UU-3′), inverted sequence duplex inv2 and also an enzymatically produced variant of S/AS with a 3 nt mismatch with the target located in the middle of the duplex (SM/ASM). The results showed that S/AS siRNA was as effective in inhibiting its cognate target as R2. Both the chemically synthesised and enzymatically produced siRNAs specifically inhibited the production of IGF1R in a dose-dependent manner. In the mutant duplex (SM/ASM), we altered the central 3 nt since it has been suggested that the target mRNA is cleaved in the centre of the region covered by siRNA and that RNA interference was sensitive to variations in this region of siRNAs (1). Although the mutant duplex SM/ASM was less effective than S/AS, it nonetheless retained noticeable inhibitory effect on the expression of IGF1R. This is in contrast to a previous report that indicated that siRNA activity is abolished by a single base pair mismatch with the target (1), and suggests that siRNA activity may not be consistently sensitive to mismatches with the target. However, a number of other reports support our observations (11,24,25). Since this issue is central to designing target-specific siRNAs, further careful experiments are needed to establish the extent of mismatches with the target tolerated in siRNA-mediated RNA interference.
Figure 2.
(A) Production of single-stranded RNAs by in vitro transcription in the presence of [α-33P]UTP and digestion with a deoxyribozyme. Transcriptions were carried out using two fully complementary oligonucleotides as templates. Complementary RNA strands were annealed to obtain siRNAs. We also performed in vitro transcriptions with DNA templates in which an oligonucleotide having the T7 promoter/leader sequence (T7ext-ATA; Table 1) was annealed to an oligonucleotide containing the complementary sequence of the T7 promoter/leader and downstream target sequence (see Fig. 1). Transcripts thus produced were used in deoxyribozyme digestion in a similar manner (data not shown). (B) Immunoblots showing inhibition of IGF1R expression by siRNA duplexes. We also treated S/AS and SM/ASM duplexes with single strand-specific RNase (Ambion), which was found to reduce the efficacy of the duplexes (lower panel). (C) Relative IGF1R levels in MDA-MB-231 cells treated with siRNA duplexes at various concentrations.
Furthermore, it is known that the 3′ termini of the RNA products are determined by the bacteriophage-encoded RNA polymerase and that T7 RNA polymerase frequently adds at the 3′ end a variable number of nucleotides that are not encoded by the DNA template. This addition, usually of a single nucleotide, is hard to predict and is also dependent on the sequence of the template (see for example 19). Therefore, this method is likely to produce siRNAs with 3 nt or longer overhangs. However, our data and reports by other groups (17,20) suggest that this may not have a considerable negative effect on the efficacy of siRNAs.
Overall, the data suggest that siRNAs produced with this method are effective in inhibiting their cognate target in cultured human cells in a manner similar to that induced by chemically synthesised siRNA.
This method provides considerable cost saving over chemical synthesis in the production of RNA duplexes, even taking into account the need to synthesise template DNA oligonucleotides and deoxyribozymes and in vitro transcription reagents. The technical advantages provided by this method are not limited to the production of siRNAs, but extended to production of any single- or double-stranded RNA of defined length and sequence. This method may also be used to produce hairpin RNA of a desired sequence (17), to remove unwanted sequence at the 3′ end of the transcript [e.g. to produce transcripts with the same 3′ end sequence (26)] or to remove unwanted sequences from both the 5′ and 3′ ends. This method is also suited to production of RNAs of specific sequences at large scale (e.g. as pharmaceutical agents) or to the production of libraries of RNA molecules of varying length and sequence (e.g. libraries of slightly varying siRNAs required for optimisation of RNAi targeting of any given gene) which would otherwise be prohibitively expensive by chemical synthesis.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to the Medical Research Council for funding M.S. and G.D., and to Cancer Research UK for supporting V.M. and J.M.
REFERENCES
- 1.Elbashir S., Martinez,J., Patkaniowska,A., Lendeckel,W. and Tuschl,T. (2001) Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J., 20, 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harborth J., Elbashir,S.M., Bechert,K., Tuschl,T. and Weber,K. (2001) Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci., 114, 4557–4565. [DOI] [PubMed] [Google Scholar]
- 3.Lewis D.L., Hagstrom,J.E., Loomis,A.G., Wolff,J.A. and Herweijer,H. (2002) Efficient delivery of siRNA for inhibition of gene expression in postnatal mice. Nature Genet., 32, 107–108. [DOI] [PubMed] [Google Scholar]
- 4.DiTullio R.A. Jr,, Mochan,T.A., Venere,M., Bartkova,J., Sehested,M., Bartek,J. and Halazonetis,T.D. (2002) 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nature Cell Biol., 12, 998–1002. [DOI] [PubMed] [Google Scholar]
- 5.Hasuwa H., Kaseda,K., Einarsdottir,T. and Okabe,M. (2002) Short 5′-phosphorylated double-stranded RNAs induce RNA interference in Drosophila. FEBS Lett., 532, 227–230.12459495 [Google Scholar]
- 6.Gitlin L., Karelsky,S. and Andino,R. (2002) Short interfering RNA confers intracellular antiviral immunity in human cells. Nature, 418, 430–434. [DOI] [PubMed] [Google Scholar]
- 7.Jacque J.M., Triques,K. and Stevenson,M. (2002) Modulation of HIV-1 replication by RNA interference. Nature, 418, 435–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang M. and Milner,J. (2002) Selective silencing of viral gene expression in HPV-positive human cervical carcinoma cells treated with siRNA, a primer of RNA interference. Oncogene, 21, 6041–6048. [DOI] [PubMed] [Google Scholar]
- 9.Qin X.F., An,D.S., Chen,I.S. and Baltimore,D. (2003) Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl Acad. Sci. USA, 100, 183–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Randall G., Grakoui,A. and Rice,C.M. (2003) Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc. Natl Acad. Sci. USA, 100, 235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holen T., Amarzguioui,M., Wiiger,M.T., Babaie,E. and Prydz,H. (2002) Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res., 30, 1757–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McManus M.T., Haines,B.B., Dillon,C.P., Whitehurst,C.E., van Parijs,L., Chen,J. and Sharp,P.A. (2002) Small interfering RNA-mediated gene silencing in T lymphocytes. J. Immunol., 69, 5754–5760. [DOI] [PubMed] [Google Scholar]
- 13.McManus M.T. and Sharp,P.A. (2002) Gene silencing in mammals by small interfering RNAs. Nature Rev. Genet., 3, 737–747. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Y., Ching,Y.P., Kok,K.H., Kung,H.F. and Jin,D.Y. (2002) Post-transcriptional suppresson of gene expression in Xenopus embryos by small interfering RNA. Nucleic Acids Res., 30, 1664–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brummelkamp T.R., Bernard,R. and Agami,R. (2001) A system for stable expression of short interfering RNAs in mammalian cells. Science, 296, 550–553. [DOI] [PubMed] [Google Scholar]
- 16.Miyagishi M. and Taira,K. (2002) U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol., 19, 497–500. [DOI] [PubMed] [Google Scholar]
- 17.Yu J.-Y., DeRuiter,S.L. and Turner,D.L. (2002) RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl Acad. Sci. USA, 99, 6047–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macaulay V.M., Salisbury,A.J., Bohula,E.A., Playford,M.P., Smorodinsky,N.I. and Shiloh,Y. (2001) Downregulation of the type 1 insulin-like growth factor receptor in mouse melanoma cells is associated with enhanced radiosensitivity and impaired activation of Atm kinase. Oncogene, 20, 4029–4040. [DOI] [PubMed] [Google Scholar]
- 19.Milligan J.F., Groebe,D.R., Witherell,G.W. and Uhlenbeck,O.C. (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res., 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donze O. and Picard,D. (2002) RNA interference in mammalian cells using siRNAs synthesized with T7 RNA polymerase. Nucleic Acids Res., 30, e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santoro S.W. and Joyce,G.F. (1997) A general purpose RNA-cleaving DNA enzyme. Proc. Natl Acad. Sci. USA, 94, 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feldman A.R. and Sen,D. (2001) A new and efficient DNA enzyme for the sequence-specific cleavage of RNA. J. Mol. Biol., 313, 283–294. [DOI] [PubMed] [Google Scholar]
- 23.Sohail M., Hochegger,H., Klotzbucher,A., Guellec,R.L., Hunt,T. and Southern,E.M. (2001) Antisense oligonucleotides selected by hybridisation to scanning arrays are effective reagents in vivo. Nucleic Acids Res., 29, 2041–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boutla A., Delidakis,C., Livadaras,I., Tsagris,M. and Tabler,M. (2001) Short 5′-phosphorylated double-stranded RNAs induce RNA interference in Drosophila. Curr. Biol., 11, 1776–1780. [DOI] [PubMed] [Google Scholar]
- 25.Amarzguioui M., Holen,T., Babaie,E. and Prydz,H. (2003) Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res., 31, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schurer H., Lang,K., Schuster,J. and Morl,M. (2002) A universal method to produce in vitro transcripts with homogenous 3′ ends. Nucleic Acids Res., 30, e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J. and Russell,D.W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.