Abstract
Ssu72 is an essential yeast protein that is involved in transcription. It physically interacts with transcription initiation and termination complexes. In this report, we provide evidence that Ssu72 is a phosphatase that physically interacts with the CTD kinase Kin28 and functionally interacts with the CTD phosphatase Fcp1. A genome-wide expression analysis of mutant ssu72-ts69 during growth in complete medium revealed a number of defects, including the accumulation of a limited number of mRNAs and the read-through transcription of small nucleolar RNAs and of some mRNAs. We hypothesize that Ssu72 plays a key role in the transcription termination of certain transcripts, possibly by promoting RNA polymerase pausing and release. The possibility that the CTD of the largest subunit of RNA polymerase II is a substrate of Ssu72 is discussed.
Keywords: CTD/microarrays/phosphatase/RNA processing/transcription
Introduction
RNA polymerase II (RNA Pol II) transcribes protein-encoding genes, small nucleolar RNA genes (snoRNAs) and several small nuclear RNA genes (snRNAs) (Kiss, 2002). The initiation of transcription requires RNA Pol II, the six general transcription factors and the mediator complex that interacts with gene-specific regulators. A number of elongation factors associate with the promoter and with the open reading frame of transcribed genes (Pokholok et al., 2002). These factors regulate the rate of transcription. Additional complexes, including the cleavage and polyadenylation complex CPF in yeast, are required for the 3′ end processing of messenger RNAs (Proudfoot et al., 2002).
A large number of general transcription factors, as well as gene-specific regulators, are phosphorylated (Kobor and Greenblatt, 2002). These include the largest subunit (Rpb1) of RNA Pol II. The phosphorylation state of the C-terminal domain (CTD) of Rpb1 is critical for the progression of the transcription cycle as it promotes the association of RNA Pol II with the pre-initiation complex and the factors responsible for elongation, pre-mRNA 5′ capping, splicing and 3′ processing (Proudfoot et al., 2002). CTD phosphorylation during transcription is highly dynamic and tightly coordinated by various kinases (Kin28, Ctk1, Bur1 and Srb10 in budding yeast), phosphatase Fcp1 and probably other phosphatases (Howe, 2002).
The different CTD phosphoacceptor sites (i.e. Ser2 and Ser5) are independently phosphorylated in response to environmental signals (Patturajan et al., 1998). This suggests that CTD kinases or phosphatases are the targets of cellular signalling pathways. This might be the case for kinase Kin28, which is phosphorylated by Cak1 on threonine 162 (Kimmelman et al., 1999), for kinase Bur1, which is phosphorylated by Cak1 on threonine 240 (Yao and Prelich, 2002), or for phosphatase Fcp1, which is activated by casein kinase 2 in Xenopus laevis (Palancade et al., 2002).
Cak1, the yeast cyclin-dependent kinase-activating kinase, is necessary not only for cell growth in rich medium but also when yeast cells experience environmental stress. We recently showed that Cak1 interacts with two other actors involved in transcription: the Paf1 complex and the Ssu72 protein (C.Ganem, C.Miled, C.Facca, J.G.Valay, G.Labesse, S.Ben Hassine, C.Mann and G.Faye, manuscript in preparation). SSU72 is an essential gene, which is phylogenetically well-conserved (Pappas and Hampsey, 2000). The Ssu72 protein interacts with the general transcription factor TFIIB (Sun and Hampsey, 1996; Wu et al., 1999; Pappas and Hampsey, 2000). Pappas and Hampsey (2000) isolated spontaneous extragenic thermoresistant revertants from a thermosensitive ssu72 mutant. One suppressor is allelic to RPB2, the gene encoding the second largest subunit of RNA Pol II. A systematic analysis of protein complexes in Saccharomyces cerevisiae revealed that Ssu72 associates with the pre-mRNA 3′ end processing complex (Gavin et al., 2002). Likewise, the identification of proteins associated with the Taf150 component of yeast TFIID by multidimensional mass spectrometry revealed Ssu72 and components of the polyadenylation complex (Sanders et al., 2002). Recently, Dichtl and co-workers demonstrated that Ssu72 is stably associated with yeast cleavage and polyadenylation factor CPF (Dichtl et al., 2002). These data suggest that Ssu72 plays an important role in transcription regulation.
In this report, we show that Ssu72 interacts genetically and physically with the Kin28 kinase, and genetically with the Fcp1 phosphatase. We provide evidence that Ssu72 is a putative tyrosine phosphatase. Using DNA microarrays and northern blot analyses, we found that only the expression of a small part of the genome is affected in an ssu72-ts mutant at the restrictive temperature. We also demonstrate that mutations in SSU72 alter the 3′ end processing of snoRNAs and of some specific pre-mRNAs. These results suggest that Ssu72 plays a specific role in the termination of transcription of snoRNA and of some specific mRNAs, possibly by altering directly or indirectly the phosphorylation state of the CTD.
Results
Mutations in Ssu72 alter the transcription of snoRNA and specific mRNAs
We were curious to know whether Ssu72 is required for the expression of most genes or just for a small sub-set during growth in complete media. We used DNA microarrays containing most of yeast open reading frames (ORFs) to compare the transcriptomes of wild-type and ssu72-ts69 cells at 28°C and after a 1 h shift at 39°C. Briefly, the expression of less than 200 genes was at least 2-fold higher and that of less than 150 genes was at least 2-fold lower in the mutant than in the wild-type, either at 28°C, after 1 h at 39°C or both (data not shown). This limited number of effects suggests that Ssu72 is implicated in the expression of particular genes.
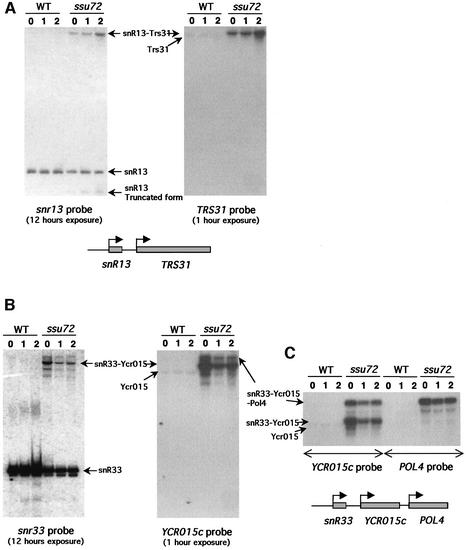
In this report, we focused on 30 genes, the mRNA of which reproducibly and strongly accumulated (>3-fold) in the mutant at 28 and 39°C (Supplementary table 1, available at The EMBO Journal Online). Nine of these 30 ORFs were immediately preceded by a snoRNA gene on the same DNA strand and two were followed by a snoRNA gene transcribed on the opposite DNA strand (convergent genes) (Table I). We wondered whether the effect observed with the microarrays was actually due to the overexpression of these 11 ORFs or to a transcriptional disorder affecting the expression of the snoRNA genes. Northern blot analyses using probes for the TRS31 ORF and for the corresponding upstream snoRNA gene (snr13, see Figure 1A) showed that the ssu72-ts69 mutant expresses a new di-cistronic transcript revealed with both probes (indicated as snr13-TRS31 on Figure 1A). Read-through transcripts were also detected for snr33-Ycr015c (Figure 1B), snr8-YTM1 and snr42-YOR272w (data not shown). In the case of YCR015c, an additional extended tri-cistronic transcript including YCR015c, the upstream snr33 gene and the downstream POL4 gene was observed (Figure 1C). These results suggest that the ssu72-ts69 mutant is defective for the correct termination of snoRNA transcription, even at the permissive temperature. Although numerous pre-snoRNA molecules appear to reach the cleavage/polyadenylation signal of the downstream ORF, a large proportion of them are correctly converted into mature species. Read-through transcription of RNA Pol II-dependent snoRNAs has already been observed in strains carrying mutations in the NRD1, NAB3 and SEN1 genes (Steinmetz et al., 2001). The proteins encoded by these genes are involved in the so-called Nrd1-dependent pathway for 3′ end formation of snoRNA and snRNA. Furthermore, Nrd1 is also able to interact with the 5′ end of the NRD1 pre-mRNA and to regulate its accumulation (Steinmetz and Brow, 1998; Steinmetz et al., 2001). Interestingly, NRD1 mRNA also accumulated in the ssu72-ts69 mutant both at 28 and 39°C, as shown by microarrays (data not shown) and confirmed by northern blot analyses (Figure 2A). All the above results suggest that Ssu72 plays a role in the 3′ termination of snoRNA transcription, possibly in conjunction with the Nrd1-dependent pathway.
Table I. Eleven ORFs whose expression is increased in ssu72-ts69 have a snoRNA gene upstream of their start codon.
| ORF | Ratio Ssu72:WT (28°C) | Ratio Ssu72:WT(1 h, 39°C) | Upstream snoRNA |
|---|---|---|---|
| YCR015c | 28.9 | 8.1 | Snr33 |
| YPR091c | 11.4 | 6.1 | Snr41, Snr70, Snr51 |
| YDR042c | 12.1 | 21.5 | Snr47 |
| YDR472w | 5.7 | 10.1 | Snr13 |
| YOL034w | 4.1 | 4.4 | Snr50 |
| YOR272w | 3.0 | 5.8 | Snr8 |
| YKR061w | 6.2 | 3.0 | Snr42a |
| YHR156c | 20.0 | 7.8 | Snr71a |
| YLR105c | 10.0 | 4.3 | Snr79 |
| YIL134w | 25.2 | No data | Snr68 |
| YGL088w | No data | 3.7 | Snr10 |
aGenes YKR061w and YHR156c are followed by a snoRNA gene transcribed convergently.
Fig. 1. Read-through transcription of snoRNAs in the ssu72-ts69 mutant. Northern blot analyses of transcripts in wild-type (WT) and ssu72-ts69 (ssu72) cells 0, 1 or 2 h after a shift to 39°C. The probes used are indicated at the bottom of each autoradiography, together with a schematic representation of the relative position of each gene analysed in the yeast genome. The different RNA transcripts are indicated with arrows. The positions of wild-type forms of YCR015c and TRS31 transcripts have been indicated as seen on overexposed autoradiographies (data not shown). (A) Probes for snr13 and TRS31 transcripts. (B) Probes for snr33 and Ycr015c transcripts. (C) Probes for POL4 and YCR015c transcripts.
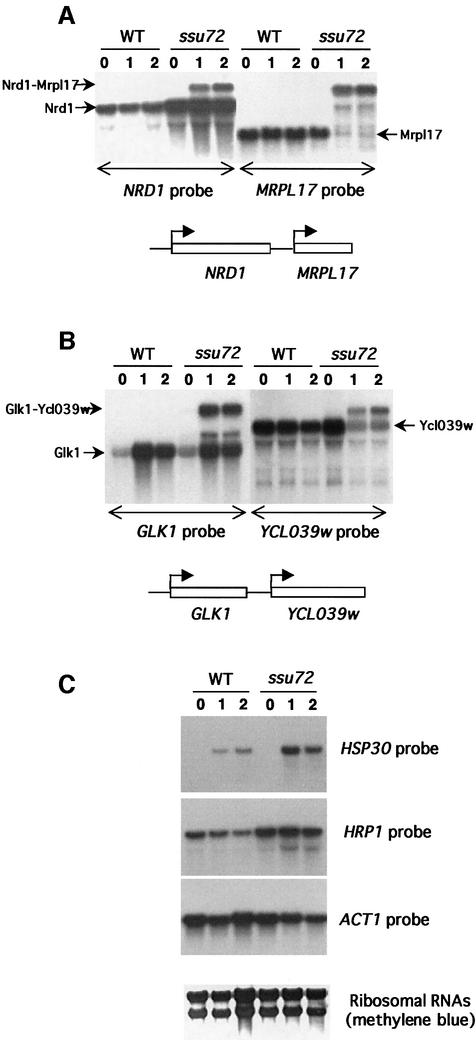
Fig. 2. Accumulation and read-through transcription of mRNAs in the ssu72-ts69 mutant. Northern blot analyses of transcripts in wild-type (WT) and ssu72-ts69 (Ssu72) cells 0, 1 or 2 h after a shift to 39°C. The probes used are indicated at the bottom (A and B) or at the right (C) of each autoradiography. For (A) and (B), a schematic representation of the relative position of each gene analyzed in the yeast genome is available. (A) Accumulation and read-through transcription of the NRD1 mRNA. (B) Read-through transcription of the GLK1 mRNA. (C) Accumulation of HRP1 and HSP30 transcripts. ACT1 probes and ribosomal RNAs are used as invariant controls.
The 19 other genes studied do not contain small RNA-encoding sequences upstream of their open reading frame. Northern blot analyses revealed that these accumulated mRNAs behaved differently. The normal NRD1 mRNA already accumulated at the permissive temperature, but an extended form of NRD1 RNA was also produced at the restrictive temperature (Figure 2A). To prove that this larger NRD1 RNA is formed by a 3′ extension of the NRD1 transcript, we used a probe for MRPL17, the ORF just downstream of NRD1 on chromosome XIV (Figure 2A). At 39°C, the rather small MRPL17 mRNA almost disappeared and was replaced by a larger form that migrated to the same position as the extended NRD1 RNA (Figure 2A). Thus, the ssu72-ts69 mutation also alters the transcription termination of the NRD1 gene. Whereas the ssu72-ts69 mutation already affected the 3′ termination of snoRNA at the permissive temperature, the read-through effect on the expression of the NRD1 mRNA appeared at 39°C only. Although the normal form of the NRD1 RNA was still produced, the level of the MRPL17 mRNA was strongly reduced. This suggests that the read-through transcription at the NRD1 cleavage/polyadenylation site inhibits the transcription initiation at the downstream MRPL17 start site. This read-through transcription of mRNAs is not specific for NRD1 as it was also found for the glucokinase-encoding gene GLK1 (Figure 2B). In this case, the normal form of GLK1 mRNA did not accumulate in the ssu72 mutant, but an additional extended form appeared at the restrictive temperature. This extended form of GLK1 mRNA cross-hybridized with a probe for the YCL039w ORF located downstream of GLK1 (Figure 2B). We also observed read-through transcription for the CUP1 transcript (data not shown). These defects that only appeared at the restrictive temperature may explain the thermosensitivity of the ssu72-ts69 mutation, because extended transcripts are not correctly translated and/or because the read-through polymerases prevent the correct expression of the downstream genes (Figure 2A and B). If the ssu72-ts69 mutation does not considerably increase the half-life of the NRD1 and GLK1 mRNA, a large proportion of transcripts produced from these genes must be efficiently cleaved and polyadenylated in the mutant cells at the restrictive temperature.
The transcriptional read-through detected in the ssu72-ts69 mutant did not affect all of the protein-coding genes as northern blot analyses showed that neither the HRP1 and HSP30 mRNAs, which accumulate in the ssu72-ts69 mutant, nor the ACT1 control displayed obvious extended forms (Figure 2C). In the case of HSP30 and HRP1, only a small, degraded band shift could be observed when the autoradiographs were overexposed (data not shown).
These results clearly show that Ssu72 plays a role in snoRNA and mRNA transcription termination, but also that not all pre-mRNAs use Ssu72 in a similar way for their correct termination.
Ssu72 interacts with the CTD kinase Kin28
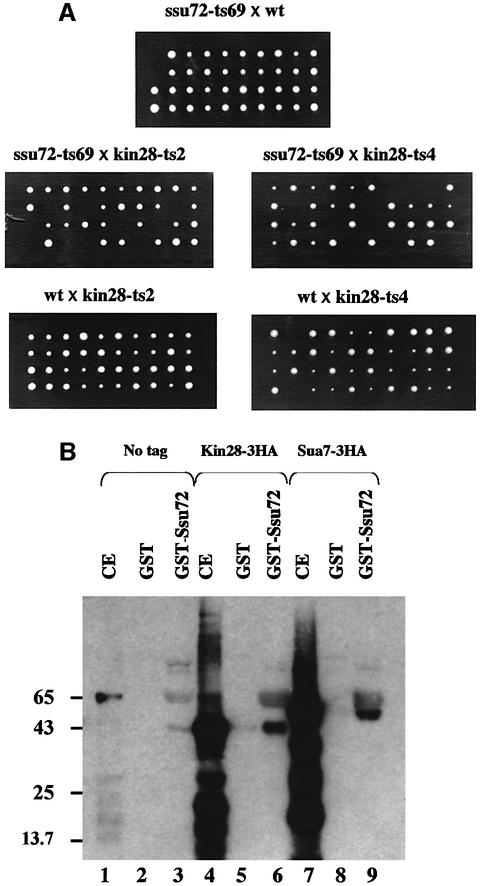
Ssu72 interacts with the general transcription factor TFIIB (Sua7) and with RNA Pol II during transcription initiation (Pappas and Hampsey, 2000). Ssu72 has also been found in the pre-mRNA 3′ end processing complex (Gavin et al., 2002; Sanders et al., 2002). We recently showed that SSU72 interacts genetically with CAK1 (C.Ganem, C.Miled, C.Facca, J.G.Valay, G.Labesse, S.Ben Hassine, C.Mann and G.Faye, in preparation). Mutations in the CAK1 gene have been found to be synthetic lethal with the kin28-ts3 (Valay et al., 1995) mutation. KIN28 encodes a cyclin-dependent kinase of the transcription factor TFIIH (Hampsey, 1998). We wondered whether SSU72 also interacts genetically with KIN28. The thermosensitive strains GF4125 (ssu72-ts52), GF3966 (ssu72-ts66), GF3969 (ssu72-ts69), GF3971 (ssu72-ts71) and GF3978 (ssu72-ts78) were crossed with GF4040 (kin28-ts2) and GF4044 (kin28-ts4). After sporulation and tetrad analysis, we observed that one quarter of the spores from each cross grew slowly or did not grow at all in the case of ssu72-ts69 (Figure 3A). This allele-specific synthetic lethality may indicate that Kin28 and Ssu72 interact physically.
Fig. 3. (A) Assay of synthetic lethality. Strains GF3969 (ssu72-ts69) was crossed with strain GF4040 (kin28-ts2) or GF4046 (kin28-ts4). As controls we crossed these mutants with wild-type strains GF1084 or GF1083. Diploids obtained were induced to sporulate. After dissection, segregants were left growing for 3 days at 28°C on YPD plates. (B) Pull-down experiments. Lanes 1, 4 and 7: 50 µg of yeast crude extracts containing no HA-tagged protein (1), protein Kin28 HA-tagged (4) and protein Sua7 HA-tagged (7) were acetone precipitated then loaded directly on the SDS–PAGE gel. Lanes 2 and 3: 50 µg of yeast crude extract containing no protein HA-tagged were incubated with glutathione–Sepharose beads coated with either GST alone (2) or GST–Ssu72 fusion protein (3), then the complexes were processed as described in Materials and methods. After acetone precipitation, the eluates were loaded on the SDS–PAGE gel. Lanes 5 and 6: 50 µg of yeast crude extract containing protein Kin28 HA-tagged were incubated with either GST alone (5) or GST–Ssu72 fusion protein (6) then processed as above. Lanes 8 and 9: 50 µg of yeast crude extract containing protein Sua7 HA-tagged were incubated with either GST alone (8) or GST–Ssu72 fusion protein (9) then processed as above. HA-tagged proteins were revealed with anti-HA 12CA5 antibody.
We used the glutathione S-transferase (GST) pull-down protocol to detect putative physical interactions between Ssu72 and Kin28. The Sua7 protein was used as a positive control, since a physical interaction between Sua7 and Ssu72 was previously described by Wu et al. (1999) in similar experiments. Crude extracts prepared from yeast strains expressing 3HA-tagged Kin28 or Sua7 proteins were incubated separately with GST or GST–Ssu72 bound to glutathione–Sepharose 4B. After elution, proteins were resolved by SDS–PAGE and detected with an anti-HA antibody (Figure 3B). It appeared that 3HA-Kin28 and 3HA-Sua7 were similarly bound to GST–Ssu72, but very poorly bound to GST alone. This in vitro binding of Ssu72 to Kin28, either directly or indirectly, is consistent with the allele-specific synthetic lethality observed between ssu72-ts69 and kin28-ts2 or kin28-ts4.
Phosphatase encoding FCP1 is a suppressor of ssu72 mutations
Strain GF3852 (ssu72-ts52) was transformed with a wild-type genomic library made with vector YEp13 (2µ, LEU2). Thermoresistant LEU+ transformants were selected on YPD plates at 35°C. The transforming plasmids were extracted and found to bear either the FCP1 gene or the SSU72 gene. FCP1 was subcloned into vector pRS425 (2µ, LEU2). The plasmid obtained, p1142, was used to transform strains GF3852 (ssu72-ts52), GF3966 (ssu72-ts66), GF3969 (ssu72-ts69), GF3971 (ssu72-ts71) and GF3978 (ssu72-ts78). LEU+ transformants were able to grow at 35°C (but not at 37°C). Fcp1 is an essential serine/threonine phosphatase that dephosphorylates the CTD of the largest subunit of RNA Pol II (Cho et al., 2001). We wondered whether the overexpression of Ssu72 could suppress fcp1 mutations. The SSU72 gene was cloned into pRS426 (2µ, URA3) and the plasmid obtained, p1467, was used to transform strains GF4698 (fcp1-2) and GF4699 (fcp1-4) (Cho et al., 2001). Neither GF4698 nor GF4699 became thermoresistant.
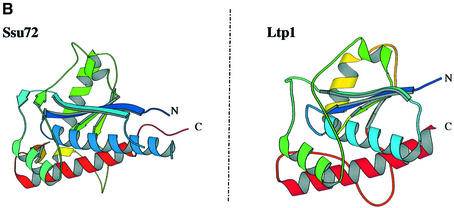
The 3-D modelling of Ssu72 reveals a phosphatase structure
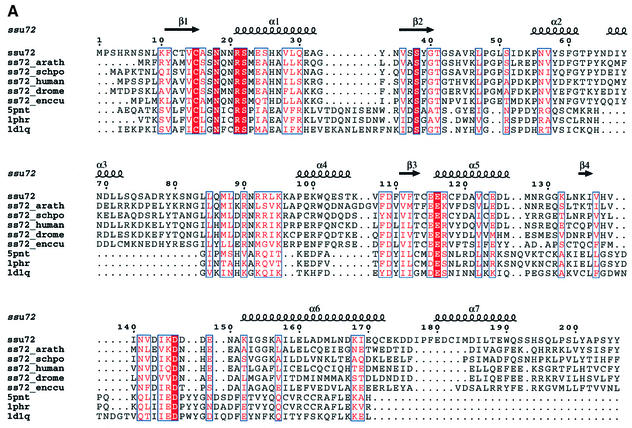
We searched protein sequence databases with the PSI-BLAST program. No putative conserved domains were detected and after two iterations the only significant hits found by the Psi-Blast searches corresponded to Ssu72 orthologues. However, these proteins present two common characteristics: they are small [160 to 210 amino acids (aa)] and contain a CxxxxxRS sequence motif at their N-terminal end (Figure 4A). This motif matches the signature of a large number of protein tyrosine phosphatases (Ramponi and Stefani, 1997) and some structurally related arsenate reductases (Bennett et al., 2001). The searches performed with ‘threaders’ identified the structures of protein phosphatases belonging to the LMW-PTP family as compatible folds, despite very weak overall sequence identity (<20%). They are PDB1PHR, PDB5PNT and PDB1D1Q (RCSB Protein Data Bank), giving a FUGUE Z-score of 5.52, a value that corresponds to >95% certainty (Shi et al., 2001). To assess the similarity, the putative homologous structures were used as templates for structural modelling. Molecular models were built using MODELLER after careful refinement of the structural alignment. The quality of the models was assessed using the PROSA, Verify3D and ERRAT programs, as described previously (Douguet et al., 2002). These models showed that Ssu72 could adopt the conserved fold of LMW-PTP, despite carrying large and variable insertions in external loops (Figure 4A). A model is presented in Figure 4B. Besides the residues of the catalytic motif, few residues appeared to be strictly conserved except for an aspartate (D146), which might be required for catalysis (Ramponi and Stefani, 1997). Thus, it is difficult to deduce the precise specificity of this new phosphatase from the proposed molecular model. In particular, the substrate-binding loops seem to lack the residues thought to be involved in phosphotyrosine recognition.
Fig. 4. (A) Structural alignment of Ssu72 of S.cerevisiae, eukaryotic orthologues and LMW-PTPs. The figure was made using the program Espript (Gouet et al., 1999). Ssu72: Ssu72 S.cerevisiae; ss72_arath: Ssu72 Arabidopsis thaliana; ss72_schpo: Ssu72 Schizosaccharomyces pombe; ss72_human: Ssu72 Homo sapiens; ss72_drome: Ssu72 Drosophila melanogaster; ss72_enccu: Ssu72 Encephalitozoon cuniculi; 5pnt: human LMW.PTP (HCPTP-A); 1phr: BPTP Bos taurus; 1d1q: LTP1: S.cerevisiae. (B) Tridimensional model of Ssu72 compared with the structure of Ltp1 obtained by X-ray diffraction (Wang et al., 2000b).
Ssu72 is a phosphatase
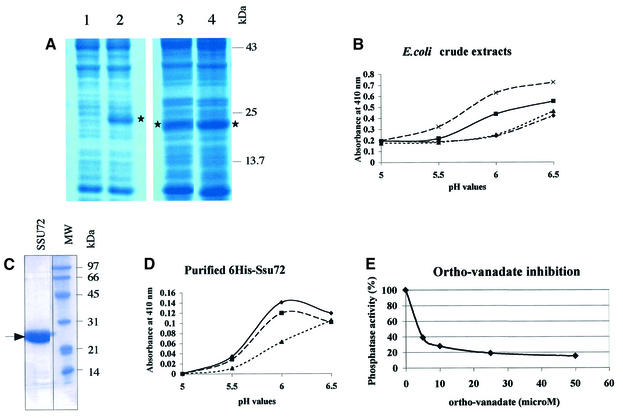
To test the phosphatase activity of Ssu72, we expressed the wild-type SSU72 borne on pET21a in Escherichia coli and tested the soluble crude extract for its ability to cleave p-nitro-phenyl phosphate (pNPP) in 100 mM citrate buffer at 28°C. The results were compared with those obtained with the soluble crude extract prepared from E.coli cells harboring pET21a without any insert. The endogenous E.coli phosphatases produced a basal level of p-nitro-phenol (pNP), which increased with the pH of the buffer (Figure 5B). However, when the wild-type Ssu72 protein was expressed, additional phosphatase activity was detected. This phosphatase activity increased the production of pNP 2-fold at pH 6.0. To confirm that this effect was actually due to the phosphatase activity of Ssu72, we conducted the same experiments with the ssu72-C15A allele, which carries a mutation in the putative catalytic site of Ssu72. This allele was unable to complement yeast cells carrying a ssu72::KanR deletion at the permissive temperature (data not shown). As expected, the ssu72-C15A allele displayed no extra pNPP hydrolysis activity compared with the empty pET21a control. Conversely, the dominant allele, ssu72-3319, which is able to suppress the thermosensitivity of a CAK1-deleted strain (GF3487, C.Ganem, C.Miled, C.Facca, J.G.Valay, G.Labesse, S.Ben Hassine, C.Mann and G.Faye, in preparation), exhibited even higher phosphatase activity than the soluble extract containing the wild-type protein. We purified the Ssu72 phosphatase and then characterized its enzymatic properties. The His-tagged Ssu72 protein was overproduced in E.coli BL21-Gold(DE3), then purified by affinity chromatography on Ni-NTA agarose and by gel filtration on a Superdex 75 column (Figure 5C). Purified His6-Ssu72 was assayed for its enzymatic activity. The fusion protein could efficiently hydrolyze pNPP at an optimum pH value of ∼6.2 at 28°C (Figure 5D). Its activity could not be increased by adding 100 µg/ml of BSA and was reduced when the temperature was increased to 37°C (Figure 5D). At 28°C and pH 6.0, the kinetic constants, Km and Vmax, were 130 mM and 0.23 mM/min per mg of protein His6-Ssu72, respectively. This suggests that the affinity of the His6-Ssu72 protein for pNPP is low but significant. Such a high Km for pNPP has already been observed with protein phosphatases, e.g. 50 mM with Cdc25 (Zhang et al., 1994) and 60 mM with Fcp1 (Kobor et al., 1999). In our conditions, the His6-Ssu72 protein was unable to hydrolyze phospho-tyrosine or phospho-serine (data not shown). Ortho-vanadate is an inhibitor of protein phosphatases, particularly of phospho-tyrosine phosphatases (Gordon, 1991). We tested the inhibitory effect of ortho-vanadate on the pNP phosphatase activity of the His6-Ssu72 protein (Figure 5E). At a concentration of 4 µM, ortho-vanadate blocked the enzyme activity of Ssu72 by 50%, at 28°C and pH 6.0.
Fig. 5. (A) Samples of soluble crude extracts prepared from E.coli cells transformed with plasmids pET21a (1), pET21a-His6-SSU72 (2), pET21a-SSU72-C15A (3) or pET21a-SSU72-3319 (4) were run on SDS–PAGE gels. The position of protein Ssu72 is indicated by an asterisk. (B) Phosphatase activities present in samples (1 mg) of soluble crude extracts prepared from E.coli cells transformed with plasmid pET21a carrying either no insert (diamond), wild-type SSU72 gene (square), mutant ssu72-C15A (triangle) or mutant ssu72-3319 (cross). Samples were assayed for their ability to cleave pNPP (20 mM) after 20 min of incubation at 28°C. (C) Purification of His6-tagged Ssu72 protein. A sample of the purified protein was run on an SDS–PAGE gel. (D) Phosphatase activity of the purified His6-Ssu72 protein. Samples of purified Ssu72 (45 µg) were incubated as described in Figure 4A (diamond), in presence of 100 µg/ml BSA (square) or at 37°C (triangle). (E) Ortho-vanadate inhibition. Samples (45 µg) of purified His6-tagged Ssu72 protein were incubated with 20 mM pNPP during 20 min at 28°C, with an increasing concentration of ortho-vanadate.
Discussion
Ssu72 is a phosphatase
Our data suggest that Ssu72 is a phosphatase. First, we constructed models of its 2D and 3D structures. The search for compatible folding identified Ltp1, a low-molecular weight tyrosine phosphatase from S.cerevisiae (Wang et al., 2000b). Ltp1 is similar to low molecular weight phosphatases from bovine heart and human placenta, but its role in cells is unknown. The sequence of Ssu72 has no evident homology with Ltp1. However, the 2D and 3D models allowed us to identify the active site signature motif (CxxNxNRSM). This motif is present at the N-terminal ends of all Ssu72 proteins sequenced so far. The proton donor residue might be aspartic acid 146 (Wang et al., 2000a).
We have overexpressed the wild-type form of SSU72, the ssu72-3319 dominant allele and the ssu72-C15A allele in E.coli. The E.coli soluble crude extract containing Ssu72 has an extra capacity to hydrolyze pNPP at pH 6.0. This activity was twice as high for the Ssu72-3319 crude extract (gain-of-function mutation), but no activity was detected in the Ssu72-C15A extract. The effect of this latter mutation is consistent with the hypothesis that cysteine 15 is an integral part of the catalytic site of Ssu72. After purification, the His-tagged Ssu72 protein was able to dephosphorylate pNPP. The inhibition of this activity by ortho-vanadate suggests that Ssu72 is indeed a phospho-tyrosine phosphatase. However, we cannot exclude the possibility that Ssu72 has the capacity to dephosphorylate phospho-serine or phospho-threonine (Ramponi and Stefani, 1997). Finally, the fact the ssu72-C15A mutant is non-viable, even at 28°C, strongly suggests that the phosphatase activity of Ssu72 is essential for its physiological function.
Ssu72 is essential for the termination of snoRNA transcription
Our microarray analyses suggested that the transcription of at least 11 snoRNA genes interferes with the expression of adjacent downstream genes. Indeed, northern blot analyses revealed that read-through transcription occurs for at least four of these 11 snoRNA genes (snr13, snr33, snr8 and snr42) into adjacent genes. These results imply that Ssu72 is required for the transcription termination of this particular group of transcripts by RNA Pol II. SnoRNAs are 60–300 nucleotides long, and metabolically stable RNAs (Kiss, 2002). Saccharomyces cerevisiae is predicted to possess up to 100 different snoRNAs. About 90% of these snoRNAs are transcribed from independent promoters. The release of individual and mature species involves both endonucleolytic cleavage and exonucleolytic trimming, but not polyadenylation (Morlando et al., 2002). The correct termination of snoRNA requires some complexes that are shared with the pre-mRNA maturation pathway [e.g. the cleavage factor IA (CFIA)] and some others that are thought to be specific for snoRNA 3′ termination (Steinmetz et al., 2001; Morlando et al., 2002). Our results suggest that the Ssu72 phosphatase interferes with the function of the Nrd1-dependent pathway for the formation of the 3′ ends of non-polyadenylated transcripts. The finding that NRD1 mRNA accumulates in the ssu72-ts69 mutant, a feature that is shared by all components of the Nrd1 pathway, strengthens this hypothesis. Indeed, Nrd1 controls the accumulation of its own mRNA, and possibly others (Steinmetz and Brow, 1998). This regulation requires the interaction of Nrd1 with both the 5′ UTR sequence of nascent NRD1 transcripts and the CTD (Steinmetz et al., 2001). Interestingly, read-through snoRNA transcripts have also been observed in strains partially lacking the CTD or deleted for the CTD kinase-encoding gene CTK1 (Steinmetz et al., 2001). Thus, Ssu72 phosphatase may participate in the Nrd1-dependent pathway by altering the phosphorylation state of the CTD either directly or indirectly. Interestingly, we found that the ssu72-ts52 ctk1Δ and ssu72-ts69 ctk1Δ double mutants were lethal (data not shown).
What is the role of Ssu72 in the mRNA transcription cycle?
Ssu72 is an intriguing protein. It was initially thought to play a general role in the initiation of transcription, on the basis of its genetic and physical interactions with Sua7 (TFIIB) (Wu et al., 1999). More recent studies have shown that Ssu72 also associates with mRNA 3′ end processing complexes (Dichtl et al., 2002; Gavin et al., 2002; Sanders et al., 2002), suggesting that Ssu72 accompanies the RNA Pol II machinery throughout the transcription process. Dichtl and co-workers found that Ssu72 is a component of the mRNA cleavage and polyadenylation complex CPF in yeast, suggesting that it is involved in general termination of transcription of mRNA (Dichtl et al., 2002). Indeed, these authors showed that the ssu72-2 mutant produced unstable read-through transcripts of the CUP1 gene. They also found that the transcription elongation inhibitor 6-azauracil suppressed ssu72-2 thermosensitivity. Thus, they hypothesized that Ssu72 balances elongation and termination by decreasing the rate of elongation of RNA Pol II (Dichtl et al., 2002).
The genetic and physical interactions that we found between Ssu72 and Kin28 on the one hand, and the genetic interactions between Ssu72 and Ctk1 on the other hand, confirm that Ssu72 acts upon the transcription machinery soon after the initiation step and during elongation. Our northern blotting studies with the ssu72-ts69 mutant also suggest that Ssu72 plays a role in the termination process. At the restrictive temperature, the ssu72-ts69 mutant exhibits stable read-through transcripts for genes NRD1, GLK1 and CUP1. As the mutant still produced significant amounts of NRD1 and GLK1 mRNA, the ssu72-ts69 mutant does not appear to be deficient for 3′ cleavage and polyadenylation. Indeed, Dichtl and co-workers have shown that extracts prepared from mutant ssu72-2 efficiently cleaved and polyadenylated CYC1 pre-mRNA (Dichtl et al., 2002). However, the transcription termination of many protein-encoding genes does not seem to require Ssu72. The HSP30, HRP1 and ACT1 genes express few or no stable extended forms in the ssu72-ts69 mutant, and the HSP82 and SSA4 genes express few or no stable extended forms in the ssu72-2 rrp6Δ double mutant (Dichtl et al., 2002).
Why is Ssu72 only required by particular genes?
Pre-snoRNA 3′ end processing is highly dependent on the activity of Ssu72. For the pre-mRNAs, this requirement seems to be more gene dependent. For instance, the NDR1 transcription termination signal is inadequately recognized in the ssu72-ts69 mutant at the restrictive temperature, whereas the MRPL17 termination signal is correctly detected. Why is the requirement for Ssu72 specific for the termination of pre-snoRNAs and of some pre-mRNAs? It is generally accepted that when an RNA Pol II elongation complex meets a cleavage/polyadenylation signal or a signal specific for the 3′ end processing of pre-snoRNAs, the elongating complex is converted into a termination-prone complex. RNA polymerase tends to pause and then to be stochastically released from the template DNA (Proudfoot et al., 2002). We hypothesize that Ssu72 is one of the components that promotes RNA polymerase pausing and release once a termination signal has been encountered. The conserved motifs that are essential for directing the 3′ end processing of pre-snoRNAs in yeast are clearly different from the cis-acting signals that direct the 3′ end formation of pre-mRNAs (Morlando et al., 2002). Furthermore, the cleavage/polyadenylation signals of pre-mRNAs in S.cerevisiae are less clearly defined than in mammalian cells (Graber et al., 1999; van Helden et al., 2000) and may have different strengths to direct termination (Proudfoot et al., 2002). This variety in the sequences directing the 3′ end processing of RNA Pol II transcripts may partly explain the different requirements for Ssu72. Another non-exclusive explanation is that different phosphatases, with non-overlapping specificities, may control the transcription termination of sub-classes of mRNA. Besides Ssu72, another candidate is phosphatase Glc7, which interacts with the cleavage and polyadenylation factor in vivo (Gavin et al., 2002).
Putative substrates of phosphatase Ssu72
Pre-mRNA 3′ end processing is dependent on the presence of RNA Pol II CTD both in vivo and in vitro (Proudfoot et al., 2002). Moreover, several subunits of termination complexes interact directly with the CTD. Our finding that Ssu72 is a phosphatase that physically and genetically interacts with CTD kinase Kin28 and genetically interacts with CTD kinase Ctk1 and with CTD phosphatase Fcp1 suggests that Ssu72 modulates the phosphorylation state of the CTD during the transcription cycle and particularly during the termination step. Tyr-1 as well as Ser-2 and Ser-5 of the YSPTSPS heptapeptide repeats are essential for the functions of RNA Pol II in vivo in S.cerevisiae (West and Corden, 1995). In mammalian cells, the CTD is known to be phosphorylated on tyrosine by the c-Abl proto-oncogene (Duyster et al., 1995). In yeast, it is not known whether Tyr-1 is phosphorylated. It would be worth testing whether the phosphorylated CTD is a substrate of Ssu72. Moreover, numerous transcription factors are also phosphorylated and could be potential substrates of Ssu72 (Kobor and Greenblatt, 2002).
In conclusion, our work shows that Ssu72 is a phosphatase. This phosphatase is an essential factor in the transcription termination of both pre-snoRNAs and pre-mRNAs, processes that are regulated by many unknown cis- and trans-acting signals. Additionally, Ssu72 is likely to have pleiotropic roles in transcription through its interactions with multiple protein factors involved in transcription initiation (TFIIB, Kin28) and elongation (Ctk1, Rpb2, Fcp1).
Materials and methods
Yeast strains, plasmids and media
Yeast strains used in this work are described in Table II. Complete medium (YPD) is 1% (w/v) yeast extract, 1% (w/v) bactopeptone, 2% (w/v) glucose. Synthetic complete medium (SC) with 2% glucose is as described by Sherman (1991). Plasmids used in this study are listed in Table III. Plasmid pET21a and E.coli strain BL21(DE3) are from Novagen.
Table II. Strains used in this study.
| Straina | Genotype |
|---|---|
| GF1047 | MATa ura3 leu2 trp1 lys2 |
| GF1083 | MATα ura3 leu2 trp1 lys2 |
| GF1084 | MATa ura3 leu2 trp1 his3 |
| GF1310 | MATα ura3 leu2 trp1 lys2 cyh2R |
| GF3721 | MATa ura3 leu2 trp1 lys2 ssu72::KanR p946(SSU72 URA3) |
| GF3852 | MATa ura3 leu2 trp1 lys2 ssu72-52 |
| GF3966 | MATα ura3 leu2 trp1 lys2 cyh2R ssu72-66 |
| GF3969 | MATα ura3 leu2 trp1 lys2 cyh2R ssu72-69 |
| GF3971 | MATα ura3 leu2 trp1 lys2 cyh2R ssu72-71 |
| GF3978 | MATα ura3 leu2 trp1 lys2 cyh2R ssu72-78 |
| GF4040 | MATa ura3 leu2 trp1 his3 kin28-ts2 |
| GF4044 | MATa ura3 leu2 trp1 his3 kin28-ts4 |
| GF4125 | MATα ura3 leu2 trp1 lys2 ssu72-52 |
| GF4698 | leu2 trp1 ura3 his3 ade2 can1 fcp1::LEU2 pRS314(fcp1-2 TRP1) |
| GF4699 | leu2 trp1 ura3 his3 ade2 can1 fcp1::LEU2 pRS314(fcp1-4 TRP1) |
| GF4790 | MATa ura3 leu2 trp1 his3 ctk1::TRP1-CYH2 |
aAll the strains are congenic, GF4698 and GF4699 excepted.
Table III. Plasmids used in this study.
| Plasmid | Vector (relevant genotype) | Insert (gene or gene fusion) |
|---|---|---|
| p912 | pRS416 (URA3, CEN) | ssu72-3319 |
| p946 | pRS416 (URA3, CEN) | SSU72 |
| p1142 | pRS425 (LEU2, 2µ) | FCP1 |
| p1349 | pGEX-6P-1 | GST-SSU72 |
| p1355 | pET21a | SSU72 |
| p1372 | pYES2 (URA3, 2µ, pGal1) | SUA7-3HA |
| p1373 | pYES2 (URA3, 2µ, pGal1) | KIN28-3HA |
| p1393 | pRS416 (URA3, CEN) | ssu72-C15A |
| p1394 | YEplac111 (LEU2, CEN) | ssu72-C15A |
| p1396 | pET21a | ssu72-3319 |
| p1399 | pET21a | ssu72-C15A |
| p1439 | pET21a | His6-SSU72 |
| p1443 | pYES2 (URA3, 2µ, pGal1) | His6-SSU72 |
| p1467 | pRS426 (URA3, 2µ) | SSU72 |
Site-directed mutagenesis of the SSU72 gene
We followed the QuikChange Site-Directed Mutagenesis method developed by Stratagene. The ssu72-C15A allele was constructed using the two complementary primers SSUCA1 CGCAATTCAAACTTGAAGTTTTGTACAGTTGCTGCATCAAACAACAATCGTTCAATGG and SSUCA2 CCATTGAACGATTGTTGTTTGATGCAGCAACTGTACAAAACTTCAAGTTTGAATTGCG and plasmid p946(SSU72) as a template. The underlined GC bases changed the TGT codon (cysteine) at position 15 into a GCT codon (alanine), whereas a BsrGI restriction site (bold letters) was introduced without changing the amino acid sequence of Ssu72. The plasmid obtained was named p1393. The mutated ssu72 gene was amplified using primers SSUNDE and DHSSUX (Supplementary table 2). The PCR product was digested with XhoI and NdeI and cloned between the XhoI and BamHI sites of pET21a creating the p1399 plasmid. The constructs were verified by sequencing.
GST pull-down analysis
The SSU72 wild-type gene was amplified by PCR using primers DHSSUE and DHSSUX and was cloned between the EcoRI and XhoI sites of vector pGEX-6P-1, yielding plasmid p1349. GST–Ssu72 was overexpressed in E.coli BL21(DE3) and crude bacterial extracts were prepared as below (see Purification of protein Ssu72). SUA7 and KIN28 wild-type genes were amplified by using primers SUA7ECOR1, SUA7PAC1, KIN28ECOR1 and KIN28PAC1 (Supplementary table 2) and were cloned EcoRI/PacI into pYES2-derived plasmid (URA3, 2µ) (p1347) allowing the fusion of 3xHA tags at the C-terminal end. The plasmids obtained were p1372 and p1373, respectively. They were introduced separately into yeast strain GF1047. After an overnight growth at 28°C in SC(–URA, +Gal, +Raf) liquid medium, cells were washed with cold water, then were crushed (Schultz, 1999). Ground cells were resuspended in PBS containing protease inhibitors (5 µg/ml leupeptin, 5 µg/ml pepstatin, 0.2 mM pefabloc). They were centrifuged at 13 000 r.p.m. for 10 min.
Crude extracts from bacteria (400 µg) containing either GST alone or GST–Ssu72, were incubated with glutathione–Sepharose 4B (100 µl) (Amersham) in 500 µl of PBS–glycerol (PBS–10% glycerol containing protease inhibitors) for 2 h at 4°C. The samples were centrifuged at 500 g for 2 min. The supernatants were removed and the beads were washed three times with 1 ml of PBS–glycerol. After that, 50 µg of yeast crude extracts containing either Sua7-3HA or Kin28-3HA were added to GST alone or GST–Ssu72 with 200 µl of PBS–glycerol. The samples were incubated overnight at 4°C. Following centrifugation, the beads were washed six times with PBS–glycerol. Proteins were eluted with 100 µl of elution buffer [10 mM reduced glutathione (Sigma), 50 mM Tris–HCl pH 8.0] for 2 h at 4°C, then were precipitated with 3 vols of acetone. Proteins were resuspended in SDS–PAGE loading buffer then run on an SDS–PAGE gel. Proteins were transferred to a nitrocellulose membrane and western analysis was performed using anti-HA 12CA5 antibody (Boehringer Mannheim).
Molecular modelling
Protein sequence database searches were performed with the PSI-BLAST version 2.0.5 program (Altschul et al., 1997) with default parameters. Compatible tridimensional structures were searched using a dedicated meta-server (Douguet and Labesse, 2001). Alignment refinement was subsequently performed using the program TITO (Labesse and Mornon, 1998) using, as templates, known structures of LMW-PTP [accession codes in the RCSB Protein Data Bank: PDB1PHR, PDB5PNT and PDB1D1Q (Su et al., 1994; Zhang et al., 1998; Wang et al., 2000b)]. Validity of the refined alignment were assessed through pseudo-energy and visual inspection. The secondary structures (α-helix, β-strand) of Ssu72 were predicted using JPRED2 (Cuff et al., 1998) and were also deduced by similarity during TITO processing. These secondary structure predictions were merged by consensus and used as additional restraints in the following modelling steps. Three-dimensional models were built using either an analogue-bound or an inactive form of LMW-PTP (PDB1D1Q and PDB1PHR, respectively) as distinct templates in MODELLER 6.1 (Sali and Blundell, 1993) and assessed using PROSA (Sippl, 1993), ERRAT (Colovos and Yeats, 1993) and Verify3D (Eisenberg et al., 1997).
Purification of protein Ssu72
Plasmids p1355 [pET21a (SSU72)], p1396 [pET21a (ssu72-3319)] or p1399 [pET21a (ssu72-C15A)] were introduced by electroporation into an E.coli BL21(DE3) strain that is able to overexpress chaperonins GroEL and GroES. Transformants were cultivated at 37°C in 200 ml of LB medium in presence of Amp (80 µg/ml) and Kan (25 µg/ml) until A600 reached 0.8, then the cultures were incubated at 16°C for 1 h before IPTG was added to a final concentration of 0.5 mM. Growth was continued overnight at 16°C. Cells were harvested by centrifugation and resuspended in 10 ml of glycerophosphate buffer [50 mM Na glycerophosphate pH 7.3, 100 mM NaCl, 20% (v/v) glycerol, 2 mM EDTA, 50 µl/500ml β-mercaptoethanol] containing proteases inhibitors (Roche, 1873580). Cells were disrupted in a One Shot Cell Disruption Model (Constant Systems Ltd, Warwick, UK) at 2.0 kBar. The resulting suspensions were centrifuged at 4°C for 1 h at 9000 r.p.m. (Sorvall, SS34) and the supernatants were kept.
Plasmid p1439 [pET21a (His6-Ssu72)] was introduced into E.coli BL21-Gold (DE3) (Stratagene) able to overexpress GroEL and GroES. Transformants were grown at 37°C in 400 ml of 2YT, then expression of Ssu72 was induced at 16°C for 8.30 h with 0.3 mM IPTG. After centrifugation, cells were suspended in 10 ml buffer A (25 mM Na phosphate pH 7.5, 150 mM NaCl, 5 mM β-mercaptoethanol). Cells were lysed by three cycles of freezing and thawing, followed by sonication. The resulting suspension was centrifuged at 4°C for 30 min at 13 000 RPM. The supernatant was loaded onto 3 ml of Ni-NTA resin (Qiagen). The column was washed with 60 ml buffer A plus 20 mM imidazol. His6-Ssu72 was eluted with 8 ml buffer A plus 100 mM imidazol. The eluate was concentrated by centrifugation (Vivaspin, Sartorius), then applied onto a Superdex 75 column equilibrated with 20 mM Tris–HCl pH 6.8, 200 mM NaCl, 5% glycerol, 10 mM β-mercaptoethanol.
Phosphatase assays
Phosphatase activity was monitored by the production of pNP from pNPP (spectroscopically assayed at 410 nm) after a reaction time of 25 min. Reactions were performed in a 100 mM citrate buffer (pH 5.0–6.5) with 20 mM pNPP at 28°C, then were stopped by adding 3.8 vols of 0.1 M NaOH. For each assay, we used either 1 mg of E.coli crude extracts or 45 µg of purified His6-Ssu72 protein.
Northern blot analyses
Strains GF3969 (ssu72-ts69) and GF1310 (SSU72) were grown at 28°C in YPD to an OD600 of 0.5. The cultures were then transferred to a 39°C water bath for 0, 1 and 2 h. Cells were processed and total cellular RNA was prepared as described previously (Valay et al., 1995). An equal amount of RNA (15 µg) from each sample was denatured, submitted to electrophoresis and transferred to a nitrocellulose sheet by capillarity. Probes were made by random priming (Nonaprimer kit) of PCR products corresponding to the ORF of interest. For snoRNA, we used oligonucleotide probes labelled with 32P by T4 kinase using a standard protocol (Sambrook et al, 1989). The sequence of these probes can be obtain from the authors (devaux@biologie.ens.fr) on request. The northern blot used to characterize the snoRNA-ORF hybrid RNAs were made as described previously (Steinmetz et al., 2001).
Microarray analysis
For microarray analyses, mRNA were prepared from total RNA used for northern blot analyses (see above) using the Micro Fast track mRNA purification kit (Invitrogen). Microarray slides containing most of the yeast open reading frames (5885 PCR products) were obtained from Hitachi Software and DNAChip Research, Inc. Four micrograms of mRNA were used for each reverse transcription reaction. Detailed protocols are at www.biologie.ens.fr/fr/genetiqu/puces/protocoles puces. html. The arrays were read by a Genepix 4000 scanner (Axon) and were analyzed with the Genepix 3.0 software. Each microarray result presented here is an average of two independent biological measurements. We excluded artefactual, saturated and low signal spots. Assuming that most of the genes have an unchanged expression, the Cy3/Cy5 ratios were normalized by use of the median of all the ratios for each experiment.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful to D.Tollervey, J.Greenblatt and C.Kane for DNA probes and yeast strains, and B.Dichtl for the communication of results before publication. We thank P.Hughes, S.Urbach, Y.Blouquit, P.Chambon, L.Cabanié, Y.Cohen, C.Craescu, C.Grangeasse, R.Chanet, M.Pierre and R.Moore for help and useful suggestions. The microarray facilities of the Ecole Normale Superieure are part of the Genopole Ile de France.
References
- Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M.S., Guan,Z., Laurberg,M. and Su,X.D. (2001) Bacillus subtilis arsenate reductase is structurally and functionally similar to low molecular weight protein tyrosine phosphatases. Proc. Natl Acad. Sci. USA, 98, 13577–13582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E.J., Kobor,M.S., Kim,M., Greenblatt,J. and Buratowski,S. (2001) Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev., 15, 3319–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colovos C. and Yeates,T.O. (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci., 2, 1511–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff J.A., Clamp,M.E., Siddiqui,A.S., Finlay,M. and Barton,G.J. (1998) JPred: a consensus secondary structure prediction server. Bioinformatics, 14, 892–893. [DOI] [PubMed] [Google Scholar]
- Dichtl B., Blank,D., Ohnacker,M., Friedlein,A., Roeder,D., Langen,H. and Keller,W. (2002) A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell, 10, 1139–1150. [DOI] [PubMed] [Google Scholar]
- Douguet D. and Labesse,G. (2001) Easier threading through web-based comparisons and cross-validations. Bioinformatics, 17, 752–753. [DOI] [PubMed] [Google Scholar]
- Douguet D., Bolla,J.M., Munier-Lehmann,H. and Labesse,G. (2002) From sequence to structure: a case study. Enzyme Microbiol. Biotech., 30, 289–294. [Google Scholar]
- Duyster J., Baskaran,R. and Wang,J.Y. (1995) Src homology 2 domain as a specificity determinant in the c-Abl-mediated tyrosine phosphorylation of the RNA polymerase II carboxyl-terminal repeated domain. Proc. Natl Acad. Sci. USA, 92, 1555–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D., Luthy,R. and Bowie,J.U. (1997) VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol., 277, 396–404. [DOI] [PubMed] [Google Scholar]
- Gavin A.C. et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]
- Gordon J.A. (1991) Use of vanadate as protein-phosphotyrosine phosphatase inhibitor. Methods Enzymol., 201, 477–482. [DOI] [PubMed] [Google Scholar]
- Gouet P., Courcelle,E., Stuart,D.I. and Metoz,F. (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics, 15, 305–308. [DOI] [PubMed] [Google Scholar]
- Graber J.H., Cantor,C.R., Mohr,S.C. and Smith,T.F. (1999) Genomic detection of new yeast pre-mRNA 3′-end-processing signals. Nucleic Acids Res., 27, 888–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M. (1998) Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev., 62, 465–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K.J. (2002) RNA polymerase II conducts a symphony of pre-mRNA processing activities. Biochim. Biophys. Acta, 1577, 308–324. [DOI] [PubMed] [Google Scholar]
- Kimmelman J., Kaldis,P., Hengartner,C.J., Laff,G.M., Koh,S.S., Young,R.A. and Solomon,M.J. (1999) Activating phosphorylation of the Kin28p subunit of yeast TFIIH by Cak1p. Mol. Cell. Biol., 19, 4774–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. (2002) Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell, 109, 145–148. [DOI] [PubMed] [Google Scholar]
- Kobor M.S. and Greenblatt,J. (2002) Regulation of transcription elongation by phosphorylation. Biochim. Biophys. Acta, 1577, 261–275. [DOI] [PubMed] [Google Scholar]
- Kobor M.S. et al. (1999) An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation. Mol. Cell, 4, 55–62. [DOI] [PubMed] [Google Scholar]
- Labesse G. and Mornon,J. (1998) A tool for incremental threading optimization (TITO) to help alignment and modelling of remote homologues. Bioinformatics, 14, 206–211. [DOI] [PubMed] [Google Scholar]
- Morlando M., Greco,P., Dichtl,B., Fatica,A., Keller,W. and Bozzoni,I. (2002) Functional analysis of yeast snoRNA and snRNA 3′-end formation mediated by uncoupling of cleavage and polyadenylation. Mol. Cell. Biol., 22, 1379–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palancade B., Dubois,M.F. and Bensaude,O. (2002) FCP1 phosphorylation by casein kinase 2 enhances binding to TFIIF and RNA polymerase II carboxyl-terminal domain phosphatase activity. J. Biol. Chem., 277, 36061–36067. [DOI] [PubMed] [Google Scholar]
- Pappas D.L. Jr, and Hampsey,M. (2000) Functional interaction between Ssu72 and the Rpb2 subunit of RNA polymerase II in S. cerevisiae. Mol. Cell. Biol., 20, 8343–8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patturajan M., Schulte,R.J., Sefton,B.M., Berezney,R., Vincent,M., Bensaude,O., Warren,S.L. and Corden,J.L. (1998) Growth-related changes in phosphorylation of yeast RNA polymerase II. J. Biol. Chem., 273, 4689–4694. [DOI] [PubMed] [Google Scholar]
- Pokholok D.K., Hannett,N.M. and Young,R.A. (2002) Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell, 9, 799–809. [DOI] [PubMed] [Google Scholar]
- Proudfoot N.J., Furger,A. and Dye,M.J. (2002) Integrating mRNA processing with transcription. Cell, 108, 501–512. [DOI] [PubMed] [Google Scholar]
- Ramponi G. and Stefani,M. (1997) Structure and function of the low Mr phosphotyrosine protein phosphatases. Biochim. Biophys. Acta, 1341, 137–156. [DOI] [PubMed] [Google Scholar]
- Sali A. and Blundell,T.L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol., 234, 779–815. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sanders S.L., Jennings,J., Canutescu,A., Link,A.J. and Weil,P.A. (2002) Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol., 22, 4723–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz M.C. (1999) Chromatin assembly in yeast cell-free extracts. Methods, 17, 161–172. [DOI] [PubMed] [Google Scholar]
- Sherman F. (1991) Getting started with yeast. Methods Enzymol., 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Shi J., Blundell,T.L. and Mizuguchi,K. (2001) FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol., 310, 243–257. [DOI] [PubMed] [Google Scholar]
- Sippl M.J. (1993) Recognition of errors in three-dimensional structures of proteins. Proteins, 17, 355–362. [DOI] [PubMed] [Google Scholar]
- Steinmetz E.J. and Brow,D.A. (1998) Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proc. Natl Acad. Sci. USA, 95, 6699–6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz E.J., Conrad,N.K., Brow,D.A. and Corden,J.L. (2001) RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature, 413, 327–331. [DOI] [PubMed] [Google Scholar]
- Su X.D., Taddei,N., Stefani,M., Ramponi,G. and Nordlund,P. (1994) The crystal structure of a low-molecular-weight phosphotyrosine protein phosphatase. Nature, 370, 575–578. [DOI] [PubMed] [Google Scholar]
- Sun Z.W. and Hampsey,M. (1996) Synthetic enhancement of a TFIIB defect by a mutation in SSU72, an essential yeast gene encoding a novel protein that affects transcription start site selection in vivo. Mol. Cell. Biol., 16, 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valay J.G., Simon,M., Dubois,M.F., Bensaude,O., Facca,C. and Faye,G. (1995) The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J. Mol. Biol., 249, 535–544. [DOI] [PubMed] [Google Scholar]
- van Helden J., del Olmo,M. and Perez-Ortin,J.E. (2000) Statistical analysis of yeast genomic downstream sequences reveals putative polyadenylation signals. Nucleic Acids Res., 28, 1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Stauffacher,C.V. and Van Etten,R.L. (2000a) Structural and mechanistic basis for the activation of a low-molecular weight protein tyrosine phosphatase by adenine. Biochemistry, 39, 1234–1242. [DOI] [PubMed] [Google Scholar]
- Wang S., Tabernero,L., Zhang,M., Harms,E., Van Etten,R.L. and Stauffacher,C.V. (2000b) Crystal structures of a low-molecular weight protein tyrosine phosphatase from Saccharomyces cerevisiae and its complex with the substrate p-nitrophenyl phosphate. Biochemistry, 39, 1903–1914. [DOI] [PubMed] [Google Scholar]
- West M.L. and Corden,J.L. (1995) Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics, 140, 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W.H., Pinto,I., Chen,B.S. and Hampsey,M. (1999) Mutational analysis of yeast TFIIB. A functional relationship between Ssu72 and Sub1/Tsp1 defined by allele-specific interactions with TFIIB. Genetics, 153, 643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S. and Prelich,G. (2002) Activation of the bur1–bur2 cyclin-dependent kinase complex by cak1. Mol. Cell. Biol., 22, 6750–6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Stauffacher,C.V., Lin,D. and Van Etten,R.L. (1998) Crystal structure of a human low molecular weight phosphotyrosyl phosphatase. Implications for substrate specificity. J. Biol. Chem., 273, 21714–21720. [DOI] [PubMed] [Google Scholar]
- Zhang Z.Y. and Dixon,J.E. (1994) Protein tyrosine phosphatases: mechanism of catalysis and substrate specificity. Adv. Enzymol. Relat. Areas Mol. Biol., 68, 1–36. [DOI] [PubMed] [Google Scholar]