Abstract
Infection with some viruses can alter cellular mRNA processing to favor viral gene expression. We present evidence that herpes simplex virus 1 (HSV-1) protein ICP27, which contributes to host shut-off by inhibiting pre-mRNA splicing, interacts with essential splicing factors termed SR proteins and affects their phosphorylation. During HSV-1 infection, phosphorylation of several SR proteins was reduced and this correlated with a subnuclear redistribution. Exogenous SR proteins restored splicing in ICP27-inhibited nuclear extracts and SR proteins isolated from HSV-1-infected cells activated splicing in uninfected S100 extracts, indicating that inhibition occurs by a reversible mechanism. Spliceosome assembly was blocked at the pre-spliceosomal complex A stage. Furthermore, we show that ICP27 interacts with SRPK1 and relocalizes it to the nucleus; moreover, SRPK1 activity was altered in the presence of ICP27 in vitro. We propose that ICP27 modifies SRPK1 activity resulting in hypophosphorylation of SR proteins impairing their ability to function in spliceosome assembly.
Keywords: HSV-1/ICP27/splicing/SRPK1/SR protein phosphorylation
Introduction
Pre-mRNA splicing is a critical step in gene expression for metazoans. While the small nuclear ribonucleoprotein particles (snRNPs) are important determinants for splice site recognition and catalysis, non-snRNP splicing factors also play essential roles in establishing and stabilizing interactions necessary for splicing (Reed and Palandjian, 1997). SR proteins are a family of highly conserved non-snRNP factors vital to constitutive and regulated splicing (Manley and Tacke, 1996; Graveley, 2000). SR proteins participate in spliceosome assembly by promoting recognition and binding of U1 snRNP to the 5′ splice site and U2AF to the 3′ splice site and by helping to recruit the U4/U6·U5 tri-snRNP (Roscigno and Garcia-Blanco, 1995; Makarova et al., 2001). SR protein structure is characterized by two domains: the N-terminus contains one or more RNA-recognition motifs, which mediate RNA binding (Mayeda et al., 1999), whereas arginine/serine-rich (RS) domains in the C-terminus mediate interactions of SR proteins with each other and other spliceosome components (Wu and Maniatis, 1993; Kohtz et al., 1994). RS domain serine residues are reversibly phosphorylated by SR protein kinases and the level of phosphorylation regulates interactions with proteins and RNA (Tacke et al., 1997; Xiao and Manley, 1997), as well as subcellular localization and recruitment to sites of transcription (Misteli, 1999). Hypo- and hyperphosphorylated SR proteins are unable to sustain their activities in splicing (Mermoud et al., 1994; Sanford and Bruzik, 1999) thus, cycles of phosphorylation and dephosphorylation regulate SR protein activities (Misteli and Spector, 1997).
Herpes simplex virus 1 protein ICP27 inhibits host cell splicing (Hardwicke and Sandri-Goldin, 1994; Hardy and Sandri-Goldin, 1994; Bryant et al., 2001; Linberg and Kreivi, 2002), which results in decreased levels of cytoplasmic cellular spliced mRNAs, thus contributing to host protein synthesis shut-off. Most HSV-1 transcripts are intronless. Because spliced mRNAs are more efficiently exported than intronless transcripts (Luo and Reed, 1999), splicing inhibition may also result in alteration of cellular splicing-dependent export pathways. In fact, ICP27 facilitates HSV RNA export by binding to viral intronless RNAs (Sandri-Goldin, 1998a) and recruiting the exon– junction complex export protein Aly/REF (Koffa et al., 2001; Chen et al., 2002).
Previous studies showed that ICP27 caused a redistribution of splicing factors (Phelan et al., 1993; Sandri-Goldin et al., 1995), co-immunoprecipitated with anti-Sm antisera and altered the phosphorylation of two co-precipitating proteins (Sandri-Goldin and Hibbard, 1996). Furthermore, ICP27 was found to interact with splicing protein SAP145 (Bryant et al., 2001). These findings suggest that ICP27 mediates inhibition by interacting with splicing factors. Here we addressed the mechanism of splicing inhibition by ICP27. We report that ICP27 interacted with SRp20 in a yeast two-hybrid screen. Co-immunoprecipitation and co-localization showed that it also interacts with other SR proteins. Significantly, ICP27 expression resulted in hypophosphorylation of SR proteins. Furthermore, ICP27 was shown to interact with SR protein kinase 1 (SRPK1), a member of a family of conserved kinases that are highly specific for arginine/serine dipeptides (Gui et al., 1994a,b). Interaction with ICP27 resulted in a compartmental redistribution of SRPK1 in vivo and altered activity in vitro. Thus, pre-mRNA splicing inhibition by ICP27 may result from its interaction with SRPK1, triggering aberrant SR protein phosphorylation.
Results
ICP27 interacts with SRp20 in vivo and in vitro
To identify cellular proteins that interact with ICP27, a HeLa cDNA library was screened by yeast two-hybrid analysis. Of 2.8 × 106 cotransformants, 32 colonies grew on His– media and produced β-galactosidase, meeting the criteria for positive interaction. Two cDNAs were identified that encode SRp20, an SR protein.
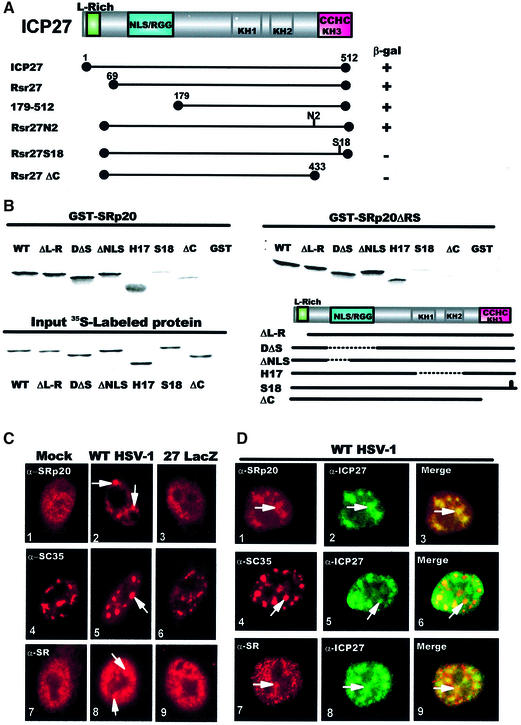
Mapping studies were performed with ICP27 mutations using yeast two-hybrid assays (Figure 1A). Interaction with SRp20 was eliminated by deleting the C-terminal 79 amino acids (Rsr27ΔC) encompassing a zinc-finger-like region. Insertion of four amino acids within this motif (Rsr27S18) also abolished interaction, whereas a mutant with a four amino acid insertion outside of this region (Rsr27N2) interacted. These results suggest that the zinc finger region of ICP27 is required for interaction with SRp20.
Fig. 1. ICP27 interacts with SRp20. (A) The ICP27 coding region is shown schematically, including a leucine-rich sequence (L-rich), NLS, RNA- binding region (RGG box), three putative KH domains and a zinc finger-like motif (CCHC). ICP27 mutations fused to the Gal4 DNA-binding domain were tested for interaction with SRp20 fused to the Gal4 activation domain as measured by β-galactosidase production. (B) GST-binding assays were performed with GST–SRp20 (left panel) or GST–SRp20ΔRS (right panel) and in vitro-translated WT ICP27 and mutants ΔL-R, D2ΔS5, ΔNLS, H17, S18 and ΔC. Input 35S-labeled proteins are shown (lower left panel) as is a schematic diagram of the mutants (lower right panel). (C) HeLa cells that were mock-infected (panels 1, 4 and 7), infected with WT HSV-1 (panels 2, 5 and 8) or 27-LacZ (panels 3, 6 and 9) for 3 h were stained with anti-SRp20 (panels 1–3), anti-SC35 (panels 4–6) or anti-SR (panels 7–9). Arrows indicate examples of coalesced SR proteins. (D) HSV-1-infected cells were double labeled with the indicated SR antibodies (red) and anti-ICP27 (green). The fields were merged to show co-localization (arrows) in yellow (panels 3, 6 and 9).
The region required for interaction was verified by in vitro binding assays using glutathione S-transferase (GST)–SRp20 and ICP27 that was translated in reticulocyte lysates. Wild-type (WT) ICP27 bound to GST–SRp20 as did mutants outside of the zinc finger region. However, binding of ΔC and S18 was detectable at background levels only (Figure 1B). ICP27 and mutants outside of the zinc finger region also bound to GST–SRp20ΔRS, which has amino acids 100–162 deleted including the RS domain. Again, ΔC and S18 failed to bind. We conclude that ICP27 interacts with SRp20 through the C-terminal zinc finger region. Significantly, the RS domain of SRp20 was not required because SRp20ΔRS interacted. Thus, SRp20 interacts with ICP27 through the N-terminal region containing the RRM, and the RS domain is not required.
Immunofluorescent staining of SRp20 was performed to determine whether ICP27 induced a coalescence similar to that reported previously for snRNPs and SC35 (Phelan et al., 1993; Sandri-Goldin et al., 1995). HeLa cells that were mock-infected, infected with WT HSV-1 or null-mutant virus, 27-LacZ were stained with anti-SRp20 (Neugebauer and Roth, 1997), anti-SC35 (Fu and Maniatis, 1990) and anti-SR (Tuma et al., 1993), which recognizes a conserved epitope present on the family of SR proteins. A striking redistribution of SRp20, SC35 and SR proteins was seen in WT HSV-1-infected cells (Figure 1C, panels 2, 5 and 8). SR proteins were coalesced into globular structures distinct from the speckled staining seen in mock and 27-LacZ-infected cells. Double-labeling experiments with WT HSV-1-infected cells (Figure 1D) showed that ICP27 was localized to coalescent SRp20, SC35 and SR protein structures in addition to being more diffusely distributed (panels 3, 6 and 9). Thus, it appears that ICP27 colocalizes with SR proteins and this results in redistribution into coalesced structures.
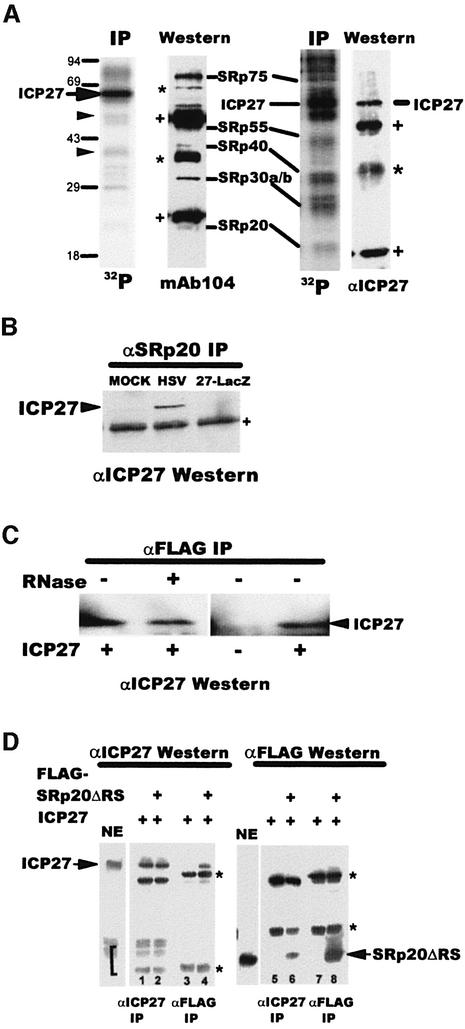
To further establish the interaction of ICP27 with SR proteins in HSV-1-infected cells, we performed co-immunoprecipitation experiments. A nuclear extract from 32P-labeled HSV-1-infected cells was immunoprecipitated with anti-ICP27 antibody and mAb104. ICP27, which has several phosphorylation sites (Zhi and Sandri-Goldin, 1999) was the predominant species seen with anti-ICP27 (Figure 2A). Other fainter bands were also seen, some of which were ICP27 degradation products as reported previously (Zhi and Sandri-Goldin, 1999), and verified by western blot analysis (data not shown). However, several of the bands were SR proteins shown by probing the blot with mAb104. SRp20, SRp30, SRp40 and SRp75 were co-immunoprecipitated with ICP27. It is not clear whether SRp55 was present because it was obscured by heavy chain IgG from the immunoprecipitation, which reacted with the secondary antibody in the immunoblot analysis. ICP27 was also co-immunoprecipitated with SR proteins recognized by mAb104 (Figure 2A, right panels) in accordance with previous findings (Sandri-Goldin, 1998b). Further, ICP27 co-precipitated with anti-SRp20 antibody (Figure 2B).
Fig. 2. ICP27 interacts with SR proteins in infected cells. (A) A nuclear extract from HSV-1-infected cells labeled with 32Pi from 1–6 h after infection was immunoprecipitated with anti-ICP27 antibody (left panels) or mAB104 (right panels). Phospho-labeled proteins were detected by autoradiography. ICP27 proteolytic degradation products are marked with arrowheads. The blots were probed with mAb104 (left) or anti-ICP27 (right). SR protein bands are indicated. Asterisks mark non-specific bands that reacted with the secondary antibody. Plus signs mark heavy and light chain IgG from the immunoprecipitation. (B) 293 cells transfected with pFlag-SRp20 were mock-infected or infected as indicated. Extracts were immunoprecipitated with anti-SRp20 antibody and the blot was probed with anti-ICP27 antibody. (C) In vitro binding was performed by mixing extracts from cells transfected with pFlag-SRp20 or pCMV-ICP27. Extracts were treated (+) or not (–) with RNase and immunoprecipitated with anti-Flag antibody. The blot was probed with anti-ICP27 antibody. (D) Extracts from cells transfected with pFlag-SRp20ΔRS or pCMV-ICP27 were mixed and immunoprecipitated with anti-ICP27 or anti-Flag antibody as indicated. The blots were probed with anti-ICP27 or anti-Flag antibody. In NE lanes, a portion of each nuclear extract was fractionated without immunoprecipitation. Asterisks mark heavy and light chain IgG. A bracket marks ICP27 degradation products (NE and lanes 1 and 2).
To confirm that the interaction between ICP27 and SRp20 was not mediated by RNA since both proteins bind RNA, we performed in vitro binding in the presence of RNase. ICP27 efficiently co-precipitated with Flag-SRp20 in the presence of RNase, indicating that the interaction was not bridged by RNA (Figure 2C). This is further supported by the finding that the ICP27 truncated protein, 179–512, which lacks the RGG motif required for RNA binding (Mears and Rice, 1996; Sandri-Goldin, 1998a) interacted with SRp20 in yeast (Figure 1A) and D2ΔS5, which also lacks the RGG RNA-binding motif interacted with GST–SRp20 (Figure 1B).
Immunoprecipitations were also performed on extracts from cells expressing Flag epitope-tagged SRp20ΔRS. ICP27 efficiently co-precipitated with Flag-SRp20ΔRS (Figure 1D, lane 4) and ∼5% of nuclear Flag-SRp20ΔRS co-precipitated with ICP27 (lane 6), further confirming the interaction of ICP27 and SRp20 in HSV-1-infected cells.
SR proteins are hypophosphorylated in the presence of ICP27
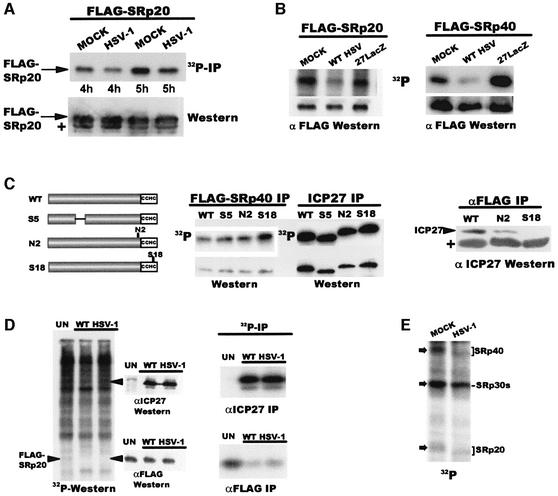
Next, we examined the phosphorylation of SR proteins. Flag-SRp20 was less phosphorylated in HSV-1-infected cells compared with mock controls, although equivalent amounts were immunoprecipitated (Figure 3A). Further, phosphorylation of Flag-SRp20 and Flag-SRp40 was decreased in WT HSV-1-infected cells compared with mock and 27-LacZ samples as seen when nuclear extracts were fractionated without immunoprecipitation (Figure 3B). To determine whether the zinc finger region is required for the effects on phosphorylation, pFlag-SRp40 was co-transfected into cells with plasmids expressing ICP27 or mutants (Figure 3C). Flag-SRp40 was more phosphorylated when co-expressed with zinc finger mutant S18 compared with WT ICP27 or mutants outside this region. Furthermore, ICP27 and N2 co-precipitated with anti-Flag antibody, and S18 did not (right panel). We conclude that the zinc finger region is required for the interaction of ICP27 with SR proteins that results in hypophosphorylation.
Fig. 3. ICP27 alters SR phosphorylation. (A) Nuclear extracts were prepared from 293 cells transfected with pFlag-SRp20, then mock- or HSV- infected for 4 and 5 h, and labeled with 32Pi. Extracts were immunoprecipitated with anti-Flag antibody. Radiolabeled proteins are shown in the upper panel. The blot was probed with anti-Flag antibody (lower panel). The plus sign marks IgG light chain from the immunoprecipitation. (B) 293 cells transfected with pFlag-SRp20 (left) or pFlag-SRp40 (right) were infected as indicated and labeled with 32Pi from 1 to 6 h after infection. Extracts were fractionated directly by SDS–PAGE. Radiolabeled proteins (upper panels) and western blot analysis with anti-Flag antibody (lower panels) are shown. (C) Nuclear extracts were prepared from 293 cells co-transfected with pFlag-SRp40 and plasmids expressing WT ICP27, S5, N2, or S18 and labeled with 32Pi for 5 h beginning 20 h after transfection. Extracts were immunoprecipitated with anti-Flag (left) or anti-ICP27 (middle). Radiolabeled proteins (upper panels) and western blot analysis with anti-Flag or anti-ICP27 (lower middle panels) are shown. Extracts containing Flag-SRp40 were mixed with WT ICP27, N2 or S18 and immunoprecipitated with anti-Flag antibody. The blot was probed with anti-ICP27 (right panel). (D) 293 cells were transfected with pFlag-SRp20. Uninfected (UN) and WT HSV-1-infected cells were labeled with 32Pi for 5 h. Nuclear extracts were fractionated directly (left panel) and the blot was later probed with anti-ICP27 and anti-Flag antibody (middle panels), or were immunoprecipitated with anti-ICP27 or anti-Flag antibody (right panels). (E) [γ-32P]ATP was added to mock or HSV-1 nuclear splicing extracts, each containing 60 µg of protein, and incubated for 3 h at 30°C. SR proteins were precipitated with MgCl2 and fractionated by SDS–PAGE.
A general dephosphorylation of cellular and viral proteins does not occur during HSV-1 infection (Figure 3D). Nuclear extracts from mock or HSV-1-infected cells expressing Flag-SRp20 were labeled with 32Pi then fractionated and transferred to nitrocellulose. Labeled protein profiles were very similar in the uninfected sample and two different HSV-1 samples (left panel). Flag-SRp20, detectable as a faint band in the UN lane, was not detectable in the HSV-1 lanes, whereas immunoblot analysis revealed that equal amounts were present (lower middle panel). Immunoprecipitation performed on portions of these extracts also showed that Flag-SRp20 was less phosphorylated in HSV-1 samples (lower right panel). These results indicate that the decrease in SRp20 phosphorylation is specific and is not due to a generalized decrease in protein phosphorylation upon HSV-1 infection.
To test whether SR proteins in splicing-incompetent extracts from HSV-1-infected cells were hypophosphorylated, SR proteins were labeled by [γ-32P]ATP and endogenous SR protein kinases in nuclear splicing extracts. SRp40 and SRp20 in the HSV-1 extract were less intensely labeled and migrated faster (Figure 3E), in accordance with reports that hypophosphorylated SR proteins migrate faster (Roth et al., 1991; Prasad et al., 1999). The band at the position of SRp30 proteins was also less intense in the HSV lane. The identity of SR protein bands was verified with mAb104 (data not shown). Thus, SRp20, SRp30 and SRp40 were less phosphorylated in splicing extracts containing ICP27.
SR proteins restore splicing activity to HSV-1 extracts
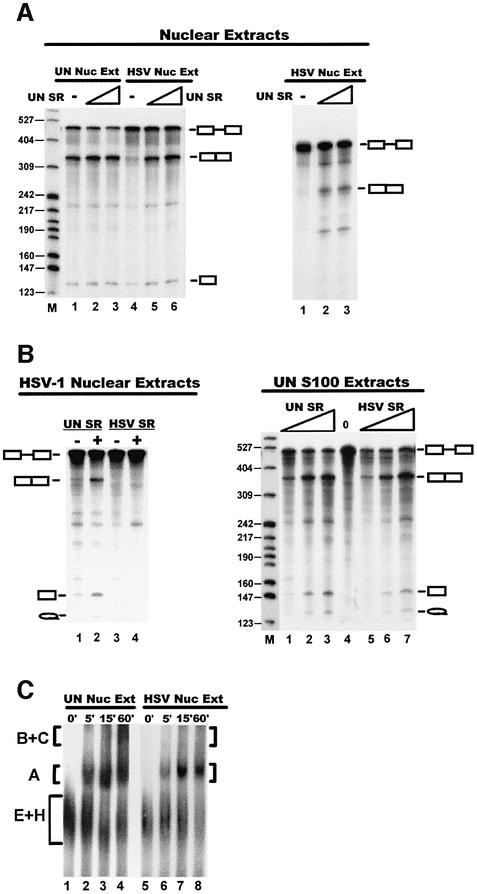
To address whether ICP27-mediated hypophosphorylation impairs SR splicing activities, purified SR proteins from uninfected cells were added to HSV-1 nuclear splicing extracts to see whether splicing activity could be restored. SR proteins from uninfected cells did restore splicing of a β-globin pre-mRNA (Figure 4A, left panel, lanes 5 and 6) and an adenovirus major late pre-mRNA (right panel). In contrast, SR proteins purified from HSV-1-infected cells were not able to restore splicing to an HSV-1 nuclear extract (Figure 4B, left panel, lane 4), whereas SR proteins from uninfected cells could restore splicing in this extract (Figure 4B, left panel, lane 2). Conversely, SR proteins from HSV-1-infected cells activated splicing when added to uninfected HeLa S100 extracts (Figure 4B, right panel, lanes 5–7). This indicates that SR proteins are not irreversibly altered in HSV-1-infected cells because S100-endogenous SR protein kinases restored activity when ICP27 was not present. We conclude that SR proteins are reversibly altered in HSV-1-infected cells and activity can be restored by phosphorylation in S100 extracts.
Fig. 4. SR proteins restore splicing to HSV-inhibited extracts. (A) 32P-labeled β-globin pre-mRNA was added to nuclear splicing extracts from uninfected (left panel, lanes 1–3) or HSV-infected (lanes 4–6) cells. Splicing extracts were not supplemented (lanes 1 and 4) or were supplemented with 200 ng (lanes 2 and 5) or 400 ng (lanes 3 and 6) of SR proteins from uninfected HeLa cells. AdML pre-mRNA was added to HSV nuclear splicing extracts (right panel, lanes 2 and 3). SR proteins from uninfected HeLa cells were added at 200 ng (lane 2) or 400 ng (lanes 3). (B) Purified SR proteins (200 ng) from uninfected (left panel, lanes 1 and 2) or HSV-1-infected (lanes 3 and 4) cells were added to nuclear splicing extracts from HSV-1-infected cells to which β-globin pre-mRNA was added. S100 extracts from uninfected cells were supplemented with 200, 400 or 800 ng of SR proteins from uninfected cells (right panel, lanes 1–3) or with the same amounts of SR proteins from HSV-1-infected cells (lanes 5–7). SR proteins were not added to lane 4. Splicing of β-globin pre-mRNA was monitored. (C) AdML pre-mRNA was added to splicing extracts from uninfected and HSV-1-infected HeLa cells that were analyzed for spliceosome complex formation on 4% non-denaturing polyacrylamide gels. Spliceosome assembly complexes A, B and C are labeled. The early complex (E) and hnRNP–pre-mRNA complex (H) co-migrated as a diffuse band labeled E+H.
Next, we analyzed splicing complex formation, which is an ordered process in which the snRNPs, SR proteins and splicing accessory factors are sequentially arranged on the pre-mRNA (Reed and Palandjian, 1997). Adenovirus major late pre-mRNA was added to nuclear splicing extracts and complexes were resolved on native polyacrylamide gels. Complexes E, A, B and C were detected in uninfected extracts (Figure 4C, lanes 1–4). In contrast, while E and A pre-spliceosomal complexes were evident in the HSV-1 extract, higher complexes representing functional spliceosomes were not seen. These results indicate that when ICP27 is present, splicing is inhibited before the first catalytic reaction because spliceosome assembly is interrupted before the entry of the tri-snRNP, which results in the formation of complex B. It has been shown that SR proteins escort the U4/U6·U5 tri-snRNP to the spliceosome, and the pre-spliceosome to spliceosome transition requires phosphorylated SR proteins (Roscigno and Garcia-Blanco, 1995). These results suggest that ICP27-induced SR protein hypophosphorylation results in a block in the transition from pre-spliceosome to spliceosome. In support of this conclusion, it has been reported that authentic B and C complexes were not formed in HSV-1 splicing extracts (Bryant et al., 2001), and that complex A to B transition was very inefficient in nuclear extracts from cells that transiently expressed ICP27 (Linberg and Kreivi, 2002).
ICP27 interacts with SRPK1
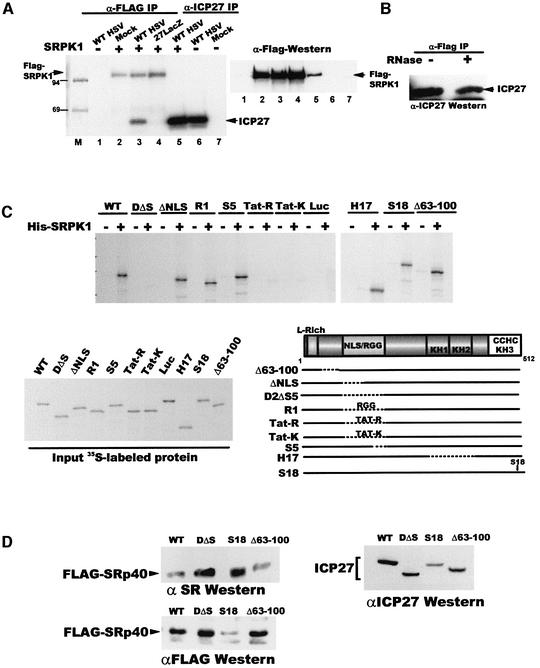
To address how ICP27 alters SR protein phosphorylation, we asked whether it interacts with an SR protein kinase, SRPK1 (Gui et al., 1994b). In HSV-1-infected cells, ICP27 was co-immunoprecipitated with Flag-SRPK1 (Figure 5A, left panel, lane 3), and Flag-SRPK1 co-immunoprecipitated with ICP27 (right panel, lane 5). To verify that the interaction was not mediated by RNA, RNase was added before mixing extracts containing each protein. Immunoblot analysis with anti-ICP27 antibody showed that ICP27 co-precipitated with Flag-SRPK1 in the presence of RNase (Figure 5B).
Fig. 5. ICP27 interacts with SRPK1. (A) Cells were transfected with pFlag-SRPK1 (lanes 2–5) or a control plasmid (lanes 1, 6 and 7) and extracts were prepared from transfected cells that were mock-infected (lanes 2 and 7), infected with WT HSV-1 (lanes 1, 3, 5 and 6) or 27-LacZ (lane 4) and labeled with 32Pi from 1 to 5 h after infection. Immunoprecipitation was performed with anti-Flag (lanes 1–4) or anti-ICP27 (lanes 5–7). Radiolabeled proteins (left) and immunoblot analysis with anti-Flag antibody (right) are shown. (B) Extracts from cells transfected with pFlag-SRPK1 or pCMV-ICP27 were treated with RNase (+) or buffer (–) and then mixed. Immunoprecipitation was performed with anti-Flag and the blot was probed with anti-ICP27 antibody. (C) In vitro binding assays were performed with His-SRPK1 and in vitro-translated WT ICP27 or mutant proteins as indicated. Input [35S]methionine-labeled proteins and a schematic showing the positions of the mutations are shown. (D) Cells were co-transfected with Flag-Srp40 and plasmids expressing WT ICP27, D2ΔS5, S18 or Δ63–100. Nuclear extracts were fractionated and western blot analysis was performed with anti-SR antibody (upper left panel), anti-Flag (lower left panel) and anti-ICP27 (right panel).
To map the region of ICP27 required for interaction with SRPK1, in vitro binding studies were performed with His-tagged SRPK1 and in vitro translated ICP27 (Figure 5C). Mutant D2ΔS5, which has a deletion encompassing the nuclear localization signal (NLS) and the RGG RNA-binding domain failed to bind to His-SRPK1, whereas ΔNLS, which lacks the NLS but has the RGG box intact, bound. Mutant R1, which has the sequence RGGRRGRRRGRGRGG inserted into the deleted region of D2ΔS5, also bound, suggesting that the RGG motif of ICP27 is required for the interaction. Furthermore, Tat-R in which the RNA binding region of HIV Tat was inserted in place of the ICP27 RGG box did not bind, and neither did Tat-K, in which lysine residues were substituted for arginine in Tat-R (Figure 5C). All other mutants tested bound, including S18. We conclude that the RGG RNA-binding motif of ICP27 is required for the interaction with SRPK1, whereas the zinc finger motif is required for the interaction of ICP27 with SRp20.
To determine whether interaction with SRPK1 is required for hypophosphorylation of SR proteins, cells were transfected with plasmids expressing Flag-SRp40 and ICP27 or mutants. SRp40 phosphorylation was analyzed by probing a western blot with anti-SR antibody, which recognizes a conserved phospho-epitope, and does not recognize unphosphorylated forms (Tuma et al., 1993). Flag-SRp40 was less phosphorylated in the presence of WT ICP27 and Δ63–100 compared with D2ΔS5, which did not bind to His-SRPK1, and mutant S18, which did not bind to SRp20 (Figure 5D, upper left panel). In fact, higher phosphorylation of Flag-SRp40 was seen despite its being less well expressed in the S18 sample as seen when probed with anti-Flag (lower left panel). We conclude that interaction with SRPK1 is necessary for ICP27-mediated hypophosphorylation of SR proteins in vivo.
ICP27 alters SRPK1 localization
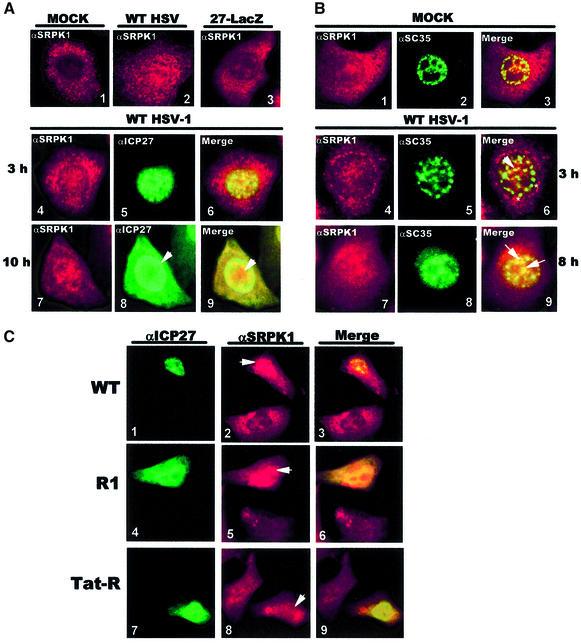
Next, immunofluorescent staining of SRPK1 was examined. SRPK1 was predominantly cytoplasmic in mock-infected cells (Figure 6A, panel 1) as reported previously (Wang et al., 1998), and a similar pattern was seen in 27-LacZ-infected cells (panel 3). Unexpectedly, SRPK1 was more concentrated in the nucleus in WT HSV-1-infected cells (panel 2). By 3 h after HSV-1 infection, SRPK1 was found in the nucleus (panel 4), ICP27 was exclusively nuclear (panel 5), and the two proteins co-localized in some areas (panel 6). By 10 h, ICP27 cytoplasmic staining was seen (panel 8), corresponding to its nucleocytoplasmic shuttling beginning ∼6 h after infection (Soliman et al., 1997; Sandri-Goldin, 1998a). SRPK1 and ICP27 nuclear localization was also altered. ICP27 showed strong nuclear rim fluorescence (panels 8 and 9), correlating with active export, as well as diffuse staining throughout the nucleus. SRPK1 was localized in cluster-like structures (panels 7 and 9). To determine whether SRPK1 clusters contained SR proteins, we examined the localization of SC35 and SRPK1 (Figure 6B). In mock-infected cells, SRPK1 was mostly cytoplasmic (panel 1) and SC35 was seen as nuclear speckles (panel 2). After HSV-1 infection, SC35 speckles coalesced into ball-like structures (panel 5), which for the most part do not appear to contain SRPK1 (asterisks, panel 6). However, by 8 h, when ICP27 was shuttling, the aggregated structures were dispersed and SC35 was redistributed in more diffuse clusters (panel 8), which appeared to contain SRPK1 (arrows, panel 9). Thus, at early times after infection SRPK1 was relocalized to the nucleus where it co-localized with ICP27, but not with SC35 coalesced structures. At later times, ICP27 was prominently cytoplasmic, while coalesced SC35 structures were dispersed and SRPK1 co-localized with SC35 in more diffuse clusters.
Fig. 6. ICP27 relocalizes SRPK1 to the nucleus. (A) Cells that were mock-, WT HSV-1- or 27-LacZ-infected for 3 h were stained with anti-SRPK1 antibody (panels 1–3). Double labeling with anti-SRPK1 (red) and anti-ICP27 (green) was performed on cells infected with WT HSV-1 for 3 (panels 4–6) and 10 h (panels 7–9). Merged images show co-localization in yellow (panels 6 and 9). ICP27 perinuclear staining is marked by an arrow (panel 8), as is SRPK1 nuclear staining (panel 9). (B) Double labeling with anti-SRPK1 (panels 1, 4 and 7) and anti-SC35 (panels 2, 5 and 8) was performed on cells that were mock-infected (panels 1–3) or infected with HSV-1 for 3 (panels 4–6) and 8 h (panels 7–9). Areas of co-localization (yellow) in the merged images are denoted by arrows. Asterisks mark regions of SC35 staining that do not contain SRPK1. (C) Cells were transfected with pCMV-ICP27, R1 or Tat-R. Double labeling was performed with anti-ICP27 (panels 1, 4 and 7) and anti-SRPK1 (panels 2, 5 and 8). The merged images are shown (lanes 3, 6 and 9).
To verify that SRPK1 is relocalized to the nucleus by ICP27, cells transfected with ICP27 or mutants R1 or Tat-R were stained with anti-SRPK1 and anti-ICP27. SRPK1 was concentrated in the nucleus of a cell that expressed ICP27, whereas it was mainly cytoplasmic in a cell that did not express ICP27 (Figure 6C, panel 2). Similarly, SRPK1 was present in the nucleus of an R1-expressing cell, but not in a cell that did not express R1 (panel 5). Note that R1, which contains the RGG box, is less efficiently localized to the nucleus because it lacks the NLS as reported previously (Hibbard and Sandri-Goldin, 1995). In contrast, SRPK1 remained cytoplasmic in a cell expressing Tat-R, which does not bind SRPK1 (panel 8). We conclude that ICP27 interacts with SRPK1 and causes it to be relocalized to the nucleus.
ICP27 alters SRPK1 activity in vitro
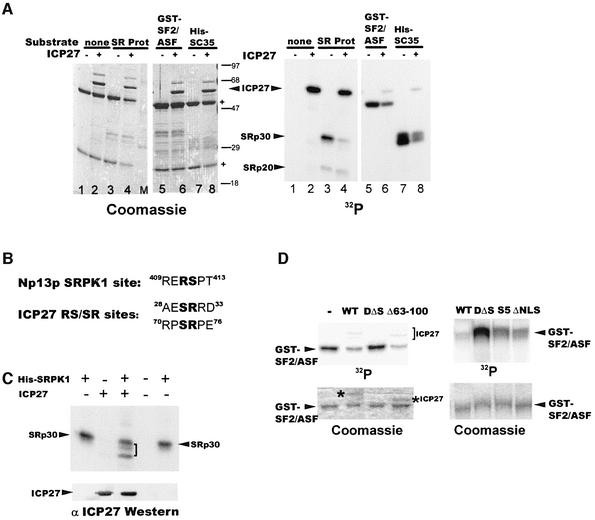
To determine the effect of the interaction of ICP27 with SRPK1 on SR protein phosphorylation, in vitro kinase assays were performed. Substrates included SR proteins purified from HeLa cells and recombinant SR proteins. His-SRPK1 was pre-incubated with immunoprecipitated ICP27 from transfected cells or with ICP27 antibody– protein A–Sepharose complexes. We used immunoprecipitated ICP27 because we (our unpublished results) and others (Mears and Rice, 1996) have been unsuccessful in obtaining homogeneous, full-length ICP27 using bacterial expression systems. SR proteins were added to the reactions and phosphorylation was measured by incorporation of a radiolabel (Figure 7A). The Coomassie Blue-stained gel (left panel) shows that equivalent amounts of substrate were present in each reaction. Several SR proteins were less phosphorylated when ICP27 was present, including SRp30 (right panel, lane 4), GST– SF2/ASF (lane 6) and His-SC35 (lane 8). Faster migrating species were seen for SRp20 (lane 4), but there was not a significant difference in labeling. Surprisingly, ICP27 incorporated the radiolabel. ICP27 has been shown to be phosphorylated by PKA and CK2 (Zhi and Sandri-Goldin, 1999). It does encode two RS/SR dipeptides (Figure 7B) that resemble the SRPK1 yeast homolog Sky1p site identified in Np13p (Nolen et al., 2001). Notably, phosphorylation of ICP27 by SRPK1 was greatly reduced in assays with unphosphorylated recombinant SR proteins (lanes 6 and 8) compared with SR proteins from HeLa cells, which were likely phosphorylated in vivo. Thus, SRPK1 apparently can act on ICP27 as an available substrate. ICP27 alone did not affect SRp30 phosphorylation (Figure 7C, lane 2), but again in its presence, SRp30 proteins from HeLa cells were less phosphorylated by SRPK1 (lane 3). Importantly, D2ΔS5, which did not bind SRPK1 (Figure 5C), also did not reduce phosphorylation of GST–SF2/ASF (Figure 7D), whereas Δ63–100, S5 and ΔNLS, which interact with SRPK1, reduced phosphorylation. Interestingly, one of the putative SR sites is deleted in Δ63–100, yet this mutant was phosphorylated to some extent by SRPK1. In contrast, D2ΔS5 was not phosphorylated by SRPK1 (data not shown). We conclude that ICP27 induced hypophosphorylation of SR proteins in vitro by interacting with SRPK1.
Fig. 7. ICP27 alters SRPK1 activity in vitro. (A) His-SRPK1 was pre-incubated with immunoprecipitated ICP27 or control antibody–protein A– Sepharose complexes. SR proteins purified from HeLa cells (lanes 3 and 4), GST–SF2/ASF (lanes 5 and 6) and His-SC35 (lanes 7 and 8) were added as was [γ-32P]ATP. Heavy and light chain IgG from antibody complexes are marked (+) in the Coomassie Blue-stained gel (left). (B) The SRPK1 homolog Sky1p site in Np13p is shown with putative SRPK1 sites in ICP27. (C) SRPK1, immunoprecipitated ICP27 or control antibody complexes were pre-incubated and SRp30 proteins from HeLa cells were added to kinase reactions. The bracket marks the hypophosphorylated species. Immunoblot analysis with anti-ICP27 antibody is shown below. (D) Kinase assays were performed with His-SRPK1 and immunoprecipitated WT ICP27 or mutants D2ΔS5, Δ63–100, S5 and ΔNLS with GST–SF2/ASF as the substrate. The Coomassie Blue-stained gels are shown below.
Discussion
We have provided evidence that ICP27, an essential HSV-1 regulatory protein, interacts with SRPK1 resulting in hypophosphorylation of SR proteins, which impairs their roles in spliceosome assembly. Alterations in SR protein phosphorylation may prove to be a general mechanism in the regulation of splicing during viral infection to redirect cellular RNA processing pathways to favor viral gene expression. Studies by Kanopka et al. (1998) showed that SR proteins isolated from adenovirus-infected cells at late times of infection were hypophosphorylated. This resulted in inactivation of SR protein splicing repressor functions so that adenovirus IIIa pre-mRNA was spliced, whereas SR proteins from uninfected HeLa cells inhibited splicing of this pre-mRNA. Further more, it was shown that adenovirus protein E4ORF4, which induces dephosphorylation of SR proteins, interacts with SR protein phosphatase 2A and with hyperphosphorylated forms of ASF/SF2 and SRp30c, but not other SR proteins (Nilsson et al., 2001). In contrast, we found that ICP27 interacts with SRPK1 and with several SR proteins, including hypophosphorylated forms (Figure 1B). Thus, adenovirus appears to modulate dephosphorylation of specific SR proteins to enhance viral gene expression by regulating alternative splicing, whereas HSV-1 uses ICP27 to inhibit pre-mRNA splicing to decrease host expression and favor export and expression of intronless HSV-1 transcripts. Interestingly, both E4ORF4 and ICP27 target SR protein phosphorylation to mediate their effects, although E4ORF4 recruits a phosphatase and ICP27 interacts with a kinase.
SR protein phosphorylation was also found to play a role in regulating splicing activity in nematode embryos (Sanford and Bruzik, 1999). SR proteins underwent dephosphorylation from an earlier stage where they were hyperphosphorylated to become active in splicing at later stages. While it is unclear to what extent active SR proteins are phosphorylated, both hyper- and hypophosphorylation inhibit SR protein activities (Prasad et al., 1999) such that an intermediate level is necessary for their critical roles in spliceosome assembly. Although ICP27 caused modest changes in SR protein phosphorylation, pre-spliceosome complex A was unable to transition into a functional spliceosome. It has been shown that phosphorylated SR proteins are required for the incorporation of the U4/U6·U5 tri-snRNP into the spliceosome (Roscigno and Garcia-Blanco, 1995). However, SR phosphorylation was not required for pre-spliceosome formation (Roscigno and Garcia-Blanco, 1995), nor was phosphorylation of ASF/SF2 required for complex E formation (Cao et al., 1997), and these complexes were able to form in HSV-1 extracts. It has been suggested that different SR protein functions require distinct phosphorylation patterns (Xiao and Manley, 1998). This could explain why SR proteins in HSV-1 splicing extracts were able to perform their roles leading to complex A formation, but were unable to recruit the tri-snRNP. Recently, Bryant et al. (2001) reported that authentic B and C complexes were not formed in HSV-1 splicing extracts and thus they concluded that splicing was inhibited before the first catalytic step. Similar results were found in extracts from cells that transiently expressed ICP27 (Linberg and Kreivi, 2002). Our findings are consistent with these studies. Although the mechanism of splicing inhibition was not probed directly in previous studies, it was shown that ICP27 interacted with SAP145 (Bryant et al., 2001), a spliceosome-associated protein that is part of a complex implicated in tethering U2 snRNP to the branch site (Champion-Arnaud and Reed, 1994). Furthermore, we found that ICP27 interacts with and alters the phosphorylation of U1 70K, which contains an RS domain (Sandri-Goldin and Hibbard, 1996). Thus, ICP27 has been reported to interact with splicing-associated factors but it was not shown previously that these interactions resulted in splicing inhibition. Here, we extend previous studies by providing evidence that ICP27, through its interaction with SRPK1 induces hypophosphorylation of SR proteins, impairing spliceosome formation.
Alteration of SR protein phosphorylation also affects subcellular localization (Misteli and Spector, 1997). Overexpression of Clk/STY kinase resulted in redistribution of splicing factors from speckles to a diffuse distribution (Misteli and Spector, 1997). In contrast, overexpression of a kinase-dead Clk-STY mutant led to an immobilization of the speckles (Sacco-Bubulya and Spector, 2002). SRPK1 was recruited to the nucleus by ICP27 where it was altered in its ability to phosphorylate SR proteins. The consequences on SR subnuclear localization were similar to overexpression of the kinase-dead mutant in that ICP27 induced the formation of coalescent structures, consistent with an alteration in phosphorylation. Significantly, coalesced SC35 structures were dispersed when ICP27 was predominantly cytoplasmic and no longer in direct contact with SRPK1. We have found that inhibition of splicing is a temporary and reversible phenomenon that occurs at early times of infection. Extracts prepared 6 h or later after infection were capable of splicing in vitro (Hardy and Sandri-Goldin, 1994). The restoration of splicing correlates with ICP27 shuttling and with dispersal of coalesced SC35 at later times. SRPK1 remains in the nucleus of infected cells at later times, however, it is not associated with ICP27 and may therefore reactivate SR proteins.
We propose that early in HSV-1 infection ICP27 recruits SRPK1 to the nucleus and interacts with SR proteins at sites of spliceosome assembly. There are several possible explanations for SR protein hypophosphorylation that occurs because of these interactions. ICP27 may compete with SR proteins for SRPK1 binding. It is also feasible that ICP27 may simultaneously interact with SR proteins and SRPK1 because different domains are involved. The interaction may alter SRPK1 conformation resulting in changes in activity, or may mask potential phosphorylation sites in SR proteins. These possibilities are not mutually exclusive. Studies are currently underway to probe these possibilities.
Materials and methods
Cells, viruses and plasmids
HeLa and 293 cells were grown as described previously (Hardy and Sandri-Goldin, 1994), as were HSV-1 strain KOS and null mutant 27-LacZ (Smith et al., 1992). Mammalian expression plasmids for ICP27 and mutants S5, N2, H17, S18 (Hardwicke et al., 1989), D2ΔS5, ΔNLS, R1, Tat-R, Tat-K (Hibbard and Sandri-Goldin, 1995) and ΔL-R (Sandri-Goldin, 1998a) were constructed by replacing the ICP27 promoter with the CMV IE promoter. Flag epitope-tagged constructs were cloned by inserting the Flag epitope sequence in frame with the 5′ ends of the cDNAs for SRp20, SRp40 (Tacke et al., 1997) and SRPK1 (Gui et al., 1994b). The RS domain (amino acids 100–164) was deleted in Flag-SRp20ΔRS. Splicing substrates were synthesized by in vitro transcription of DNA templates for pSP64-HβΔ6 and pAdML (Michaud and Reed, 1993). pRSETb-SRPK1 encoding His-tagged SRPK1 was provided by X.-D.Fu. ICP27 yeast expression plasmids were constructed as described previously (Zhi et al., 1999). In pDBD-SRPK, a 2.59 Kb BamHI–EcoRI fragment from pRSETb-SRPK1 was inserted into pGBT9(5).
Yeast two-hybrid assay
Plasmid pDBD-Rsr27 was cotransformed with a HeLa cDNA library fused to the Gal4 activation domain in pGADGH (Clontech) into Saccharomyces cerevisiae HF7c carrying Gal1-lacZ/HIS3 reporter genes. Transformants were plated on media lacking tryptophan, leucine and histidine and were screened for β-galactosidase production by colony- lift filter-assay. Blue colonies, scored as a positive interaction, were re-streaked for purification and amplified.
Recombinant proteins
GST–SRp20 and GST–SRp20ΔRS were purified from Escherichia coli Rosetta extracts with glutathione–Sepharose beads. His-SRPK1 was purified by Ni2+-NTA chromatography from E.coli BL21 extracts. The protein concentration of His-SRPK1 (9 mg/ml) was determined by the Bradford assay, and kinase activity (450 units/ml) was determined as described previously (Gui et al., 1994b). Purified His-SC35 was provided by K.Hertel and purified GST–SF2/ASF was provided by X.-D.Fu.
Virus infection, transfection and radiolabeling
Cells were infected with HSV-1 at an m.o.i. of 10 for the times indicated. When cells were radiolabeled, 32Pi (NEN Dupont) was added as described previously (Zhi et al., 1999). Cells were transfected using lipofectamine (Life Technologies).
Immunofluorescent staining
HeLa cells grown on coverslips were mock-infected or infected with HSV-1 as indicated. Immunofluorescent staining was performed as described previously (Sandri-Goldin et al., 1995). Primary antibodies included anti-ICP27 monoclonal antibody H1119 (Goodwin Institute), used at a dilution of 1:500; anti-SRp20 (Zymed Laboratories) at 2 µg/ml, anti-SC35 hybridoma supernatant at 1:50; anti-SR (Zymed Laboratories) at 3 µg/ml, and anti-SRPK1 (PharMingen) at 1:1000. In double-staining experiments, cells were treated with the first antibody and detected with FITC-conjugated goat anti-mouse IgG secondary antibody (Jackson Immunoresearch) at 7.5 µg/ml. Cells were blocked with normal mouse serum (500 µg/ml) and then with unconjugated goat anti-mouse Fab fragments (Jackson Immunoresearch) at 7 µg/ml to prevent cross-reactions when labeling with primary antibodies from the same species. Cells were probed with the second monoclonal antibody and Texas Red-conjugated goat anti-mouse IgG used at 7.5 µg/ml. Cells were viewed at 100× magnification with a Zeiss Axiovert S100 microscope.
Cellular extracts and purification of SR proteins
Nuclear extracts for immunoprecipitation of ICP27 were prepared in extraction buffer containing 500 mM NaCl as described previously (Sandri-Goldin et al., 1996). A phosphatase inhibitor, 5 mM β-glycerolphosphate, was added to all buffers (Zahler et al., 1992). For immunoprecipitation of SR proteins, extracts were prepared in SR isolation buffer (Zahler et al., 1992) containing 5 mM potassium fluoride and 5 mM β-glycerolphosphate. Nuclear splicing extracts and cyto plasmic S100 extracts were prepared from mock and HSV-1-infected HeLa cells as described previously (Hardy and Sandri-Goldin, 1994). SR proteins were purified from HeLa cells as described previously (Zahler et al., 1992).
Splicing reactions
In vitro splicing reactions were performed as described previously (Hertel and Maniatis, 1999), as was native gel electrophoresis of splicing complexes (Konarska and Sharp, 1986), except that heparin (1 mg/ml) was added before gel fractionation.
In vitro binding assays
GST-binding assays were performed at room temperature by combining 20 µl glutathione–Sepharose-bound GST proteins with 10 µl 35S-labeled proteins from in vitro translation reactions in 200 µl TBST buffer (10 mM Tris–HCl pH 7.5, 200 mM NaCl, 0.2% Tween 20 and 0.5% BSA) for 1 h. Beads were washed three times in TBST and bound proteins were fractionated by SDS–PAGE. For in vitro binding immunoprecipitation assays, extracts from transfected cells were cleared by centrifugation at 14 000 r.p.m. at 4°C for 20 min, then incubated in the presence of 4 mM pefabloc, 0.1 µg/ml leupeptin, 10 mM β-glycerolphosphate, 2.5 units of RNase A and 200 units of RNase T1 (Ambion) at 37°C for 60 min. Equal volumes of each extract were combined and rotated overnight at 4°C before immunoprecipitation.
Immunoprecipitation and immunoblotting
Immunoprecipitation was performed with anti-ICP27, mAb104, anti-SRp20, anti-SRPK1 or anti-Flag (M2, Sigma) antibodies as described previously (Sandri-Goldin and Hibbard, 1996). Extracts were pre-cleared with rabbit anti-mouse IgG (Pierce) bound to protein A–Sepharose before primary antibody was added. For immunoblot blot analysis, anti-ICP27 was used at a dilution of 1:10 000; anti-SRp20 at 1:500; anti-SRPK1 at 1:2000; and mAb104 hybridoma supernatant was undiluted. Horseradish peroxidase-conjugated sheep anti-mouse Ig secondary antibody (Jackson ImmunoResearch) was used at 1:100 000. Proteins were detected by ECL (Amersham Pharmacia).
In vitro phosphorylation
SR protein labeling by endogenous kinases was performed by mixing 50% (vol/vol) nuclear splicing extract with buffer containing 50 mM Tris pH 8.5, 4 mM MgCl2, 2 mM DTT, 0.2% NP-40, 1 µM ATP and 0.1 mCi [γ-32P]ATP for 3 h at 30°C. SR proteins were precipitated by adding 10 mM MgCl2 and incubating on ice for 60 min, followed by centrifugation at 14 000 r.p.m. for 20 min at 4°C. Pellets were washed with 20 mM MgCl2 and resuspended before fractionation by SDS–PAGE. SRPK1 kinase assays were performed as described previously (Gui et al., 1994b). Immunoprecipitated ICP27 bound to protein A–Sepharose was incubated with 5 units (180 ng) of recombinant SRPK1 in 15 µl of kinase assay mixture for 30 min at 30°C. SR proteins (500 ng) were added to the reactions for 15 min followed by boiling for 2 min in SDS–PAGE loading buffer.
Acknowledgments
Acknowledgements
We thank R.Tacke, J.Manley, C.Y.Yun, X.-D.Fu and K.Hertel for cDNAs, plasmids, recombinant proteins and helpful suggestions, and K.Hertel for critical review of the manuscript. This work was supported by NIH grant AI21515 to R.M.S.-G. K.S.S. was supported by NIAID T32-AI-07319.
References
- Bryant H.E., Wadd,S., Lamond,A.I., Silverstein,S.J. and Clements,J.B. (2001) Herpes simplex virus IE63 (ICP27) protein interacts with spliceosome-associated protein 145 and inhibits splicing prior to the first catalytic step. J. Virol., 75, 4376–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W., Jamison,S.F. and Garcia-Blanco,M.A. (1997) Both phosphoryl ation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA, 3, 1456–1467. [PMC free article] [PubMed] [Google Scholar]
- Champion-Arnaud P. and Reed,R. (1994) The prespliceosome components SAP 49 and SAP 145 interact in a complex implicated in tethering U2 snRNP to the branch site. Genes Dev., 8, 1974–1983. [DOI] [PubMed] [Google Scholar]
- Chen I.B., Sciabica,K.S. and Sandri-Goldin,R.M. (2002) ICP27 interacts with the export factor Aly/REF to direct herpes simplex virus 1 intronless RNAs to the TAP export pathway. J. Virol., 76, 12877–12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X.D. and Maniatis,T. (1990) Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature, 343, 437–441. [DOI] [PubMed] [Google Scholar]
- Graveley B.R. (2000) Sorting out the complexity of SR protein functions. RNA, 6, 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui J.-F., Lane,W.S. and Fu,X.-D. (1994a) A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature, 369, 678–682. [DOI] [PubMed] [Google Scholar]
- Gui J.-F., Tronchere,H., Chandler,S.D. and Fu,X.-D. (1994b) Purification and characterization of a kinase specific for the serine- and arginine-rich pre-mRNA splicing factors. Proc. Natl Acad. Sci. USA, 91, 10824–10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwicke M.A. and Sandri-Goldin,R.M. (1994) The herpes simplex virus regulatory protein ICP27 can cause a decrease in cellular mRNA levels during infection. J. Virol., 68, 4797–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwicke M.A., Vaughan,P.J., Sekulovich,R.E., O’Conner,R. and Sandri-Goldin,R.M. (1989) The regions important for the activator and repressor functions of the HSV-1 α protein ICP27 map to the C-terminal half of the molecule. J. Virol., 63, 4590–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy W.R. and Sandri-Goldin,R.M. (1994) Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J. Virol., 68, 7790–7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel K.J. and Maniatis,T. (1999) Serine-arginine (SR)-rich splicing factors have an exon-independent function in pre-mRNA splicing. Proc. Natl Acad. Sci. USA, 96, 2651–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbard M.K. and Sandri-Goldin,R.M. (1995) Arginine-rich regions succeeding the nuclear localization region of the HSV-1 regulatory protein ICP27 are required for efficient nuclear localization and late gene expression. J. Virol., 69, 4656–4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanopka A., Muhlemann,O., Petersen-Mahrt,S., Estmer,C., Ohrmalm,C. and Akusjarvi,G. (1998) Regulation of adenovirus alternative splicing by dephosphorylation of SR proteins. Nature, 393, 185–187. [DOI] [PubMed] [Google Scholar]
- Koffa M.D., Clements,J.B., Izaurralde,E., Wadd,S., Wilson,S.A., Mattaj,I.W. and Kuersten,S. (2001) Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J., 20, 5769–5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohtz J.D., Jamison,S.F., Will,C.L., Zuo,P., Luhrmann,R., Garcia-Blanco,M.A. and Manley,J.L. (1994) Protein–protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature, 368, 119–124. [DOI] [PubMed] [Google Scholar]
- Konarska M.M. and Sharp,P.A. (1986) Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell, 46, 845–855. [DOI] [PubMed] [Google Scholar]
- Linberg A. and Kreivi,J.P. (2002) Splicing inhibition at the level of spliceosome assembly in the presence of herpes simplex virus protein ICP27. Virology, 294, 189–198. [DOI] [PubMed] [Google Scholar]
- Luo M.J. and Reed,R. (1999) Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl Acad. Sci. USA, 96, 14937–14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova O.V., Makarov,E.M. and Luhrmann,R. (2001) The 65 and 110 kDa SR-related proteins of the U4/U6.U5 tri-snRNP are essential for the assembly of mature spliceosomes. EMBO J., 20, 2553–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley J.L. and Tacke,R. (1996) SR proteins and splicing control. Genes Dev., 10, 1569–1579. [DOI] [PubMed] [Google Scholar]
- Mayeda A., Screaton,G.R., Chandler,S.D., Fu,X.D. and Krainer,A.R. (1999) Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Mol. Cell. Biol., 19, 1853–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears W.E. and Rice,S.A. (1996) The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J. Virol., 70, 7445–7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermoud J.E., Cohen,P.T.W. and Lamond,A.I. (1994) Regulation of mammalian splicesome assembly by a protein phosphorylation mechanism. EMBO J., 13, 5679–5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud S. and Reed,R. (1993) A functional association between the 5′ and 3′ splice sites is established in the earliest prespliceosome complex (E) in mammals. Genes Dev., 7, 1008–1020. [DOI] [PubMed] [Google Scholar]
- Misteli T. (1999) RNA splicing: what has phosphorylation got to do with it? Curr. Biol., 9, R198–R200. [DOI] [PubMed] [Google Scholar]
- Misteli T. and Spector,D.L. (1997) Protein phosphorylation and the nuclear organization of pre-mRNA splicing. Trends Cell Biol., 7, 135–138. [DOI] [PubMed] [Google Scholar]
- Neugebauer K.M. and Roth,M.B. (1997) Distribution of pre-mRNA splicing factors at sites of RNA polymerase II transcription. Genes Dev., 11, 1148–1159. [DOI] [PubMed] [Google Scholar]
- Nilsson C.E., Petersen-Mahrt,S., Durot,C., Shtrichman,R., Krainer,A.R., Kleinberger,T. and Akusjarvi,G. (2001) The adenovirus E4-ORF4 splicing enhancer protein interacts with a subset of phosphorylated SR proteins. EMBO J., 20, 864–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen B., Yun,C.Y., Wong,C.F., McCammon,J.A., Fu,X.D. and Ghosh,G. (2001) The structure of Sky1p reveals a novel mechanism for constitutive activity. Nat. Struct. Biol., 8, 176–183. [DOI] [PubMed] [Google Scholar]
- Phelan A., Carmo-Fonseca,M., McLauchlan,J., Lamond,A.I. and Clements,J.B. (1993) A herpes simplex virus type 1 immediate-early gene product, IE63, regulates small nuclear ribonucleoprotein distribution. Proc. Natl Acad. Sci. USA, 90, 9056–9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad J., Colwill,K., Pawson,T. and Manley,J.L. (1999) The protein kinase Clk/Sty directly modulates SR protein activity: both hyper- and hypophosphorylation inhibit splicing. Mol. Cell. Biol., 19, 6991–7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. and Palandjian,L. (1997) Spliceosome assembly. In Krainer,A.R. (ed.), Eukaryotic mRNA Processing. Oxford University Press, New York, NY, pp. 103–129.
- Roscigno R.F. and Garcia-Blanco,M.A. (1995) SR proteins escort the U4/U6-U5 tri-snRNP to the spliceosome. RNA, 1, 692–706. [PMC free article] [PubMed] [Google Scholar]
- Roth M.B., Zahler,A.M., and Stolk,J.A. (1991) A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J. Cell Biol., 115, 587–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco-Bubulya P. and Spector,D.L. (2002) Disassembly of inter chromatin granule clusters alters the coordination of transcription and pre-mRNA splicing. J. Cell Biol., 156, 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri-Goldin R.M. (1998a) ICP27 mediates herpes simplex virus RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev., 12, 868–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri-Goldin R.M. (1998b) Interactions between an HSV regulatory protein and cellular mRNA processing pathways. Methods, 16, 95–104. [DOI] [PubMed] [Google Scholar]
- Sandri-Goldin R.M. and Hibbard,M.K. (1996) The herpes simplex virus type 1 regulatory protein ICP27 coimmunoprecipitates with anti-Sm antiserum and the C-terminus appears to be required for this interaction. J. Virol., 70, 108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri-Goldin R.M., Hibbard,M.K. and Hardwicke,M.A. (1995) The C-terminal repressor region of HSV-1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J. Virol., 69, 6063–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford J.R. and Bruzik,J.P. (1999) Developmental regulation of SR protein phosphorylation and activity. Genes Dev., 13, 1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith I.L., Hardwicke,M.A. and Sandri-Goldin,R.M. (1992) Evidence that the herpes simplex virus immediate early protein ICP27 acts post-transcriptionally during infection to regulate gene expression. Virology, 186, 74–86. [DOI] [PubMed] [Google Scholar]
- Soliman T.M., Sandri-Goldin,R.M. and Silverstein,S. (1997) Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J. Virol., 71, 9188–9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke R., Chen,Y. and Manley,J.L. (1997) Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: creation of an SRp40-specific splicing enhancer. Proc. Natl Acad. Sci. USA, 94, 1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuma R.S., Stolk,J.A. and Roth,M.B. (1993) Identification and characterization of a sphere organelle protein. J. Cell Biol., 122, 767–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.-Y., Lin,W., Dyck,J.A., Yeakley,J.M., Songyang,Z., Cantley,L.C. and Fu,X.D. (1998) SRPK2, a differentially expressed SR protein-kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J. Cell Biol., 140, 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.Y. and Maniatis,T. (1993) Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell, 75, 1061–1070. [DOI] [PubMed] [Google Scholar]
- Xiao S.-H. and Manley,J.L. (1997) Phosphorylation of the ASF/SF2 RS domain affects both protein–protein and protein–RNA interactions and is necessary for splicing. Genes Dev., 11, 334–344. [DOI] [PubMed] [Google Scholar]
- Xiao S.-H. and Manley,J.L. (1998) Phosphorylation–dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J., 17, 6359–6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler A.M., Lane,W.S., Stolk,J.A. and Roth,M.B. (1992) SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev., 6, 837–847. [DOI] [PubMed] [Google Scholar]
- Zhi Y. and Sandri-Goldin,R.M. (1999) Analysis of the phosphorylation sites of the herpes simplex virus type 1 regulatory protein ICP27. J. Virol., 73, 3246–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi Y., Sciabica,K.S. and Sandri-Goldin,R.M. (1999) Self interaction of the herpes simplex virus type 1 regulatory protein ICP27. Virology, 257, 341–351. [DOI] [PubMed] [Google Scholar]