Abstract
Pro-survival Bcl-2-related proteins, critical regulators of apoptosis, contain a hydrophobic groove targeted for binding by the BH3 domain of the pro-apoptotic BH3-only proteins. The solution structure of the pro-survival protein Bcl-w, presented here, reveals that the binding groove is not freely accessible as predicted by previous structures of pro-survival Bcl-2-like molecules. Unexpectedly, the groove appears to be occluded by the C-terminal residues. Binding and kinetic data suggest that the C-terminal residues of Bcl-w and Bcl-xL modulate pro-survival activity by regulating ligand access to the groove. Binding of the BH3-only proteins, critical for cell death initiation, is likely to displace the hydrophobic C-terminal region of Bcl-w and Bcl-xL. Moreover, Bcl-w does not act only by sequestering the BH3-only proteins. There fore, pro-survival Bcl-2-like molecules probably control the activation of downstream effectors by a mechanism that remains to be elucidated.
Keywords: apoptosis/Bcl-2/binding/NMR/protein structure
Introduction
Apoptosis, the physiological process of killing and removing damaged, unwanted or surplus cells during development, tissue homeostasis, or in response to stress or damage signals, is conserved between organisms as diverse as worms and man. The Bcl-2 family of proteins (Cory and Adams, 2002) are critical regulators of this process and function to regulate cell fate by controlling the activation of caspases, cysteine proteases that cause cellular destruction by cleaving vital substrates. Some members of the Bcl-2 family, including Bcl-2 itself, Bcl-xL, Bcl-w, Mcl-1 and A1, promote cell survival and their absence, such as that observed in mice lacking Bcl-w (Print et al., 1998), results in tissue degeneration.
The pro-survival proteins display a high degree of sequence identity and generally contain four conserved Bcl-2 homology domains (BH1–4), as well as a C-terminal hydrophobic region. Solution structures of Bcl-xL (Muchmore et al., 1996), Bcl-2 (Petros et al., 2001) and the Kaposi sarcoma herpes virus (KSHV) Bcl-2 homolog (Huang et al., 2002), reveal that the BH1-3 domains are in close proximity to each other and form a hydrophobic groove that is the docking site for BH3-only proteins (Sattler et al., 1997; Petros et al., 2000). In contrast, the BH3-only proteins, including mammalian Bik/Nbk, Bid, Bad, Bim/Bod and Bmf, have distinct sequences, but contain a conserved 9–16 residue BH3 domain required for binding to the pro-survival proteins (Huang and Strasser, 2000). The BH3-only proteins seem to act as sensors of cellular well-being, normally held in check unless stress signals cause their activation to initiate killing (Huang and Strasser, 2000). Precisely how these proteins induce killing is unclear, but their action is dependent on a second group of pro-apoptotic Bcl-2-like proteins, the multi-domain Bax subfamily (Bax, Bak and Bok/Mtd) (Cheng et al., 2001; Zong et al., 2001). The Bax sub-family of proteins also contain BH1–3 and surprisingly, the overall structure of full-length Bax resembles that of pro-survival Bcl-xL (Suzuki et al., 2000). However, in contrast to the structures of C-terminally truncated Bcl-xL or Bcl-2, the hydrophobic groove formed by the BH1–3 domains in Bax is occluded by its C-terminus. Translocation of Bax from the cytosol to intracellular membranes, particularly the outer mitochondrial membrane (Nechushtan et al., 1999, 2001), is an early step in its damage signal-induced activation and exposure of the hydrophobic C-terminal region may be important for this process.
Pro-survival Bcl-w is functionally indistinguishable from Bcl-2 and Bcl-xL (Gibson et al., 1996; O’Reilly et al., 2001; J.Wilson-Annan et al., in preparation), although it appears to be located exclusively on the mitochondrion, whereas a significant proportion of Bcl-2 (∼90%) (Krajewski et al., 1993; Lithgow et al., 1994) and Bcl-xL (∼50%) (Gonzalez-Garcia et al., 1994; Hsu et al., 1997) are present on the outer nuclear and contiguous endoplasmic membranes. Surprisingly, Bcl-w is loosely associated with membranes in healthy cells; tight membrane association is only triggered in dying cells (J.Wilson-Annan et al., in preparation) upon binding of a BH3-only protein. This suggests that a conformational change might occur upon induction of death. However, no such change was apparent from the structures of C-terminally truncated Bcl-xL in complex with either Bak or Bad BH3 peptides (Sattler et al., 1997; Petros et al., 2000). Instead, it appears that the BH3-binding groove on pro-survival molecules pre-exists and ligand binding does not cause major conformational alteration (Muchmore et al., 1996; Sattler et al., 1997). To resolve the issue of conformational change, and to elucidate the potential role of the C-terminal residues in pro-survival Bcl-2 proteins, we have studied Bcl-w.
We report here the solution structure of Bcl-w that is missing only 10 residues at the C-terminus (Bcl-wΔC10), and show that Bcl-wΔC5, which is fully functional, has an identical structure. Although the overall structure of Bcl-w resembles those reported previously for Bcl-2 and Bcl-xL, the C-terminal region of Bcl-w, like that in pro-apoptotic Bax, surprisingly is bound in the hydrophobic groove formed by the BH1-3 domains. Moreover, binding experiments suggest that the C-terminal residues restrict access to the binding groove in Bcl-w and Bcl-xL. Thus, the hydrophobic groove in these proteins may not be normally accessible, and displacement of the C-terminal residues by the BH3-only proteins is likely to be a critical early step in neutralizing the activity of pro-survival Bcl-2-like proteins.
Results
Solution structure of Bcl-w
Our initial attempts to solve the structure of Bcl-w were hampered by its poor solubility and the propensity of full-length protein to aggregate when expressed in Escherichia coli. To obtain a molecule that was amenable to study by NMR, a series of truncated Bcl-w proteins were generated. The most complete sequence that was highly soluble and could be purified in large quantities was one lacking the last 10 residues. However, acquisition of high quality spectral data from this protein was not possible due to its broad line widths. To resolve this, hydrophobic residues predicted to be solvent accessible (based on homology to Bcl-xL) were mutated. One such mutation, A128E, improved the solution properties without disrupting either the structural integrity of the protein, as demonstrated by the similarity of the 2D-NOESY spectrum, or its ability to bind BH3-only proteins (data not shown). Although deletion of the 10 C-terminal residues, but not the A128E mutation itself, abolished the activity of Bcl-w, longer active proteins were found to have indistinguishable structures (see below). Since the recombinant longer forms of Bcl-w were not readily amenable for structural studies, we focused on characterizing Bcl-wΔC10 (A128E), subsequently referred to as Bcl-wΔC10.
Essentially complete, sequence-specific, backbone and side-chain assignments for Bcl-wΔC10 were determined using a series of heteronuclear 3D NMR experiments (Sattler et al., 1999). Structures were calculated using a total of 3871 constraints (Table I). Figure 1A shows the superposition of the final 20 lowest-energy structures over the backbone atoms (N, Cα, C′) of residues 8–183. The structural statistics for the ensemble are shown in Table I and demonstrate that the NMR structures are both energetically reasonable and have acceptable covalent geometry. The N-terminal 13 residues, including the five cloning artefacts (GPLGS), lack any long-range distance constraints and are disordered in solution. In addition, the amide protons for residues 59, 114 and 115 are in short solvent-accessible loops that exchange rapidly with solvent and are not observable.
Table I. Summary of restraints and structural statistics for the 20 lowest energy structures of Bcl-wΔC10 in aqueous solution, at pH 6.7 and 30°C, following minimization in water using CNS.
| Experimental constraints | ||
|---|---|---|
| Total | 3871 | |
| Intraresidue | 694 | |
| Sequential (|i – j| = 1) | 843 | |
| Short range (1 < |i – j| < 5) | 912 | |
| Long range (|i – j| ≥ 5) | 993 | |
| Hydrogen bonds | 64 | |
| Dihedral angles (φ, 136; ψ, 101; χ1, 34; χ2, 30) | 301 | |
| R.m.s.d. from experimental distance restraints (Å) | 0.0201 ± 0.0005 | |
| R.m.s.d. from experimental dihedral restraints (°) |
0.3765 ± 0.0226 |
|
| R.m.s.d. from idealized covalent geometry |
|
|
| Bonds (Å) | 0.0034 ± 0.0001 | |
| Angles (°) | 0.4867 ± 0.0092 | |
| Impropers (°) |
0.3793 ± 0.0101 |
|
| Measures of structural quality |
|
|
| ELJ (kcal mol–1) | –864.1 ± 10.8 | |
| Procheck % residues in region of Ramachandran plot (residues with S(φ) and S(ψ) ≥ 0.9) | ||
| most favorable | 84.7 (87.7) | |
| additionally allowed | 14.2 (11.6) | |
| generously allowed | 0.8 (0.7) | |
| disallowed | 0.3 (0.1) | |
| Angular order residues | ||
| with S(φ) ≥ 0.9 | 178 | |
| with S(ψ) ≥ 0.9 |
173 |
|
| Coordinate precision |
|
|
| Mean pairwise r.m.s.d. (Å) | Cα, C, N | All heavy atoms |
| Residues 8–183 | 0.52 ± 0.09 | 1.14 ± 0.11 |
| α1–α9 | 0.36 ± 0.07 | 0.97 ± 0.10 |
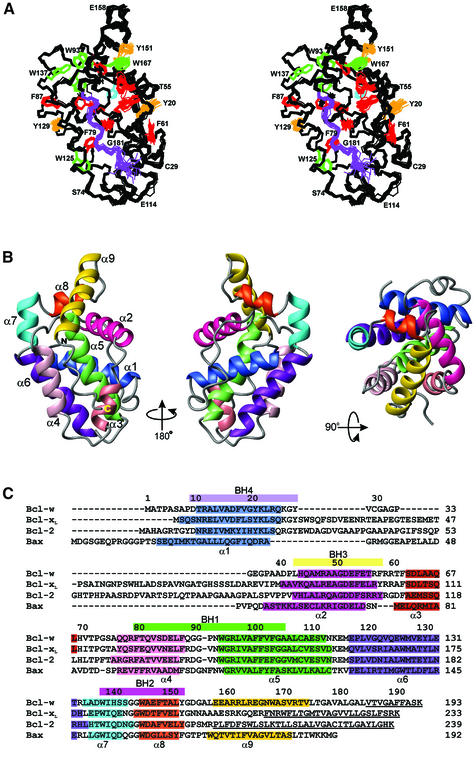
Fig. 1. Sequence and structure of Bcl-w. (A) A stereoview of the backbone (N, Cα, C) superposition of the 20 NMR-derived structures of Bcl-wΔC10 (residues 8–183). Aromatic side chains are shown for residues with <25% solvent accessibility in different colors: Trp (green), Phe (red), His (cyan) and Tyr (orange). The region of extended structure at the C-terminus is shown in purple. (B) Ribbon diagram of the structure closest to the mean (residues 8–183). The helices are indicated in different colors and are labeled. The view on the left has the same orientation as (A) while the middle view has been rotated 180° about the vertical axis and the right view 90° about the horizontal axis. (C) Structure-based sequence alignment of Bcl-w, Bcl-xL, Bcl-2 and Bax. The BH domains are indicated by colored bars above the sequence and the helical residues in all the structures are indicated in colors identical to those used in (B). The C-terminal residues, missing from the structures, are underlined.
Bcl-wΔC10 is an α-helical protein containing a well-defined core formed by a central hydrophobic helix, α5 (residues 93–111), and flanking amphipathic helices α1 (residues 10–24), α2 (residues 43–56), α3 (residues 62–68), α4 (residues 76–87) and α6 (residues 116–132) (Figure 1). The helices are connected by a series of well-defined loops. As predicted, residue 128 occupies a solvent-accessible position in α6 making it unlikely that mutation to glutamic acid has influenced the structure. Helix α7 (residues 134–141) is essentially continuous with α6 except for a sharp bend, indicated by a change in the coupling constants at residue 133, that disrupts the two helices. At the base of α7 lies helix α8 (residues 144–150) which primarily contacts α2. A sharp turn containing two glycine residues connects α8 to helix α9 (residues 157–173). As a consequence of this turn α9 is folded back onto the structure and the C-terminus of α9 makes a series of hydrophobic contacts with residues at the N-terminus of α5 and the α4–α5 loop (Figure 2A). Following α9 is a region (residues 174–183) of extended but ordered structure. This extended region lies in a groove that is principally formed by residues located in α3, α4 and the N-terminus of α5. The position of the extended region is stabilized by a series of hydrophobic interactions between the tail and residues in α3 and α4. Analysis of preliminary heteronuclear relaxation experiments (15N-R1, R2 and 1H-15N NOE; data not shown) on Bcl-wΔC10 shows all helices have similar relaxation properties. This suggests that, within the time scale sensitivity of these experiments, there is little difference in the backbone mobility of α9 compared with the core. Residues 176–183 are more mobile according to the NMR relaxation data.
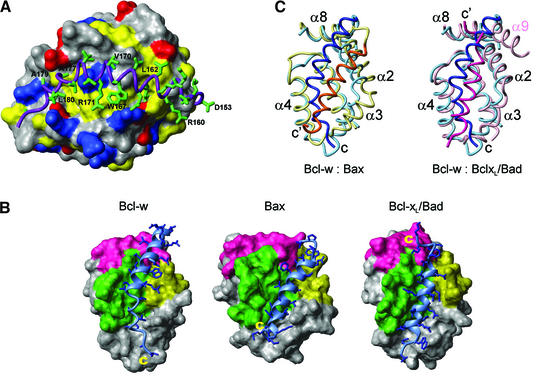
Fig. 2. The hydrophobic binding grooves in Bcl-2 family members. (A) A close-up view of the C-terminal residues of Bcl-w. Residues 8–152 are shown as a surface with the side chains of basic, acidic and hydrophobic residues colored blue, red and yellow, respectively. The C-terminal residues (153–183) are shown as a ribbon (purple) and the side chains of these residues are shown in stick representation (green). (B) Comparison of the hydrophobic binding grooves from Bcl-w, Bax and Bcl-xL. In all three structures residues equivalent to 8–152 in Bcl-w are shown as a surface representation with the BH domains indicated (BH1, green; BH2, pink; BH3, yellow). The residues that lie in the groove (Bcl-w residues 153–181, Bax residues 166–192 and the Bad peptide) are shown as a ribbon (light blue) with the side chains as sticks (blue). The atomic coordinates of Bax (1f16) and the Bcl-xL/Bad (1g5j) peptide complex were obtained from the Protein Data Bank. (C) Comparison of the binding groove in Bcl-w with those in Bax and Bcl-xL. On the left, the ribbon diagram representing Bcl-w (pale blue) is superimposed with Bax (yellow). The C-terminal residues are shown in dark blue (Bcl-w) and orange–yellow (Bax). On the right Bcl-w (pale blue, dark blue) is superimposed with the Bcl-xL (pink):Bad (dark pink) complex. The C-terminus of Bcl-w (C) and the C-termini of Bax and the Bad peptide (C′) are labeled. The structures were superimposed using TOP (Lu, 2000) and the equivalent view is shown for all of them.
The presence of the C-terminal residues in the hydrophobic groove means that Bcl-wΔC10 is a compact globular molecule with no significant hydrophobic surface attributes. The most distinct surface feature of Bcl-w is a region of negative electrostatic potential formed by residues from α1, α1–α2 loop, α5–α6 loop, α6 and α7. A smaller region of positive potential, which is largely formed by basic residues in α9, is seen on the opposite face of the molecule. The presence of a much longer C-terminal tail in Bcl-w, when compared with the published structures of pro-survival Bcl-2 proteins, led us to a detailed comparison with those structures.
Structural comparison with other Bcl-2 family proteins
The overall topology of Bcl-w is very similar to that observed for other Bcl-2 family members. These include the pro-survival proteins Bcl-xL (Muchmore et al., 1996; Aritomi et al., 1997), Bcl-2 (Petros et al., 2001) and the viral Bcl-2 homolog from KSHV (Huang et al., 2002) as well as the pro-apoptotic proteins Bax (Suzuki et al., 2000) and Bid (Chou et al., 1999; McDonnell et al., 1999). The position of the helices for some of the pro-survival proteins and Bax is indicated in Figure 1C. Notably, helices α1–α7 occupy similar positions in all structures and the hydrophobic core is conserved. When compared with the other mammalian pro-survival molecules over the core of the protein (Cα, N and C′ atoms of helices α1–α7 as defined for Bcl-w), the r.m.s.d. lies between 1.39 and 2.02 Å. In addition, the conserved BH domains present comparable surfaces in all pro-survival Bcl-2 proteins and pro-apoptotic Bax (Figure 2B). Despite these common features a number of significant differences exist.
Bcl-w differs from both Bcl-2 and Bcl-xL in that the α1–α2 loop is both shorter and structurally well defined (Figure 1). This 13 residue loop packs against both α1 and the N-terminus of α2 in Bcl-w and is relatively immobile according to the 15N-relaxation data. In contrast, the equivalent loop in Bcl-xL and Bcl-2 is longer (∼58 residues), and in the structures of Bcl-xLΔC24 where the full loop is present, it is disordered as indicated by both the lack of electron density in the X-ray structure (Protein Data Bank code 1maz) and 1H-15N NOE data (Protein Data Bank code 1lxl) (Muchmore et al., 1996). A consequence of the short, well-defined α1–α2 loop in Bcl-w is that it reduces the solvent accessibility of residues 15–20 in α1 that form part of the BH4 domain. Since this domain appears essential for pro-survival activity (Borner et al., 1994; Huang et al., 1998), the α1–α2 loop might control this by modulating access to the BH4 region in Bcl-w.
The most important difference between the structure of Bcl-w and those of Bcl-xL and Bcl-2 is the conformation of the C-terminal residues. The structures of Bcl-xL and Bcl-2 have been determined using proteins that not only contain deletions in the α1–α2 loop but are also missing C-terminal residues; hereafter these molecules are referred to as, Bcl-xLΔC24 (truncated at position 209; Muchmore et al., 1996; Petros et al., 2000) and Bcl-2ΔC32 (truncated at position 207; Petros et al., 2001). In addition, both proteins contained a C-terminal His6 tag and the residues after the last helix, α8 (Figure 1C), are disordered. However, an additional helix (α9), that is displaced from the core of the protein and does not make any contacts with the rest of the structure, is present in Bcl-xL complexed to BH3 peptides (Protein Data Bank codes 1bxl and 1g5j; Sattler et al., 1997; Petros et al., 2000). This suggests that, in Bcl-xL, residues beyond α8 have some helix forming ability but the location of these residues in the groove may have been destabilized by the C-terminal truncation.
Although Bcl-w is a pro-survival Bcl-2 protein, the general location of the C-terminal residues is most similar to that seen for the pro-apoptotic protein Bax (Suzuki et al., 2000; Figure 2B and C). While the C-terminal residues in both proteins occupy the hydrophobic groove formed by residues from α2–α5 a detailed comparison reveals a number of differences. The C-terminal tail of Bax is shorter and forms a single α-helix that lies in the center of the hydrophobic groove (Figure 2C). In contrast, the region beyond α8 in Bcl-wΔC10 is considerably longer and, unlike the continuous helix seen in Bax, only residues 157–173 have a helical conformation. Beyond α9 in Bcl-w is a region of irregular structure (residues 174–183), as indicated by the φ and ψ angles. This region contains a number of hydrophobic residues (V173, L174, A177, V178, A179 and L180; Figure 2A) that contact hydrophobic residues in α3 and α4. In contrast, the N-terminus of α9 in Bcl-w only contacts L150 and Y151 while the C-terminus interacts with the α4–α5 loop. This differs from the situation in Bax, where in addition to hydrophobic contacts, S184 in α9 makes specific contacts in the binding groove. These contacts appear to be required for interaction of α9 with the binding groove as a mutation, S184V, results in a protein that constitutively associates with mitochondria (Nechushtan et al., 1999). The C-terminal helix in Bax, α9, also lies deeper in the groove and makes a number of contacts to residues at the base of the groove (Figure 2B and C). The position of α9 in Bax is accommodated by the displacement of helices α2 and α3 relative to Bcl-w, although α4–α8 occupy similar positions in both structures (Figure 2C). The position of the tail in Bcl-w suggests that, as has been proposed for Bax (Nechushtan et al., 1999; Suzuki et al., 2000), exposure of its C-terminal residues could well regulate binding of Bcl-w to membranes.
Structural comparison of Bcl-wΔC10 and Bcl-xLΔC24 in complex with BH3 peptides from either Bak or Bad (r.m.s.d. of 1.58 Å over Cα, N and C′ atoms of helices α1–α7 between Bcl-wΔC10 and Bcl-xLΔC24; Figure 2B and C) suggests a mechanism for regulating the position of the C-terminal hydrophobic residues. The BH3-domain peptide ligand and the C-terminal residues in Bcl-w have similar locations, although they have opposite orientations with respect to the direction of the protein chain (Figure 2C). However, the helices forming the hydrophobic groove (α2–α5) have very similar positions in Bcl-wΔC10 and the Bcl-xLΔC24 complex structures (Figure 2C). In particular, when the structures are superimposed to give the overall best agreement α2, α4 and α5 overlay closely (r.m.s.d. 1.14 Å over Cα, N and C′ atoms) and many of the corresponding side chains that contact the ligand in Bcl-xLΔC24 have similar rotamer conformations in both structures. Only in α3 are differences in the position of the helices and the associated side chains seen. Thus, binding of BH3-only ligands to Bcl-w probably requires displacement of the C-terminal residues from the groove but only small local movements of interacting residues. Once displaced, the hydrophobic C-terminal tail of Bcl-w would be free to tightly integrate into membranes.
The C-terminal residues of Bcl-w influence BH3-domain binding
To test the idea that the C-terminal residues of Bcl-w might restrict access to the hydrophobic groove, we examined the ability of Bim to interact with Bcl-w. The BH3-only protein Bim is a potent initiator of apoptotic cell death and all isoforms, including BimL and BimEL used here, have identical BH3 domains (O’Connor et al., 1998). The BH3 domain is necessary for binding to and neutralizing pro-survival Bcl-2 proteins, and in Bim mutation of a highly conserved residue in this domain (L94A) reduced binding to Bcl-w (or other pro-survival molecules) and killing activity (Figure 3; data not shown). Due to difficulties associated with producing full-length BimL, C-terminally truncated BimL (BimLΔC27) and a mutant version (BimLΔC27-L94A) were used to examine the binding properties of Bcl-w by surface plasmon resonance measurements on a BIAcore and GST pull-down experiments.
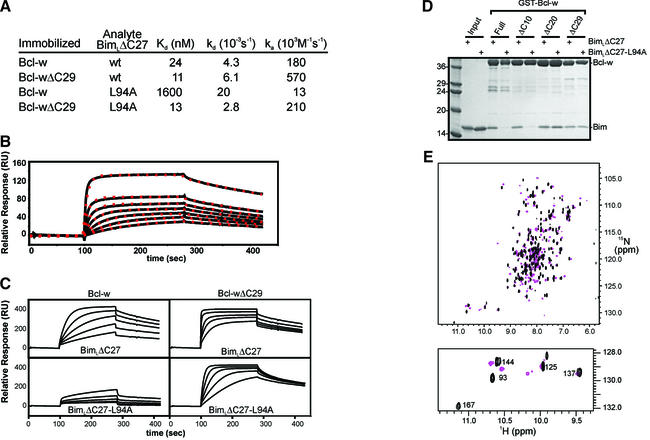
Fig. 3. Binding properties of Bcl-w proteins. (A) The binding constants were determined using biosensor experiments as described in Materials and methods. (B) Binding of Bcl-w to BimLΔC27 fits a 1:1 model. Samples of serially diluted Bcl-w (2 µM–62.5 nM) were analyzed on a BimLΔC27 sensor surface. The experimental data (solid line) and the suggested fit to a 1:1 Langmuir binding model (red dots) are shown. (C) Interaction kinetics for Bcl-w binding to BimLΔC27. Serial dilutions of Bcl-w or Bcl-wΔC29 were analyzed on parallel sensor surfaces that had been derivatized at comparable densities with either BimLΔC27 or BimLΔC27-L94A. Relative responses of samples between 1 µM and 62.5 nM are shown. (D) GST pull-down assay to assess the binding capacity of Bcl-w proteins. Approximately equivalent amounts of the indicated GST–Bcl-w proteins were mixed with either soluble wild-type BimLΔC27 or soluble BimLΔC27-L94A. The intensity of the Bim band indicated the amount of protein that bound to Bcl-w. Molecular weight standards in kDa are indicated. (E) 1H-15N-HSQC spectra of 15N Bcl-w in the presence (pink) and absence (black) of a 26 residue Bim-BH3 domain peptide. Inset shows the indole region.
As expected from other studies (O’Connor et al., 1998; J.Wilson-Annan et al., in preparation), biosensor and pull-down experiments demonstrated tight binding of wild-type Bim to Bcl-w (Figure 3). Global analysis of the BIAcore binding data revealed unambiguously 1:1 Langmuir interactions between surface-immobilized BimL and Bcl-w in solution (Figure 3B). This analysis also suggested a nanomolar affinity for the interaction between full-length Bcl-w and BimLΔC27 (Kd = 24 nM). In contrast, Bim containing the L94A mutation bound with significantly reduced affinity (Kd = 1600 nM) and the >60-fold reduction was accounted for by an ∼14-fold decrease in the association rate and a 5-fold increase in the dissociation rate of the mutant protein (Figure 3A). In agreement with the notion that the C-terminal region of Bcl-w restricts access to the groove, we observed a 3-fold increase in the association rate between BimLΔC27 and Bcl-w truncated by 29 residues at the C-terminus (Bcl-wΔC29), resulting in an improved affinity for this interaction (Kd = 11 nM; Figure 3B and C). In contrast to the results obtained with full-length protein, Bcl-wΔC29 also bound BimLΔC27-L94A with a comparable affinity (Kd = 13 nM). Similar results were obtained when a corresponding mutation (L138A) was introduced into another BH3-only protein, Bmf (data not shown). The significantly increased affinity of the Bim and Bmf BH3 point mutants for the truncated Bcl-w proteins suggests that the conserved leucine in BH3-domains is required for displacement of the C-terminus, yet is largely dispensible for tight binding to the groove of truncated Bcl-w. Although further studies are required to define the mechanistic details, these results support a role for the C-terminal residues regulating the binding of BH3-only proteins to Bcl-w.
To identify the C-terminal region that occluded the binding groove in Bcl-w, the ability of Bcl-w, or C-terminal truncations of it, to bind wild-type BimL or the L94A mutant was assessed by GST pull-down experiments (Figure 3D). Bcl-wΔC10 behaved like full-length Bcl-w and only bound wild-type BimLΔC27, whereas Bcl-wΔC20 and Bcl-wΔC29 bound equally well to BimLΔC27 and BimLΔC27-L94A (Figure 3D). This suggests that residues 173–183, those that distinguish ΔC10 from ΔC20, have an essential role in preventing the interaction of BimLΔC27-L94A with Bcl-w. These data are consistent with the NMR structure, as residues 173–183, although dynamically more mobile, make a number of contacts with residues in the hydrophobic groove (Figure 2A). As α9 only has a limited set of contacts, it is likely that residues 173–183 play a role in stabilising the position of α9.
To demonstrate directly that the C-terminal residues are displaced upon binding of a BH3 domain, we compared the 1H-15N-HSQC spectra of Bcl-wΔC10 in the presence and absence of a 26 residue peptide encompassing the Bim-BH3 domain (Figure 3E). As expected, resonances of residues in α3 and α4, and resonances of residues C-terminal to G152 have moved, suggesting a new chemical environment upon binding the peptide. This is most clearly observed for the indole Hε1Nε1 resonances. The indole HN of W167 has undergone a significant shift while the other Trp indole resonances are essentially unchanged (Figure 3E, inset). These data are consistent with a conformational change in the C-terminal residues upon binding Bim-BH3 peptides as would be expected if the C-terminus was displaced by the ligand. The details of the interaction between Bcl-w and Bim, and the nature of the conformational change are the focus of ongoing studies.
Access to the surface groove of Bcl-w and Bcl-xL is normally restricted in vivo
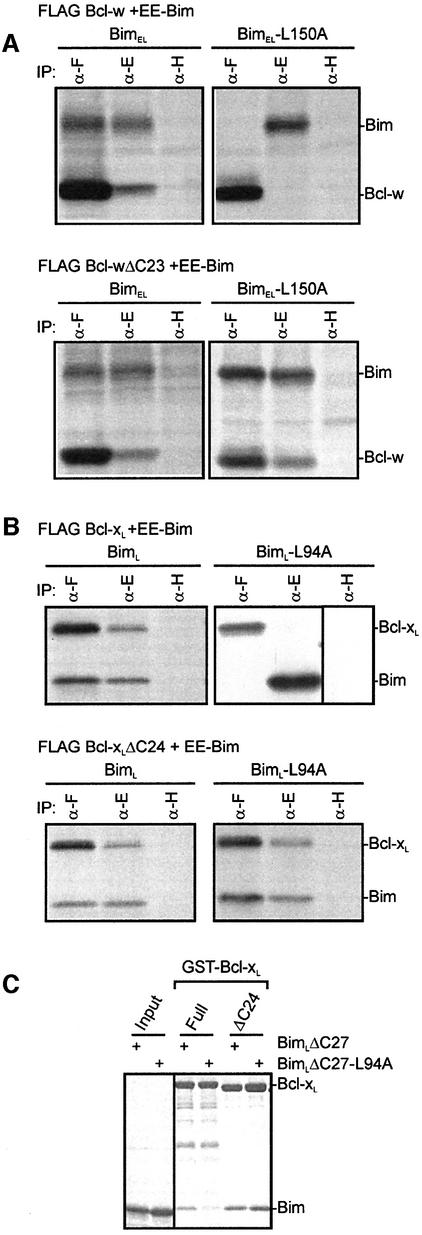
Our structural and binding studies suggest that access to the hydrophobic groove on Bcl-w may normally be restricted by its C-terminal residues. To determine whether Bcl-w adopts a similar conformation in vivo we tested the ability of FLAG-tagged full-length or C-terminally truncated Bcl-w, to bind to EE-tagged Bim when overexpressed in 293T cells. Depending on which could be more readily distinguished by its size, either full-length BimL or BimEL was used in these experiments. Interactions between wild-type or mutant Bim (L94A in BimL or L150A BimEL) and Bcl-w were assessed by the ability of these proteins to be co-immunoprecipitated from 293T cell lysates (Figure 4A). In agreement with our findings with purified recombinant proteins (Figure 3), Bcl-wΔC23 bound both wild-type and the L150A mutant BimEL equally, while full-length Bcl-w only bound wild-type BimEL (Figure 4A).
Fig. 4. In vivo and in vitro binding properties of Bcl-xL resemble those of Bcl-w. (A) The C-terminal residues of Bcl-w restrict access to the binding groove in vivo. Equivalent 35S-labeled 293T lysates obtained from cells co-expressing FLAG Bcl-w or Bcl-wΔC23, and EE-BimEL or BimEL-L150A, were immunoprecipitated using the anti-FLAG M2 (α-F), anti-EE (α-E) or control anti-HA (α-H) monoclonal antibodies. The immunoprecipitates were fractionated on SDS–PAGE gels. (B) The C-terminal residues of Bcl-xL restrict access to the binding groove in vivo. Co-precipitation experiments similar to those described in (A) using lysates from cells co-expressing FLAG Bcl-xL or Bcl-xLΔC24, and EE-BimL or BimL-L94A. (C) GST pull-down experiment as for Figure 3D, except in this case GST–Bcl-xL proteins were mixed with either soluble wt BimLΔC27 or soluble BimLΔC27-L94A.
Bcl-xL and Bcl-2 also contain hydrophobic residues at their C-termini, similar to those found in Bcl-w (Figure 1C), yet the low level of sequence identity following α8 and the absence of 3D structural information, makes prediction of their conformation difficult. Since Bcl-xL, like Bcl-w, is only partially membrane bound in healthy cells (Hsu et al., 1997), we compared the ability of FLAG-tagged full-length Bcl-xL or a C-terminally truncated mutant (ΔC24) to bind to BimL or the L94A BH3 mutant. Like Bcl-w, full-length Bcl-xL associates only with wild-type BimL in cell extracts (Figure 4B) and more tightly with wild-type BimL in GST pull-down experiments (Figure 4C). However, Bcl-xLΔC24 behaved like Bcl-wΔC29 since it bound wild-type BimL and the L94A mutant equally. Given that Bcl-xL also becomes tightly associated with membranes in response to apoptotic stimuli, presumably due to binding of BH3-only proteins as suggested for Bcl-w (J.Wilson-Annan et al., in preparation), our results suggest similar roles for the C-terminal residues in both proteins.
The C-terminal residues in Bcl-w are required for its pro-survival activity
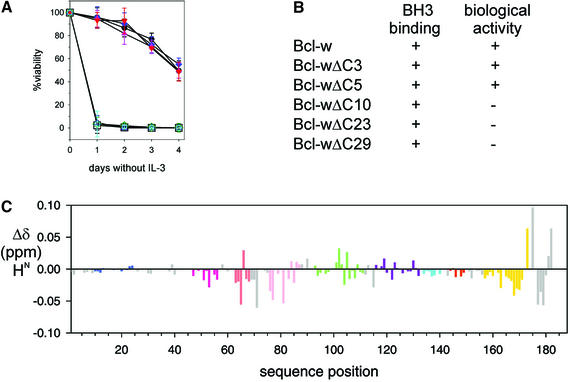
Like its cousins Bcl-2 and Bcl-xL, Bcl-w overexpression protects cells from diverse death stimuli, including cytokine deprivation and γ-irradiation (Gibson et al., 1996). Since C-terminal truncation of Bcl-w did not affect binding to BH3-only proteins such as Bim, we explored whether BH3-binding alone is sufficient for the pro-survival activity of Bcl-w by comparing the functionality of full-length Bcl-w with C-terminal truncated variants when overexpressed in FDC-P1 myeloid cells. The survival of cells expressing comparable levels of FLAG-tagged proteins in response to IL-3 withdrawal, γ-irradiation or cytotoxic drugs was monitored (Figure 5A and B). Surprisingly, only the smallest deletions (ΔC5 and ΔC3) were fully active. Expression of the other deletion mutants failed to afford the cells any protection, even though they were indistinguishable from full-length Bcl-w in their ability to bind to the BH3-only proteins Bim, Bad, Bik/Nbk or Bmf (Figures 3 and 4; data not shown).
Fig. 5. Bcl-wΔC10 is functionally inert but is structurally similar to biologically active Bcl-wΔC5. (A) Bcl-w cannot tolerate extensive C-terminal deletions. The viability of parental FDC-P1 cells (black squares) or representative clones expressing different Bcl-w constructs [Bcl-w (black circles); Bcl-w(A128E) (blue); Bcl-wΔC3 (pink); Bcl-wΔC5 (red); Bcl-wΔC10 (green); Bcl-wΔC23 (blue); Bcl-wΔC29 (cyan)] deprived of IL-3 were determined by PI-staining analyzed flow cytometrically. Data shown are means ± 1 SD of at least three experiments. (B) Summary of the binding properties and biological activity of full-length or C-terminal truncated mutants of Bcl-w. (C) Comparison of the 2D 1H-15N-HSQC spectra for Bcl-wΔC10 and Bcl-wΔC5. Backbone amide proton chemical shift differences plotted for residues in 15N-labeled Bcl-wΔC10 relative to those for Bcl-wΔC5 are indicated. Colors for the helices correspond to those used in Figure 1.
As Bcl-wΔC10 appeared biologically inert, while Bcl-wΔC5 behaved like the full-length protein, we next compared the spectra of these two molecules to determine whether there was a structural basis for the marked functional difference. The 1H-15N-HSQC spectra for Bcl-wΔC5 (A128E), the longest protein that we could purify in sufficient quantities for NMR analysis, was compared with that of Bcl-wΔC10 (Figure 5C). Only small differences in the position of resonances were seen, consistent with addition of five residues to the C-terminus of Bcl-wΔC10. In addition, analysis of a 15N-edited NOESY spectrum obtained for Bcl-wΔC5 indicated that the additional five residues were disordered and all other residues that were ordered in Bcl-wΔC10 had a similar pattern of NOEs. Thus, despite its impaired biological activity, Bcl-wΔC10 is structurally equivalent to the functional Bcl-wΔC5 molecule (Figure 5A and C). Together, our findings demonstrate that the solution structure of Bcl-w reported here is that of a biologically relevant molecule and represents the most complete model of a pro-survival Bcl-2 protein.
Since the structure and binding properties of functional Bcl-wΔC5 and non-functional Bcl-wΔC10 appear indistinguishable, we are currently investigating other possible differences between these proteins to explain their contrasting activities. One possible explanation is their localization. Full-length Bcl-w is located exclusively on the mitochondria, and as the C-terminal residues are important for localization of other Bcl-2 proteins, it is possible that in Bcl-w the most C-terminal residues have a critical role in mediating membrane association.
Discussion
Our recent work suggests that binding of pro-apoptotic BH3-only proteins, which initiate cell death, mediates a change in Bcl-w enabling it to interact more tightly with membranes (J.Wilson-Annan et al., in preparation). Based on the solution structure of Bcl-w we present a possible mechanism for this C-terminally mediated process. The most remarkable feature of the Bcl-w structure is the conformation and location of the C-terminal residues. The C-terminus of Bcl-w, composed of a helical region (α9) followed by a region of ordered extended structure, lies in the hydrophobic groove formed by helices α2–α4, thereby allowing tail residues to form a series of close contacts with residues from the protein core (Figures 1 and 2). The position of the C-terminal residues is similar to that occupied by BH3 peptides when bound to Bcl-xL (Figure 2; Sattler et al., 1997; Petros et al., 2000). This implies that binding of pro-apoptotic BH3-only proteins to Bcl-w requires displacement of the C-terminal residues.
Structures of ligand-free and bound forms of Bcl-xL indicated that only minor conformational changes were associated with ligand binding and showed the pre-existing nature of the binding groove (Muchmore et al., 1996; Sattler et al., 1997; Petros et al., 2000). However, this simple description of binding is complicated by the absence of the C-terminal residues from the molecules studied. The presence of residues shielding the binding groove in Bcl-w would be expected to influence ligand binding, and consistent with this we observed a reduction in both the Kd and association rates in the presence of these residues (Figures 3 and 4). Accordingly, our data, including titration experiments of Bcl-w with Bim (Figure 3E), support a model in which the C-terminal residues gate access to the binding site and are displaced to allow binding. Such a model is not evident from the available structures. Furthermore, such a gating mechanism is also likely to occur with Bcl-xL, where we have observed a similar reduction in binding in the presence of C-terminal residues (Figure 4).
How is Bcl-w targeted to the outer mitochondrial membrane? In the case of Bcl-2 the C-terminus has been proposed to function as a membrane anchor (Chen-Levy et al., 1989) and recent studies suggest it has a role in membrane targeting (Motz et al., 2002). Notably, peptides based on the C-terminal sequence form transmembrane (TM) helices (del Mar Martinez-Senac et al., 2000), and we have observed that the C-termini of Bcl-w, Bcl-xL and Bcl-2 contain motifs (Senes et al., 2000; Kleiger et al., 2002) commonly found in TM helices. In contrast to Bcl-2, which appears to always be associated with membranes (Chen-Levy et al., 1989), Bcl-xL and Bcl-w are loosely membrane associated in healthy cells and tighter association is triggered by damage signals, suggesting that there is more than one mode of membrane interaction (Hsu et al., 1997; Hausmann et al, 2000; J.Wilson-Annan et al., in preparation). In its loosely attached mode Bcl-w may interact directly with the membrane or with a mitochondrial membrane associated protein. Although further studies are required, the latter seems more likely since the largely negatively charged surfaces present on the pro-survival Bcl-2 proteins, including Bcl-w, are unlikely to favor direct membrane binding. Upon induction of death, we envisage that binding of a BH3 domain would release the hydrophobic C-terminal residues from their extended conformation in the solvent-protected environment of the groove allowing tight membrane association, possibly as a TM helix.
While the precise functional role of the C-terminal hydrophobic residues in pro-survival Bcl-2 proteins remains unclear, our results show that the most C-terminal residues are required for Bcl-w’s activity (Figure 5A). Importantly, our data support a model whereby these proteins do not act solely to sequester the BH3-only proteins, since the ability to bind BH3-only proteins does not appear sufficient for biological activity (Figure 5). As mutants of pro-survival Bcl-2 proteins that cannot bind BH3-only proteins are also inactive (Cory and Adams, 2002), we conclude that both the ability to bind BH3 domains and the presence of the C-terminal residues are necessary, but not individually sufficient, for biological activity. Taken together, these results suggest that Bcl-w may control the availability of a factor required for downstream events, such as Bax/Bak activation (Cheng et al., 2001; Zong et al., 2001). Given the evolutionary conservation of the cell death pathways, it seems plausible that mammalian pro-survival molecules function like CED-9, the Bcl-2 homolog in the nematode Caeno rhabditis elegans (Cory and Adams, 2002). CED-9 sequesters the adapter protein CED-4, activator of the caspase CED-3, until the BH3-only protein EGL-1 displaces it. The identity of such a mammalian CED-4-like target remains elusive since recent work suggests that its mammalian counterpart, Apaf-1, is neither the direct nor the sole target for Bcl-2’s action (Moriishi et al., 1999; Hausmann et al., 2000; Marsden et al., 2002).
A structural alignment of pro-survival Bcl-w and pro-apoptotic Bax (Suzuki et al., 2000) shows they closely resemble each other, but the location and detailed interactions of their C-terminal residues differ. BH3 binding readily displaces the tail of Bcl-w to trigger tight membrane association and its inactivation, but it is unclear if Bax is activated by a similar mechanism (Cheng et al., 2001). Intriguingly, Bax translocation and consequent oligomerization, steps critical in its activation, appears to be linked to its C-terminal residues (Nechushtan et al., 1999; Suzuki et al., 2000). Thus, understanding at atomic level how the C-terminal tails of pro-survival Bcl-w and pro-apoptotic Bax are regulated may be important for understanding their opposing biological activities.
The data we have presented support a model for coordinate regulation of Bcl-w whereby displacement of the C-terminal residues from the hydrophobic binding groove, by high-affinity interaction of a BH3-domain, brings about tight membrane association and possibly the release of a cell death effector. Coordinate regulation of ligand binding and membrane attachment may represent a general regulatory mechanism as both Bcl-w and Bcl-xL have comparable binding and membrane association properties. Since numerous proteins contain α-helices with a hydrophobic face, all that is required for binding to the surface of pro-survival molecules (Sattler et al., 1997; Petros et al., 2000), restricting access to the hydrophobic binding groove may prevent inappropriate cell deaths. In this context, the C-terminal tail in Bcl-w may act as a ligand selection filter.
Materials and methods
Production of Bcl-w and Bim proteins
Human Bcl-w, mouse BimL and human Bcl-xL were expressed as GST fusion proteins in E.coli BL21(DE3) and purified by affinity chromatography. All purified proteins have five additional N-terminal residues (GPLGS) as a result of cloning. Isotopically labeled proteins were prepared as described previously (Day et al., 1999). Samples of Bcl-wΔC10 used for NMR contained ∼1.0 mM protein in 50 mM sodium phosphate (pH 6.7), 70 mM NaCl, 2 mM Tris–(2-carboxyethyl) phosphine and 0.04% sodium azide in H2O:2H2O (95:5). The Bcl-wΔC5 sample contained 0.4 mM protein in the same buffer. Site specific mutants of BimL and Bcl-w were generated using a PCR-based strategy as described previously (Day et al., 1999) and confirmed by sequencing.
NMR spectroscopy and spectral assignments
Spectra were recorded at 30°C on a Bruker DRX-600 spectrometer equipped with triple resonance probes and pulsed field gradients. A series of heteronuclear 3D NMR experiments were recorded using either 15N or 13C, 15N double-labeled protein (Sattler et al., 1999). Experiments recorded on 15N-labeled Bcl-wΔC10 included a 15N-edited NOESY at mixing times of 50 and 150 ms, HSQC, HNHA and 15N–edited TOCSY. A series of 1H-15N-HSQC spectra were recorded as Bim BH3 peptide (sequence: DLRPEIRIAQELRRIGDEFNETYTRR) was titrated into a solution of 15N-Bcl-w to follow resonance changes on binding ligand. 15N-R1,-R2 and 1H-15N NOE spectra were recorded on 15N-Bcl-w. Triple resonance experiments recorded on a 13C, 15N-labeled Bcl-wΔC10 sample included a HNCA, CBCA(CO)NH and 13C-edited NOESY at mixing times of 50 and 150 ms, a 3D HCCH-TOCSY was also recorded on this sample. A 150 ms mixing time 2D NOESY was acquired on unlabeled Bcl-wΔC10. A 1H-15N-HSQC and 150 ms 15N-edited NOESY were recorded on 15N-Bcl-wΔC5. Spectra were processed using XWINNMR (Bruker AG) and analyzed using XEASY (Bartels et al., 1995).
Distance and dihedral angle restraints
Distance restraints were measured from the 3D 15N-edited NOESY and 3D 13C-edited NOESY as well as the 2D NOESY spectra. Hydrogen bond constraints were applied at a late stage of the structure calculation where there existed the characteristic low 3JHNHα coupling constant and NOE patterns observed for α-helices as described previously (Day et al., 1999). No hydrogen bond constraints were employed outside α-helices.
Dihedral angle restraints for φ, ψ, χ1 and χ2 angles were used as summarized in Table I. 3JHNHα were derived from a 3D HNHA spectrum (Vuister and Bax, 1993). Additional φ and ψ backbone torsion angles plus uncertainties for these values were derived from 13Cα chemical shifts according to procedures we have reported previously (Day et al., 1999). Stereospecific assignments, χ1 and χ2 restraints were derived using HABAS and GLOMSA routines in DYANA (Güntert et al., 1997).
Structure calculation and analysis
Initial structures were calculated using DYANA 1.5 (Güntert et al., 1997) and refined with CNS 1.1 (Brünger et al., 1998) before minimisation in a box of water with the OPLSX force field (Linge and Nilges, 1999). Structural statistics for the final set of 20 structures, chosen on the basis of their stereochemical energies, are presented in Table I. PROCHECK_NMR (Laskowski et al., 1996) and MOLMOL (Koradi et al., 1996) were used for the analysis of structure quality. The final structures had no experimental distance violations >0.25 Å or dihedral angle violations >5°. Structural figures were generated in MOLMOL. Coordinates for the final set of 20 structures have been deposited in the Protein Data Bank under accession number 1o0l.
Binding measurements
Direct interactions between Bcl-w and BimL were monitored using GST pull-down experiments as described previously in Day et al. (1999).
Analysis of protein interactions by surface plasmon resonance were performed on a BIAcore 2000 biosensor. For immobilization to CM 5 sensorchips (BIAcore), wild-type or mutant Bim proteins were buffer exchanged into 20 mM Na-acetate pH 4.5. N-hydroxysuccinimide, coupling and binding analysis were as described previously (Lackmann et al., 1997). Kinetic experiments were performed at 20 µl/min to minimize mass-transport mediated effects. Bcl-w binding at concentrations between 2 and 0.03 µM in running buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 3.4 mM EDTA, 0.005% Tween 20) was performed on sensorchip surfaces derivatized on parallel channels with a non-relevant protein, BimLΔC27 and BimLΔC27-L94A. The binding kinetics were derived from the sensorgrams, following subtraction of baseline responses, by ‘global analysis’ using the BIA Evaluation software (v3.02; BIAcore). The surface of the chip was regenerated with 50 mM 1,2-diethylamine containing 0.1% Triton X-100, followed by two washes with running buffer.
Tissue culture, transfection and immunoprecipitation
Cell culture, stable transfections into FDC-P1, transient transfections into 293T human embryonic kidney cells, metabolic labeling with [35S]methionine/cysteine (NEN) and co-immunoprecipitation have been described previously (Huang et al., 1998; O’Connor et al., 1998; J.Wilson-Annan et al., in preparation). Mammalian pEF-based expression vectors for Bcl-w, Bcl-xL, Bim, Bmf, Bad and Bik were transiently transfected using liposome-mediated transfection (LipofectAMINE™, Invitrogen). After transfection (48 h), equivalent trichloroacetic acid precipitable 35S counts were immunoprecipitated using the anti-FLAG M2 (Sigma), anti-EE (CRP) or anti-HA.11 (CRP) mouse monoclonal antibodies. The immunoprecipitates were resolved by SDS–PAGE (Novex), transferred onto nitrocellulose membranes (APB) and the proteins detected by fluorography (Amplify, APB).
Survival assays
Survival assays were performed as described previously (Huang et al., 1998; O’Connor et al., 1998; J.Wilson-Annan et al., in preparation). Briefly, cells (2–5 × 104 per time point) were left untreated, deprived of their essential growth factor IL-3, exposed to 10 Gy γ-irradiation (provided by a 60Co source) or 1–100 nM staurosprorine (Sigma). Cell viability was quantified by flow cytometric analysis of cells excluding 5 µg/mL PI (Sigma) using a FACScan (Becton Dickinson). Each time point was performed in triplicate on at least three independent clones of each genotype and the experiments repeated at least three times.
Acknowledgments
Acknowledgements
We thank J.Beaumont, L.Parma and H.Yang for excellent technical assistance; and J.Adams, P.Colman, S.Cory, T.Lithgow, H.Puthalakath, A.Strasser and D.Vaux for gifts of reagents and numerous discussions. Fellowships and grants from the Marsden Fund (NZ), NHMRC (Canberra), the US NCI (CA80188), the Leukemia and Lymphoma Society, and the Sylvia and Charles Viertel Charitable Foundation supported this work.
References
- Aritomi M., Kunishima,N., Inohara,N., Ishibashi,Y., Ohta,S. and Morikawa,K. (1997) Crystal structure of Rat Bcl-xL. J. Biol. Chem., 272, 27886–27892. [DOI] [PubMed] [Google Scholar]
- Bartels C., Xia,T.H., Billeter,M., Güntert,P. and Wüthrich,K. (1995) The program XEASY for computer-supported nmr spectral-analysis of biological macromolecules. J. Biomol. NMR, 6, 1–10. [DOI] [PubMed] [Google Scholar]
- Borner C., Martinou,I., Mattmann,C., Irmler,M., Schaerer,E., Martinou,J.C. and Tschopp,J. (1994) The protein Bcl-2α does not require membrane attachment, but two conserved domains to suppress apoptosis. J. Cell Biol., 126, 1059–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D., 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Chen-Levy Z., Nourse,J. and Cleary,M.L. (1989) The bcl-2 candidate proto-oncogene product is a 24-kilodalton integral-membrane protein highly expressed in lymphoid cell lines and lymphomas carrying the t(14;18) translocation. Mol. Cell. Biol., 9, 701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng E.H., Wei,M.C., Weiler,S., Flavell,R.A., Mak,T.W., Lindsten,T. and Korsmeyer,S.J. (2001) Bcl-2, Bcl-XL sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol. Cell, 8, 705–711. [DOI] [PubMed] [Google Scholar]
- Chou J.J., Li,H., Salvesen,G.S., Yuan,J. and Wagner,G. (1999) Solution structure of BID, an intracellular amplifier of apoptotic signaling. Cell, 96, 615–624. [DOI] [PubMed] [Google Scholar]
- Cory S. and Adams,J.M. (2002) The Bcl-2 family: regulators of the cellular life-or-death switch. Nat. Rev. Cancer, 2, 647–677. [DOI] [PubMed] [Google Scholar]
- Day C.L., Dupont,C., Lackmann,M., Vaux,D.L. and Hinds,M.G. (1999) Solution structure and mutagenesis of the caspase recruitment domain (CARD) from Apaf-1. Cell Death Differ., 6, 1125–1132. [DOI] [PubMed] [Google Scholar]
- del Mar Martinez-Senac M., Corbalan-Garcia,S. and Gomez-Fernandez,J.C. (2000) Study of the secondary structure of the C-terminal domain of the antiapoptotic protein Bcl-2 and its interaction with model membranes. Biochemistry, 39, 7744–7752. [DOI] [PubMed] [Google Scholar]
- Gibson L. et al. (1996) Bcl-w, a novel member of the Bcl-2 family, promotes cell survival. Oncogene, 13, 665–675. [PubMed] [Google Scholar]
- Gonzalez-Garcia M., Perez-Ballestero,R., Ding,L., Duan,L., Boise,L.H., Thompson,C.B. and Núñez,G. (1994) bcl-xL is the major bcl-x mRNA form expressed during murine development and its product localizes to mitochondria. Development, 120, 3033–3042. [DOI] [PubMed] [Google Scholar]
- Güntert P., Mumenthaler,C. and Wüthrich,K. (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol., 273, 283–298. [DOI] [PubMed] [Google Scholar]
- Hausmann G., O’Reilly,L.A., van Driel,R., Beaumont,J.G., Strasser,A., Adams,J.M. and Huang,D.C.S. (2000) Pro-apoptotic apoptosis protease-activating factor 1 (Apaf-1) has a cytoplasmic localization distinct from Bcl-2 or Bcl-xL. J. Cell Biol., 149, 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y.T., Wolter,K.G. and Youle,R.J. (1997) Cytosol-to-membrane redistribution of Bax and Bcl-XL during apoptosis. Proc. Natl Acad. Sci. USA, 94, 3668–3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.C.S. and Strasser,A. (2000) BH3-only proteins-essential initiators of apoptotic cell death. Cell, 103, 839–842. [DOI] [PubMed] [Google Scholar]
- Huang D.C.S., Adams,J.M. and Cory,S. (1998) The conserved N-terminal BH4 domain of Bcl-2 homologues is essential for inhibition of apoptosis and interaction with CED-4. EMBO J., 17, 1029–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Petros,A.M., Virgin,H.W., Fesik,S.W. and Olejniczak,E.T. (2002) Solution structure of a Bcl-2 homolog from Kaposi sarcoma virus. Proc. Natl Acad. Sci. USA, 99, 3428–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiger G., Grothe,R., Mallick,P. and Eisenberg,D. (2002) GXXXG and AXXXA: common α-helical interaction motifs in proteins, particularly in extremophiles. Biochemistry, 41, 5990–5997. [DOI] [PubMed] [Google Scholar]
- Koradi R., Billeter,M. and Wüthrich,K. (1996) MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph., 14, 51–55. [DOI] [PubMed] [Google Scholar]
- Krajewski S., Tanaka,S., Takayama,S., Schibler,M.J., Fenton,W. and Reed,J.C. (1993) Investigation of the subcellular distribution of the Bcl-2 oncoprotein: residence in the nuclear envelope, endoplasmic reticulum, and outer mitochondrial membranes. Cancer Res., 53, 4701–4714. [PubMed] [Google Scholar]
- Lackmann M. et al. (1997) Ligand for EPH-related kinase (LERK) 7 is the preferred high affinity ligand for the HEK receptor. J. Biol. Chem., 272, 16521–16530. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Rullmann,J.A.C., MacArthur,M.W., Kaptein,R. and Thornton,J.M. (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR, 8, 477–486. [DOI] [PubMed] [Google Scholar]
- Linge J.P. and Nilges,M. (1999) Influence of non-bonded parameters on the quality of NMR structures: a new force field for NMR structure calculation. J. Biomol. NMR, 13, 51–59. [DOI] [PubMed] [Google Scholar]
- Lithgow T., van Driel,R., Bertram,J.F. and Strasser,A. (1994) The protein product of the oncogene Bcl-2 is a component of the nuclear envelope, the endoplasmic reticulum, and the outer mitochondrial membrane. Cell Growth Differ., 5, 411–417. [PubMed] [Google Scholar]
- Lu G.G. (2000) TOP: a new method for protein structure comparisons and similarity searches. J. Appl. Crystallogr., 33, 176–183. [Google Scholar]
- Marsden V.S. et al. (2002) Apoptosis initiated by Bcl-2 regulated caspase activation independently of the cytochrome c/Apaf-1/caspase-9 apoptosome. Nature, 419, 634–637. [DOI] [PubMed] [Google Scholar]
- McDonnell J.M., Fushman,D., Milliman,C.L., Korsmeyer,S.J. and Cowburn,D. (1999) Solution structure of the proapoptotic molecule BID: a structural basis for apoptotic agonists and antagonists. Cell, 96, 625–634. [DOI] [PubMed] [Google Scholar]
- Moriishi K., Huang,D.C.S., Cory,S. and Adams,J.M. (1999) Bcl-2 family members do not inhibit apoptosis by binding the caspase activator Apaf-1. Proc. Natl Acad. Sci. USA, 96, 9683–9688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motz C., Martin,H., Krimmer,T. and Rassow,J. (2002) Bcl-2 and porin follow different pathways of tom-dependent insertion into the mitochondrial outer membrane. J. Mol. Biol., 323, 729–738. [DOI] [PubMed] [Google Scholar]
- Muchmore S.W. et al. (1996) X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature, 381, 335–341. [DOI] [PubMed] [Google Scholar]
- Nechushtan A., Smith,C.L., Hsu,Y.T. and Youle,R.J. (1999) Conform ation of the Bax C-terminus regulates subcellular location and cell death. EMBO J., 18, 2330–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechushtan A., Smith,C.L., Lamensdorf,I., Yoon,S.H. and Youle,R.J. (2001) Bax and Bak coalesce into novel mitochondria-associated clusters during apoptosis. J. Cell Biol., 153, 1265–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor L., Strasser,A., O’Reilly,L.A., Hausmann,G., Adams,J.M., Cory,S. and Huang,D.C.S. (1998) Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J., 17, 384–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly L.A., Print,C., Hausmann,G., Moriishi,K., Cory,S., Huang,D.C.S. and Strasser,A. (2001) Tissue expression and subcellular localization of the pro-survival molecule Bcl-w. Cell Death Differ., 8, 486–494. [DOI] [PubMed] [Google Scholar]
- Petros A.M. et al. (2000) Rationale for Bcl-xL/Bad peptide complex formation from structure, mutagenesis, and biophysical studies. Protein Sci., 9, 2528–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petros A.M., Medek,A., Nettesheim,D.G., Kim,D.H., Yoon,H.S., Swift,K., Matayoshi,E.D., Oltersdorf,T. and Fesik,S.W. (2001) Solution structure of the antiapoptotic protein Bcl-2. Proc. Natl Acad. Sci. USA, 98, 3012–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Print C.G. et al. (1998) Apoptosis regulator Bcl-w is essential for spermatogenesis but appears otherwise redundant. Proc. Natl Acad. Sci. USA, 95, 12424–12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler M. et al. (1997) Structure of Bcl-xL–Bak peptide complex: recognition between regulators of apoptosis. Science, 275, 983–986. [DOI] [PubMed] [Google Scholar]
- Sattler M., Schleucher,J. and Griesinger,C. (1999) Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Magn. Reson. Spectrosc., 34, 93–158. [Google Scholar]
- Senes A., Gerstein,M. and Engelman,D.M. (2000) Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with β-branched residues at neighboring positions. J. Mol. Biol., 296, 921–936. [DOI] [PubMed] [Google Scholar]
- Suzuki M., Youle,R.J. and Tjandra,N. (2000) Structure of Bax: coregulation of dimer formation and intracellular localization. Cell, 103, 645–654. [DOI] [PubMed] [Google Scholar]
- Vuister G.W. and Bax,A. (1993) Quantitative J correlation: a new approach for measuring homonuclear three-bond J(HNHα) coupling constants in 15N-enriched proteins. J. Am. Chem. Soc., 115, 7772–7777. [Google Scholar]
- Zong W.X., Lindsten,T., Ross,A.J., MacGregor,G.R. and Thompson,C.B. (2001) BH3-only proteins that bind pro-survival Bcl-2 family members fail to induce apoptosis in the absence of Bax and Bak. Genes Dev., 15, 1481–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]