Abstract
Integration of biochemical and biophysical data on the lactose permease of Escherichia coli has culminated in a molecular model that predicts substrate–protein proximities which include interaction of a hydroxyl group in the galactopyranosyl ring with Glu269. In order to test this hypothesis, we studied covalent modification of carboxyl groups with carbodiimides using electrospray ionization mass spectrometry (ESI-MS) and demonstrate that substrate protects the permease against carbodiimide reactivity. Further more, a significant proportion of the decrease in carbodiimide reactivity occurs specifically in a nanopeptide containing Glu269. In contrast, carbodiimide reactivity of mutant Glu269→Asp that exhibits lower affinity is unaffected by substrate. By monitoring the ability of different substrate analogs to protect against carbodiimide modification of Glu269, it is suggested that the C-3 OH group of the galactopyranosyl ring may play an important role in specificity, possibly by H-bonding with Glu269. The approach demonstrates that mass spectrometry can provide a powerful means of analyzing ligand interactions with integral membrane proteins.
Keywords: bioenergetics/H+ symport/lactose permease/ligand binding/membrane proteins
Introduction
The lactose permease (LacY) of Escherichia coli is typical of membrane transport proteins from Archaea to the mammalian central nervous system that transduce free energy stored in electrochemical ion gradients into solute concentration gradients and vice versa (Konings et al., 1996). LacY is a 12 transmembrane-helix bundle with the N- and C-termini on the cytoplasmic face of the membrane (Foster et al., 1983; Calamia and Manoil, 1990; Kaback and Wu, 1997), and the 417-residue polypeptide has been solubilized, purified, reconstituted into proteoliposomes and shown to be solely responsible for the galactoside-H+ symport (reviewed in Viitanen et al., 1984). Several lines of evidence indicate that LacY is both functionally (Sahin-Tóth et al., 1994) and structurally (Costello et al., 1987; Sun and Kaback, 1997; Guan et al., 2001) a monomer. Analysis of an extensive library of mutants, particularly Cys replacement mutants (Frillingos et al., 1998), with a battery of site-directed biophysical and biochemical techniques has led to the formulation of a tertiary-structure model (Sorgen et al., 2002), as well as a proposed mechanism for lactose-H+ symport (reviewed in Kaback et al., 2001).
LacY is selective for d-galactopyranosides including d-galactose (Sandermann, 1977; Olsen and Brooker, 1989; Sahin-Tóth et al., 2000a, 2001), but has no affinity for d-glucopyranosides (Wu and Kaback, 1994; Sahin-Tóth et al., 2000a). Specificity is directed towards the galactopyranosyl ring of LacY substrates, and although the C-4 OH is clearly the major determinant for ligand binding, the C-2, C-3 and C-6 OH groups (C-3 OH ≥ C-6 OH > C-2OH) also participate (Sahin-Tóth et al., 2000a, 2001).
Cys148 (helix V) is not irreplaceable with respect to activity, but interacts directly with substrate (Bieseler et al., 1985; Jung et al., 1994; Wu and Kaback, 1994; Frillingos and Kaback, 1996; le Coutre et al., 2000). Alkylation with N-ethylmaleimide (NEM) abolishes transport and ligand binding, and d-galactopyranosides afford protection against alkylation (Kaback et al., 2001). Furthermore, replacement of Cys148 with small hydrophobic residues such as Ala or Val decreases the Km for lactose, while hydrophilic replacements such as Ser or Thr lead to an increase in Km. In addition, hydrophilic replacements decrease transport of d-galactose relative to disaccharide substrates (Jung et al., 1994). These and other observations (Guan et al., 2002a) indicate that Cys148 interacts weakly and hydrophobically with the galactopyranosyl moiety of substrate.
Site-directed NEM-labeling of single-Cys mutants in helices IV and V (Kwaw et al., 2001) reveals that a Cys residue in place of Ala122 (helix IV) exhibits properties similar to those of native Cys148. Thus, NEM inactivates lactose transport and the ligand protects against inactivation as well as alkylation by NEM. Since Ala122 is located at about the same depth in the membrane as Cys148 (Wolin and Kaback, 2000), the findings indicate that Ala122 may be a component of the substrate binding site (Figure 1). Strikingly, site-directed alkylation of A122C or replacement with Phe or Tyr selectively inactivates binding and transport of disaccharides with little or no effect on d-galactose transport, leading to the hypothesis that Ala122 abuts the non-galactosyl portion of disaccharide substrates (Guan et al., 2002a). This conclusion receives strong support from recent studies with methanethiosulfonyl-β-d-galactopyranosides which act as affinity inactivators of mutant A122C (Guan et al., 2002b).
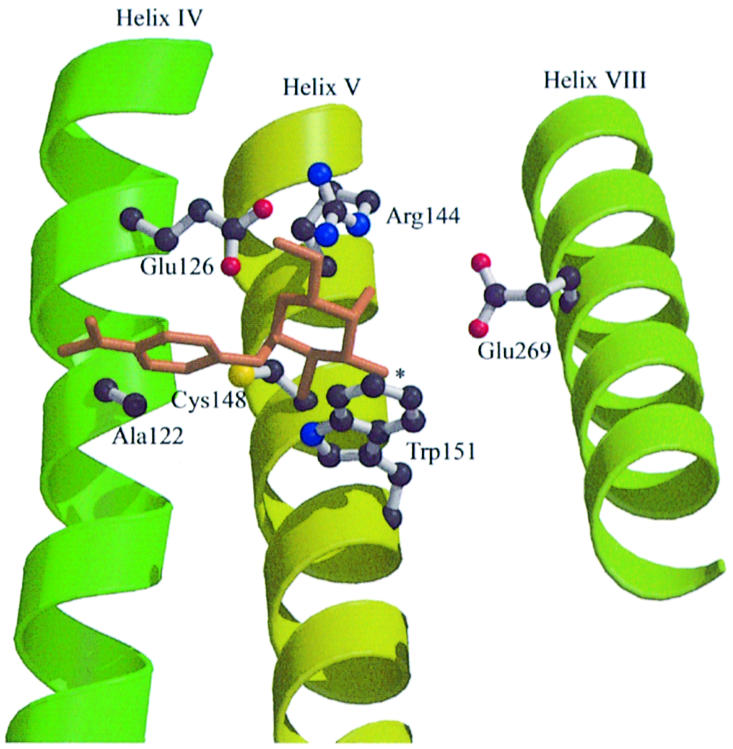
Fig. 1. Proposed LacY binding site with NPGal as ligand. The galactopyranosyl ring, which contains all the determinants for specificity, is shown with a nitrophenyl group at the anomeric position in the α configuration. The C-4 OH is most important with respect to specificity (Sahin-Tóth et al., 2000a, 2001). Cys148 (helix V) interacts weakly and hydrophobically with the galactopyranosyl ring, and Ala122 (helix IV) is in close proximity to the nitrophenyl moiety (Guan et al., 2002a). Trp151 stacks with the hydrophobic face of the galactopyranosyl ring (Guan et al., 2003), placing it at right angles to helix IV and abutting Cys148 near the 1-position. In this orientation, the C-4 OH can H-bond directly with either of the primary amines of Arg144. Since the C-3 OH (indicated by an asterisk) is close to Glu269 (helix VIII) but at an angle, a water molecule(s) may mediate this interaction.
LacY with Tyr or Phe in place of Trp151, which is one turn of helix V removed from Cys148, exhibits at least a 20- or a 50-fold decrease in affinity, respectively, despite relatively minor effects on the kinetic parameters of transport (Guan et al., 2003). Therefore, it is likely that Trp151 stacks with the hydrophobic face of the galactopyranosyl ring, orienting the ring at a right angle with helix V and abutting Cys148 near the 1 position (Figure 1). In this orientation, the C-4 OH can H-bond with either of the primary amines of Arg144 (helix V) and the C-6 OH can H-bond with Glu126 (helix IV), interactions that are consistent with the obligatory requirements for a guanidinium group at position 144 and a carboxyl group at position 126 for ligand binding (Frillingos et al., 1997a; Venkatesan and Kaback, 1998; Kaback et al., 2001).
Glu269 (helix VIII) is another irreplaceable residue that is located at the interface between helices VIII and V (Wu et al., 1995, 1996; Sorgen et al., 2002). This residue is also important for substrate recognition and may also be involved in H+ translocation (Franco and Brooker, 1994; Ujwal et al., 1994; He and Kaback, 1997; Venkatesan and Kaback, 1998; Sahin-Tóth et al., 2000b). Neutral replacements abolish binding and transport, while replacement with Asp leads to decreased affinity for β-d-galactopyranosyl-1-thio-β-d-galactopyranoside (TDG) (Sahin-Tóth et al., 2000b), as well as a marked increase in stoichiometry between H+ and sugar transport (Franco and Brooker, 1994; Ujwal et al., 1994). Finally, Gly-scanning mutagenesis of mutant E269D indicates that it can be rescued with respect to all translocation reactions, suggesting that positioning of the carboxyl group at position 269 in the binding site is critical (Weinglass et al., 2002).
In this communication, we present a novel approach for probing the role of Glu269 in substrate binding. By applying electrospray ionization mass spectrometry (ESI-MS) to purified full-length LacY, we observe that substrate reduces carbodiimide modification. Furthermore, analysis of a CNBr fragment of LacY containing Glu269 demonstrates that this residue reacts specifically with hydrophobic carbodiimides in a manner that is protected by substrate. Interestingly, replacement of Glu269 with Asp abolishes substrate protection against carbodiimide reactivity. These and other considerations are consistent with a model in which the C-3 OH of the d-galactopyranosyl ring may H-bond with Glu269.
Results
Substrate binding alters the modification profile of LacY by diisopropylcarbodiimide
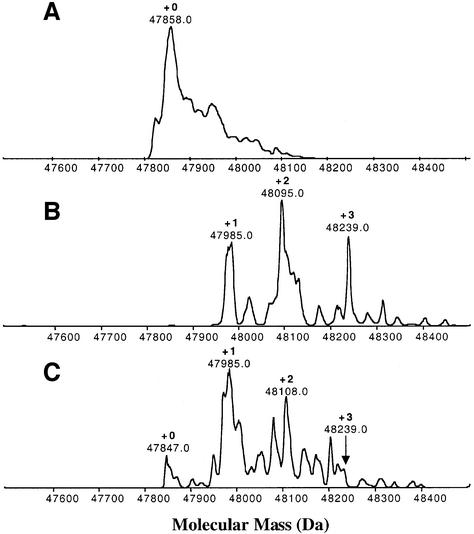
Molecular weight reconstruction of the uncharged permease with a His10 tag reveals that the LacY gene product is homogeneous and the experimentally measured mass (47 858 Da) is precisely that calculated from the DNA sequence with a formyl group on the initiating Met residue and 10 His residues at the C-terminus (Whitelegge et al., 1999) (Figure 2A). Following treatment of purified LacY with the hydrophobic carbodiimide diisopropylcarbodiimide (DiPC), the computed mass of the protein changes by the addition of one, two or three DiPC molecules within 0.03% of the calculated mass (Figure 2B): 47 985 (+1), 48 095 (+2) and 48 239 (+3), respectively, where each DiPC molecule is 126 Da. Pre-incubation with substrate leads to a loss of approximately one DiPC group, as indicated by an increase in the amount of unmodified material [47 847 (+0)], an increase in the magnitude of the +1 relative to the +2 peak and a large decrease in the +3 peak [48239 (+3)] (Figure 2C).
Fig. 2. DiPC modification and substrate protection by NPGal. ESI-MS is used to quantitate the addition of DiPC groups to Asp and Glu residues in the purified protein. Each DiPC addition increases the mass of the protein by 126 Da. (A) Unmodified permease. (B) Thirty-minute labeling of purified wild-type His10 LacY (∼170 µM) with 20 mM DiPC at pH 6.0 and 30°C. (C) Thirty-minute labeling of purified wild-type His10 LacY (∼170 µM) with 20 mM DiPC at pH 6.0 and 30°C after pre-incubation with a saturating concentration (20 mM) of the substrate analog NPGal. Pre-incubation with substrate reveals differential protection of these Asp/Glu residues against modification with DiPC. The molecular mass of the uncharged full-length protein was reconstructed using BioMultiView (Perkin–Elmer, Sciex, Applied Biosystems). Under the conditions described the accuracy of mass measurement is of the order of 0.03% (300 p.p.m.).
Identification of CNBr fragments by ESI-MS
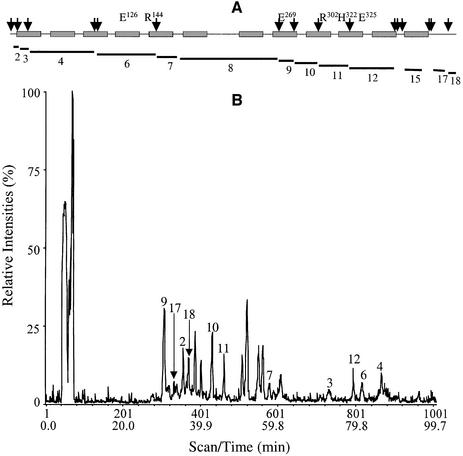
Since LacY has 17 carboxyl-group-containing side-chains, to dissect the effect of substrate on carbodiimide modification of Glu269, the protein was fragmented with CNBr and subsequently resolved by either reverse-phase (RP) or size-exclusion (SE) high-performance liquid chromatography (HPLC) in-line with an electrospray ionization mass spectrometer as described in Materials and methods. Pseudo-wild-type single Cys-148 LacY was used for these experiments. Of the 18 predicted peptides (Figure 3A and Table I; compare with Venter et al., 2002), 11 peptides including those containing the six essential residues [Glu126 (helix IV), Arg144 (helix V), Glu269 (helix VIII), Arg302 (helix IX), His322 and Glu325 (helix X)] are resolved and identified by RP-HPLC coupled with ESI-MS (Figure 3B; Table I). In addition, solubilization of the pellet with 90% formic acid followed by SE-HPLC and ESI-MS allows resolution of an additional two large hydrophobic peptides (11 636 Da and 9097.3 Da, Table I). Five small peptides (Table I, peptides 1, 5, 13, 14 and 16) at or below the mass range scanned were not detected. However, the peptides recovered represent 97% coverage of the full-length molecule. Notably, the total ion current profile also contains additional peaks that are not numbered. Based on the finding that size exclusion HPLC in-line with ESI-MS identifies a single protein species corresponding to LacY (data not shown), these peaks most likely arise from either cleavage of an acid labile bond or a Trp residue (Savige and Fontana, 1977). Mass analysis indicates that the peaks do not correspond to formylation of hydroxyamino acids, non-cleavage at Met sulfoxide or Met sulfone, incomplete cleavage at Met-Ser/Thr/Cys bonds or oxidation of Trp residues.
Fig. 3. ESI-MS of CNBr peptides from LacY. CNBr peptides from 100 µg of purified single-Cys148 LacY were separated as described in the Materials and methods. (A) Schematic representation of single-Cys148 LacY. The 12 transmembrane α-helices (rectangles), six irreplaceable residues and CNBr cleavage sites (arrows) are shown. Peptides resolved by either RP- or SE-HPLC are indicated by lines. (B) Identification of fragments. The total ion current following ESI-MS is shown. Individual CNBr fragments, numbered as in (A), were assigned by comparing the expected molecular masses obtained from the program Peptide Mass in the Expasy Molecular Biology Server (http://www.expasy.org) with the measured molecular masses.
Table I. Summary of mass spectrometry data for CNBr peptides from single-Cys148 LacY.
| Residue | Observed massa (Da) | Calculated massa (Da) | Δmass (observed – calculated) (Da) |
|---|---|---|---|
| 1 | ND | 102.1 | ND |
| 2–13 | 1331.4 | 1331.6 | 0.2 |
| 12–23 | 1578.0 | 1577.9 | 0.1 |
| 24–83 | 6810.6 | 6811.1 | 0.5 |
| 84–86 | ND | 313.4 | ND |
| 87–145 | 6503.4 | 6504.6 | 1.2 |
| 146–161 | 1572.9 | 1574.8 | 1.9 |
| 162–267 | 11 657.0b | 11 637.3 | 19.7 |
| 268–276 | 899.7 | 899.5 | 0.2 |
| 277–299 | 2396.0 | 2396.9 | 0.9 |
| 300–323 | 2525.1 | 2526.0 | 0.9 |
| 324–362 | 4541.2 | 4542.3 | 1.1 |
| 363–365 | ND | 361.4 | ND |
| 366–372 | ND | 642.7 | ND |
| 373–466 | 9095.3b | 9096.5 | 1.2 |
| 467–468 | ND | 229.3 | ND |
| 469–498 | 2922.9 | 2924.3 | 1.4 |
| 499–517 | 2077.0 | 2077.2 | 0.2 |
aThe masses of peptides under 2300 Da are reported as the monoisotopic values [M + H+]. Peptides of over 2300 Da are reported as average values [Mav + H+].
bThe insoluble material left after solubilizing CNBr fragments in 60% formic acid was solubilized in 90% formic acid and separated by SE-HPLC.
ND, these masses were not observed after resolving CNBr peptides by either RP- or SE-HPLC.
Glu269 is in a hydrophobic environment
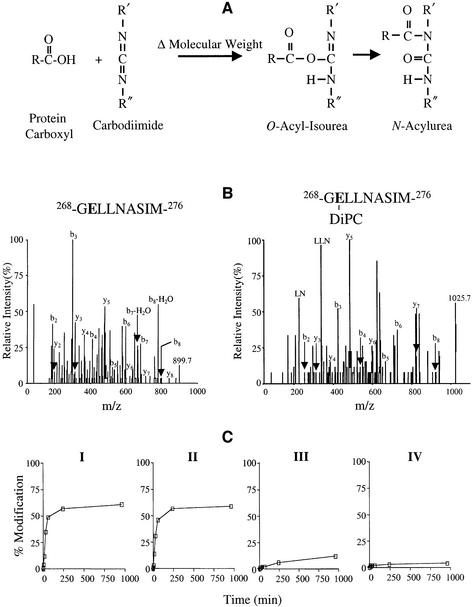
Carbodiimides react with protonated carboxyl groups (Khorana, 1953; Hoare and Koshland, 1967). The reaction takes place in two steps: (i) reaction of the carboxyl group to form a protonated O-acyl-isourea; (ii) rearrangement to form a stable N-acyl-isourea (Figure 4A). By utilizing carbodiimides with different solubility properties, it is possible to probe the environment of a carboxyl group within a protein by monitoring the increase in mass of CNBr fragments containing a reactive carboxyl side-chain(s).
Fig. 4. Modification of Glu269 by carbodiimides. (A) Reaction of carbodiimide with carboxyl side-chain. Nucleophilic attack of a carbodiimide on a protonated carboxyl side-chain leads to the formation of O-acyl-isourea with a change in molecular weight (Δ molecular weight) followed by rearrangement to form an N-acylurea with no further change in mass. (B) Collision-induced dissociation spectrum of 268-GELLNASIM-276 and 268-GELLNASIM-276 modified with DiPC. Annotated peaks in the spectra include the singly charged parents (m/z 899.7 and 1025.7, respectively) and the b- and y-ion series. (C) Time course of 268-GELLNASIM-276 modification by carbodiimides with different solubilities. Purified LacY (∼20 µM) in DDM at pH 6.0 was incubated with 20 mM of the following carbodiimides at 30°C leading to the mass shifts shown in parentheses: DiPC (Δ126), DCCD (Δ206), CMC (Δ251) and EDC (Δ155). The total ion current from peptide 268-GELLNASIM-276 modified by carbodiimide divided by the total ion current from unmodified and modified 268-GELLNASIM-276 was used to calculate the percentage modification of Glu269. No measurable difference in the ionization efficiency of the unmodified and modified peptides was detected. Each point represents the average of two independent experiments. Maximum peak height in the absence of modification was ∼5 × 105 counts.
Following CNBr cleavage, peptide 9 (268-GELLNA SIM-276) which contains Glu269 displays the isotopic distribution typical of a singly charged peptide with an experimentally determined m/z value of 899.7 in agreement with the expected m/z of 899.5. Sequencing of 268-GELLNASIM-276 by tandem mass spectometry (MS/MS) yields both a b-type and a y-type series of fragment ions that confirm the identity of the nanopeptide (Figure 4B, left panel). Following reaction with DiPC, a new peptide with an m/z value of 1025.7 is generated, and sequencing by MS/MS identifies this as 268-GEL LNASIM-276 modified at Glu269 by DiPC (Figure 4B, right panel). It is noteworthy that modification with DiPC leads to internal fragmentation of the peptide during MS/MS.
The local environment of Glu269 was probed by modification with each of four different carbodiimides with hydrophobic [DiPC or dicyclohexylcarbodiimide (DCCD)] or hydrophilic [1-cyclohexyl-3-[2-morpholinoethyl]carbodiimide (CMC) or 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide (EDC)] properties. In clear-cut fashion, both hydrophobic carbodiimides (DiPC and DCCD), react relatively rapidly with 268-GELLNA SIM-276 in a linear manner for ∼1 h (Figure 4C, panels I and II, respectively), after which the reaction terminates at ∼60% modification primarily because of the instability of carbodiimides in aqueous solution. In contrast, the hydrophilic carbodiimides CMC and EDC react slowly, if at all, over the time period tested (Figure 4C, panels III and IV, respectively).
Substrate protection against carbodiimide modification of Glu269
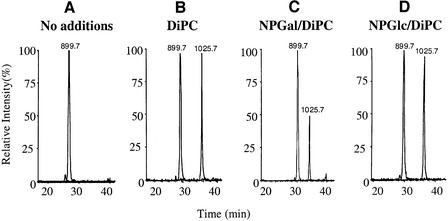
At pH 6.0, DiPC covalently modifies ∼48% of LacY at position 269 in 30 min as determined by analysis of the peptide 268-GELLNASIM-276 (Figure 5A, compare with B). Pre-incubation with the high-affinity ligand p-nitrophenyl-α-d-galactopyranoside (p-NPGal) reduces the rate of modification of Glu269 to ∼26% (Figure 5C). The effect is substrate specific since p-nitrophenyl-α-d-gluco pyranoside (NPGlc), which has no measurable affinity for LacY, exerts no significant effect on the rate of DiPC modification (Figure 5D).
Fig. 5. DiPC modifies Glu269 in a substrate-dependent manner. Purified single-Cys148 LacY (∼20 µM) in DDM at pH 6.0 was incubated for 30 min at 30°C with 2% dimethylsulfoxide (DMSO) (A) and 20 mM DiPC (B), 20 mM DiPC in the presence of p-NPGal (C) or 20 mM DiPC in the presence of NPGlc (D). Data are displayed as selected ion chromatograms of unmodified and DiPC-modified peptide 268-GELLNASIM-276 (m/z 899.7 and 1025.7, respectively). The chromatograms are representative of three independent experiments. Maximum peak height in the absence of modification was typically 5 × 105 counts.
Glu269 may interact with the C-3 OH of the galactopyranosyl ring
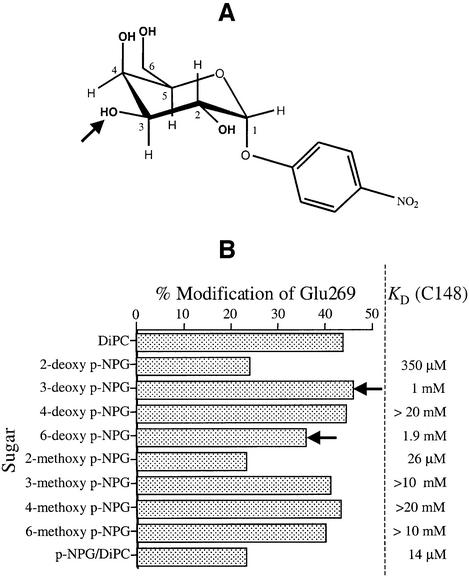
In an effort to identify the position(s) of the galactopyranosyl ring in p-NPGal which interacts with Glu269, a series of deoxy and methoxy analogs of p-NPGal were analyzed with respect to their ability to protect against DiPC modification of Glu269 (Figure 6). As shown by experiments in which protection against alkylation of Cys148 by N-ethylmaleimide was studied (Sahin-Tóth et al., 2001), the 2-deoxy analog binds with 25-fold lower affinity, the C-3 deoxy analog exhibits ∼70-fold decreased binding, affinity for 6-deoxy NPG is diminished by at least 130-fold and, remarkably, the 4-deoxy derivative binds with almost 1500-fold reduced affinity. No significant substrate protection is afforded by NPG analogs with methoxy substitutions at positions C-3, C-4 and C-6, but the C-2 methoxy analog binds almost normally.
Fig. 6. Ability of NPGal substrate analogs to protect against DiPC reactivity of Glu269. (A) Structure of NPGal. The arrow indicates the position of the C3 hydroxyl group on the galactopyranosyl ring. (B) Purified C148 (∼20 µM) in DDM at pH 6.0 was incubated for 30 min at 30°C with either 20 mM DiPC alone or 20 mM DiPC in the presence of given deoxy- or methoxy p-NPGal derivatives at pH 6.0. Selected ion chromatograms of unmodified and DiPC-modified 268-GELLNASIM-276 (m/z 899.7 and 1025.7, respectively) were used to determine the percentage of modification. Each point represents the average of two independent experiments. Maximum peak height in the absence of modification was ∼5 × 105 counts.
At saturating concentrations, the 2-deoxy or 2-methoxy derivatives of p-NPGal protect against DiPC modification of 268-GELLNASIM-276 as well as the parent ligand p-NPG; these findings are consistent with previous studies (Sahin-Tóth et al., 2001). Also consistent with Cys148 protection experiments, the 4-deoxy analog exhibits no ability to protect Glu269 against DiPC reactivity, the 6-deoxy derivative exhibits small but significant protection and the 3-, 4- and 6-methoxy compounds do not protect. Although 3-deoxy-NPGal with a KD of 1 mM is expected to protect as well as or better than the 6-deoxy analog (KD ≈ 2 mM), there is no protection relative to an identical reaction carried out in the absence of the substrate analog.
Ligand binding with Asp in place of Glu269
Mutant E269D exhibits decreased affinity for ligands of LacY (Ujwal et al., 1994; He and Kaback, 1997; Sahin-Tóth et al., 2000b). However, unlike TDG which binds with extremely poor affinity to mutant E269D/single-Cys148 relative to the pseudo-wild-type single-Cys148 LacY (KD values of 4 mM and 14 µM, respectively; Sahin-Tóth et al., 2000b), mutant E269D/single-Cys148 retains reasonable affinity for p-NPGal (KD values of 280 µM and 14 µM, respectively) (Figure 7; Table II).
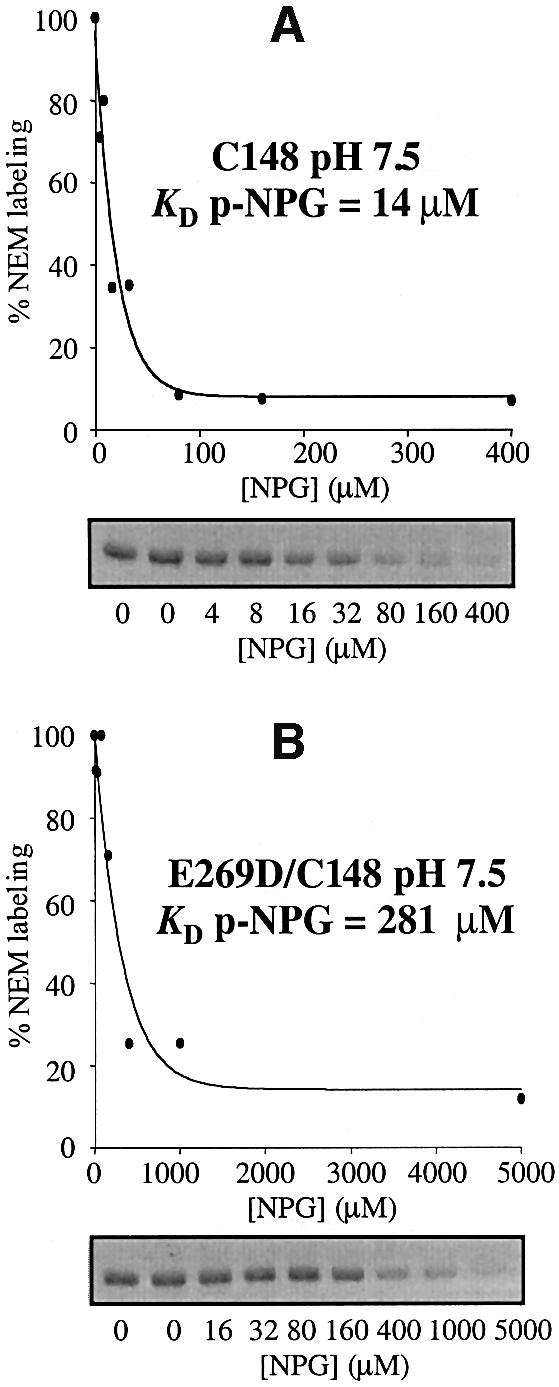
Fig. 7. NPGal protection against [14C]NEM labeling of (A) pseudo-wild-type single-Cys148 LacY and (B) mutant E269D/single-Cys148. RSO membrane vesicles containing single-Cys148 LacY with a biotin acceptor domain at the C-terminus were incubated in 100 mM KPi pH 7.5 with 0.5 mM [14C]NEM (40 mCi/mmol) for 5 min in the absence or presence of the indicated concentrations of p-NPGal. Reactions were quenched with DTT, and biotinylated LacY was solubilized and purified by affinity chromatography on monomeric avidin. Samples were separated on a sodium dodecylsulfate–12% polyacrylamide gel and labeled protein was visualized by autoradiography. Labeling was quantitated with a Storm 860 PhosphorImager, and labeling in the presence of given concentrations of NPG is expressed as percent labeling observed in the absence of sugar.
Table II. Effect of altering the position of the nitro moiety in NPGal on the apparent affinity constants KD of single Cys148 and E269D/C148 LacYa.
| Ligand | C148 LacY | E269D/C148 LacY |
|---|---|---|
| p-NPGal | 14 µM | 280 µM |
| o-NPGal | 45 µM | 6.3 mM |
| m-NPGal | 67 µM | >20 mM |
| p-NPGlc | >20 mM | >20 mM |
aNEM labeling experiments were carried out at pH 7.5, and the KD were determined as described in Materials and methods. Each value represents the average of two independent experiments where the KD did not differ by >10%.
Although specificity of LacY for substrate is directed exclusively towards the galactopyranosyl ring, adducts at the 1-position can cause a marked increase in affinity, particularly if hydrophobic and/or in the α configuration (Sahin-Tóth et al., 2002). Moreover, with respect to NPGal, the highest affinity is observed with p-NPGal (KD = 14 µM), followed by o-NPGal (KD = 45 µM) and m-NPGal (KD = 67 µM) (Table II). In contrast, with Asp in place of Glu269, affinity for o-NPGal or m-NPGal decreases by a factor of 25 or >75, respectively (KD values of 6.3 mM and >20 mM, respectively) (Table II). Thus, replacement of Glu269 with Asp leads to changes in both affinity and specificity, consistent with the notion that this carboxyl group is involved in substrate binding and recognition and that precise geometry is critical for optimal interaction.
Replacement of Glu269 with Asp abolishes substrate protection
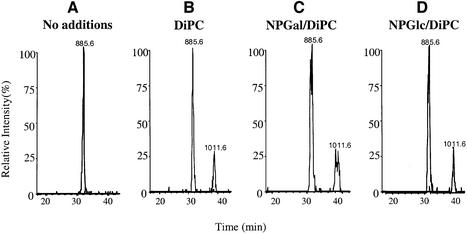
The nanopeptide containing Asp in place of Glu269 from mutant E269D/single-Cys148 (268-GDLLNASIM-276) displays the expected m/z value of 885.6 as a result of removing a single methylene group from position 269. Furthermore, mutant E269D/single-Cys148 is modified by DiPC or DCCD in a manner similar to that observed with the pseudo-wild-type single-Cys148, although the rate of modification is approximately half of that observed with the nanopeptide containing Glu269 (Figure 8A and B). Reactivity with either hydrophilic carbodiimide did not increase (data not shown). Strikingly, pre-incubation with either p-NPGal or NPGlc has no effect whatsoever on DiPC modification (Figure 8C and D), and nor does pre-incubation with TDG (data not shown).
Fig. 8. Replacement of Glu269 with Asp abolishes substrate protection against DiPC reactivity. Purified E269D/C148 LacY (∼20 µM) in DDM at pH 6.0 was incubated for 30 min at 30°C with 2% DMSO (A) and 20 mM DiPC (B), 20 mM DiPC in the presence of NPGal (C) or 20 mM DiPC in the presence of NPGlc (D). Data are displayed as selected ion chromatograms of unmodified and DiPC-modified peptide 268-GDLLNASIM-276 (m/z 885.6 and 1011.6, respectively). The chromatograms are representative of three independent experiments. Maximum peak height in the absence of modification was typically 5 × 105 counts.
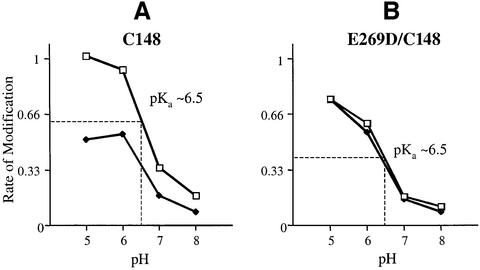
Reduced reactivity of the E269D mutant with DiPC in the absence of p-NPGal may reflect either a change in the local environment of Asp269 which alters accessibility to DiPC or a decrease in pKa. To distinguish between these possibilities, DiPC reactivity was monitored as a function of pH (Figure 9). Both the pseudo-wild-type single-Cys148 LacY and the E269D/single-Cys148 mutant exhibit higher rates of reactivity at low pH, as expected, and both exhibit perturbed apparent pKa values of ∼pH 6.5, which is consistent with the position of the carboxyl group within the permease in dodecyl-β-d-maltopyranoside (DDM) micelles (Figure 6). In addition, p-NPGal protection is observed at all pH values with the pseudo wild type, and no protection is observed with the E269D mutant (Figure 9).
Fig. 9. pH-dependent DiPC reactivity of Glu or Asp at position 269. Purified single-Cys148 LacY (A) or mutant E269D/C148 (B) (∼20 µM) in DDM at pH 6.0 was incubated for 60 min at 30°C with either 2% DMSO and 20 mM DiPC or 20 mM DiPC in the presence of p-NPGal at pH 5.0, 6.0, 7.0 or 8.0. Selected ion chromatograms of unmodified and DiPC-modified 268-GELLNASIM-276 (m/z 899.7 and 1025.7, respectively) or 268-GDLLNASIM-276 (m/z 885.6 and 1011.6, respectively) were used to determine the rate of modification (% modification/min). Each point represents the average of two independent experiments. Maximum peak height in the absence of modification was ∼5 × 105 counts.
Discussion
Currently there are no atomic-resolution structures for symporters, but only two-dimensional structures of OxlT (Hirai et al., 2002), NhaA (Williams, 2000) and MelB (Hacksell et al., 2002) obtained from electron crystallography. In contrast, insight into the structure of LacY at a resolution of ∼4 Å has come from the application of site-directed biochemical and biophysical techniques (Sorgen et al., 2002). Integrating the structural information with functional insights obtained from characterization of site-directed mutants has led to a working hypothesis for the role of the six irreplaceable residues involved in the mechanism of lactose-H+ symport (reviewed in Kaback et al., 2001). Glu269 is strongly implicated in both H+ and substrate translocation. However, problems in interpreting signals generated from multiple reaction sites restrict substrate-dependent studies on Glu269 reactivity with radioactive or fluorescent carboxylate modifying reagents. Furthermore, replacement with Cys in a background devoid of native Cys residues renders LacY completely inactive and unable to bind ligand (Frillingos et al., 1997b), thereby precluding studies with thiol-specific reagents on single-Cys269 LacY. To overcome this obstacle, the covalent modification of full-length purified LacY by carboxyl-specific carbodiimides is monitored by mass spectrometry. Since the pKa of a carboxyl group is typically ∼4.3, by using the hydrophobic carbodiimide DiPC at pH 6 a limited subset of the 17 carboxyl side-chains in LacY is modified, most likely those embedded in the membrane which includes Glu269. Clearly, substrate binding reduces the reactivity of at least one carboxyl side-chain (Figure 2), despite heterogeneity arising from ESI-MS of large polytopic membrane proteins. Such heterogeneity is generally due to: (i) formylation of hydroxylated amino acids; (ii) maintenance of structure during ESI-MS leading to non-stoichiometric ion binding of certain species (Whitelegge et al., 1999); and/or (iii) reduction in mass accuracy due to a decrease in signal-to-noise ratio and an increase in heterogeneity (Figure 2B and C) (le Coutre et al., 2000; Gomez et al., 2002). The properties of Glu or Asp at position 269 have been examined by following the reaction of LacY with hydrophobic carbodiimides followed by CNBr fragmentation and in-line RP-HPLC ESI-MS, and it has been shown that Glu269 interacts directly with substrate.
As shown previously (Franco and Brooker, 1994; Ujwal et al., 1994; He and Kaback, 1997; Sahin-Tóth et al., 2000b; Weinglass et al., 2002), neutral replacements at position 269 lead to a complete loss of substrate binding and translocation, while mutant E269D binds and translocates substrate poorly. Furthermore, if the packing and/or dynamics of helix VIII are altered with Asp in place of Glu269, both binding and transport are rescued, suggesting that interaction with substrate is perturbed (Weinglass et al., 2002). The exaggerated differences between the binding affinities of p-, o- or m-NPGal derivatives with mutant E269D/single-Cys148 relative to the pseudo-wild-type single-Cys148 LacY support the contention that this carboxyl group plays an important role in both affinity and substrate specificity. Therefore, changes in specificity observed with Asp in place of Glu269 are probably due to alterations in the positioning of the galactosyl moiety in the substrate binding site secondary to subtle changes in interactions of the non-galactopyranosyl end of the ligands. Regardless of the precise explanation for the phenomena, since p-NPGlc exhibits no protection, it is clearly the galactopyranosyl ring in the ligand that is responsible for protection of Glu269 against DiPC reactivity. The observation that the DiPC reactivity of an Asp residue in place of Glu269, which leads to a marked decrease in affinity (Sahin-Tóth et al., 2000b), is unaffected by p-NPGal is consistent with the interpretation that the carboxyl group at position 269 plays an important role in substrate recognition.
There are at least two explanations for the decrease in carbodiimide reactivity of the carboxyl group when Glu269 is replaced with Asp. Either the accessibility of the carbodiimide to the carboxyl group is reduced when the side-chain is shortened by 1.5 Å, or the carboxyl group is in a more hydrophilic environment leading to a decrease in pKa and reduced reactivity at pH 6.0. Since carbodiimides react specifically with protonated carboxyl groups, an apparent pKa can be approximated by studying reactivity as a function of pH (Kluge and Dimroth, 1993; Velazquez et al., 1997; Valiyaveetil et al., 2002). As shown in Figure 6, a perturbed apparent pKa of ∼6.5 is observed with either a Glu or an Asp residue at position 269, but no effect on the apparent pKa is observed in the presence of p-NPGal. The finding is consistent with the exclusive reactivity of Glu269 with hydrophobic carbodiimides. Moreover, the reduced rate of DiPC reaction with an Asp residue at position 269 is unlikely to reflect a change in pKa. It is noteworthy in this regard that the present studies were carried out with purified LacY solubilized in dodecyl-β-d-maltopyranoside (DDM) and that the pKa of the carboxyl group may be even higher when LacY is embedded in a phospholipid bilayer. Left unresolved by this work is the possibility that the pKa may be perturbed in the presence of ligand when LacY is in the membrane. These possibilities are currently being pursued.
In an effort to identify the position in the galactopyranosyl ring that might interact with Glu269, a series of p-nitrophenyl-α-d-deoxy- and methoxy-galactopyranoside analogs were tested for their ability to protect against DiPC reactivity of Glu269 relative to the ability of the same ligands to protect single-Cys148 LacY against N-ethylmaleimide reactivity (Sahin-Tóth et al., 2001). The studies indicate that the C-2, C-4 and C-6 derivatives of p-NPGal have the same relative ability to protect Glu269 or Cys148 qualitatively. That is, the C-2 analogs protect against modification of both side-chains, the C-4 analogs do not protect either Glu269 or Cys148 and the C-6 analog protects both side-chains relatively weakly. Therefore, it seems unlikely that either the C-2 or the C-6 OH is responsible for protection of Glu269 against DiPC reactivity. Since the C-4 OH binds with little or no affinity, as judged by protection of Cys148 against NEM reactivity, it cannot be concluded that the OH at this position plays a role in the protection of Glu269. However, the affinity of the C-3 deoxy derivative is twice as high as that of the C-6 deoxy analog (KD values of 1.0 mM and ∼2.0 mM, respectively) and the latter exhibits reproducible, albeit weak, protection of Glu269. Therefore, the C-3 deoxy analog should protect at least as well, but no protection is observed.
As depicted in the model of the binding site in LacY (Figure 1), Cys148 (helix V) interacts weakly and hydrophobically with the galactosyl moiety of substrate, and Ala122 (helix IV) abuts the non-galactosyl moiety (Guan et al., 2002a; Sorgen et al., 2002). Importantly, Trp151 (helix V) stacks with the hydrophobic face of the galactopyranosyl ring, placing it at right angles to helix V approximating Cys148 near the 1-position (Guan et al., 2003). In this orientation, the C-4 OH can H-bond directly with either of the primary amines of Arg144 and the C-6 OH can H-bond with Glu126 (helix IV). Taking the model together with the data presented in this paper, it seems reasonable to suggest that C-3 OH is the most likely candidate to interact with Glu269 (helix VIII). Furthermore, since C-3 OH is close to Glu269 but appears to be at an angle, a water molecule(s) may mediate the interaction.
In conclusion, by engineered replacement of non-essential residues with Met, it should be possible to create CNBr fragments such that each contains a single reactive species. This will allow studies on the role of other important residues in substrate and H+ translocation with appropriate group-specific reagents. Thus, while ultimate verification requires an atomic structure, this study shows that accurate mass measurements in conjunction with group-specific chemical reagents can provide a new level of insight into substrate binding interactions in a specific membrane transport protein, LacY. Furthermore, by monitoring the protonation state of residues implicated in H+ translocation in mutants conformationally locked at different stages in the transport cycle, it should be possible to gain insight into the pathway of H+ flux. Finally, it is apparent that the general approach may be broadly applicable to a wide variety of membrane proteins, as well as to transporters.
Materials and methods
Materials
Oligodeoxynucleotides were synthesized by Sigma-Genosys (The Woodland, TX). Restriction endonucleases, T4 DNA ligase and Vent polymerase were obtained from New England Biolabs (Beverly, MA). All other materials were reagent grade and obtained from commercial sources.
Growth of cells and preparation of right-side-out (RSO) membrane vesicles
Escherichia coli T184 was grown in Luria–Bertani broth, and RSO membrane vesicles were prepared as described (Kaback, 1971; Short et al., 1975), except that 5.0 mM dithiothreitol (DTT) was included. At the end of the preparation, the vesicles were washed with 100 mM potassium phosphate (KPi, pH 7.5) to remove DTT, resuspended in the same buffer at a protein concentration of 13–18 mg/ml, frozen in liquid nitrogen and stored at –80°C until use.
NEM labeling
Reactivity of single-Cys148 LacY with [14C]NEM was determined in situ in the absence or presence of given concentrations of ligand (Sahin-Tóth et al., 2000b). Labeling was initiated by the addition of 10 µl of [14C]NEM (40 mCi/mmol) to a final concentration of 0.5 mM, and the vesicles were incubated for 5 min at 25°C. Reactions were quenched by the addition of 10 mM DTT. After sodium dodecylsulfate–12% polyacrylamide electrophoresis, the gels were dried and [14C]NEM labeling was visualized and quantitated with a Storm 860 PhosphorImager. Apparent affinity constants (KDapp) were determined with the MicroCalTMOrigin TM computer program using non-linear least-squares curve fitting to a user-defined equation as described (Sahin-Tóth et al., 2000b).
Purification of LacY
Escherichia coli T184 [lacI+O+Z–Y–(A), rpsL, met–, thr, recA, hsdM, hsdR/F′, lacIqO+ZD118(Y+A+)] (Teather et al., 1978) was grown in Luria–Bertani medium. Flasks were innoculated with an overnight culture of freshly transformed cells bearing the pT7-5/cassette lacY encoding given to LacY mutants with a C-terminal biotin acceptor domain (Consler et al., 1993) or a His10 tag. Cells were induced with 0.5 mM i-propyl-1-thio-β,d-galactopyranoside at an OD600 of 0.6 and harvested at an OD of ∼2. Concentrated cells were washed once in cold 50 mM KPi pH 7.5 and a second time in the same buffer plus 10 mM MgSO4. To prepare membranes, cells were resuspended in a cold mixture containing 50 mM KPi pH 7.5, 10 mM MgSO4, 0.5 mM Pefabloc, 10 mM β-mercaptoethanol (BME) and 30 µg/ml DNAse and disrupted by passage through a French pressure cell (15 000–20 000 psi). Unbroken cells were removed by centrifugation (15 min at 10 000 g) and the membrane fraction was obtained from the supernatant by centrifugation (3 h at 150 000 g). To remove peripheral membrane proteins, the membranes were washed with a mixture containing 5 M urea, 50 mM KPi pH 7.5, 0.5 mM Pefabloc and 10 mM BME, stirred for 30 min on ice and centrifuged overnight at 170 000 g. The supernatant was discarded, and the membranes were resuspended in a mixture containing 50 mM sodium phosphate (NaPi) pH 7.5, 0.1 M NaCl, 10 mM BME and 0.5 mM Pefabloc, and DDM was added to a final concentration of 2% while stirring on ice for 30 min. After removal of insoluble material by centrifugation at 150 000 g for 30 min, the supernatant was combined with either washed monomeric avidin coupled to Sepharose (Pierce, Rockford, IL) (0.25 ml resin per 1.0 g starting cells) or a cobalt Talon resin (Invitrogen, Palo Alto, CA) (0.35 ml resin per 1.0 g starting cells) and rotated for 1 h at 4°C. The loaded avidin resin was transferred to a glass column, washed exhaustively in a mixture containing 50 mM NaPi pH 7.5, 10 mM BME, 0.5 mM Pefabloc, 0.15 M NaCl and 0.02% DDM. LacY was then eluted with 5 mM biotin in the same buffer. The loaded cobalt resin was transferred to a glass column and washed with 50 mM NaPi pH 7.0, 300 mM NaCl, 5 mM imidazole and 0.01% DDM until the baseline was reached. To remove the remaining low-affinity contaminants the resin was further washed with 50 mM NaPi pH 7.0, 300 mM NaCl, 50 mM imidazole and 0.01% DDM. Finally, LacY was eluted with 50 mM NaPi pH 7.0, 200 mM imidazole and 0.01% DDM. For both purifications, the eluted protein was dialyzed overnight against a mixture containing 50 mM NaPi pH 7.5, 0.15 M NaCl, 0.02% DDM and 10% glycerol, and for carbodiimide modification experiments the pH of the buffer was adjusted.
Carbodiimide labeling and sample preparation for SE-HPLC of full-length protein or RP-HPLC of CNBr fragments
Carbodiimides were freshly prepared as stocks of 1 M DiPC or DCCD dissolved in dimethylsulfoxide or 1 M EDC and 200 mM CMC dissolved in water. Following reaction of a given carbodiimide with purified membrane protein at 30°C for a given time, the sample was immediately precipitated with CHCl3-MeOH, as described (Wessel and Flügge, 1984). For this, an aliquot (100 µl) of the aqueous protein solution at a concentration of 1–2 mg/ml was diluted 1:3 (v/v) with MeOH and mixed briefly. CHCl3 (100 µl) was added and mixed, yielding a single phase. Phase separation was accomplished by adding water (200 µl) and mixing vigorously. The phases were separated by centrifugation (10 000 g for 2 min), yielding a precipitate at the interface. The bulk of the upper aqueous methanol phase was then carefully aspirated, and methanol (300 µl) was added. After gentle mixing, insoluble protein in the single-phase mixture was recovered by centrifugation at 10 000 g for 1 min. For SE-HPLC of the full length protein, the pellet was air dried for 1–2 min with the tube inverted and resuspended in 90% formic acid. For cleavage of the protein, the dried pellet was resuspended in a saturated solution of CNBr in 90% formic acid (Witkop, 1961) and left for 2 h in the dark. Formic acid was removed in a vacuum centrifuge, and the pellet was left in 0.5 ml of 0.1% trifluoroacetic acid (TFA) in water overnight. The sample was dried again and resuspended in 60% formic acid immediately prior to RP-HPLC.
High-performance liquid chromatography
For final purification of lactose permease either before or after covalent modification with carbodiimides, HPLC was used before ESI-MS in an in-line set-up (LCMS). The permease was purified by using isocratic elution (0.25 ml/min) from a size exclusion column (Super SW2000, ToSo BioSep, Montgomeryville, PA; 4 µM, 300 mm × 4.6 mm) in an aqueous organic solvent mixture (Whitelegge et al., 1999) using degassed CHCl3-MeOH-1% aqueous formic acid (4:4:1 v/v) (Fearnley and Walker, 1996) as eluant. To separate the CNBr peptides generated from permease, a polystyrene–divinylbenzene column (PLRP/S, Polymer Labs; 5 µM, 300 Å, 150 mm × 2.1 mm) at 40°C was used for reverse-phase LCMS (Whitelegge et al., 1992). Following equilibration in 95% solvent A and 5% solvent B for 5 min, the percentage of solvent B was increased to 40% over the next 25 min and then to 100% over the subsequent 120 min at a flow rate of 0.1 ml/min [solvent A, 0.1% TFA in water; solvent B, 0.1% TFA in CH3CN-isopropanol (1:1)]. All chromatographic separations were performed at 40°C using a modified ABI 120A dual-syringe pump HPLC equipped with a post-detector (A280) splitter for back-pressure regulation.
Electrospray ionization mass spectrometry
ESI-MS analysis was performed using a Perkin–Elmer Sciex API III triple-quadrupole instrument operating in the positive ion mode as described (Whitelegge et al., 1998). The optimal orifice potential was 75 V for full-length lactose permease and ramped from 60 to 120 V across the mass range (600–2300) for CNBr fragments. MS/MS fragment ion spectra were obtained using an orifice potential of 80 V and a collision gas (99.999% argon) thickness of 180. The nomenclature used for fragment ions is N-terminal fragments (b type) and C-terminal fragments (y type) (Roepstorff and Fohlman, 1984; Biemann, 1990).
Acknowledgments
Acknowledgements
We thank Jose Luis Vazquez-Ibar for help with purification and Vladimir Kasho, Irina Smirnova, Miklós Sahin Tóth and Andy Norris for helpful discussions. This work was supported in part by NIH grant DK51131:07 to H.R.K. The Pasarow Family and the W.M.Keck foundation provided partial funds for instrument purchase.
References
- Biemann K. (1990) Appendix 5. Nomenclature for petide fragment ions (positive ions). Methods Enzymol., 193, 886–887. [DOI] [PubMed] [Google Scholar]
- Bieseler B., Prinz,H. and Beyreuther,K. (1985) Topological studies of lactose permease of Escherichia coli by protein sequence analysis. Ann. N.Y. Acad. Sci., 456, 309–325. [DOI] [PubMed] [Google Scholar]
- Calamia J. and Manoil,C. (1990) Lac permease of Escherichia coli: topology and sequence elements promoting membrane insertion. Proc. Natl Acad. Sci. USA, 87, 4937–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consler T.G., Persson,B.L., Jung,H., Zen,K.H., Jung,K., Prive,G.G., Verner,G.E. and Kaback,H.R. (1993) Properties and purification of an active biotinylated lactose permease from Escherichia coli. Proc. Natl Acad. Sci. USA, 90, 6934–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello M.J., Escaig,J., Matsushita,K., Viitanen,P.V., Menick,D.R. and Kaback,H.R. (1987) Purified lac permease and cytochrome o oxidase are functional as monomers. J. Biol. Chem., 262, 17072–17082. [PubMed] [Google Scholar]
- Fearnley I.M. and Walker,J.E. (1996) Analysis of hydrophobic proteins and peptides by electrospray ionization MS. Biochem. Soc. Trans, 24, 912–917. [DOI] [PubMed] [Google Scholar]
- Foster D.L., Boublik,M. and Kaback,H.R. (1983) Structure of the lac carrier protein of Escherichia coli. J. Biol. Chem., 258, 31–34. [PubMed] [Google Scholar]
- Franco P.J. and Brooker,R.J. (1994) Functional roles of Glu-269 and Glu-325 within the lactose permease of Escherichia coli. J. Biol. Chem., 269, 7379–7386. [PubMed] [Google Scholar]
- Frillingos S. and Kaback,H.R. (1996) Probing the conformation of the lactose permease of Escherichia coli by in situ site-directed sulfhydryl modification. Biochemistry, 35, 3950–3956. [DOI] [PubMed] [Google Scholar]
- Frillingos S., Gonzalez,A. and Kaback,H.R. (1997a) Cysteine-scanning mutagenesis of helix IV and the adjoining loops in the lactose permease of Escherichia coli: Glu126 and Arg144 are essential. Biochemistry, 36, 14284–14290. [DOI] [PubMed] [Google Scholar]
- Frillingos S., Ujwal,M.L., Sun,J. and Kaback,H.R. (1997b) The role of helix VIII in the lactose permese of Escherichia coli. I. Cys-scanning mutagenesis. Protein Sci., 6, 431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frillingos S., Sahin-Tóth,M., Wu,J. and Kaback,H.R. (1998) Cys-scanning mutagenesis: a novel approach to structure–functions relationships in polytopic membrane proteins. FASEB J., 12, 1281–1299. [DOI] [PubMed] [Google Scholar]
- Gomez S.M., Nishio,J.N., Faull,K.F. and Whitelegge,J.P. (2002) The chloroplast grana proteome defined by intact mass measurements from liquid chromatography mass spectrometry. Mol. Cell Proteomics, 1, 46–59. [DOI] [PubMed] [Google Scholar]
- Guan L., Weinglass,A.B. and Kaback,H.R. (2001) Helix packing in the lactose permease of Escherichia coli: localization of helix VI. J. Mol. Biol., 312, 69–77. [DOI] [PubMed] [Google Scholar]
- Guan L., Sahin-Tóth,M. and Kaback,H.R. (2002a) Changing the lactose permease of Escherichia coli into a galactose-specific symporter. Proc. Natl Acad. Sci. USA, 99, 6613–6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L., Sahin-Tóth,M., Kálai,T., Hideg,K. and Kaback,H.R. (2002b) Probing the mechanism of a membrane transport protein with affinity inactivators. J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- Guan L., Hu,L. and Kaback,H.R. (2003) Aromatic stacking in the sugar binding site of the lactose permease. Biochemistry, 42, 1377–1382. [DOI] [PubMed] [Google Scholar]
- Hacksell I., Rigaud,J.L., Purhonen,P., Pourcher,T., Hebert,H. and Leblanc,G. (2002) Projection structure at 8 Å resolution of the melibiose permease, an Na-sugar co-transporter from Escherichia coli. EMBO J., 21, 3569–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M. and Kaback,H.R. (1997) Interaction between residues Glu269 (helix VIII) and His322 (helix X) of the lactose permease of Escherichia coli is essential for substrate binding. Biochemistry, 36, 13688–13692. [DOI] [PubMed] [Google Scholar]
- Hirai T., Heymann,J.A., Shi,D., Sarker,R., Maloney,P.C. and Subramaniam,S. (2002) Three-dimensional structure of a bacterial oxalate transporter. Nat. Struct. Biol., 9, 597–600. [DOI] [PubMed] [Google Scholar]
- Hoare D.G. and Koshland,D.E., Jr (1967) A method for the quantitative modification and estimation of carboxylic acid groups in proteins. J. Biol. Chem., 242, 2447–2453. [PubMed] [Google Scholar]
- Jung H., Jung,K. and Kaback,H.R. (1994) Cysteine 148 in the lactose permease of Escherichia coli is a component of a substrate binding site. I. Site-directed mutagenesis studies. Biochemistry, 33, 12160–12165. [DOI] [PubMed] [Google Scholar]
- Kaback H.R. (1971) Bacterial membranes. Methods Enzymol., 22, 99–120. [Google Scholar]
- Kaback H.R. and Wu,J. (1997) From membrane to molecule to the third amino acid from the left with the lactose permease of Escherichia coli. Q. Rev. Biophys., 30, 333–364. [DOI] [PubMed] [Google Scholar]
- Kaback H.R., Sahin-Tóth,M. and Weinglass,A.B. (2001) The kamikaze approach to membrane transport. Nat. Rev. Mol. Cell Biol., 2, 610–622. [DOI] [PubMed] [Google Scholar]
- Khorana H.G. (1953) The chemistry of carbodiimides. Chem. Rev., 53, 145–166. [Google Scholar]
- Kluge C. and Dimroth,P. (1993) Specific protection by Na+ or Li+ of the F1F0-ATPase of Propionigenium modestum from the reaction with dicyclohexylcarbodiimide. J. Biol. Chem., 268, 14557–14560. [PubMed] [Google Scholar]
- Konings W.N., Kaback,H.R. and Lolkema,J.S. (1996) Handbook of Biological Physics: Transport Processes in Eukaryotic and Prokaryotic Organisms. Elsevier, Amsterdam, The Netherlands.
- Kwaw I., Zen,K.-C., Hu,Y. and Kaback,H.R. (2001) Site-directed labeling of the lactose permease of Escherichia coli: helices IV and V which contain the major determinants for substrate binding. Biochemistry, 40, 10491–10499. [DOI] [PubMed] [Google Scholar]
- le Coutre J., Whitelegge,J.P., Gross,A., Turk,E., Wright,E.M., Kaback,H.R. and Faull,K.F. (2000) Proteomics on full-length membrane proteins using mass spectrometry. Biochemistry, 39, 4237–4242. [DOI] [PubMed] [Google Scholar]
- Olsen S.G. and Brooker,R.J. (1989) Analysis of the structural specificity of the lactose permease toward sugars. J. Biol. Chem., 264, 15982–15987. [PubMed] [Google Scholar]
- Roepstorff P. and Fohlman,J. (1984) Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed. Mass Spectrom., 11, 601. [DOI] [PubMed] [Google Scholar]
- Sahin-Tóth M., Lawrence,M.C. and Kaback,H.R. (1994) Properties of permease dimer, a fusion protein containing two lactose permease molecules from Escherichia coli. Proc. Natl Acad. Sci. USA, 91, 5421–5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin-Tóth M., Akhoon,K.M., Runner,J. and Kaback,H.R. (2000a) Ligand recognition by the lactose permease of Escherichia coli: specificity and affinity are defined by distinct structural elements of galactopyranosides. Biochemistry, 39, 5097–5103. [DOI] [PubMed] [Google Scholar]
- Sahin-Tóth M., Karlin,A. and Kaback,H.R. (2000b) Unraveling the mechanism of lactose permease of Escherichia coli. Proc. Natl Acad. Sci. USA, 97, 10729–10732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin-Tóth M., Lawrence,M.C., Nishio,T. and Kaback,H.R. (2001) The C-4 hydroxyl group of galactopyranosides is the major determinant for ligand recognition by the lactose permease of Escherichia coli. Biochemistry, 43, 13015–13019. [DOI] [PubMed] [Google Scholar]
- Sahin-Tóth M., Gunawan,P., Lawrence,M.C., Toyokuni,T. and Kaback,H.R. (2002) Binding of hydrophobic d-galactopyranosides to the lactose permease of Escherichia coli. Biochemistry, 41, 13039–13045. [DOI] [PubMed] [Google Scholar]
- Sandermann H. Jr (1977) β-d-Galactoside transport in Escherichia coli: substrate recognition. Eur. J. Biochem., 80, 507–515. [DOI] [PubMed] [Google Scholar]
- Savige W.E. and Fontana,A. (1977) Cleavage of the tryptophanyl peptide bond by dimethyl sulfoxide–hydrobromic acid. Methods Enzymol., 47, 459–469. [DOI] [PubMed] [Google Scholar]
- Short S.A., Kaback,H.R. and Kohn,L.D. (1975) Localization of d-lactate dehydrogenase in native and reconstituted Escherichia coli membrane vesicles. J. Biol. Chem., 250, 4291–4296. [PubMed] [Google Scholar]
- Sorgen P.L., Hu,Y., Guan,L., Kaback,H.R. and Girvin,M.E. (2002) An approach to membrane protein structure without crystals. Proc. Natl Acad. Sci. USA, 99, 14037–14040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J. and Kaback,H.R. (1997) Proximity of periplasmic loops in the lactose permease of Escherichia coli determined by site-directed cross-linking. Biochemistry, 36, 11959–11965. [DOI] [PubMed] [Google Scholar]
- Teather R.M., Müller-Hill,B., Abrutsch,U., Aichele,G. and Overath,P. (1978) Amplification of the lactose carrier protein in Escherichia coli using a plasmid vector. Molec. Gen. Genet., 159, 239–248. [DOI] [PubMed] [Google Scholar]
- Ujwal M.L., Sahin-Tóth,M., Persson,B. and Kaback,H.R. (1994) Role of glutamate-269 in the lactose permease of Escherichia coli. Mol. Membr. Biol., 11, 9–16. [DOI] [PubMed] [Google Scholar]
- Valiyaveetil F., Hermolin,J. and Fillingame,R.H. (2002) pH dependent inactivation of solubilized F1F0 ATP synthase by dicyclohexyl carbodiimide: pKa of detergent unmasked aspartyl-61 in Escherichia coli subunit c. Biochim. Biophys. Acta, 1553, 296–301. [DOI] [PubMed] [Google Scholar]
- Velazquez I., Martinez,F. and Pardo,J.P. (1997) Inactivation of the Kluyveromyces lactis H+-ATPase by dicyclohexylcarbodiimide: binding stoichiometry and effect of nucleophiles. Arch. Biochem. Biophys., 346, 294–302. [DOI] [PubMed] [Google Scholar]
- Venkatesan P. and Kaback,H.R. (1998) The substrate binding site in the lactose permease of Escherichia coli. Proc. Natl Acad. Sci. USA, 95, 9802–9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter H., Ashcroft,A.E., Keen,J.N., Henderson,P.J. and Herbert,R.B. (2002) Molecular dissection of membrane-transport systems: mass spectrometry and sequence determination of the galactose-H+ symport protein, GalP, of Escherichia coli and quantitative assay of the incorporation of [ring-2–13]histidine and (15)HN(3). Biochem. J., 363, 243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viitanen P., Garcia,M.L. and Kaback,H.R. (1984) Purified reconstituted lac carrier protein from Escherichia coli is fully functional. Proc. Natl Acad. Sci. USA, 81, 1629–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinglass A.B., Sondej,M. and Kaback,H.R. (2002) Manipulating conformational equilibria in the lactose permease of Escherichia coli. J. Mol. Biol., 315, 561–571. [DOI] [PubMed] [Google Scholar]
- Wessel D. and Flügge,U.I. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem., 138, 141–143. [DOI] [PubMed] [Google Scholar]
- Whitelegge J.P., Jewess,P., Pickering,M.G., Gerrish,C., Camilleri,P. and Bowyer,J.R. (1992) Sequence analysis of photoaffinity-labelled peptides derived by proteolysis of photosystem-2 reaction centres from thylakoid membranes treated with [14C]azidoatrazine. Eur. J. Biochem., 207, 1077–1084. [DOI] [PubMed] [Google Scholar]
- Whitelegge J.P., Gundersen,C.B. and Faull,K.F. (1998) Electrospray-ionization mass spectrometry of intact intrinsic membrane proteins. Protein Sci., 7, 1423–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelegge J.P., le Coutre,J., Lee,J.C., Engel,C.K., Privé,G.G., Faull,K.F. and Kaback,H.R. (1999) Toward the bilayer proteome, electrospray ionization-mass spectrometry of large, intact transmembrane proteins. Proc. Natl Acad. Sci. USA, 96, 10695–10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K.A. (2000) Three-dimensional structure of the ion-coupled transport protein NhaA. Nature, 403, 112–115. [DOI] [PubMed] [Google Scholar]
- Witkop B. (1961) Nonenzymatic methods for the preferential and selective cleavage and modification of proteins. Adv. Protein Chem., 16, 221–321. [DOI] [PubMed] [Google Scholar]
- Wolin C.D. and Kaback,H.R. (2000) Thiol cross-linking of transmembrane domains IV and V in the lactose permease of Escherichia coli. Biochemistry, 39, 6130–6135. [DOI] [PubMed] [Google Scholar]
- Wu J. and Kaback,H.R. (1994) Cysteine 148 in the lactose permease of Escherichia coli is a component of a substrate binding site. 2. Site-directed fluorescence studies. Biochemistry, 33, 12166–12171. [DOI] [PubMed] [Google Scholar]
- Wu J., Perrin,D., Sigman,D. and Kaback,H. (1995) Helix packing of lactose permease in Escherichia coli studied by site-directed chemical cleavage. Proc. Natl Acad. Sci. USA, 92, 9186–9190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Voss,J., Hubbell,W.L. and Kaback,H.R. (1996) Site-directed spin labeling and chemical crosslinking demonstrate that helix V is close to helices VII and VIII in the lactose permease of Escherichia coli. Proc. Natl Acad. Sci. USA, 93, 10123–10127. [DOI] [PMC free article] [PubMed] [Google Scholar]