Abstract
λ Exonuclease is a highly processive 5′→3′ exonuclease that degrades double-stranded (ds)DNA. The single-stranded DNA produced by λ exonuclease is utilized by homologous pairing proteins to carry out homologous recombination. The extensive studies of λ biology, λ exonuclease enzymology and the availability of the X-ray crystallographic structure of λ exonuclease make it a suitable model to dissect the mechanisms of processivity. λ Exonuclease is a toroidal homotrimeric molecule and this quaternary structure is a recurring theme in proteins engaged in processive reactions in nucleic acid metabolism. We have identified residues in λ exonuclease involved in recognizing the 5′-phosphate at the ends of broken dsDNA. The preference of λ exonuclease for a phosphate moiety at 5′ dsDNA ends has been established in previous studies; our results indicate that the low activity in the absence of the 5′-phosphate is due to the formation of inert enzyme–substrate complexes. By examining a λ exonuclease mutant impaired in 5′-phosphate recognition, the significance of catalytic efficiency in modulating the processivity of λ exonuclease has been elucidated. We propose a model in which processivity of λ exonuclease is expressed as the net result of competition between pathways that either induce forward translocation or promote reverse translocation and dissociation.
INTRODUCTION
λ Exonuclease (λ exo) is a 5′→3′ double-stranded (ds)DNA exonuclease that processively degrades one chain of the duplex to yield mononucleotides and single-stranded (ss)DNA (1–3). The ssDNA thus generated by λ exo serves as the substrate for pairing enzymes that promote homologous recombination. During bacteriophage λ recombination, λ exo acts in conjunction with its molecular partner, β, a ssDNA annealing factor (4). λ exo and β carry out homologous recombination, either with or without the aid of host proteins such as RecA, to generate linear concatemers of the λ genome, which get cleaved and packaged to produce viable phage particles (4,5).
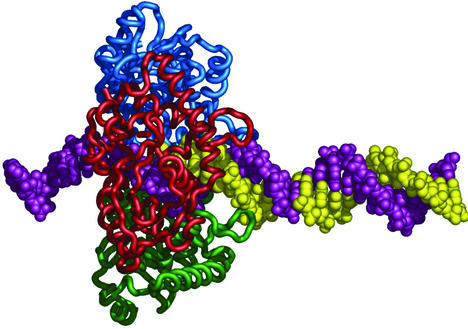
The X-ray crystallographic structure of λ exo shows it to be a homotrimeric ring-shaped molecule with a central funnel-shaped channel. This channel is wide enough to encircle dsDNA at one end but can accommodate only ssDNA at the other end (6). Docking models of partially ssDNA suggest that the ssDNA product of dsDNA resection gets threaded through the central channel (6). This mode of substrate binding, whereby an oligomeric, ring-shaped protein molecule or complex encircles its string-shaped nucleic acid substrate, has been implicated in many processive reactions in DNA and RNA metabolism. For instance, the sliding-clamp β, an accessory factor for DNA polymerase III of Escherichia coli, is thought to enhance the processivity of DNA polymerization by tethering the polymerase to the DNA via topological linkage (7). DnaB is a processive, hexameric, ring-shaped DNA helicase essential for E.coli DNA replication. Fluorescence resonance energy transfer experiments indicate that the ssDNA gets threaded through the DnaB ring (8). Similarly, electron microscopic studies of the phage T7 helicase/primase and E.coli RuvB helicases indicate that the ssDNA passes through the inner channel formed by the oligomeric rings (9,10). The tapered shape of the central channel in λ exo, the nature of the enzymatic reaction, in which dsDNA is converted to ssDNA, and the recurrence of oligomeric ring-shaped proteins in processive enzymatic transactions suggest that the processivity of λ exo is also derived from topological linkage between the enzyme and the ssDNA produced as a result of digestion of dsDNA (Fig. 1). The observation that digestion of ssDNA by λ exo is distributive rather than processive (11; our unpublished results) supports the structural model for processivity above, since partially digested dsDNA, but not ssDNA, has the undigested 3′-ending strand that is proposed to thread through the enzyme (6) (Fig. 1).
Figure 1.
Proposed model for topological linkage between λ exo and partially digested dsDNA. The model was created by assembling the structures of λ exo and DNA manually, using the program O (30) and subsequently rendered in PyMOL (31). The model is not a result of energetic docking, but is presented here for illustrative purposes. The monomers of λ exo are shown in green, red and blue. The 5′-ending strand that gets digested by λ exo is shown in yellow and the 3′-ending strand that is proposed to thread through the enzyme is shown in purple.
The putative active site of λ exo has been identified by multiple sequence alignment of viral proteins with known or suspected λ exo-like properties and by structural similarities with type II restriction enzymes (12–15). The putative active site of λ exo consists of a catalytic triad formed by two acidic residues, D119 and E129, and a positively charged residue, K131. The two acidic residues chelate a divalent cation, magnesium in the case of λ exo, which is essential for the hydrolysis of the phosphodiester bond. The positively charged lysine residue is proposed to stabilize the negatively charged reaction intermediate that is thought to arise during hydrolytic cleavage of DNA as hypothesized for restriction endonucleases (16). There are three active sites in λ exo, one in each monomer (Fig. 2). The three active sites lie on the same plane on the inner surface of the channel, and the channel is ∼33 Å wide in that plane. The central channel is ∼23 Å wide at the end through which dsDNA is proposed to be taken up and ∼15 Å at the narrow end, through which the ssDNA is proposed to be extruded during processive resection. The varying dimensions of the channel invoke conformational changes in the λ exo–dsDNA complex, so that the 5′ end of the dsDNA can access the active site and be positioned there stably during the hydrolysis of hundreds of nucleotides, one nucleotide at a time.
Figure 2.
Position of the three catalytic sites in λ exo. The monomers of λ exo are represented in green, red and blue. The catalytic triad (D119, E129 and K131) is shown in violet. The phosphate ion that we propose to be analogous to the 5′ phosphoryl terminus of the DNA substrate is represented in space filling form in orange. The conserved arginine 28 residue is depicted in space filling form in yellow. This figure was generated using PyMOL (31).
In a previous study, λ exo was stored under high phosphate conditions and subsequently crystallized in acetate buffer (6). Three residual phosphate ions remained in the trimeric molecule after crystallization, and they were coordinated by identical residues in each monomer. These residues, R28, S35, S117 and Q157, are all highly conserved in the λ exo family of recombination nucleases (13) in the SynExo family of two-component recombinases (12,17). Based on the proximity of the phosphate ion to the putative catalytic site and the preference of λ exo for a phosphate at the 5′ ends of dsDNA, we propose that the phosphate ion represents the phosphate group at the 5′ end of dsDNA and that the conserved residues interacting with the phosphate are involved in positioning the 5′ end in the λ exo–dsDNA complex to facilitate DNA digestion. We tested this hypothesis by comparing the digestion of DNA with either 5′-phosphate (5′-P) or 5′-hydroxyl (5′-OH) ends and by mutating one of the conserved phosphate-interacting residues, arginine 28, to an alanine to test the activity of the mutant (R28A) in vivo and in vitro. The results are consistent with our model for 5′ end recognition by λ exo. Our results also demonstrate that binding of the 5′-phosphorylated end of the dsDNA substrate by residues proximal to the catalytic site is a crucial step in the processive resection of linear dsDNA molecules by λ exo.
MATERIALS AND METHODS
Plasmids and bacterial strains
In the pET28a+::exo plasmid, the N-terminus of the coding sequence for λ exo has been fused to the hexa-histidine tag sequence in the pET28a+ vector, so that λ exo is expressed as a fusion protein with an N-terminal hexa-histidine tag. The pUC18::his.exo plasmid was constructed by subcloning the BamHI–BglII fragment from pET28a+::exo containing the T7 promoter region and the his.exo gene fusion into the BglII site of pUC18. The exoR28A mutant was constructed by synthesizing full-length plasmid DNA with Pfu polymerase (Stratagene) using pUC18::his.exo as template and mutagenic oligonucleotide primers, R28A1 (5′-GGCACAAATTAGC GCTCGGCGTCATC-3′) and R28A2 (5′-GATGACGCC GAGCGCTAATTTGTGCC-3′) (purchased from Operon), encoding the arginine to alanine amino acid change (altered nucleotides are shown underlined). PCR cycling conditions were as follows: 1 cycle of 2 min at 95°C, 4 min at 45°C and 6 min at 68°C followed by 18 cycles of 30 s at 95°C, 1 min at 55°C and 8 min at 68°C and one final cycle of 12 min at 68°C. The products of the PCR were transformed into E.coli DH5α by electroporation (18) and plasmid DNA was prepared from the transformants using the alkaline lysis method (19). The plasmid DNA was sequenced (at the DNA Core Laboratory, University of Miami School of Medicine) to confirm the presence of the mutation and named pUC18::his.exoR28A. The pET28a+::his.exoR28A plasmid used for expression of the R28A mutant of λ exo protein was constructed by subcloning the BamHI–NcoI fragment of pUC18::his. exoR28A into pET28a+::exo restricted with BamHI and NcoI. All bacterial strains used in this study were derivatives of E.coli K12 except E.coli BL21(DE3) which is a hybrid strain (20). The restriction enzymes used in the cloning procedures were purchased from New England Biolabs.
Complementation test
The pUC18::his.exo plasmids carrying wild-type or mutant exo genes were used to test complementation of the exo– λ phage growth defect. Escherichia coli CC118 (su° recA1) was transformed with the respective plasmids by electroporation (18). λ Plating cultures of these strains and stocks of λ exo– phage MMS1682 (χA Δb527 redα329 γ210 cI857 Rts129) and an isogenic λ exo+ phage MMS1687 (χA Δb527 γ210 cI857 Rts129) were prepared using the protocols in Arber et al. (21) by growth on an amber suppressor strain of E.coli, C600, at 30°C, since the phage used in the study carried the γ210 amber mutation and the Rts129 temperature-sensitive allele of the lysozyme gene (21).
Expression and purification of proteins
His-tagged wild-type λ exo (λ exoWT) and the his-tagged R28A mutant form of λ exo (λ exoR28A) were expressed from pET28a+::exo derivatives in E.coli BL21(DE3)[pLysS] as described (6) with the following changes. Pure his-tagged λ exo proteins were obtained by one-step purification of the clarified cell lysates by affinity chromatography over TALON resin (Clontech) and elution with an imidazole gradient (5–200 mM). The λ exo-containing fractions were pooled and dialyzed into storage buffer (0.3 M potassium phosphate, 1 mM EDTA, 1 mM dithiothreitol and 50% glycerol) and stored at –80°C. A large amount of phosphate in the storage buffer is necessary to maintain solubility of λ exo concentrates (R.Kovall, personal communication). Proteins were purified to homogeneity (as detected by Coomasie staining on SDS–polyacrylamide gels) and quantified using UV absorbance spectroscopy using a molar extinction coefficient determined by amino acid sequence (ε280 nm of 46 140/M/cm) [ProtParam primary sequence analysis tool at www.expasy.org, using the method described in Gill and von Hippel (22)].
Protein gel electrophoresis
Purified λ exoWT and λ exoR28A were subjected to SDS and native polyacrylamide gel electrophoresis to assess their purity and aggregation state. SDS–polyacrylamide gel electrophoresis was conducted using precast 12% Tris–glycine gels (Bio-Rad) and a mini-gel protein electrophoresis system (Bio-Rad). Native polyacrylamide gel electrophoresis was conducted using the PHAST gel system (8–25% gradient gels; Pharmacia). BSA and chicken egg albumin were used as molecular weight markers for native gels (Native PAGE Markers Kit; Sigma).
Preparation of long linear dsDNA substrates
DNA substrates were prepared by purification of pUC19 plasmid DNA from overnight cultures of E.coli DH5α in TB culture medium (DIFCO) containing 100 µg/ml of ampicillin by the alkaline lysis method (19). The plasmid preparation was linearized by restriction with PstI (New England Biolabs) and cleared of proteins by extraction with phenol:chloroform: isoamyl alcohol mixture at 25:24:1. The DNA was then ethanol precipitated twice and resuspended in T0.1E buffer (10 mM Tris pH 8.0, 0.1 mM EDTA). The DNA solution was quantified using UV absorbance spectrophotometry at 260 nm. DNA substrates with 5′-OH ends, prepared by treating the above DNA with calf intestinal phosphatase, were a generous gift from T. Vellani.
Preparation of 32P-labeled oligonucleotide substrates
Oligonucleotides WL65 (5′-GCCTGACCATGTACAGAG TGCTGATCTCCTCATTCTGGTATCGTCTAGATGGAG AAAACTAGTAG-3′) and WL65C (5′-CTACTAGTTTT CTCCATCTAGACGATACCAGAATGAGGAGATCAGC ACTCTGTACATGGTCAGGC-3′) (Sigma Genosys) were gel purified from 20% acrylamide/urea sequencing gels followed by DNA extraction with DNA elution buffer (0.5 M ammonium acetate, 10 mM magnesium acetate, 1 mM EDTA pH 8.0 and 0.1 M SDS), phenol/chloroform extraction and ethanol precipitation.
Aliquots of 16 pmol of WL65 were 3′ end-labeled by treating with 60 µCi of [α-32P]ddATP (Amersham Pharmacia Biotech) and terminal deoxynucleotidyl transferase (New England Biolabs) at 37°C for 2 h. The terminal transferase was inactivated at 90°C for 7 min to obtain 3′ end-labeled WL65 (WL65*). A portion of WL65* (8 pmol) was treated with T4 polynucleotide kinase (New England Biolabs) and 1 mM ATP at 37°C for 30 min to obtain 5′-phosphorylated WL65* (5′-P WL65*). Phosphorylated WL65C (5′-P WL65C) was prepared by treating 32 pmol of gel purified WL65C as described above. The kinase reactions were inactivated by heating at 90°C for 10 min and cleared of proteins using phenol/chloroform extraction. The oligonucleotides were ethanol precipitated and resuspended in T0.1E buffer. Radioactively labeled dsDNA substrates with 5′-P and 5′-OH ends were obtained by annealing 5′-P WL65* with a 4-fold excess of 5′-P WL65C and WL65* with a 4-fold excess of WL65C, respectively. The preceding mixture of the two complementary oligonucleotides, containing 100 nM labeled ends, was heated at 90°C for 4 min and cooled slowly to room temperature to allow annealing to produce dsDNA substrates. The formation of labeled dsDNA oligonucleotides was confirmed by ascertaining the susceptibility of annealed substrates to cleavage by restriction enzymes (data not shown).
Fluorescence-based exonuclease assays
The exonuclease activity of purified his-tagged λ exo proteins was quantified using the differential fluorescence output of Picogreen dye (Molecular Probes, OR) bound to dsDNA versus ssDNA. Picogreen dye has a high fluorescence output when bound to dsDNA and weaker fluorescence when bound to ssDNA (nucleotides do not alter the fluorescence of the dye; data not shown). Hence, DNA digestion can be monitored by following the decrease in fluorescence over time. The Picogreen assay method is discontinuous because the presence of the dye was found to inhibit DNA digestion by λ exo (data not shown). In a typical assay, the nuclease reactions were performed with 1 nM dsDNA ends of linear pUC19 DNA in assay buffer (20 mM CHES pH 9.4, 2.5 mM MgCl2 and 1 mM DTT) in 10 µl reactions. Fixed amounts of substrate were titrated with varying amounts of λ exo to calculate the fractional activity and DNA hydrolysis rates at saturation of the enzyme preparations, assuming that the active species is a trimer (6,23). In each reaction, the enzymes were diluted in dilution buffer (0.3 M potassium phosphate, 1 mM EDTA, 1 mM dithiothreitol and 10% glycerol) and an equal volume (1 µl) of diluted enzyme was added to the tube and the samples were incubated at room temperature for 2 min, followed by addition of MgCl2 to 2.5 mM to initiate the reaction. The reactions were incubated at 37°C for the desired amount of time and quenched with EDTA added to 25 mM. Then 2 ml of Picogreen dye, diluted 1:1000 in TEK buffer (10 mM Tris–HCl pH 8, 25 mM EDTA), was allowed to bind the DNA in the dark for 3 min and steady-state fluorescence spectra were collected (TimeMaster; Photon Technology International) by exciting the samples at 484 nm and scanning the emission from 510 to 540 nm, with a 5 nm bandpass. Each fluorescence spectrum was collected four times in succession, averaged, integrated and the integrated data were plotted against enzyme concentration. Since the limit digest converts 1 mol of dsDNA to 0.5 mol of ssDNA and 0.5 mol of nucleotides (1), the percentage DNA digested can be calculated by treating the fluorescence output of thermally melted DNA equivalent of half the initial concentration of dsDNA used in the assay as 100% digestion and referencing the output of each sample to the difference in fluorescence intensity between undigested dsDNA and the ssDNA control. Initial rates of digestion were determined by applying this formula: initial rate = (f × 2686)/t, where f is the fraction of substrate digested, 2686 is the number of nucleotides per strand in linear pUC19 and t is the incubation time (30 s). The fraction of substrate digested under saturating conditions was between 0.13 (for λ exoWT acting on 5′-OH DNA) and 0.26 (for λ exoWT acting on 5′-P DNA). The limiting rate, Vmax, was calculated by fitting the data to a hyperbolic rate equation (GraphPad Prism 3.0; GraphPad Software Inc., San Diego, CA). The turnover number, kcat, or rate per active λ exo trimer per dsDNA end, was calculated by dividing Vmax by 2, since at saturation we expect two active λ exo molecules to be acting on each molecule of linear dsDNA. To determine the suitability of the single time point chosen for performing the titrations described above and to determine the maximum extent of digestion carried out by the enzymes on the DNA, a time course was performed as follows. Linear pUC19 DNA at 1 nM ends was treated with a slight excess (4–8 nM trimers) of λ exo trimers in assay buffer in a 10 µl reaction for varying lengths of time. The reactions were quenched by addition of EDTA to 25 mM and diluted Picogreen dye was added and fluorescence spectra were collected and analyzed as decribed above. The data were fitted to a hyperbolic equation, in which the asymptote represents the maximum extent of digestion.
Digestion of linear dsDNA by λ exoWT
Linear pUC19 DNA (11.4 nM ends) with either 5′-P or 5′-OH ends was treated with 11 nM active trimers of λ exoWT in the assay buffer (same as in the fluorescence-based assay) in a reaction volume of 10 µl for varying lengths of time. The reactions were quenched by addition of gel-loading buffer (50% glycerol, 50 mM EDTA, 0.2% bromophenol blue, 0.2% xylene cyanol and 1% SDS). The samples were loaded onto a 1% agarose gel and electrophoresed at 4.5–5 V/cm. Gels were stained using ethidium bromide and images recorded using a CCD camera, a UV transilluminator and AlphaImager 2000 (Alpha Innotech Corp.). The gel was scanned and the band intensity was converted into percentage DNA digested by normalizing the 0 time point to 0% digestion in each case. The percentage DNA digested was plotted and the curves were fitted to hyperbolae (GaphPad Prism 3.0). The asymptote of the hyperbola gave the extent of DNA digested under saturating conditions. The rate of digestion was estimated from the first derivative of the curve close to 0 and converting that to rate in nucleotides digested per second using the formula, rate of hydrolysis = (S × 2686)/2, where S is the differential expressed in percentage DNA digested per unit time, 2686 is the number of nucleotides per strand in linear pUC19 and 2 is the number of ends per molecule of linear pUC19.
Digestion of 32P-labeled oligonucleotide substrates
Radioactively labeled dsDNA oligonucleotides (10 nM labeled ends) were treated with λ exo (at a ratio of active trimers to dsDNA ends varying from 1:1 to 1:50) in BSA (50 µg/ml) in exonuclease assay buffer (described above) at 37°C for varying lengths of time. The reactions were quenched with 2× urea/TTE buffer (16 M urea, 180 mM Tris, 58 mM taurine, 1 mM EDTA, 0.5% bromophenol blue and 0.5% xylene cyanol). Samples were reheated (90°C for 3 min), separated by electrophoresis through a 20% polyacrylamide gel containing 8 M urea and the species on the gel were imaged using a Storm PhosphorImager System (Molecular Dynamics) (24).
DNase I protection of 32P-labeled 5′-OH oligonucleotide substrates
Radioactively labeled dsDNA oligonucleotides (10 nM labeled ends) with 5′-OH ends were incubated with λ exo (87 nM trimers) in BSA (50 µg/ml) in exonuclease assay buffer (without MgCl2) in a 50 µl reaction at 37°C for 10 min in duplicate. Fifty microliters of DNase I reaction mixture containing DNase I (2 U/µl) (USB), MgCl2 and CaCl2 was added to one tube to bring the final concentrations to 0.2 U/100 µl DNase I, 0.5 mM CaCl2 and 20 µM MgCl2. The DNase I reactions were incubated on the bench at room temperature for 10 s and quenched with 12 µl of 1% SDS, 50 mM EDTA. Samples were then centrifuged under vacuum to dryness and resuspended in 10 µl TE buffer. Magnesium chloride (to 2.5 mM) was added to the other tube containing the 5′-OH DNA and λ exo and the reaction was incubated at 37°C. Ten microliters was withdrawn from this tube at various time points and quenched in 10 µl 2× urea/TTE buffer. Ten microliters of 2× urea/TTE buffer was also added to the resuspended DNase I samples. Both the λ exo-treated and DNase I-treated samples were electrophoresed through a 20% polyacrylamide gel containing 8 M urea and processed and analyzed as described above.
Redigestion of the products of the λ exoR28A reaction with fresh enzyme
Radioactively labeled dsDNA oligonucleotides (10 nM labeled ends) were treated with 20 nM active trimers of λ exoR28A under the assay conditions described above for 1 h. At the end of the 1 h incubation at 37°C, the products of the reaction were subjected to another round of digestion by addition of 20 nM active trimers of λ exoWT or λ exoR28A. A portion of the reaction was withdrawn at different times after addition of the second enzyme and quenched with 2× urea/TTE buffer. The samples were then resolved by electrophoresis and analyzed further as described above.
Processivity assay using heparin trap
The average processivities of λ exoWT and λ exo R28A enzymes were determined by using heparin to trap the excess enzyme in the fluoresence-based exonuclease assay. Reaction conditions were as described in the section on the fluoresence-based exonuclease assay with the following modification. Sufficient λ exo was added to saturate the dsDNA ends in the reaction (determined by titration). Heparin to a final concentration of 9.6 mg/ml was added to the reaction either after the reaction was quenched at the desired time point or after the reaction was initiated and had progressed for 10 s. The samples were treated and the data analyzed as described before to determine the percentage DNA digested. The nucleotides digested per active λ exo complex was determined by multiplying the value of the percentage DNA digested by 1343, which is the number of nucleotides/strand available for digestion per active λ exo complex on linear pUC DNA upon saturation of both ends.
RESULTS
Complementation test
To assess the in vivo function of λ exo gene variants, we employed a complementation test that utilizes a conditional requirement for exo function during λ phage growth. Escherichia coli recA strains are unable to support the growth of λ phage that are defective in Gam and Exo functions, therefore gam– exo– λ phage are unable to form plaques on such strains. The plaque-forming ability was restored if λ exo was produced from a plasmid source, pUC18::his.exo, transformed into the non-permissive recA host (Table 1). The pUC18::his.exoR28A plasmid, expressing the R28A mutant of λ exo, was unable to restore the plaque-forming ability of λ exo-defective phage, indicating that the highly conserved R28 residue is important for in vivo function of λ exo (Table 1).
Table 1. The λ exoR28A mutant does not support phage growth in vivo.
| Plasmid | Expressed version of λ exo | exo+ phage | exo– phage |
|---|---|---|---|
| pUC18 | None | + | – |
| pUC18::his.exoWT | WT | + | + |
| pUC18::his.exoR28A | R28A | + | – |
Growth of exo+ phage (λ MMS 1687) and exo– phage (λ MMS 1689) was followed by monitoring the appearance of plaques after 10 h of growth at 30°C. Escherichia coli recA mutants support low λ plating efficiencies in general, therefore the data were used as qualitative indicators of the ability of exo genes to complement the growth defect in exo– phage.
Oligomeric state of λ exoWT and λ exoR28A
The complementation test suggested that λ exoR28A is impaired for exonuclease function in vivo. We purified λ exoR28A protein to test its activity in vitro and compare it to that of λ exoWT protein. The proteins were purified to apparent homogeneity as judged by SDS–polyacrylamide gel analysis (detected using Coomassie staining; data not shown). Native polyacrylamide gel electrophoresis indicates that migration of λ exoWT and λ exoR28A proteins is consistent with an oligomeric state larger than a dimer (Fig. 3). The structure determined by X-ray crystallography indicates that λ exo is a trimer, with a molecular weight of 85 350 kDa (6). The λ exoR28A mutant migrates faster than λ exoWT, probably because of the removal of three positive charges per trimer. The behavior of λ exoR28A during purification and its migration on the native gel suggests that the tertiary and quaternary structure of the protein resembles that of λ exoWT. Therefore, the defect of the mutant evident in the complementation test is unlikely to be a result of gross folding problems as we were able to purify the mutant enzyme and study its enzymatic activity (see below).
Figure 3.

Oligomeric state of λ exo proteins. The oligomeric state of λ exoR28A was compared to that of λ exoWT by native polyacrylamide gel electrophoresis as described in Materials and Methods. Lane 1 contains molecular weight markers consisting of 1 µg each of BSA (migrates as two species, a 66 kDa monomer and a 132 kDa dimer) and chicken egg albumin (45 kDa). Lane 2 contains λ exoWT (1.46 µg). Lane 3 contains λ exoR28A (1.28 µg). The migration of both proteins is consistent with formation of trimers (his-tagged λ exo trimers have a molecular weight of 85 350 kDa).
The R28 residue of λ exo is involved in recognition of the 5′-P at the end of dsDNA
The exonuclease activities of pure λ exoWT and λ exoR28A were assayed as described in Materials and Methods. An example of fluorescence spectra (raw data) is shown in Figure 4. The titration of linear dsDNA with λ exoWT indicates that the activity saturates at a 1:3 ratio of dsDNA ends:λ exo trimers, irrespective of the state of phosphorylation of the DNA ends. This is consistent with 33% fractional activity, assuming that λ exo is a trimer, as determined by X-ray crystallography and gel filtration (6,23). The titration of linear dsDNA with λ exoR28A indicates that the activity saturates at 1:4 and 1:3 dsDNA ends:λ exo trimers on 5′-P and 5′-OH DNA, consistent with 25 and 33% fractional activities, respectively. λ exoWT degrades the pUC19 dsDNA substrate containing 5′-P 2-fold faster than that with a 5′-OH moiety; the kcat for dsDNA with 5′-P is 11.7 nt degraded/s (nt/s), whereas that for DNA with 5′-OH is 5.9 nt/s (Fig. 5). These results are qualitatively similar to those obtained in a previous report (25). In the same previous report, the Kmapp values reported for dsDNA substrates that differ only in the phosphorylation state of the 5′ end were the same (25), and our assays were performed under saturating DNA conditions (determined empirically), well above the Kmapp determined in that report. The reduced turnover number, as measured by the fluorescence- based assay under saturating conditions, suggests that the reduced rate of hydrolysis of 5′-OH substrates by λ exoWT enzyme is due to an inherent catalytic defect. A more detailed view of the reduced activity of λ exoWT on 5′-OH substrates is presented at the end of this section. The λ exoR28A enzyme has a measurable but not significantly reduced turnover number in vitro. However, the kcat for exonuclease activity of λ exoR28A is similar for dsDNA substrates with or without a 5′-P (8.9 and 7.8 nt/s) (Fig. 5) (P > 0.05, unpaired t-test performed using GraphPad InStat 3.05; GraphPad Software Inc.). The apparent insensitivity of λ exoR28A to the phosphorylation state of the ends of dsDNA suggests that the arginine residue in λ exoWT is indeed responsible for recognizing the 5′-phosphate at the ends of native dsDNA.
Figure 4.
Examples of fluorescence spectra used to follow digestion of dsDNA by λ exoWT. Linear dsDNA was digested with λ exoWT enzyme under standard reaction conditions and fluorescence spectra were collected (see Materials and Methods). The dye fluoresces with a higher intensity when bound to dsDNA than to single-stranded DNA or to nucleotides, therefore a decrease in fluorescence reflects digestion of DNA. The titrations were performed with various concentrations of enzyme against 1 nM dsDNA ends. The cartoon representations on the right depict the form of the DNA substrate in the reaction at the time the fluorescence spectrum was collected, the ring represents λ exo and the lines represent DNA strands. The curve labeled ‘dsDNA’ is the fluorescence spectrum of the sample in which the reaction was quenched with EDTA as described in Materials and Methods prior to the addition of enzyme. The sample used in the ‘ssDNA’ curve contained 0.5 equivalents of thermally denatured input DNA to imitate limit digestion conditions. Curves 1–5 contained 1 nM dsDNA ends treated with 0.6, 1.2, 2.5, 5 and 10 nM λ exoWT trimers for 30 s, respectively.
Figure 5.
λ exoR28A is impaired for 5′ end recognition. Double-stranded DNA (1 nM ends) in the form of linear pUC19 was titrated with the indicated amounts of λ exo; λ exoWT on 5′-P DNA is represented by open circles, λ exoWT on 5′-OH DNA is given by closed circles, open squares represent the activity of λ exoR28A on 5′-P DNA and λ exoR28A on 5′-OH DNA is given by closed squares. The data were fitted to a hyperbolic rate equation to obtain the solid lines and the turnover number (kcat) indicated in the text. Each data set is derived from two trials, and the error bars represent standard errors.
The limiting rate of DNA digestion at saturating λ exoWT concentration is lower when the 5′ end of the substrate lacks a phosphate (11.7 versus 5.9 nt/s), even though the assay measures the net result of several successive cleavage events. The difference in limiting rates suggests one of the following. (i) The turnover of the first nucleotide at the 5′ end is rate-limiting and is slower for the 5′-OH terminated substrate than for the 5′-P terminated substrate. Once the first nucleotide is cleaved, all other subsequent nucleotides are turned over at the same rate on both 5′-OH and 5′-P substrates. (ii) Only a fraction of enzyme–substrate complexes acting on 5′-OH DNA are competent to turn the substrate over and the measured rate represents the digestion of that sub-population of DNA molecules. (iii) A combination of the preceding two models, such that only a fraction of enzyme–substrate complexes acting on 5′-OH DNA are competent to turn the substrate over and this active fraction performs the first hydrolytic turnover at a rate that is slower than the rate of hydrolysis of 5′-P DNA.
These possibilities were addressed by monitoring the digestion of linear pUC19 DNA on an agarose gel (see Fig. 7 and associated text).
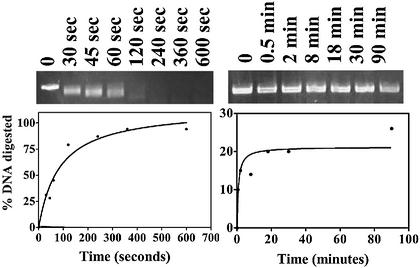
Figure 7.
Digestion of linear dsDNA by λ exoWT. Linear pUC19 DNA (11.4 nM ends) with either 5′-P or 5′-OH ends was treated with 11 nM active trimers of λ exoWT for the lengths of time indicated above each lane. The gel was scanned and the data plotted as described in Materials and Methods. The data on the left correspond to the digestion of 5′-P DNA and the data on the right correspond to the digestion of 5′-OH DNA. The extent of digestion of 5′-P and 5′-OH substrates were 100 and 21%, respectively. The rates of hydrolysis of 5′-P and 5′-OH substrates were 17 and 3.3 nt/s, respectively.
Recognition of 5′ ends of DNA is necessary for efficient resection
The digestion of linear dsDNA by λ exo enzymes under saturating conditions was followed over time to estimate the extent of digestion. DNA is 100% digested given sufficient time only in the case of λ exoWT acting on 5′-P DNA (Fig. 6). λ exoWT acting on 5′-OH DNA digests only 21% of the total DNA, whereas λ exoR28A manages to digest only 15 and 18% of the total 5′-P and 5′-OH DNA, respectively. The data suggest that end-recognition is essential for efficient resection of DNA, by affecting either the number of active complexes formed by λ exo on dsDNA ends or the processivity of the enzyme–substrate complexes formed in the reaction. If the number of active complexes is altered, then the same fraction of total DNA should be digested on a linear dsDNA substrate that is shorter or longer than pUC19. If the processivity of the enzyme is altered, then a greater extent of the DNA should become digestible when a shorter dsDNA substrate is treated with the enzyme. These two models were addressed by monitoring the digestion of linear dsDNA on agarose and polyacrylamide gels (following sections).
Figure 6.
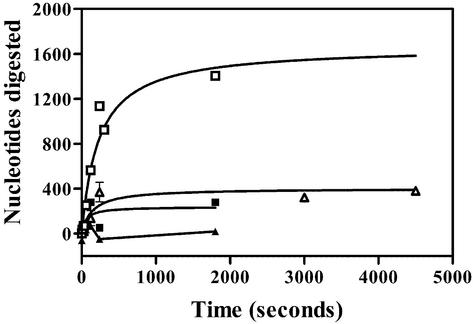
Recognition of 5′ ends is necessary for efficient DNA resection. Linear pUC19 DNA (1 nM ends) with either 5′-P or 5′-OH ends was treated with a slight excess (4–8 nM active trimers) of λ exoWT or λ exoR28A for varying lengths of time. The percentage DNA digested and the rates in each case were calculated from the fluorescence spectra as described in Materials and Methods. The symbols used represent the enzyme–substrate pairs as in Figure 5. The extents of digestion by λ exoWT acting on 5′-P and 5′-OH DNA were 120 and 21%, respectively. The extents of digestion by λ exoR28A acting on 5′-P and 5′-OH DNA were 15 and 18%, respectively. The inset is a truncated version of the time course graph that depicts the early portion of the progress curve in better detail.
The λ exoWT enzyme forms inert complexes on 5′-OH dsDNA
Linear pUC19 DNA with 5′-P or 5′-OH ends was treated with saturating amounts of λ exoWT enzyme for varying lengths of time. Linear dsDNA with 5′-P ends was digested efficiently (Fig. 7). The extent and rate of digestion of 5′-P DNA were 100% and 17 nt/s. The extent and rate of digestion of 5′-OH DNA, on the other hand, were 21% and 3.3 nt/s. The lower extent of digestion of 5′-OH DNA suggests that inert complexes are formed, since a fraction of the DNA (∼79%) appears to be refractory to digestion by λ exoWT enzyme. If the reduction in rate can be accounted for by the formation of inert complexes alone, then the rate of hydrolysis of 5′-OH DNA, after correcting for the fraction of active complexes, should be similar to the rate of hydrolysis of the 5′-P DNA. If, however, there is a delay in the turnover of the first nucleotide at the end of the 5′-OH DNA, and only a fraction of DNA–enzyme complexes can carry out this first turnover, then the corrected rate of hydrolysis of the 5′-OH end will be lower than the rate of hydrolysis of the 5′-P end. The rate of DNA hydrolysis on 5′-OH DNA, after accounting for the fraction of DNA–enzyme complexes apparently active in the reaction (0.21), is 16 nt/s, which is comparable to the DNA hydrolysis rate of 5′-P DNA. The results are consistent with the former model, which is also model (ii) described in the previous section, i.e. only a fraction of enzyme–substrate complexes acting on 5′-OH DNA are competent to turn the substrate over and the measured rate represents the digestion of that sub-population of DNA molecules.
DNA end recognition influences catalytic efficiency of λ exo
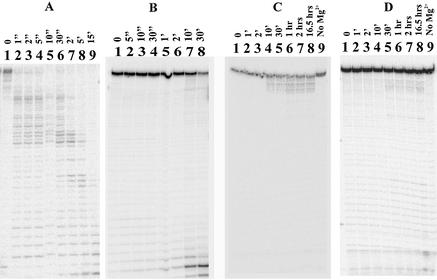
To determine the origin of the reduced activity of λ exo on linear dsDNA when end recognition is perturbed, we followed the digestion of 32P-labeled short oligonucleotide substrates by λ exo. If dsDNA molecules are digested by a processive mechanism, the following predictions will hold true: (i) when dsDNA is treated with saturating amounts of enzyme, then full-length DNA should disappear before limit digestion products appear; (ii) when DNA ends are present in vast excess over enzyme molecules, undigested DNA should persist and limit digestion products should increase over the time course; (iii) under these sub-saturating conditions a distribution of partially digested products should not coexist with the full-length substrate in a time window that is long enough to allow for digestion of an individual DNA molecule. The results indicate that λ exoWT digests 5′-P DNA rapidly and processively, and the DNA is digested to completion within minutes (Figs 8A and 9). The λ exoWT enzyme degrades 65 bp 5′-OH DNA processively but poorly, and the limit digestion products are the same length as those appearing in digestion of 5′-P DNA under sub-saturating conditions (Fig. 8B). A similar fraction of the short radiolabeled 5′-OH DNA (∼28%; data not shown) appears to be susceptible to digestion as in the assays on 5′-OH linear pUC19 DNA (preceding section). These results lead us to propose that a large fraction of complexes formed by λ exoWT at 5′-OH DNA ends are inert and that hence for any length of the substrate the same fraction of the total DNA is digestible. We propose that, with low probability, the enzyme–substrate complex with 5′-OH DNA is able to make the transition to a hydrolytically competent complex, giving rise to the low rates and extents of digestion measured in the assays described in this study.
Figure 8.
DNA end recognition influences catalytic efficiency of λ exo. In each set of experiments, the 65 bp dsDNA substrate (10 nM labeled ends) was treated with λ exo at a 1:1 ratio of DNA ends to λ exo trimers for varying lengths of time. (A) λ exoWT digests 5′-P dsDNA rapidly (lanes 1–9 are 0, 1, 2, 5, 10 and 30 s and 2, 5, 15 min, respectively). (B) λ exoWT digests 5′-OH dsDNA slowly and poorly (lanes 1–8 are 0, 5, 10 and 30 s and 1, 2, 10 and 30 min, respectively). (C) λ exoR28A digests 5′-P dsDNA distributively and poorly (lanes 1–9 are 0, 1, 2, 10 and 30 min and 1, 2 and 16.5 h and a control without Mg2+, respectively). (D) λ exoR28A digests 5′-OH dsDNA distributively and poorly (lanes 1–9 are 0, 1, 2, 10 and 30 min and 1, 2 and 16.5 h and a control without Mg2+, respectively). Note the persistence of full-length substrate even after prolonged exposure to λ exo in (B), (C) and (D) in contrast to the rapid disappearance of full-length substrate in (A).
Figure 9.
Distribution of digestion products generated by λ exoWT and λ exoR28A. Assay conditions were the same as described in Figure 4. The radiolabeled oligonucleotide 5′-P substrate was treated with sub-saturating amounts of λ exo for varying lengths of time. dsDNA was treated with λ exoWT at a ratio of 1:50, 1:10 and 1:2 active λ exo trimers to dsDNA ends in sets 1, 3 and 5, respectively. In sample sets 2, 4 and 6, dsDNA was treated with λ exoR28A at a ratio of 1:50, 1:10 and 1:2 active λ exo trimers to dsDNA ends, respectively. In 1 and 2, the samples were incubated at 37°C for 0, 15, 45 and 60 min, from left to right, respectively. In set 3 samples were incubated for 0, 5, 15, 60 and 120 min, respectively. In sets 4–6, samples were incubated for 0, 5, 15, 30, 60 and 120 min, respectively.
The λ exoR28A mutant digests DNA poorly, irrespective of the phosphorylation state of the 5′ end of DNA (Fig. 8C and D), even when the reaction is incubated for 1 h or longer, at which times we expect the DNA to have been digested to completion (time expected for complete digestion is calculated by dividing the number of nucleotides in the substrate by the turnover number. In the case of λ exoR28A acting on both ends of oligonucleotide substrates that are 66 bp long, this time is ∼8 s). Moreover, digestion products generated by λ exoR28A (46–49 nt long, see Fig. 11B) never approach the length of the limit digestion products seen in λ exoWT reactions (8–10 nt long). λ exoR28A exhibits compromised processivity, since it carries out a limited number of turnovers on dsDNA and generates a similar distribution of partially digested DNA molecules under sub-saturating conditions (Fig. 9).
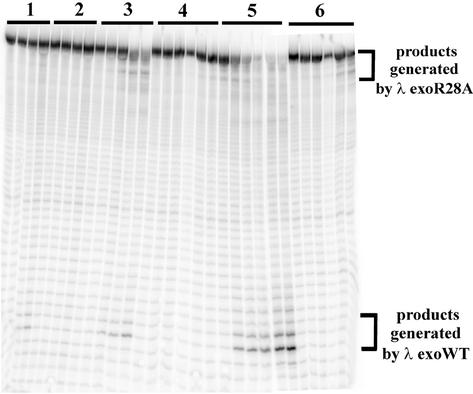
Figure 11.
λ exoR28A has limited processivity. A reaction containing λ exoR28A and dsDNA was challenged with a fresh aliquot of either λ exoWT or λ exoR28A protein. (A) After a 1 h incubation of the 65 bp dsDNA substrate with λ exoR28A, a fresh aliquot of 20 nM active trimers of λ exoWT (lanes 1–5) or λ exoR28A (lanes 6–10) was added to the tube and 10 µl aliquots were withdrawn at the various time points (lanes 1–5 and lanes 6–10 represent samples taken at 0, 1, 2, 30 and 60 min, respectively) and quenched in an equal volume of 2× urea/TTE buffer and further processed and analyzed as described in Materials and Methods. (B) The size distribution of limit digestion products generated by λ exoR28A. Lane 5 contains the same sample as lane 10 in (A); the gel was run longer to allow for better resolution of the digested DNA species; lanes 1–4 contain molecular weight markers generated by restriction digestion of the radiolabeled dsDNA substrate used in the assay.
In contrast to the relative insensitivity of λ exoR28A rates of DNA hydrolysis to the phosphorylation state of the 5′ ends of dsDNA in the assay conducted on much longer substrates (Figs 5 and 6), λ exoR28A appears to digest 5′-P DNA more efficiently than 5′-OH DNA in this sensitive assay (compare Fig. 8C and D). This suggests that other residues that appear to bind the phosphate ion in the crystal structure also contribute to recognition of the phosphate at the 5′ end.
These data suggest that the defect in catalysis that is evident in the wild-type enzyme in cleavage of the first nucleotide of 5′-OH DNA is reiterated at every step of hydrolysis in the λ exoR28A mutant. Therefore the digestion observed in the fluorescence-based assay, in which the initial rates are plotted, is a result of a limited number of turnovers, and hence limited processivity, of λ exoR28A on dsDNA.
λ exoWT enzyme binds to 5′-OH DNA
The formation of λ exoWT–5′-OH DNA complexes was monitored by performing a DNase I protection assay. Radioactively labeled short oligonucleotide substrates with 5′-OH ends were incubated with λ exoWT enzyme under saturating conditions in the absence of magnesium and subsequently treated with DNase I as described in Materials and Methods. The formation of λ exoWT–5′-OH DNA complexes was indicated by the protection of DNA from DNase I action (Fig. 10). The protection extends to about 10–12 nt on the 5′ strand. The footprint of λ exoWT on 5′-OH DNA detected by us is similar to, albeit shorter than, its footprint on 5′-P DNA reported in a previous study (25). The nature of products formed on λ exoWT treatment of 5′-OH DNA under these conditions was similar to those described in the previous section (data not shown). Furthermore, the 5′-OH DNA used in these assays was susceptible to digestion by T7 gene 6 exonuclease (data not shown), which is a non-processive 5′→3′ dsDNA exonuclease that, unlike λ exo, does not have specificity for the 5′-P (3). Therefore, the limited digestion of 5′-OH DNA by λ exoWT is due to a catalytic defect in the enzyme–substrate complex and is not due to an inability of λ exoWT to bind the DNA substrate, nor is it due to an unspecified block at the ends of a large fraction of the substrate.
Figure 10.
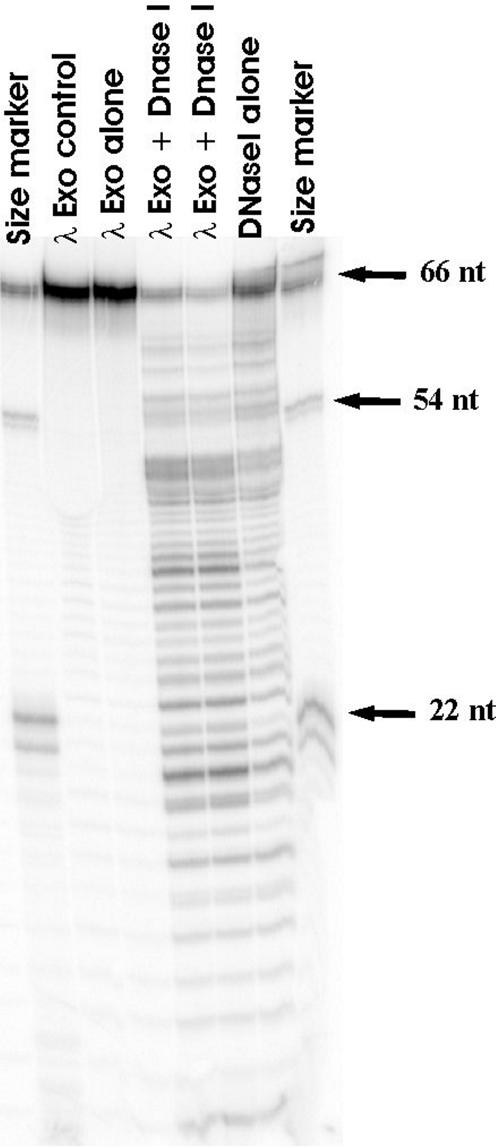
λ exoWT protects 5′-OH DNA from DNase I. Radiolabeled 5′-OH dsDNA was incubated with λ exoWT enzyme and the complexes formed were probed by DNase I treatment. The lanes marked as size marker contain molecular weight markers generated by restriction digestion of the radiolabeled dsDNA substrate used in the assay. The λ exo control lane contains only DNA and λ exoWT enzyme as reaction components. The λ exo alone lane contains all components of the reaction except DNase I. The two lanes marked λ exo + DNase I are duplicate samples containing all reaction components. The DNase I alone lane contains all reaction components except λ exoWT. λ exoWT protects a region spanning ∼10 bp from the 5′ end of the DNA from digestion by DNase I.
λ exoR28A enzyme has limited processivity
The limit digestion products resulting from digestion of 5′-P dsDNA by saturating amounts of λ exoR28A appear around 1 h and persist through overnight incubation. The persistence of the partially digested DNA molecules might reflect the inherently reduced processivity of λ exo R28A or might result from inactivation of the λ exoR28A enzyme during the course of the experiment. To distinguish between these two possibilities, we challenged an ongoing λ exoR28A reaction with saturating amounts of either λ exoWT or λ exoR28A enzyme. The results, shown in Figure 11, indicate that the partially digested DNA molecules produced by λ exoR28A are susceptible to further digestion by both enzymes. However, whereas the challenge with fresh λ exoWT results in the digestion of the partially digested molecules to completion (the formation of limit digestion products), the addition of fresh λ exoR28A results in the formation of limit digestion products of the same length as were observed in the absence of challenge (compare last lane of set 6 in Fig. 9 with lane 5 in Fig. 11B). The results suggest that although λ exoR28A appears to undergo some inactivation during the course of the reaction, the enzyme has a greatly reduced processivity, and is only able to cleave 17–20 nt from dsDNA substrates before arresting.
The average processivities of λ exoWT and λ exoR28A enzymes were measured directly using a single-turnover assay utilizing heparin to trap excess enzyme. In an exonuclease reaction, when an ongoing λ exo reaction is challenged with heparin, an amount of digestion corresponding to the average amount of digestion carried out by a λ exo–DNA complex will be observed. The results, shown in Figure 12, indicate that λ exoWT has an average processivity of 400 nt, whereas the λ exo R28A enzyme appears distributive in this assay (digestion was not detectable when the heparin trap was added before or after initiation of the reaction).
Figure 12.
λ exoR28A is a distributive exonuclease. The processivity of λ exoWT and λ exoR28A was determined using heparin to trap excess enzyme. Linear pUC19 DNA (1.1 nM ends) was treated with saturating amounts of λ exo as described in the fluoresence-based exonuclease assay and the reaction was incubated for varying amounts of time. Heparin to 9.6 mg/ml was added either after quenching in the control reactions (open squares for λ exoWT; closed squares for λ exoR28A) or after 10 s post-initiation of the reaction with MgCl2 (open triangles for λ exoWT; closed triangles for λ exoR28A). Control reactions in which the heparin was added prior to the addition of λ exo enzymes were performed to confirm that the amount of heparin used was sufficient to trap the enzyme and prevent digestion of DNA (data not shown).
DISCUSSION
Processivity is a property common to many enzyme systems that carry out critical functions in nucleic acid metabolism. The mechanism by which processivity is imparted in these systems has benefited from a structural point of view. More specifically, the structural view of processivity is largely derived from work on toroidal proteins that have been demonstrated to encircle their substrates (see for example 7–10). The topological linkage model provides an elegant mechanism for balancing the forces that enable enzymes to remain stably bound to their substrates during catalysis and also allow for freedom of movement (reviewed in 26,27). λ exo is a highly processive nuclease that shares this toroidal structure with the processive enzyme systems described above. Moreover, the distributive action of λ exo on ssDNA and the tapered channel in the enzyme point to the same structural basis for processivity.
In this study we have used λ exo as a model system to identify components of the dynamic enzyme–substrate complex that facilitate access of the DNA to the active site of the enzyme during processive digestion. The active site of λ exo was identified independently by several laboratories and unpublished preliminary studies in our laboratory confirmed the active site residues, indicated in Figure 2. In the structural studies (6), λ exo was stored under high phosphate conditions and subsequently crystallized in acetate buffer. Residual phosphate ions remained in the molecule after crystallization, and they were coordinated by specific residues in each monomer. These residues, R28, S35, S117 and Q157, are all highly conserved in the λ exo family of recombination nucleases (13) in the SynExo family of two-component recombinases (12,17). The proximity (∼7–8 Å) of the phosphate ion to the catalytic triad, the conservation of the residues that chelate the phosphate ion and the preference of λ exo and other enzymes in the SynExo family for a 5′-P to a 5′-OH, taken together, lead us to the following model. We propose that the phosphate ion in the crystal structure represents the phosphate at the 5′ ends of broken dsDNA, the natural substrate for λ exo. Moreover, we propose that the residues involved in binding the phosphate ion (Fig. 13) are responsible for recognizing the 5′ end of DNA and facilitating DNA positioning near the catalytic site for hydrolysis of the scissile phosphodiester bond between the ultimate and the penultimate nucleotides on the 5′ ending strand of the dsDNA substrate.
Figure 13.
Residues interacting with the phosphate ion. The residues in λ exo interacting with the phosphate ion are shown. The distances indicated are derived from the X-ray crystallographic structure (PDB no. 1AVQ) (6) for monomer chain A.
To test this hypothesis we mutated one of the highly conserved residues, arginine 28, to alanine to abrogate its ability to interact with the phosphate. The inability of this R28A mutant to support phage growth in vivo suggests that it is important for function. The reduced extent of DNA digestion by λ exoR28A in vitro and its relative insensitivity to the phosphorylation state of the 5′ ends of dsDNA, as evident from the fluorescence-based exonuclease assay, are both consistent with the hypothesis that it participates in 5′ end recognition during DNA digestion by λ exo.
The low activity of λ exo on 5′-OH substrates has been described before (1,23). In this study we have expanded our understanding of the origin of this low activity. The digestion profile of 5′-OH linear dsDNA by λ exoWT indicates that only a fraction of the substrate gets turned over even after prolonged exposure to the enzyme under saturation conditions. The remaining fraction of λ exoWT–5′-OH DNA complexes appear to be inert, trapped in a form unable to carry out catalysis. Topological linkage of the DNA through the enzyme imposes conformational constraints on the enzyme–DNA complex. We propose that a large fraction of the complexes formed when the enzyme encounters 5′-OH ends are unable to satisfy these constraints and the DNA is presented to the active site in a sub-optimal manner, leading to inert complex formation.
The effect of end recognition on processivity: model for behavior of λ exoR28A
The λ exoR28A mutant, besides being impaired in the ability to discern the phosphorylation state of the 5′ end of dsDNA, has limited processivity on dsDNA. We propose that λ exoR28A represents a class of mutants in which the ability to hydrolyze DNA processively is affected by changing the presentation of the substrate to the enzyme. We propose that R28, and other conserved residues involved in chelating the phosphate ion, aid in positioning the 5′ end during catalysis and facilitate the transition from a non-specific complex to a specific, catalytically competent complex. When the productive interaction between the 5′ end and λ exo is perturbed, either by changing the phosphorylation state of the 5′ end of the dsDNA or mutating residues that interact with the 5′ end, the enzyme fails to make the transition to the productive catalytic complex.
In any processive enzyme system, the processivity factor (ρ, defined in this case as the average number of nucleotides digested per binding event by λ exo), is governed by the relative probabilities of moving forward (Pf), moving backward (Pb) and dissociation from the substrate (Poff). This can be represented as follows:
processivity factor (ρ) ≡ Pf/(Pb + Poff)
In the topological model for processivity, processivity is enhanced by decreasing Poff, since the diffusion of the substrate away from the enzyme is limited. The probability of moving forward on the substrate, Pf, is modulated by the catalytic efficiency of the enzyme, which is directly related to the turnover number, kcat and the forward translocation rate constant, kf. The probability of moving backwards, Pb, is influenced by the relative affinity of λ exo for dsDNA versus ssDNA and the backward translocation rate constant, kb. We propose that in λ exoR28A, the contribution to processivity from topological linkage remains unchanged since the quaternary structure of the mutant enzyme resembles that of λ exoWT enzyme, but processivity is reduced by decreasing the ratio between Pf and Pb. In other words, since the efficiency of recognizing and holding on to the 5′ end of the digested strand is reduced in λ exoR28A, the enzyme translocates forward with a lower efficiency, and hence cleaves DNA with lower efficiency and is now distracted to a greater extent by the ssDNA produced as a result of the digestion. The model predicts that the relative affinity for dsDNA versus ssDNA would be lower in λ exoR28A when compared to λ exoWT. Characterization of the binding of the enzymes to various DNA substrates are the focus of our future studies.
Processivity in λ exo can be thought to originate from many aspects of the enzyme–substrate complex, including topological linkage of the protein ring around the ssDNA product of dsDNA digestion and the stable positioning of the 5′ end of the digested DNA chain through specific interactions with the 5′-P at the end of dsDNA. This study, which focuses on the in vitro characterization of λ exoR28A, a distributive mutant of λ exo, highlights the importance of end recognition in modulating the partition of the enzyme between a productive forward-translocating form and a non-productive backward- translocating form. The influence of specific attributes of a dynamic substrate–enzyme complex on processivity, via changes in catalytic efficiency and changes in propensity of the enzyme to translocate forward preferentially, is likely to be important in other processive enzyme systems as well. One such example is evident in the behavior of stalled complexes formed by E.coli RNA polymerase. The upstream or reverse translocation of E.coli RNA polymerase induced by stalling, for example by NTP starvation, has been proposed to cause the RNA polymerase to loose its grip on the 3′ end of the nascent transcript, thus causing the polymerase to be inappropriately positioned for catalyzing RNA synthesis and results in the formation of arrested transcription complexes (28,29). Although the direct effect of such slippage on processivity of transcription by RNA polymerase has not been described, and this repositioning event is reversible in RNA polymerase (28,29), reverse translocation followed by arrest would result in shorter transcripts, which is reminiscent of limited processivity. Such features may be apparent in DNA polymerases and helicases as well, both of which overlap with the set of processive enzymes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Brian Matthews and Rhett Kovall for pET::his.exo plasmid DNA and for personal communications, the DNA core facility of the Sylvester Cancer Center, University of Miami, for sequencing the pET::his.exoR28A plasmid, and Arun Malhotra for guiding us in the use of his Phastgel system and for review of the manuscript. We thank Peter von Hippel and Paul Mitsis for critically reviewing the manuscript and for suggesting future experiments. We are grateful to all the members of the Myers laboratory for discussions and commentary on the manuscript. Tia Vellani kindly gifted us dephosphorylated substrate DNA and the protocol for the processivity assay. We are also grateful to Tristan Fiedler, Yuhong Zuo and Gökhan Tolun who facilitated the generation of the structural models. This work was supported by the American Cancer Society RPG-00-100-01-MBC and the Florida Biomedical Research Program BM032 (to R.S.M.), NIH grant AI-39973 from the NIAID (to W.S.) and a pre-doctoral fellowship from the American Heart Association (to W.R.).
REFERENCES
- 1.Little J.W. (1967) An exonuclease induced by bacteriophage λ. II. Nature of the enzymatic reaction. J. Biol. Chem., 242, 679–686. [PubMed] [Google Scholar]
- 2.Carter D.M. and Radding,C.M. (1971) The role of exonuclease and β protein of phage λ in genetic recombination. II. Substrate specificity and the mode of action of λ exonuclease. J. Biol. Chem., 246, 2502–2512. [PubMed] [Google Scholar]
- 3.Thomas K.R. and Olivera,B.M. (1978) Processivity of DNA exonucleases. J. Biol. Chem., 253, 424–429. [PubMed] [Google Scholar]
- 4.Hendrix R.W., Roberts,J.W., Stahl,F.W. and Weisberg,R.A. (eds) (1983) Lambda II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 5.Black L.W. (1989) DNA packaging in dsDNA bacteriophages. Annu. Rev. Microbiol., 43, 267–292. [DOI] [PubMed] [Google Scholar]
- 6.Kovall R. and Matthews,B.W. (1997) Toroidal structure of λ-exonuclease. Science, 277, 1824–1827. [DOI] [PubMed] [Google Scholar]
- 7.Stukenberg T.P., Studwell-Vaughan,P.S. and O’Donnell,M. (1991) Mechanism of the sliding β-clamp of DNA polymerase III holoenzyme. J. Biol. Chem., 273, 10515–10529. [PubMed] [Google Scholar]
- 8.Jezewska M.J., Rajendran,S., Bujalowska,D. and Bujalowski,B. (1998) Does single-stranded DNA pass through the inner channel of the protein hexamer in the complex with the Escherichia coli DnaB helicase? Fluorescence energy transfer studies. J. Biol. Chem., 266, 11328–11334. [DOI] [PubMed] [Google Scholar]
- 9.Egelman E.H., Yu,X., Wild,R., Hingorani,M.M. and Patel,S.S. (1995) Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggest a general structure for hexameric helicases. Proc. Natl Acad. Sci. USA, 92, 3869–3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stasiak A., Tsaneva,I.R., West,S.C., Benson,C.J.B., Yu,X. and Egelman,E.H. (1994) The Escherichia coli RuvB branch migration protein forms double hexameric rings around DNA. Proc. Natl Acad. Sci. USA, 91, 7618–7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sriprakash K.S., Lundh,N., Huh,M.M. and Radding,C.M. (1975) The specificity of λ exonuclease. Interactions with single-stranded DNA. J. Biol. Chem. 250, 5438–5445. [PubMed] [Google Scholar]
- 12.Myers R.S. and Rudd,K.E. (1998) Mining sequences for molecular enzymology: the Redα superfamily defines a set of recombination nucleases. In Proceedings of the 1998 Miami Nature Biotechnology Symposium. Oxford University Press, USA, Vol. 9, pp. 49–50.
- 13.Kovall R.A. and Matthews,B.W. (1998) Structural, functional and evolutionary relationships between λ-exonuclease and the type II restriction endonucleases. Proc. Natl Acad. Sci. USA, 95, 7893–7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aravind L., Makarova,K.S. and Koonin,E.V. (2000) Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res., 28, 3417–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bujnicki J.M. and Rychlewski,L. (2001) The herpesvirus alkaline exonuclease belongs to the restriction endonuclease PD-(D/E)XK superfamily: insight from molecular modeling and phylogenetic analysis. Virus Genes, 22, 219–230. [DOI] [PubMed] [Google Scholar]
- 16.Pingoud A. and Jeltsch,A. (2001) Structure and function of type II restriction endonucleases. Nucleic Acids Res., 29, 3705–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vellani T. and Myers,R.S. (2003) Bacteriophage SPP1 Chu is an alkaline exonuclease in the SynExo family of viral two-component recombinases. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan D., Jessee,J. and Bloom,F.R. (1991) Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol., 204, 63–113. [DOI] [PubMed] [Google Scholar]
- 19.Maloy S. (1989) Experimental Techniques in Bacterial Genetics. Jones & Bartlett, Boston, MA, p. 83.
- 20.Studier F.W., Rosenberg,A.H., Dunn,J.J. and Dubendorff,J.W. (1990) Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol., 185, 60–89. [DOI] [PubMed] [Google Scholar]
- 21.Arber W., Enquist,L., Hohn,B., Murray,N.E. and Murray,K. (1983) In Hendrix,R.W., Roberts,J.W., Stahl,F.W. and Weisberg,R.A. (eds), Lambda II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 433–466.
- 22.Gill S.C. and von Hippel,P.H. (1989) Calculation of protein extinction coefficients, from amino acid sequence data. Anal. Biochem., 182, 319–326. [DOI] [PubMed] [Google Scholar]
- 23.Little J.W., Lehman,I.R. and Kaiser,A.D. (1967) An exonuclease induced by bacteriophage λ. I. Preparation of the crystalline enzyme. J. Biol. Chem., 242, 672–678. [PubMed] [Google Scholar]
- 24.Meyer P.R., Matsuura,S., So,A.G. and Scott,W.A. (1998) Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc. Natl Acad. Sci. USA, 95, 13471–13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitsis P.G. and Kwagh,J.G. (1999) Characterization of the interaction of lambda exonuclease with the ends of DNA. Nucleic Acids Res., 27, 3057–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hingorani M. and O’Donnell,M. (1998) Toroidal proteins: running rings around DNA. Curr. Biol., 8, R83–R86. [DOI] [PubMed] [Google Scholar]
- 27.Breyer W.A. and Matthews,B.W. (2001) A structural basis for processivity. Protein Sci., 10, 1699–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee D.N., Feng,G. and Landick,R. (1994) GreA-induced transcript cleavage is accompanied by reverse translocation to a different transcription complex conformation. J. Biol. Chem. 269, 22295–22303. [PubMed] [Google Scholar]
- 29.Wang D. and Landick,R. (1997) Nuclease cleavage of the upstream half of the non-template strand DNA in an Escherichia coli transcription elongation complex causes upstream translocation and transcriptional arrest. J. Biol. Chem. 272, 5989–5994. [DOI] [PubMed] [Google Scholar]
- 30.Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr., A47, 110–119. [DOI] [PubMed] [Google Scholar]
- 31.DeLano W.L. (2002) The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA.