Abstract
Determining how chromatin is remodelled during early development, when totipotent cells begin to differentiate into specific cell types, is essential to understand how epigenetic states are established. An important mechanism by which chromatin can be remodelled is the replacement of major histones with specific histone variants. During early mammalian development H2A.Z plays an essential, but unknown, function(s). We show here that undifferentiated mouse cells of the inner cell mass lack H2A.Z, but upon differentiation H2A.Z expression is switched on. Strikingly, H2A.Z is first targeted to pericentric hetero chromatin and then to other regions of the nucleus, but is excluded from the inactive X chromosome and the nucleolus. This targeted incorporation of H2A.Z could provide a critical signal to distinguish constitutive from facultative heterochromatin. In support of this model, we demonstrate that H2A.Z can directly interact with the pericentric heterochromatin binding protein INCENP. We propose that H2A.Z functions to establish a specialized pericentric domain by assembling an architecturally distinct chromatin structure and by recruiting specific nuclear proteins.
Keywords: early mammalian development/H2A.Z/inactive X chromosome/inner centromere protein/pericentric heterochromatin
Introduction
It has become clear that eukaryotic gene regulation, at the level of transcription, is connected to the structural organization of the genome. The basis for this structural organization is chromatin. Although it has been established that chromatin plays a fundamental role in maintaining regulated gene expression, very little is known about the chromatin structural and compositional changes that occur in early mammalian development when active and silenced chromosomal domains are established.
The fundamental unit of chromatin, the nucleosome core, is a multi-subunit structure consisting of four different histone types. Each histone has the potential to be differentially post-translationally altered by a number of different types of modifications (Strahl and Allis, 2000). This enables the structure and function of the nucleosome, and higher-order structures, to be regulated in a very precise way. It has been proposed that histone modifications represent an important epigenetic mechanism for the establishment and the long-term maintenance of specialized chromatin structures (Lachner and Jenuwein, 2002).
One such modification, methylation at Lys9 of histone H3, has been shown to be an essential signal for the formation of heterochromatin from yeast to mammals (Lachner and Jenuwein, 2002). In fission yeast, a stepwise model has been proposed for the formation of heterochromatin where Lys9 is first deacetylated and then methylated (Nakayama et al., 2001). Fundamental to histone H3–K9 methylation providing a stable epigenetic mark is its ability to recruit the nuclear protein HP1 (Lachner et al., 2001). Another type of histone modification, histone acetylation, appears to have a dual role. It can modulate chromatin structure by inhibiting the formation of the 30 nm fibre (Tse et al., 1998) and by facilitating the recruitment of remodelling complexes (Horn and Peterson, 2001). Whether other forms of chromatin modifications can have such a dual role is not known.
Another equally important way to control chromatin function is to alter the biochemical make-up of the nucleosome by replacing an individual histone with a histone variant. Interestingly, most histone variants belong to the histone H2A family, suggesting that H2A plays a unique role in the nucleosome. Modulating nucleosomal and higher-order chromatin structure through variation in H2A will have an impact on all nuclear functions.
Several different H2A histone variants with specialized functions have been described. One histone variant, H2AX, appears to play a fundamental role in the repair of DNA double-strand breaks (Paull et al., 2000). Another member of the histone H2A variant family, macroH2A1.2, is concentrated in the inactive X chromosome of adult female mammals. Recent evidence suggests that macroH2A1.2 plays an important role in X chromosome transcriptional silencing in extra-embryonic lineages, since accumulation of this histone variant in pre-implantation embryos is the earliest marker of the inactive X chromosome (Costanzi et al., 2000). On the other hand, in embryonic lineages, macroH2A1.2 enrichment is a relatively late event (Rasmussen et al., 2000).
The H2A.Z variant family of proteins is highly conserved (across species the amino acid sequence of H2A.Z is more conserved than the amino acid sequence of major H2A), suggesting that H2A.Z plays an important role in chromosome function (van Daal et al., 1988; White and Gorovsky, 1988; Iouzalen et al., 1996). Consistent with this proposal, H2A.Z has been shown to be essential in Drosophila (van Daal and Elgin, 1992), Tetrahymena (Liu et al., 1996) and mice (Faast et al., 2001). H2A.Z-deficient mouse embryos die early in development, around the time of implantation. It is therefore clearly established that H2A.Z performs a unique role(s) in metazoans, which cannot be substituted for by H2A (Jackson and Gorovsky, 2000). This essential function(s) of H2A.Z remains to be elucidated.
Previously, we performed studies in Drosophila and identified the docking domain of H2A.Z, located in its C-terminus, as being the region required for its specialized role(s) (Clarkson et al., 1999). The crystal structure of a nucleosome containing H2A.Z revealed that a significant portion of this altered docking domain of H2A.Z is exposed on the surface of the nucleosome (Suto et al., 2000). We therefore proposed that one important function of H2A.Z is to recruit a specific nuclear factor(s) and/or to modulate nucleosome–nucleosome interactions and the subsequent formation of high-order chromatin structures. Supporting this hypothesis, we recently demonstrated that H2A.Z could facilitate nucleosome–nucleosome interactions within the chromatin fibre to assemble a higher-order compacted domain in vitro (Fan et al., 2002).
The present study investigates the function of H2A.Z by examining whether it is involved in the formation of compacted chromosomal domains during early mouse development at a time when H2A.Z–/– mice die. Strikingly, major chromatin remodelling events occur during the earliest stages of differentiation since H2A.Z is absent from the nuclei of totipotent inner cell mass (ICM) cells, but its expression is switched on upon differentiation. Interestingly, as H2A.Z expression is switched on, it is first targeted to pericentric heterochromatin before localizing to other regions in the nucleus. In contrast, the inactive X chromosome is depleted in H2A.Z. This result raises the possibility that H2A.Z provides a specific signal for the assembly of an architecturally distinct structure at pericentric heterochromatin. Supporting the proposal that H2A.Z marks pericentric heterochromatin, we demonstrate that H2A.Z interacts directly with the pericentric heterochromatin-binding protein INCENP (a protein critical for chromosome segregation).
Results
H2A.Z expression is switched on during differentiation
H2A.Z null mouse embryos die shortly after hatching and implantation, suggesting a critical role of H2A.Z at this stage (Faast et al., 2001). To begin to understand the role of H2A.Z during early development, the cellular and nuclear distribution of H2A.Z was investigated employing immunofluorescent techniques. At 3.5 days post-coitum (d.p.c.) mouse blastocysts were collected and cultured under conditions where they hatched and attached onto coverslips. During in vitro culture, the trophoblast spreads out to form an adherent monolayer of cells while the undifferentiated cells of the ICM expand and outgrow to form a dense mound of cells (Abbondanzo et al., 1993). Mimicking an early differentiating step during mouse development, the cells located at the outer edge of the ICM mound differentiate into a layer of extraembryonic endoderm (Handyside and Barton, 1977; Nichols and Gardner, 1984).
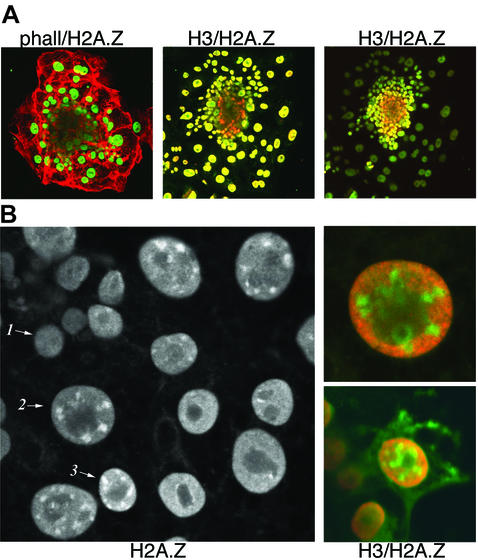
Immunofluorescent staining of these early mouse embryos revealed the presence of H2A.Z in the nuclei of trophoblast cells and in endoderm nuclei located around the edge of the ICM. Surprisingly, H2A.Z was not detected in ICM cells (Figure 1A). Co-immunostaining of H2A.Z with phallodium (which binds to filamentous actin) revealed trophoblast nuclei containing H2A.Z and a ‘black hole’ lacking H2A.Z at the centre of the ICM (Figure 1A). On the other hand, di-acetylated histone H3 is present in ICM nuclei as well as in the nuclei of trophoblast and endoderm cells (Figure 1A).
Fig. 1. H2A.Z expression is switched on when the ICM begins to differentiate. (A) Early mouse embryos were stained with phallodium (red) and immunostained with H2A.Z antibodies (green) or co-immunostained with diacetylated histone H3 (red) and H2A.Z (green) antibodies. Both antibodies were used at a 1:1000 dilution. The centre and right panels display two different confocal views highlighting trophoblast nuclei, and the ICM plus endoderm nuclei, respectively. The absence of H2A.Z at the ICM (red) and the co-localization of H2A.Z and diacetylated histone H3 at trophoblast and endoderm cells (yellow) can be seen clearly. (B) Immunostaining of ICM and endoderm nuclei at the periphery of the ICM with H2A.Z antibodies. An ICM nucleus (cell-1), an endoderm nucleus (cell-3) and an apparent differentiation intermediate between these two cell types (cell-2) are shown. Other cells in this image, which are also located at the edge of the ICM mound, are not in the same confocal plane. The right panels show cell-2, and a similar cell type, co-immunostained with H2A.Z (green) and diacetylated histone H3 (red) antibodies.
To confirm this observation, cells of the ICM and trophoblast cells were isolated, RNA extracted and semi-quantitative RT–PCRs were carried out. Consistent with the immunostaining data, H2A.Z transcripts are ∼12-fold reduced in the ICM compared with trophoblasts (Figure 2). As controls, we also confirmed that endo-β-N-acetyl glucosaminidase and fibroblast growth factor transcript levels are significantly higher in trophoblast cells and the ICM, respectively (Brison and Schultz, 1996). Taken together, these results indicate that H2A.Z is not, or poorly, expressed in ICM cells but its expression is swtiched on when ICM cells begin to differentiate.
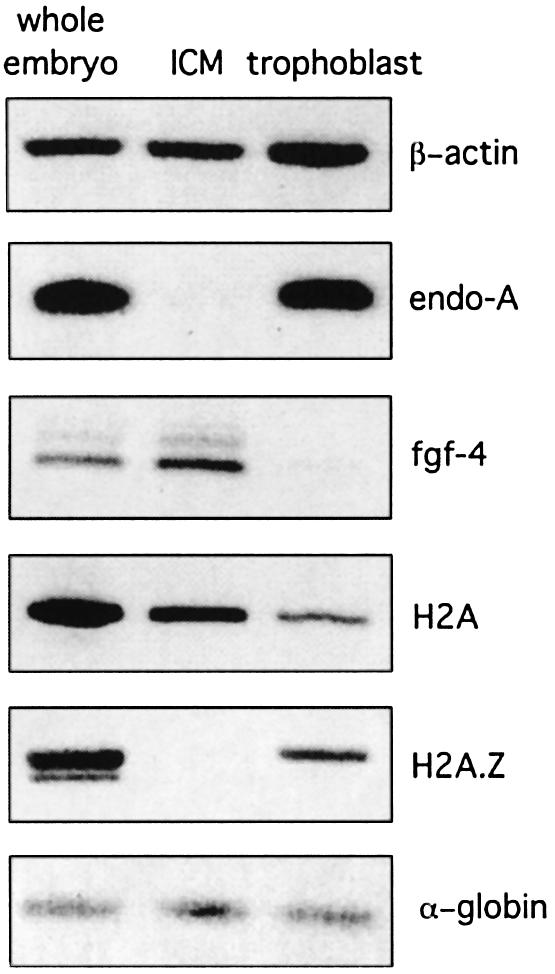
Fig. 2. Analysis of H2A.Z and H2A transcript levels in the ICM. Transcript levels of β-actin, endo-β-N-acetylglucosaminidase, fibroblast growth factor, H2A and H2A.Z were determined by semi-quantitative RT–PCR as described in Materials and methods. Approximately 32 whole embryos, 90 ICMs and 130 trophoblast samples were used to yield approximately the same total number of cells. Rabbit α-globin mRNA was used as an internal control to correct for differences between samples in RNA recovery and efficiency of RT–PCR.
Closer examination of trophoblast and endoderm cells revealed a second novel observation. H2A.Z is not only located throughout the nucleus in these cells types, but it is also enriched at specific foci (Figures 1B and 3). Figure 1B shows an ICM cell lacking H2A.Z (cell-1), and an endoderm cell containing H2A.Z throughout the nucleus (excluding the nucleolus) as well as located at distinct foci (cell-3). These cells were located at the outer edge of the ICM, where endoderm cells form (Handyside and Barton, 1977; Nichols and Gardner, 1984). Intriguingly, another cell type was identified displaying a different H2A.Z nuclear localization pattern. In these cell types, H2A.Z was only located at these distinct foci and not throughout the nucleus (cell-2). This is seen more clearly when H2A.Z and acetylated histone H3 are co-localized in cell-2 (Figure 1B, right panel). In contrast to H2A.Z, acetylated H3 is excluded from these foci but is distributed throughout the rest of the nucleus. Therefore, cell-2 may be a differentiation intermediate in the formation of an endoderm cell.
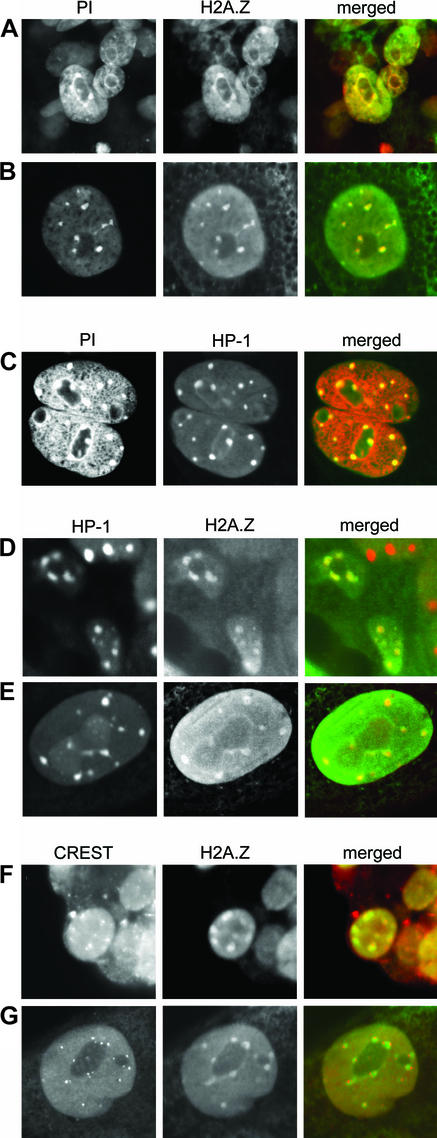
Fig. 3. H2A.Z and HP1-α co-localize at pericentric heterochromatin. Early mouse embryos were immunostained with H2A.Z (1:1000), HP1-α (1:250), and CREST (1:500) antibodies or stained with propidium iodide (PI). The images show extraembryonic endoderm cells (A, D and F) and trophoblast cells (B, C, E and G). For the merged panel, the first and second panels are artificially coloured red and green, respectively. Undifferentiated cells of the ICM are also seen next to endoderm cells in (D, upper right).
H2A.Z is targeted specifically to pericentric heterochromatin
H2A.Z was located at distinct foci not only in endoderm nuclei (Figure 3A, D and F), but also in the nuclei of trophoblast cells (Figure 3B, E and G). The location of H2A.Z at these foci suggested that it was performing an important function at a specialized chromosomal domain. This function was unrelated to transcription because these foci lacked acetylated histone H3 (Figure 1B) and acetylated H4 (data not shown).
Further immunofluorescence co-localization experiments unequivocally demonstrated that these foci were pericentric heterochromatin, the heterochromatin domain that surrounds the centromere. First, identified by their dense staining with propidium iodide, the domains of pericentric heterochromatin co-localize with H2A.Z (Figure 3A and B). Secondly, the specific heterochromatin binding protein HP1-α (Minc et al., 1999) co-localized precisely with H2A.Z (Figure 3D and E) and with propidium iodide (Figure 3C). Interestingly, in the nuclei of ICM cells that lack H2A.Z, HP1-α foci are still observed (Figure 3D). Finally, co-immunostaining experiments were carried out using H2A.Z and CREST antibodies, an autoimmune serum that recognizes components of the inner kinetochore region including CENPA (Van Hooser et al., 1999). Most importantly, these results show clearly that H2A.Z is not located at the centromere with CENPA but in the heterochromatin domain adjacent to the inner kinetochore region (Figure 3G). The inner kinetochore region can be seen as a doublet, indicating that these trophoblast cells were in the G2 phase of the cell cycle.
H2A is depleted in pericentric heterochromatin
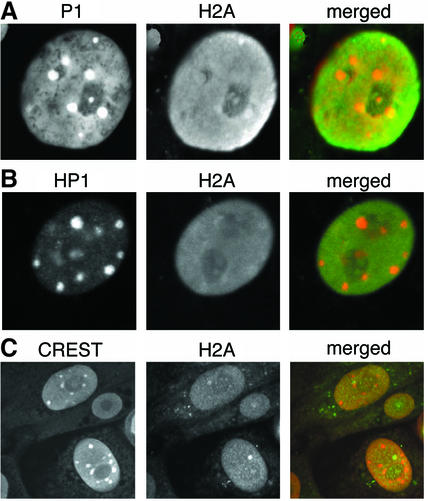
To address the question of whether H2A.Z is actually enriched in pericentric heterochromatin, co-immunostaining experiments were repeated using H2A affinity-purified antibodies. In contrast to H2A.Z, H2A displays a much more uniform distribution in the nuclei of trophoblast (Figure 4) and endoderm cells (data not shown). Surprisingly, very little co-localization is observed with propidium iodide (Figure 4A) and HP1-α (Figure 4B), which is consistent with the absence of H2A foci located between inner kinetochore regions (Figure 4C). Therefore, H2A is either depleted in pericentric heterochromatin or H2A nucleosomal arrays are buried deeper than H2A.Z in pericentric heterochromatin, thus being inaccessible to antibodies. In any event, H2A.Z is indeed enriched in pericentric heterochromatin and its location is distinct from that of H2A.
Fig. 4. H2A is not enriched at pericentric heterochromatin. Mouse trophoblast cells were immunostained with H2A (1:1000), HP1-α (1:250) and CREST (1:500) antibodies, and the DNA was stained with PI. For the merged panel, the first and second panels are artificially coloured red and green, respectively.
The inactive X chromosome is depleted in H2A.Z
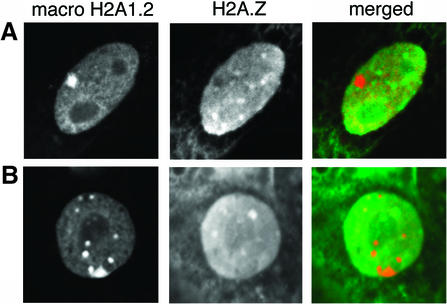
Next, we wanted to investigate whether H2A.Z is located in other heterochromatin domains. It has been shown that in extraembryonic interphase cells, the inactive X chromosome is identified as a large macroH2A1.2 dense domain referred to as macrochromatin body (MCB) (Costanzi et al., 2000). Figure 5A and B shows clearly the location of a single MCB, which was observed in ∼50% of embryos. Most significantly, H2A.Z does not co-localize with macroH2A1.2 at MCBs (Figure 5A and B) or, interestingly, at any other minor foci of macroH2A1.2 (Figure 5B). We conclude that H2A.Z is absent from facultative heterochromatin, at least at a global level, which supports our proposal that H2A.Z performs a specialized function unique to pericentric heterochromatin.
Fig. 5. H2A.Z and macroH2A1.2 do not co-localize. Mouse trophoblast cells were co-immunostained with H2A.Z (1:1000) and macroH2A1.2 (1:100) (Costanzi et al., 2000) antibodies. For the merged panel, the first and second panels are artificially coloured red and green, respectively.
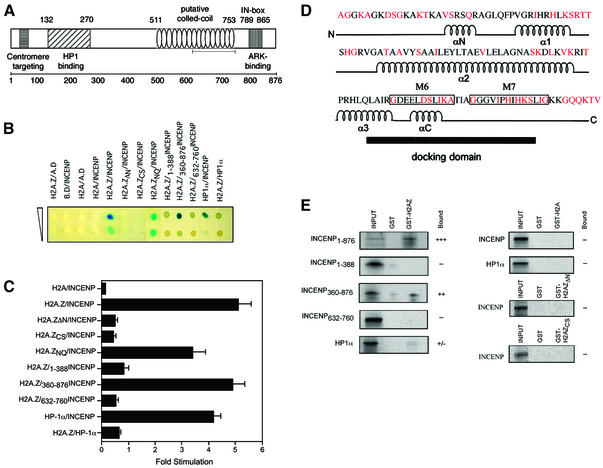
H2A.Z interacts with the pericentric heterochromatin-binding protein INCENP
Since H2A.Z is enriched in constitutive heterochromatin (but depleted in facultative heterochromatin), H2A.Z might provide a distinctive mark, analogous to histone tail modifications, important for pericentric heterochromatin structure and/or function. The identification of a pericentric heterochromatin binding that specifically interacts with H2A.Z would provide strong support for this hypothesis. To identify novel H2A.Z-interacting proteins, we used a GAL4-based yeast two-hybrid system. Mouse H2AZ cDNA was fused with the DNA binding domain of GAL4 to create the ‘bait’ construct. The ‘prey’ construct was a mouse embryo cDNA library fused with the activation domain of GAL4. Four positive colonies were ultimately identified; two of these clones encoded a truncated version of mouse INCENP. When an identical experiment was carried out using mouse H2A as the bait, no meaningful positive clones were identified.
To confirm this in vivo interaction between H2A.Z and INCENP, full-length INCENP (Figure 6A) was cloned into the prey construct and the yeast two-hybrid analysis was repeated. Following selection on minimal media, the in vivo interaction between H2A.Z and INCENP was analysed using two different assays. This interaction was visualized using a β-galactosidase agarose overlay assay (Figure 6B) and quantified by measuring the induction of the β-galactosidase reporter gene using a liquid culture assay (Figure 6C). Whereas no interaction between H2A and INCENP was observed, Figure 6B and C clearly shows an interaction between H2A.Z and full-length INCENP. We also confirmed the interaction reported previously between the specific pericentric heterochromatin binding protein HP1-α (Minc et al., 1999) and INCENP (Ainsztein et al., 1998) (Figure 6B and C). This interaction between H2A.Z and INCENP is significant, since the strength of this association is similar to that of the interaction between INCENP and HP1-α (Figure 6C).
Fig. 6. H2A.Z interacts with the pericentric heterochromatin binding protein INCENP. (A) Schematic showing the regions of INCENP shared by vertebrates (Uren et al., 2000; Adams et al., 2001b,c). (B) The interaction between H2AZ and INCENP, and various mutations of these two proteins, were analysed using a yeast two-hybrid approach and visualized using an agarose overlay assay. Undiluted cultures and cultures diluted 1 in 10 are shown. (C) The quantification of protein–protein interactions using a β-galactosidase liquid culture assay. The relative fold stimulation was calculated from the β-galactosidase activities of individual co-transformants using the equation [(pGBT7X + pACT2Y) – (pGBT7 + pACT2Y)] / (pGBT7X+pACT2), where X is H2AZ and Y is INCENP protein. The average of three independent experiments are shown. ‘Bait’ constructs included the GAL4 DNA binding domain alone (B.D.) or fused with H2A, H2A.Z, H2A.ZΔN (the N-terminus removed), H2A.ZCS (the C-terminal α-helix of H2A.Z swapped with the equivalent region of H2A), or with H2A.ZNQ (the two evolutionarily conserved histidine residues at positions 112 and 114 changed to asparagine and glutamine residues, which are the identically positioned amino acid residues in H2A). ‘Prey’ constructs included the activation domain by itself (A.D.) or fused with full-length INCENP, or with different deletion mutants. A ‘prey’ construct was also generated that contained the activation domain fused with HP1-α. (D) The amino acid sequence and secondary structure of mouse H2A.Z are shown. Amino acid residues that differ from H2A are shown in red. The docking domain, encompassing the essential M6 and M7 regions, is also shown. (E) Either GST, GST–H2A.Z, GST–H2A.ZΔN, GST–H2A.ZCS or GST–H2A were expressed in E.coli, bound to glutathione–agarose and, following purification, incubated with in vitro-labelled translation products of INCENP and various deletion mutants. Histone H2B was included in all binding reactions.
To determine which region of INCENP interacts with H2A.Z in vivo, various deletion mutants of INCENP were cloned into the ‘prey’ vector, and the interaction of these INCENP mutants with H2A.Z were also analysed using β-galactosidase overlay and liquid assays (Figure 6B and C). Whereas the N-terminus of INCENP (amino acids 1–388, which encompass the binding region for HP1-α and the poorly conserved centromere targeting domain) is not required for H2A.Z interaction, amino acids 360–876 are needed. Within this C-terminal region, the microtubule-binding domain (amino acids 632–760) (Mackay et al., 1998) is not responsible for the interaction with H2A.Z.
Amino acid swap experiments in Drosophila revealed that a region (M6 and M7) in the C-terminus of H2A.Z, which overlaps with docking domain (Luger et al., 1997; Suto et al., 2000), is essential for adult fly survival (Clarkson et al., 1999) (Figure 6D). The M6 sub-region, which encompasses the C-terminal α-helix of H2A.Z, is especially important because it plays a crucial role in early Drosophila development. Most significantly, swapping the M6 sub-region of H2A.Z with the identically positioned amino acids in H2A (H2A.ZCS) does inhibit the interaction between H2A.Z and INCENP (Figure 6B and C). On the other hand, the two highly conserved histidine residues in the M7 sub-region are not required for this interaction (H2A.ZNQ). We therefore conclude that the same region identified in flies as being essential for Drosophila development is required for the interaction between H2A.Z and INCENP. Interestingly, removal of the N-terminus of H2A.Z also prevents the interaction with INCENP (Figure 6B and C). We propose that the N-terminus may play a stabilizing role in this association, perhaps mediated by a non-specific ionic interaction (Ren and Gorovsky, 2001).
To verify this interaction between H2A.Z and INCENP in the yeast nucleus, in vitro pull-down assays were performed. Glutathione S-transferase (GST)–H2A, GST–H2A.Z, GST–H2A.ZΔN and GST–H2A.ZCS fusion proteins were expressed and purified from Escherichia coli. Full-length INCENP, and various INCENP mutant proteins, were expressed and labelled in vitro using reticulocyte lysates. The results presented in Figure 6E demonstrate that, under stringent binding conditions, H2A.Z and INCENP can interact with each other in vitro. In addition, these data confirm the results from the yeast two-hybrid analysis, which showed that amino acids 360–876 of INCENP (but not the microtubule binding domain by itself), and the N-terminus and the C-terminal α-helix of H2A.Z, are required for the interaction. Interestingly, although no interaction between H2A.Z and HP1-α was observed in the yeast two-hybrid analysis (Figure 6B and C), a weak but reproducible interaction was observed between GST–H2A.Z and HP1-α in vitro (Figure 6E). The ability of H2A.Z to interact with a nuclear protein, INCENP, provides a new example of how histone variants can potentially modify the structure and function of chromatin and supports our proposal that H2A.Z may be a marker that distinguishes constitutive heterochromatin form facultative heterochromatin.
Discussion
H2A.Z performs an essential function during early mammalian development but this role(s) has not yet been elucidated. Our previous structural studies suggested that H2A.Z might be involved in establishing specialized higher-order chromatin domains (Suto et al., 2000; Fan et al., 2002). Immunofluorescence analyses performed here demonstrate that H2A.Z is enriched in pericentric heterochromatin. Moreover, the specific targeting of H2A.Z to pericentric heterochromatin and the depletion of H2A.Z in the inactive X chromosome, argue that H2A.Z may provide an important signal for the establishment of a condensed pericentric domain when cells of the ICM begin to differentiate. Consistent with a model where H2A.Z marks pericentric heterochromatin is the novel finding that H2A.Z, but not H2A, can interact with the pericentric heterochromatin-binding protein INCENP.
The composition of pericentric heterochromatin changes during development becoming enriched with H2A.Z
A remarkable finding is that H2A.Z is absent in cells of the ICM and that its expression is switched on as these totipotent cells differentiate into extraembryonic endoderm cells. This novel observation shows that major chromatin remodelling events occur during early differentiation and raises the distinct possibility that incorporation of H2A.Z into chromatin, especially constitutive heterochromatin may be one of the steps that leads to the loss of totipotency.
Interestingly, in addition to ICM cells, two other cells types were observed that differed in their nuclear localization of H2A.Z. Endoderm cells have H2A.Z located throughout the nucleus (except for the nucleolus) and enriched at pericentric heterochromatin. The third cell type identified had H2A.Z located only at pericentric heterochromatin. Since this cell type was located between ICM cells and the endoderm layer, we suggest that it may be a differentiation intermediate, leading to the formation of endoderm cells. Clearly, during early differentiation, incorporation of H2AZ into different chromosomal regions occurs as a staged process.
The incorporation of H2A.Z into pericentric heterochromatin before other regions of the nucleus, and the finding that H2A is depleted in pericentric heterochromatin, argues that it performs an important function at this domain (see below). In fission yeast, it is proposed that the formation of pericentric heterochromatin is a sequential process, with H3-K9 being first deacetylated and then methylated. Methylated H3-K9 provides an important epigenetic mark by subsequently recruiting HP1, which establishes and maintains pericentric heterochromatin in a condensed state (Nakayama et al., 2001). In mouse embryonic stem cells (which are derived from cells of the ICM), histone H4 in pericentric heterochromatin is acetylated and the DNA within this domain is undermethylated (Keohane et al., 1996). It will be of interest to determine the timing of events, and the relationship between H2A.Z incorporation, deacetylation of histone H4, methylation of histone H3 and DNA methylation during the formation of pericentric heterochromatin when the ICM begins to differentiate. Interestingly, we found that HP1 is associated with ICM pericentric heterochromatin despite this domain being acetylated.
As discussed above, H2A.Z is not present in ICM pericentric heterochromatin. This indicates that the structure and function of pericentric heterochromatin is different in totipotent cells of the ICM. Perhaps, in the absence of H2A.Z, this heterochromatin domain is less condensed, which may be necessary for the rapid rate of cell division observed for the ICM (Abbondanzo et al., 1993). Interestingly, in a human condition known as ICF immuno deficiency syndrome, the replication timing of pericentric heterochromatin is advanced (Hassan et al., 2001). Peri centric heterochromatin associated with this syndrome is less condensed and its DNA is hypomethylated.
H2A.Z might be an important signal distinguishing constitutive heterochromatin from facultative heterochromatin
The critical signals that distinguish the formation and maintenance of pericentric heterochromatin and the inactive X chromosome are far from being understood. For example, both of these heterochromatin types display methylation of K9 in histone H3, yet HP1 only binds to pericentric heterochromatin. Clearly, other signals are required for the recruitment of HP1 and the assembly of a condensed pericentric heterochromatin structure.
Our observation that H2A.Z is targeted to pericentric heterochromatin during early development and that it is absent from the inactive X chromosome, raises the possibility that H2A.Z may be an important signal for the formation of constitutive heterochromatin. In vitro experiments are in progress to investigate whether a H2A.Z-containing nucleosomal array is a more suitable scaffold for HP1 recruitment. Further support for our proposal that H2A.Z may provide a mark to distinguish constitutive heterochromatin from facultative heterochromatin is the observation that H2A.Z can directly interact with INCENP, a pericentric heterochromatin binding protein. The basis of the ‘histone code’ hypothesis is the ability of histone tail modifications to recruit different specific nuclear proteins.
The inactive X chromosome is enriched with macroH2A. An unanswered question is whether different histone H2A types can co-exist in the same nucleosome. The observation that macroH2A and H2A.Z do not co-localize suggests that these histone variants cannot be assembled into the same nucleosome. Although other mechanisms may operate, such as the existence of specific chaperones, we propose that these two histone variants are unable to co-exist because of structural considerations. Within the same nucleosome, pairing of one H2A.Z and one macroH2A polypeptide chain could lead to a steric clash in the L1 loop-L1′ loop region (Suto et al., 2000).
The role of H2AZ at pericentric heterochromatin
Pericentric heterochromatin plays at least two important roles in chromosome segregation and cytokinesis. First, the condensed nature of pericentric heterochromatin provides mechanical strength necessary to withstand the forces associated with the chromosome segregation process (Rieder and Salmon, 1998). Secondly, it serves as a recruitment area for many chromosomal binding proteins involved in chromosome cohesion, segregation and cytokinesis such as cohesin (Nonaka et al., 2002) and INCENP (Ainsztein et al., 1998; Adams et al., 2001a).
Our recent biophysical study on the folding properties of H2A.Z-containing nucleosomal arrays is consistent with H2A.Z having a structural role at pericentric heterochromatin (Fan et al., 2002). H2A.Z facilitated intra-nucleosome interactions within the nucleosomal fibre to generate a more compacted structure. Although compacted, H2A.Z prevented the subsequent formation of highly condensed structures by inhibiting inter-nucleo somal fibre–fibre interactions. In other words, H2A.Z generated a fibre that was a conformational intermediate in the chromatin-folding pathway. Such an intermediate would still be accessible to the binding of chromosomal proteins. In addition, compared with H2A nucleosomal arrays, this study also found that H2A.Z nucleosomes were positioned more regularly along the DNA template. Such regular positioning is an important characteristic of pericentric heterochromatin (Wallrath and Elgin, 1995). Totally consistent with this proposed role of H2A.Z, a recent study employing analytical sucrose centrifugation demonstrated that pericentric heterochromatin isolated from living cells was more condensed than bulk chromatin, but less condensed than centromeric heterochromatin containing CENPA (Gilbert and Allan, 2001). It is also worth noting that in budding yeast, it was shown that H2A.Z is involved in the establishment of a silenced domain at the HMR loci (Dhillon and Kamakaka, 2000).
In addition to having a structural role, pericentric heterochromatin also functions as a docking site for a number of different nuclear proteins such as INCENP. INCENP is a chromosomal passenger protein required for proper chromosome segregation and cytokinesis (Terada, 2001). Recent studies have shown that INCENP is in a macromolecular complex with Aurora B kinase, which is responsible for mitotic histone H3 phosphorylation, and Survivin (Adams et al., 2000, 2001a). How INCENP is targeted to pericentric heterochromatin at the onset of metaphase (Ainsztein et al., 1998; Adams et al., 2001a) is not known. We propose that, in addition to having a structural function, H2A.Z may have a second role in creating a docking site for INCENP. The finding that both HP1-α and H2A.Z co-localize at pericentric heterochromatin, and that both of these proteins interact with different regions of INCENP, raises the interesting possibility that they may form a combined docking pad for the recruitment of the INCENP complex. Consistent with this hypothesis, the fidelity of minichromosome segregation in fission yeast decreased markedly when the H2A.Z gene was deleted (Carr et al., 1994).
Finally, an important question is whether H2A.Z has other functions. The work of others has suggested that H2A.Z may facilitate the transcription process in budding yeast (Santisteban et al., 2000; Adam et al., 2001) and in Tetrahymena (Allis et al., 1986; Stargell et al., 1993). Whether H2A.Z functions to activate or silence transcription may depend upon the nature of the nuclear factor(s) recruited to H2A.Z-containing nucleosomes.
Materials and methods
Immunofluorescence and culture of early mouse embryos
H2A and H2A.Z antisera were raised in sheep using a synthetic peptide equivalent to the unique C-terminal 19 amino acids of mouse H2A and H2A.Z, respectively. Importantly, the observed immunostaining patterns of H2A.Z and H2A were blocked when affinity purified antibodies where first incubated with recombinant H2A.Z and H2A, respectively (data not shown). Di-acetylated histone H3 antibodies were obtained from Upstate Biotechnology. Approximately 3.5 d.p.c. mouse blastocysts were collected from superovulated C57BL6 female mice and cultured in Dulbecco’s modified Eagle’s medium (DMEM; for ∼5 days) until the embryos hatched, attached and out-grew on the coverslip (Abbondanzo et al., 1993). The embryos were fixed in 4% paraformaldehyde for 20 min, permeabilized in 0.1% SDS/PBS/1% BSA for 15 min, then blocked in PBS/1% BSA for 1 h. All antibodies were incubated overnight at 4°C. Bound antibodies were visualized using Texas Red- or FITC-labelled secondary antibodies (Jackson Immunoresearch). Cells were examined using a Leica (Germany) confocal fluorescence microscope. For propidium iodide (PI) staining, embryos were incubated for 1 h with freshly prepared RNase (200 µg/ml) and PI (20 µg/ml). Between steps, embryos were washed with PBS/1% BSA.
RT–PCR-based method to semi-quantiate the expression of the H2AZ gene in early mouse embryos
ICM and trophoblast cells were isolated as described previously (Temeles et al., 1994). Special care was taken to ensure that ICM cells were free from contaminating trophoblast and endoderm cells. Following washing of embryos grown in culture for 5 days, with DMEM media containing 3 mg/ml polyvinylpyrrolidone, only the central half of the ICM cone was surgically removed. For isolation of trophoblast cells, the remaining ICM cone was completely removed surgically before isolation of adherent trophoblast cells by gently scraping with siliconized fine-grade glass needles. Whole embryos were also removed by scraping. All samples were washed once with DMEM containing 3 mg/ml polyvinylpyrrolidone before being frozen immediately and stored at –70°C.
Isolation of mRNA, RT–PCR conditions, and the primers to detect α-globin, β-actin, endo-β-N-acetylglucosaminidase and fibroblast growth factor transcript levels, have been described previously (Brison and Schultz, 1996). Rabbit α-globin mRNA (0.1 pg; BRL) was added following lysis of the different cell types, which served as an internal control to correct for differences between samples in RNA recovery and efficiency of RT–PCR. Both the α-globin transcript and the mRNA of interest were co-amplified. The forward primers for H2A.Z and H2A, respectively, were 5′-ATGGCTGGCGGTAAGGCTGGAAAG-3′ and 5′-ATGTCTGGACGCGGCAAGCAGGGTG-3′. The reverse primers for H2A.Z and H2A, respectively, were 5′-AAACAGTCTTCTGTTGTCC-3′ and 5′-TTATTTTCCCTTGGCCTTGTGG-3′.
Yeast two-hybrid analysis and in vitro binding assays
Mouse H2A.Z and H2A cDNA were fused with the DNA binding domain of GAL4 by cloning into the NdeI–BamHI sites of pGBT7, and these ‘bait’ constructs were transformed into the yeast strain, AH109 (MATa). The ‘prey’ construct, a 17 d.p.c. mouse embryo cDNA library fused with the activation domain of GAL4 (pACT2), was transformed into yeast strain Y187 (MATα). Following a yeast mating, ∼2 × 106 yeast transformants were screened on selective conditions. Four positive colonies were isolated on selective minimal media (SD/–Ade/–His/– Trp/–Leu/40 mM 3-amino-1, 2,4-triazole) and identified using the methods described by the supplier (Clontech). Following the identification of INCENP from this screen, full-length and truncations of INCENP cDNA (generated by PCR) where cloned into the NcoI–XhoI sites of pACT2. Following site-directed mutagenesis of H2A.Z cDNA (Stratagene) and truncation of amino acid residues 1–16, these mutated H2AZ constructs (in pET15b) were cloned into NdeI–BamHI sites of pGBT7. The in vivo interaction between these ‘prey’ and ‘bait’ constructs were visualized using a β–galactosidase agarose overlay assay and quantitated employing a β–galactosidase liquid culture assay as described by Clontech. Chlorophenol red-β-d-galactopyranoside was used as the substrate. For in vitro binding assays, GST, GST–H2A.Z, GST–H2A.ZΔN, GST–H2A.ZCS or GST–H2A fusion proteins (wild-type and mutated cDNA clones were inserted into pGEX-4T-1) were expressed and purified from E.coli BL21. INCENP and truncations thereof were inserted into pET15b. In vitro transcription and translation reactions were performed with [35S]methionine using the TNT T7 system (Promega). GST pull-down assays were carried out as described previously (Ainsztein et al., 1998), except that the beads were washed five times with more stringent TSA buffer (10 mM Tris–HCl, pH 8.0, 140 mM NaCl, 0.025% NaN3, 0.05% Triton X-100) and bound fractions were visualized and quantitated on a phosphoimager. It is worth noting that purified H2B was added to these binding reactions, since this enhanced the interaction between INCENP and H2A.Z (core histones, including H2A.Z, remain unfolded until they dimerize) (data not shown).
Acknowledgments
Acknowledgements
We thank the following for providing important reagents: Michael Clarkson for generating H2A.Z and H2A antibodies; Jeff Craig and Andy Choo for HP1-α and CREST antibodies; John Pehrson for macroH2A1 antibodies; Lori Wallrath for the cDNA for HP1-α; and David Vaux for giving us the full-length INCENP cDNA clone. We are also grateful to Klaus Matthaei and Helen Taylor for providing us with mouse blastocysts. This work was supported by a Human Frontiers Science program grant to D.J.T., and a NHMRC grant to P.R. and D.J.T.
References
- Abbondanzo S.J., Gadi,I. and Stewart,C.L. (1993) Derivation of embryonic stem cell lines. Methods Enzymol., 225, 803–823. [DOI] [PubMed] [Google Scholar]
- Adam M., Robert,F., Larochelle,M. and Gaudreau,L. (2001) H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol. Cell. Biol., 21, 6270–6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams R.R., Wheatley,S.P., Gouldsworthy,A.M., Kandels-Lewis,S.E., Carmena,M., Smythe,C., Gerloff,D.L. and Earnshaw,W.C. (2000) INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol., 10, 1075–1078. [DOI] [PubMed] [Google Scholar]
- Adams R.R., Carmena,M. and Earnshaw,W.C. (2001a) Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol., 11, 49–54. [DOI] [PubMed] [Google Scholar]
- Adams R.R., Eckley,D.M., Vagnarelli,P., Wheatley,S.P., Gerloff,D.L., Mackay,A.M., Svingen,P.A., Kaufmann,S.H. and Earnshaw,W.C. (2001b) Human INCENP colocalizes with the Aurora-B/AIRK2 kinase on chromosomes and is overexpressed in tumour cells. Chromosoma, 110, 65–74. [DOI] [PubMed] [Google Scholar]
- Adams R.R., Maiato,H., Earnshaw,W.C. and Carmena,M. (2001c) Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction and chromosome segregation. J. Cell Biol., 153, 865–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainsztein A.M., Kandels-Lewis,S.E., Mackay,A.M. and Earnshaw,W.C. (1998) INCENP centromere and spindle targeting: identification of essential conserved motifs and involvement of heterochromatin protein HP1. J. Cell Biol., 143, 1763–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis C.D., Richman,R., Gorovsky,M.A., Ziegler,Y.S., Touchstone,B., Bradley,W.A. and Cook,R.G. (1986) hv1 is an evolutionarily conserved H2A variant that is preferentially associated with active genes. J. Biol. Chem., 261, 1941–1948. [PubMed] [Google Scholar]
- Brison D.R. and Schultz,R.M. (1996) RT–PCR-based method to localize the spatial expression of genes in the mouse blastocyst. Mol. Reprod. Dev., 44, 171–178. [DOI] [PubMed] [Google Scholar]
- Carr A.M., Dorrington,S.M., Hindley,J., Phear,G.A., Aves,S.J. and Nurse,P. (1994) Analysis of a histone H2A variant from fission yeast: evidence for a role in chromosome stability. Mol. Gen. Genet., 245, 628–635. [DOI] [PubMed] [Google Scholar]
- Clarkson M.J., Wells,J.R., Gibson,F., Saint,R. and Tremethick,D.J. (1999) Regions of variant histone His2AvD required for Drosophila development. Nature, 399, 694–697. [DOI] [PubMed] [Google Scholar]
- Costanzi C., Stein,P., Worrad,D.M., Schultz,R.M. and Pehrson,J.R. (2000) Histone macroH2A1 is concentrated in the inactive X chromosome of female preimplantation mouse embryos. Development, 127, 2283–2289. [DOI] [PubMed] [Google Scholar]
- Dhillon N. and Kamakaka,R.T. (2000) A histone variant, Htz1p and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol. Cell, 6, 769–780. [DOI] [PubMed] [Google Scholar]
- Faast R. et al. (2001) Histone variant H2A.Z is required for early mammalian development. Curr. Biol., 11, 1183–1187. [DOI] [PubMed] [Google Scholar]
- Fan J.Y., Gordon,F., Luger,K., Hansen,J.C. and Tremethick,D.J. (2002) The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat. Struct. Biol., 9, 172–176. [DOI] [PubMed] [Google Scholar]
- Gilbert N. and Allan,J. (2001) Distinctive higher-order chromatin structure at mammalian centromeres. Proc. Natl Acad. Sci. USA, 98, 11949–11954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handyside A.H. and Barton,S.C. (1977) Evaluation of the technique of immunosurgery for the isolation of inner cell masses from mouse blastocysts. J. Embryol. Exp. Morphol., 37, 217–226. [PubMed] [Google Scholar]
- Hassan K.M., Norwood,T., Gimelli,G., Gartler,S.M. and Hansen,R.S. (2001) Satellite 2 methylation patterns in normal and ICF syndrome cells and association of hypomethylation with advanced replication. Hum. Genet., 109, 452–462. [DOI] [PubMed] [Google Scholar]
- Horn P.J. and Peterson,C.L. (2001) The bromodomain: a regulator of ATP-dependent chromatin remodeling? Front. Biosci., 6, D1019–D1023. [DOI] [PubMed] [Google Scholar]
- Iouzalen N., Moreau,J. and Mechali,M. (1996) H2A.ZI, a new variant histone expressed during Xenopus early development exhibits several distinct features from the core histone H2A. Nucleic Acids Res., 24, 3947–3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J.D. and Gorovsky,M.A. (2000) Histone H2A.Z has a conserved function that is distinct from that of the major H2A sequence variants. Nucleic Acids Res., 28, 3811–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keohane A.M., O’Neill L,P., Belyaev,N.D., Lavender,J.S. and Turner,B.M. (1996) X-inactivation and histone H4 acetylation in embryonic stem cells. Dev. Biol., 180, 618–630. [DOI] [PubMed] [Google Scholar]
- Lachner M. and Jenuwein,T. (2002) The many faces of histone lysine methylation. Curr. Opin. Cell Biol., 14, 286–298. [DOI] [PubMed] [Google Scholar]
- Lachner M., O’Carroll,D., Rea,S., Mechtler,K. and Jenuwein,T. (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature, 410, 116–120. [DOI] [PubMed] [Google Scholar]
- Liu X., Bowen,J. and Gorovsky,M.A. (1996) Either of the major H2A genes but not an evolutionarily conserved H2A.F/Z variant of Tetrahymena thermophila can function as the sole H2A gene in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 2878–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Mader,A.W., Richmond,R.K., Sargent,D.F. and Richmond,T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- Mackay A.M., Ainsztein,A.M., Eckley,D.M. and Earnshaw,W.C. (1998) A dominant mutant of inner centromere protein (INCENP), a chromosomal protein, disrupts prometaphase congression and cytokinesis. J. Cell Biol., 140, 991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minc E., Allory,Y., Worman,H.J., Courvalin,J.C. and Buendia,B. (1999) Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma, 108, 220–234. [DOI] [PubMed] [Google Scholar]
- Nakayama J., Rice,J.C., Strahl,B.D., Allis,C.D. and Grewal,S.I. (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science, 292, 110–113. [DOI] [PubMed] [Google Scholar]
- Nichols J. and Gardner,R.L. (1984) Heterogeneous differentiation of external cells in individual isolated early mouse inner cell masses in culture. J. Embryol. Exp. Morphol., 80, 225–240. [PubMed] [Google Scholar]
- Nonaka N., Kitajima,T., Yokobayashi,S., Xiao,G., Yamamoto,M., Grewal,S.I. and Watanabe,Y. (2002) Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol., 4, 89–93. [DOI] [PubMed] [Google Scholar]
- Paull T.T., Rogakou,E.P., Yamazaki,V., Kirchgessner,C.U., Gellert,M. and Bonner,W.M. (2000) A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol., 10, 886–895. [DOI] [PubMed] [Google Scholar]
- Rasmussen T.P., Mastrangelo,M.A., Eden,A., Pehrson,J.R. and Jaenisch,R. (2000) Dynamic relocalization of histone MacroH2A1 from centrosomes to inactive X chromosomes during X inactivation. J. Cell Biol., 150, 1189–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Q.H. and Gorovsky,M.A. (2001) H2A.Z acetylation modulates an essential charge patch. Mol. Cell, 7, 1329–1335. [DOI] [PubMed] [Google Scholar]
- Rieder C.L. and Salmon,E.D. (1998) The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol., 8, 310–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santisteban M.S., Kalashnikova,T. and Smith,M.M. (2000) Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell, 103, 411–422. [DOI] [PubMed] [Google Scholar]
- Stargell L.A., Bowen,J., Dadd,C.A., Dedon,P.C., Davis,M., Cook,R.G., Allis,C.D. and Gorovsky,M.A. (1993) Temporal and spatial association of histone H2A variant hv1 with transcriptionally competent chromatin during nuclear development in Tetrahymena thermophila. Genes Dev., 7, 2641–2651. [DOI] [PubMed] [Google Scholar]
- Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Suto R.K., Clarkson,M.J., Tremethick,D.J. and Luger,K. (2000) Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat. Struct. Biol., 7, 1121–1124. [DOI] [PubMed] [Google Scholar]
- Temeles G.L., Ram,P.T., Rothstein,J.L. and Schultz,R.M. (1994) Expression patterns of novel genes during mouse preimplantation embryogenesis. Mol. Reprod. Dev., 37, 121–129. [DOI] [PubMed] [Google Scholar]
- Terada Y. (2001) Role of chromosomal passenger complex in chromosome segregation and cytokinesis. Cell Struct. Funct., 26, 653–657. [DOI] [PubMed] [Google Scholar]
- Tse C., Sera,T., Wolffe,A.P. and Hansen,J.C. (1998) Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol., 18, 4629–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren A.G., Wong,L., Pakusch,M., Fowler,K.J., Burrows,F.J., Vaux,D.L. and Choo,K.H. (2000) Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr. Biol., 10, 1319–1328. [DOI] [PubMed] [Google Scholar]
- van Daal A. and Elgin,S.C. (1992) A histone variant, H2AvD, is essential in Drosophila melanogaster. Mol. Biol. Cell, 3, 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Daal A., White,E.M., Gorovsky,M.A. and Elgin,S.C. (1988) Drosophila has a single copy of the gene encoding a highly conserved histone H2A variant of the H2A.F/Z type. Nucleic Acids Res., 16, 7487–7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hooser A.A., Mancini,M.A., Allis,C.D., Sullivan,K.F. and Brinkley,B.R. (1999) The mammalian centromere: structural domains and the attenuation of chromatin modeling. FASEB J., 13, Suppl. 2, S216–S220. [DOI] [PubMed] [Google Scholar]
- Wallrath L.L. and Elgin,S.C. (1995) Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev., 9, 1263–1277. [DOI] [PubMed] [Google Scholar]
- White E.M. and Gorovsky,M.A. (1988) Localization and expression of mRNA for a macronuclear-specific histone H2A variant (hv1) during the cell cycle and conjugation of Tetrahymena thermophila. Mol. Cell. Biol., 8, 4780–4786. [DOI] [PMC free article] [PubMed] [Google Scholar]