Abstract
The essential response regulator CtrA controls the Caulobacter crescentus cell cycle and phosphorylated CtrA∼P preferentially binds target DNA in vitro. The CtrA aspartate to glutamate (D51E) mutation mimics phosphorylated CtrA∼P in vivo and rescues non-viable C.crescentus cells. However, we observe that the CtrA D51E and the unphosphorylated CtrA wild-type proteins have identical DNA affinities and produce identical DNase I protection footprints inside the C.crescentus replication origin. There fore, D51E promotes essential CtrA activities separate from increased DNA binding. Accordingly, we argue that CtrA protein recruitment to target DNA is not sufficient to regulate cell cycle progression.
INTRODUCTION
CtrA (cell cycle transcription regulator) is a globally acting response regulator in Caulobacter crescentus (1) and presumably many related bacteria (2). Caulobacter crescentus provides a model cell cycle where flagellated ‘swarmer cells’ first differentiate into replicating ‘stalked cells’, and then divide asymmetrically to produce new stalked and swarmer cells. This cellular differentiation requires the cell cycle timing of flagellar synthesis, stalk formation, cell division and chromosome replication. CtrA was originally identified in a genetic screen for flagellar transcription activators, and purified CtrA protein binds flagellar gene promoters (1). CtrA also represses chromosome replication, and purified CtrA protein binds five sites that span the autonomously replicating DNA (3,4). This repression may result from CtrA blocking transcription from an exceptionally strong promoter inside the replication origin, but it is also likely that CtrA productively interacts with other replication proteins (4). Complex regulatory interactions are also implied by recent whole-genome transcription analysis demonstrating that CtrA controls over a quarter of the cell cycle regulated genes of C.crescentus (5). It is not known how CtrA mediates precise cell cycle timing, and it is not known how CtrA activates transcription in one context while repressing transcription in another. However, appropriate levels of CtrA activity are clearly required, both for cell viability and for cell cycle progression (1,3,6).
CtrA activity is modulated during the cell cycle by phosphorylation and by proteolysis (6). CtrA wild-type is first degraded at the start of S-phase, and then replenished and phosphorylated during mid S-phase. Yet, like other response regulator proteins, CtrA phosphorylation is the dominant modulator of CtrA activity. For example, a non-proteolyzable CtrAΔ3 mutant protein is abundantly present throughout the cell cycle, and it is the periodic phosphorylation of CtrAΔ3 that drives cell cycle progression (6). Also, like other two-component response regulator proteins, CtrA is phosphorylated by cognate histidine kinases, such as the cell cycle kinase CckA (7).
We wish to understand how CtrA phosphorylation alters CtrA cell cycle activity. Like other response regulators, CtrA binds specific DNA motifs and phosphorylated CtrA∼P has substantially higher affinity for the same target DNA (8,9). Therefore, in principle, CtrA protein recruitment to its target DNA may sufficiently explain in vivo CtrA activity. Regulation by recruitment, defined as localizing proteins to where they are more likely to interact, is a dominant principle in eukaryotic (10) and bacterial regulatory systems (11). For example, this recruitment principle is apparently sufficient to explain how Bacillus subtilis response regulator Spo0A activates genetic transcription (12). However, CtrA phosphorylation might also significantly alter CtrA protein conformation and this could alter CtrA contacts with RNA polymerase and other proteins. This second principle is illustrated by the nitrogen-response regulator NtrC. Phosphorylation alters NtrC protein conformation, increasing an intrinsic ATPase, contacts between NtrC proteins, and transcription by RNA polymerase (13).
The CtrA aspartate to glutamate (D51E) mutation may suggest how phosphorylation alters CtrA activity. CtrA is exclusively phosphorylated at the conserved position 51 aspartate inside the receiver domain (8). The analogous aspartate to glutamate mutation (D54E) at the phosphorylation site of NtrC mimics aspartyl-phosphate and partially activates NtrC (14). Likewise, the Escherichia coli OmpR D55E mutation mimics aspartyl-phosphate activation, since appropriate genetic expression depends on adjusting the in vivo abundance of OmpR D55E which bypasses the requirement for signaling through protein kinase EnvZ (15). Previous in vivo studies demonstrated that CtrA D51E also mimics aspartyl-phosphate (6). Since the CtrA D51E protein cannot be phosphorylated, CtrA D51E is apparently active in vivo independent of its cognate kinase (6). However, the biochemical basis of this activation is unknown.
In this study, we compare the binding of unphosphorylated CtrA wild-type and CtrA D51E to the five binding sites inside the C.crescentus replication origin (Cori). We accurately measured binding constants and rigorously controlled for protein molar activity. Unexpectedly, we observe essentially identical DNA binding by both unphosphorylated CtrA wild-type and CtrA D51E proteins that is distinct from strong and cooperative binding by phosphorylated CtrA∼P (9). We propose that CtrA D51E separates DNA binding from other biochemical properties that constitute CtrA activation, and we discuss the implications for cell cycle control.
MATERIALS AND METHODS
Protein preparation
Plasmids pTRC7.4 and pXD51E, containing hexa-histidine tagged CtrA wild-type and the CtrA D51E, were used to express and affinity purify these proteins from E.coli BL21 (Novagen), essentially as recommended by Novagen, and as detailed previously (8,9). Plasmids pTRC7.4 and pXD51E were derived from pTrcHisC (Invitrogen), and are identical except for the mutation in the CtrA coding sequences that changes aspartate 51 to glutamate 51 (8). Protein concentrations were measures from the molar extinction coefficients derived by the method of Gill and von Hippel (16). CtrA was phosphorylated using purified MBP-EnvZ kinase as previously described (8). MBP-EnvZ kinase was prepared from BL21 E.coli cells containing plasmid pKJH5, as described (8). To measure the amount of CtrA phosphorylation the standard kinase reaction (0.4 mM ATP) was supplemented with 10 µCi/ml [γ-32P]ATP (Amersham), as described previously (9).
DNA binding assays
DNase I protection, ‘footprinting’ experiments. DNase I footprinting experiments were based on the method of Galas and Schmitz (17), with minor modification as described before (9). The DNA substrate was prepared as described before (9), using plasmid pGM1877 which was 32P 5′ end-labeled at a unique HindIII site and digested with XhoI to yield a 600 bp fragment containing all five a–e CtrA binding sites from the C.crescentus replication origin (3). The band intensities presented in Figure 3, and in similar footprint experiments (data not shown), were measured using phosphorimaging and the IQuant program (Molecular Dynamics).
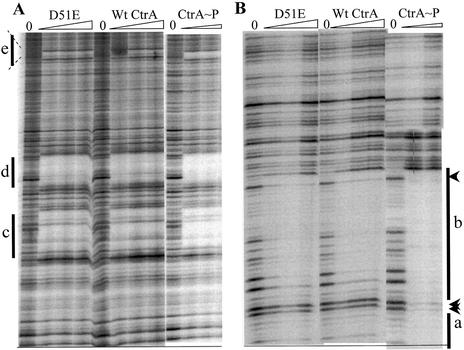
Figure 3.
DNase I footprinting. (A) Upper and (B) lower parts of the same autoradiograms are shown. End-labeled 32P Cori DNA (∼20 000 c.p.m.) was incubated, as described in Materials and Methods, with increasing concentrations (0.2–1 µM) of unphosphorylated CtrA wild-type and CtrA D51E. Lower concentrations (0.01–0.03 µM) of phosphorylated CtrA∼P were used as controls. Binding sites designated a, b, c, d, e are marked by the bars. The arrowheads mark selected bands that are only partially protected by unphosphorylated CtrA and CtrA D51E, but that are fully protected by CtrA∼P.
Band mobility shift assays. The DNA substrate, a 24 bp double-stranded oligonucleotide (based on the sequence of CtrA site e) was prepared by annealing 5′-GTG GTT AAG CAA CCG TTA ACG GAT-3′ (top strand) and 5′-ATC CGT TAA CGG TTG CTT AAC CAC-3′ (bottom strand), prior to end-labeling with T4 polynucleotide kinase and [γ-32P]ATP (Amersham). 10 000 c.p.m. of DNA substrate was incubated with different concentrations of CtrA and CtrA D51E protein (and for Figure 2, unlabeled DNA substrate was added, as indicated). Total 20 µl reactions [20 mM Tris (pH 8), 50 mM KCl, 5 mM MgCl2, 10% glycerol, 10 µg/ml poly(dI–dC); Pharmacia Biotech] were incubated at 25°C for 20 min, then immediately loaded on a 6% Tris–glycine polyacrylamide gel, and run for 2 h at 150 V and 4°C. These gels were blotted to Whatman 3MM paper, dried, and exposed to phosphorimaging screens (Molecular Dynamics).
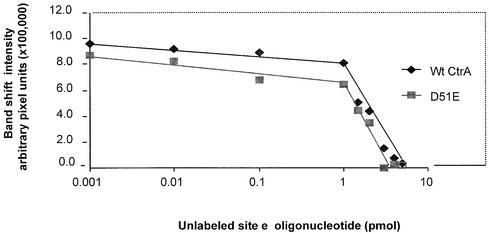
Figure 2.
Equal activities of CtrA wild-type and CtrA D51E protein preparations. Titration of CtrA proteins by the 24 bp unlabeled site e oligonucleotide in a band mobility shift assay to determine molar protein activity. The bound band intensities were measured using the phosphorimager (arbitrary pixel units) and plotted against the concentration of unlabeled oligonucleotide.
RESULTS
Accurate titration of CtrA wild-type and CtrA D51E molar binding activities
We standardized protein purification to ensure equal molar activities of both our CtrA wild-type and CtrA D51E protein preparations. Both proteins were prepared from the same E.coli strain (BL21), employing the same reagents and otherwise processed under identical conditions. As demonstrated previously, this CtrA wild-type preparation was efficiently phosphorylated (>90% CtrA phosphorylation) by EnvZ, the cognate histidine kinase for OmpR. Under these conditions, the CtrA D51E protein remained unphosphorylated (8) (data not shown).
Band mobility shift assays demonstrated that unphosphorylated CtrA wild-type and CtrA D51E have essentially identical Kd values and that both protein preparations have equal molar activities. Unlike footprint experiments that require a molar excess of protein over DNA, band mobility shift assays can be performed in molar DNA excess, and all active protein molecules can be driven to bind DNA.
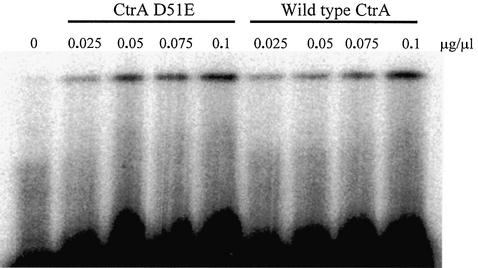
To determine the binding affinity of CtrA wild-type and CtrA D51E, equal protein concentrations were incubated with a 24 bp 32P 5′ end-labeled double-stranded oligonucleotide, based on CtrA Cori binding site e. The binding affinity of the CtrA D51E was only slightly, and not significantly, increased when compared with the wild-type protein (Fig. 1).
Figure 1.
Band mobility shift assays of CtrA and CtrA D51E. As described in Materials and Methods, 10 000 c.p.m. of a 24 bp double-stranded CtrA site e oligonucleotide, 5′-GTG GTT AAG CAA CCG TTA ACG GAT-3′ (top strand), was incubated with increasing concentrations of CtrA and CtrA D51E, as indicated above each lane.
Titration of CtrA protein by the unlabeled target site e oligonucleotide confirmed the equal molar binding activities of both protein preparations. Equal concentrations (0.05 µM) of CtrA wild-type and CtrA D51E were incubated with 10 000 c.p.m. of 32P end-labeled site e oligonucleotide and increasing concentrations of unlabeled site e oligonucleotide. Band shift intensities were measured and plotted against the concentration of unlabeled oligonucleotide (Fig. 2). The band shift intensities were not significantly affected, until the molarity of the oligonucleotide approached the molarity of the active (able to bind) protein. The virtually identical breakpoints at ∼1 pmol of oligonucleotide indicate virtually identical concentrations of active protein in both the CtrA wild-type and the CtrA D51E preparations. Without this control experiment, it was possible that the D51E mutation promotes protein denaturation and perhaps fewer CtrA D51E molecules were available for binding. This titration control also implies that D51E creates a very subtle change of protein structure, since both proteins behave essentially identically when subjected to the same binding analysis and protein preparation protocols.
CtrA wild-type and CtrA D51E proteins have equal DNA binding affinities
We employed DNase I footprinting (17) to compare the binding characteristics of both wild-type and D51E proteins with the C.crescentus replication origin (Cori). As the substrate, a 600 bp Cori DNA fragment, containing all five a–e CtrA binding sites, was 5′ end-labeled with 32P. Equal concentrations of unphosphorylated CtrA wild-type and CtrA D51E were added to the footprint reactions. Both proteins bound identically to all five Cori binding sites (Fig. 3). The band intensities presented in Figure 3, and in similar footprint experiments (data not shown), were measured using phosphorimaging and the IQuant program (Molecular Dynamics). The dissociation binding constant (Kd) values were calculated from the half maximal binding concentrations (Table 1). The Kd values of the CtrA D51E protein were essentially identical to Kd values of CtrA wild-type unphosphorylated protein (Table 1). As a control, we also bound phosphorylated CtrA∼P protein to the same target DNA (Fig. 3), and this confirmed our published results that phosphorylated CtrA∼P protein gains substantial DNA affinity (9). The CtrA D51E clearly does not resemble the wild-type phosphorylated protein, which shows 40–100-fold lower Kd values at Cori sites a–e (Table 1).
Table 1. Dissociation constant (Kd) values of CtrA D51E, CtrA wild-type and phosphorylated CtrA∼P proteins at Coria.
| Cori binding site | Kd values (µM) Ctr D51E | CtrA wild-type | CtrA∼P |
|---|---|---|---|
| a | 0.5 ± 0.2 | 0.6 ± 0.3 | 0.015 ± 0.005 |
| b | 0.3 ± 0.1 | 0.4 ± 0.1 | 0.004 ± 0.002 |
| c | 0.4 ± 0.1 | 0.6 ± 0.1 | 0.008 ± 0.002 |
| d | 0.2 ± 0.1 | 0.2 ± 0.1 | 0.006 ± 0.003 |
| e | 0.3 ± 0.1 | 0.2 ± 0.1 | 0.006 ± 0.002 |
aAs described in the text, these Kd values were derived from footprint assays, such as those presented in Figure 3.
Unphosphorylated CtrA and CtrA D51E lack cooperative binding inside Cori
CtrA wild-type and CtrA D51E produce identical footprint patterns that are significantly different from the CtrA∼P footprint pattern (9). For example, phosphorylated CtrA∼P footprints have wider zones of complete protection than unphosphorylated CtrA (9). This qualitative appearance is also interpreted as tighter cooperative binding by CtrA∼P. Tight cooperative binding is most apparent at adjacent sites a and b. Figure 3 also illustrates the binding patterns of unphosphorylated CtrA, CtrA∼P and CtrA D51E to sites a and b. The unphosphorylated CtrA wild-type and the CtrA D51E footprints are essentially identical. However, the binding of phosphorylated CtrA∼P to sites a and b is distinctly cooperative (9). One indication of cooperative binding is blocked DNase I digestion between a and b (Fig. 3). Presumably CtrA∼P binding to site b aids CtrA∼P binding to site a through a direct protein bridge that blocks DNase I (9). This blocked DNase I digestion is clearly seen with CtrA∼P in Figure 3 (arrowheads), but not with unphosphorylated CtrA wild-type and CtrA D51E. Even at the highest allowable protein concentrations (∼5 µM), where our footprint assays are limited by protein precipitation (data not shown), both unphosphorylated CtrA wild-type and CtrA D51E fail to block this DNase I cutting between adjacent sites a and b. Therefore, the D51E mutation does not increase DNA binding affinity and it does not promote cooperative binding between sites a and b that characterizes CtrA∼P binding.
DISCUSSION
Despite the essential requirement of CtrA for cell cycle progression in C.crescentus (1) and presumably in many related bacteria (2,4), we still do not know how phosphorylated CtrA∼P regulates transcription and replication. Since phosphorylated CtrA∼P has substantially higher affinity for DNA binding sites, CtrA recruitment from the cytoplasm to its target DNA has dominated cell cycle models (4).
Our present work indicates that essential biochemical properties, besides DNA binding, are altered upon CtrA∼P phosphorylation. We demonstrated that CtrA D51E does not alter DNA binding. Since CtrA D51E clearly provides essential activities in vivo (as discussed below) the D51E mutation must separate DNA binding from other essential activities. Phosphorylated CtrA∼P binds five TTAA-N7-TTAA target sites a–e inside Cori with substantially greater affinity than the unphosphorylated CtrA (9). Upon phosphorylation, the dissociation binding constant (Kd) values are lowered 40–100-fold by two types of cooperative CtrA protein interactions. First, phosphorylation stimulates cooperative CtrA binding between two TTAA half-sites. For example, phosphorylation does not stimulate CtrA binding to only one half-site if the other half-site is mutated. Secondly, phosphorylation stimulates cooperative CtrA binding between adjacent whole sites a and b, and phosphorylation promotes a uniform zone (a ‘tight zone’) of DNase I protection. For example (Fig. 3), phosphorylated CtrA∼P blocks DNase I cuts between adjunct Cori sites a and b. Since CtrA D51E binds identically, both quantitatively (by Kd values) and qualitatively (by footprint appearance) to unphosphorylated CtrA wild-type, the D51E mutation cannot promote either of the two cooperative protein interactions that characterize phosphorylated CtrA∼P.
Nonetheless, ctrA D51E is clearly a gain-of-function allele that mimics aspartyl-phosphate in vivo. This was first demonstrated by the synergy between CtrAΔ3 and CtrA D51E mutations. CtrA represses chromosome replication and its C-terminus mutation (CtrAΔ3) blocks proteolysis (6). However, when CtrAΔ3 is oversupplied, CtrAΔ3 is abundantly present, but it does not block chromosome replication, because CtrAΔ3 phosphorylation is still regulated during the cell cycle. Presumably decreased CtrAΔ3 phosphorylation releases CtrAΔ3 from the replication origin prior to S-phase (3,6). In contrast, when CtrAΔ3 D51E is oversupplied, only this double mutant protein arrests chromosome replication, presumably because D51E mimics the CtrA phosphorylation.
CtrA D51E also provides essential biological activities that uniquely characterize CtrA∼P. CtrA, as well as its kinase (CckA) are both required for cell viability (7), presumably because they regulate essential cell cycle events. For defective cckA kinase mutants, oversupply of CtrA D51E, but not an oversupply of CtrA wild-type, uniquely restores cell viability (7). Protein abundance does not explain extra CtrA D51E activity under these in vivo conditions. For example, an oversupply of CtrAΔ3 also does not complement defective cckA kinase mutants even when the in vivo abundance of CtrAΔ3 exceeds CtrA D51E. Although it cannot be proteolysed, CtrAΔ3 is otherwise fully functional, since a relatively low abundance of CtrAΔ3 fully complements defective ctrA mutants, and such cells exhibit a normal cell cycle program (3). In contrast, a relatively high oversupply of CtrA D51E is required to complement defective ctrA mutants. This argues that CtrA D51E is only partially active. In light of our results, this also argues that extra CtrA D51E protein is needed to drive CtrA onto its DNA targets. Although our DNA binding results were unexpected, they are nonetheless consistent with previous in vivo results, if D51E provides alternative in vivo activities.
Response regulator proteins NtrC (14), OmpR (15), as well as CtrA (6) are phosphorylated at one conserved aspartate, and for each of these proteins an aspartate to glutamate mutation produces an active protein that bypasses requirements for the cognate kinase in vivo. CtrA (8,9) and OmpR (18,19) binding affinities are enhanced upon protein phosphorylation, while NtrC and NtrC∼P bind equally well to a single binding site (20). However, NtrC phosphorylation promotes cooperative binding at two adjacent sites and the consequent transcription activation of the glnA promoter (20,21). Likewise, phosphorylation promotes CtrA (9) and OmpR (18,19) cooperative binding at adjacent binding sites. In a band mobility shift experiment with NtrC D54E, the cooperative binding was not enhanced by D54E when compared with unphosphorylated wild-type NtrC (14). Instead, NtrC D54E increased an intrinsic ATPase activity and its ability to activate transcription (14).
If CtrA D51E resembles CtrA∼P in vivo but does not enhance DNA binding in vitro, then CtrA D51E must acquire other biochemical activities. Since CtrA is primarily a transcription regulator (1), CtrA D51E most likely has altered contacts with RNA polymerase that at least partially resemble those of CtrA∼P. CtrA also interacts with the ClpX/ClpP chaperone/protease system that degrades CtrA during the cell cycle (22). Since the CtrA D51E protein is more stable in vivo (6), CtrA D51E may have altered interactions with ClpX/ClpP. Also, the five CtrA binding sites inside Cori present an exceptionally high concentrate of CtrA binding sites that span this unusual replication origin (4). This implies that CtrA contacts many proteins besides RNA polymerase. Perhaps CtrAD51E has altered interactions with replication proteins such as DnaA that are likewise essential and required to drive the cell cycle (23).
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported in part by Le Fonds pour la Formation de Chercheurs et l’Aide à la Recherche as a PhD scholarship to R.S. and by the Canadian Institutes for Health Research (CIHR) Grant MT-13453, and an MRC Scholarship Award SH-50791-AP007403 to G.T.M.
REFERENCES
- 1.Quon K.C., Marczynski,G.T. and Shapiro,L. (1996) Cell cycle control by an essential bacterial two-component signal transduction protein. Cell, 84, 83–93. [DOI] [PubMed] [Google Scholar]
- 2.Barnett M.J., Hung,D.Y., Reisenauer,A., Shapiro,L. and Long,S.R. (2001) A homolog of the CtrA cell cycle regulator is present and essential in Sinorhizobium meliloti. J. Bacteriol., 183, 3204–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quon K.C., Yang,B., Domian,I.J., Shapiro,L. and Marczynski,G.T. (1998) Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc. Natl Acad. Sci. USA, 95, 120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marczynski G.T. and Shapiro,L. (2002) Control of chromosome replication in Caulobacter crescentus. Annu. Rev. Microbiol., 56, 625–656. [DOI] [PubMed] [Google Scholar]
- 5.Laub M.T., McAdams,H.H., Fraser,C.M. and Shapiro,L. (2000) Global analysis of the genetic network controlling a bacterial cell cycle. Science, 290, 2144–2148. [DOI] [PubMed] [Google Scholar]
- 6.Domian I.J., Quon,K.C. and Shapiro,L. (1997) Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1 to S transition in a bacterial cell cycle. Cell, 90, 415–424. [DOI] [PubMed] [Google Scholar]
- 7.Jacobs C., Domian,I.J., Maddock,J.R. and Shapiro,L. (1999) Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell, 97, 111–120. [DOI] [PubMed] [Google Scholar]
- 8.Reisenauer A., Quon,K. and Shapiro,L. (1999) The CtrA response regulator mediates temporal control of gene expression during the Caulobacter cell cycle. J. Bacteriol., 181, 2430–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siam R. and Marczynski,G.T. (2000) Cell cycle regulator phosphorylation stimulates two distinct modes of binding at a chromosome replication origin. EMBO J., 19, 1138–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ptashne M. and Gann,A. (1997) Transcription activation by recruitment. Nature, 386, 569–577. [DOI] [PubMed] [Google Scholar]
- 11.Hochschild A. and Dove,S.L. (1998) Protein–protein contacts that activate and repress prokaryotic transcription. Cell, 92, 597–600. [DOI] [PubMed] [Google Scholar]
- 12.Baldus J.M., Green,B.D., Youngman,P. and Moran,C.P.,Jr (1994) Phosphorylation of Bacillus subtilis transcription factor Spo0A stimulates transcription from the spoIIG promoter by enhancing binding to weak 0A boxes. J. Bacteriol., 176, 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hwang I., Thorgeirsson,T., Lee,J., Kustu,S. and Shin,Y.-K. (1999) Physical evidence for a phosphorylation-dependent conformational change in the enhancer-binding protein NtrC. Proc. Natl Acad. Sci. USA, 96, 4880–4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klose K.E., Weiss,D.S. and Kustu,S. (1993) Glutamate at the site of phosphorylation of nitrogen-regulatory protein NTRC mimics aspartyl-phosphate and activates the protein. J. Mol. Biol., 232, 67–78. [DOI] [PubMed] [Google Scholar]
- 15.Lan C.-Y. and Igo,M.M. (1998) Differential expression of the OmpF and OmpC proteins in Escherichia coli K-12 dependent upon the level of active OmpR. J. Bacteriol., 180, 171–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gill S.C. and von Hippel,P.H. (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem., 182, 319–326. [DOI] [PubMed] [Google Scholar]
- 17.Galas D.J. and Schmitz,A. (1978) DNAse footprinting: a simple method for the detection of protein–DNA binding specificity. Nucleic Acids Res., 5, 9357–9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang K.-J., Lan,C.-Y. and Igo,M.M. (1997) Phosphorylation stimulates the cooperative DNA-binding properties of transcription factor OmpR. Proc. Natl Acad. Sci. USA, 94, 2828–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bergstrom L.C., Qin,L., Harlocker,S.L., Egger,L.A. and Inouye,M. (1998) Hierarchical and co-operative binding of OmpR to a fusion construct containing the ompC and ompF upstream regulatory sequences of Escherichia coli. Genes Cells, 3, 777–788. [DOI] [PubMed] [Google Scholar]
- 20.Weiss V., Claverie-Martin,F. and Magasanik,B. (1992) Phosphorylation of nitrogen regulator I of Escherichia coli induces strong cooperative binding to DNA essential for activation of transcription. Proc. Natl Acad. Sci. USA, 89, 5088–5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wyman C., Rombel,I., North,A.K., Bustamante,C. and Kustu,S. (1997) Unusual oligomerization required for activity of NtrC, a bacterial enhancer-binding protein. Science, 275, 1658–1661. [DOI] [PubMed] [Google Scholar]
- 22.Jenal U. and Fuchs,T. (1998) An essential protease involved in bacterial cell cycle control. EMBO J., 17, 5658–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorbatyuk B. and Marczynski,G.T. (2001) Physiological consequences of blocked Caulobacter crescentus DnaA expression, an essential DNA replication gene. Mol. Microbiol., 40, 485–497. [DOI] [PubMed] [Google Scholar]