Abstract
Capping protein, a heterodimeric protein composed of α and β subunits, is a key cellular component regulating actin filament assembly and organization. It binds to the barbed ends of the filaments and works as a ‘cap’ by preventing the addition and loss of actin monomers at the end. Here we describe the crystal structure of the chicken sarcomeric capping protein CapZ at 2.1 Å resolution. The structure shows a striking resemblance between the α and β subunits, so that the entire molecule has a pseudo 2-fold rotational symmetry. CapZ has a pair of mobile extensions for actin binding, one of which also provides concomitant binding to another protein for the actin filament targeting. The mobile extensions probably form flexible links to the end of the actin filament with a pseudo 21 helical symmetry, enabling the docking of the two in a symmetry mismatch.
Keywords: actin filament/capping protein/CapZ/cytoskeleton/2-fold rotational symmetry
Introduction
Capping protein (Isenberg et al., 1980; Schafer and Cooper, 1995), also known as CapZ in muscle (Casella et al., 1987; Caldwell et al., 1989; Maruyama et al., 1990), is a heterodimeric protein composed of α and β subunits that is present in a wide variety of tissues and organisms. This protein is one of the key components that regulate the dynamic properties and organization of actin filaments, both temporally and spatially, in a variety of important biological processes. The actin filament is polar, so that the barbed end (fast-growing end) and the pointed end (slow-growing end) are distinct from each other in both their structural and kinetic properties. Capping protein binds to the barbed end with a high affinity (Kd ≈ 1 nM) and a 1:1 stoichiometry (Caldwell et al., 1989; Schafer et al., 1993), thus preventing the addition and loss of actin monomers at the end.
In terms of temporal control of the actin filament dynamics, capping protein is crucial for rapid filament elongation in response to cell signaling. Capping protein blocks a large fraction of the barbed ends, thus maintaining a high steady state concentration of actin monomers in the cell. Therefore, upon activation of the cell, actin polymerization is confined to a few non-capped filaments which individually grow faster than those in the system without capping protein (Pantaloni et al., 2001), resulting in cell motility such as the lamellipodial protrusion. The importance of capping protein has been experimentally demonstrated in that the absence of this protein prevented the reconstitution of the motility of Listeria and Shigella in vitro (Loisel et al., 1999). The functions of capping protein might also be regulated in vivo (Pantaloni et al., 2001), and one possible regulatory molecule is phosphatidylinositol-4,5-bisphosphate (PIP2) (Czech, 2000), which dissociates capping protein from the barbed ends in vitro (Schafer et al., 1996). Capping protein can also promote the nucleation of actin polymerization in vitro (Isenberg et al., 1980; Caldwell et al., 1989).
In terms of spatial control of the dynamics, capping protein plays important roles in targeting the actin filament ends to other structural components. The sarcomeric isoform of capping protein (CapZ α1β1) is localized at the Z-line in muscle (Casella et al., 1987; Schafer et al., 1994b), possibly through an interaction with α-actinin at the Z-line (Papa et al., 1999), whereas the non-sarcomeric isoforms are localized at the sites of membrane–actin contact (Amatruda and Cooper, 1992; Schafer et al., 1992, 1994b). These findings suggest that capping protein binds not only to actin but also to another component, thereby targeting and orienting the barbed ends of the actin filament at the proper place. Capping protein also interacts with twinfillin (Palmgren et al., 2001) and with the CARMIL protein, which further interacts with the Arp2/3 complex and myosin I (Jung et al., 2001), both of which are key players in actin-based cell motility. Capping protein also caps the Arp1 minifilament in the dynactin complex (Schafer et al., 1994a), an activator of the microtubule motor dynein.
Here we describe the crystal structure of chicken skeletal muscle CapZ at 2.1 Å resolution. The CapZ structure must be a good representative of the highly conserved capping protein family. The structure shows striking 2-fold symmetry, with a pair of mobile functional extensions that are well suited for binding to the end of the actin filament.
Results and discussion
Structure determination
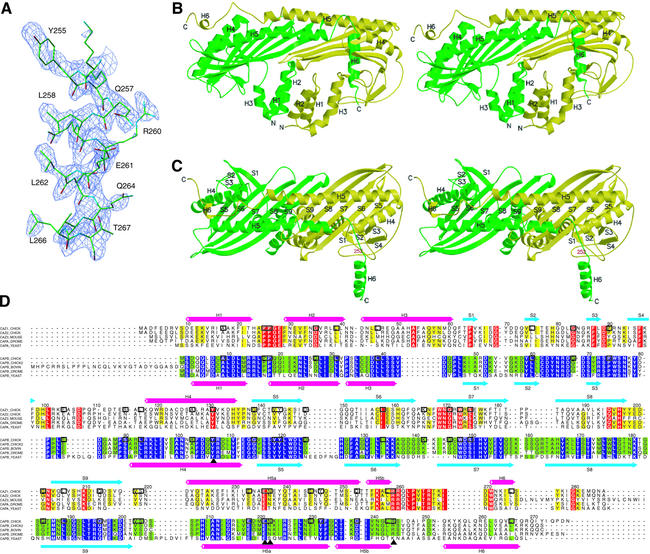
The recombinant chicken CapZ α1β1 heterodimer was crystallized at neutral pH (∼pH 6.5) and the structure was determined by the multiwavelength anomalous dispersion method, mainly using a gold-derivative crystal and adjunctly a platinum-derivative crystal (Table I). The electron density map obtained from the crystal at neutral pH showed no clear density at the C-terminal part of the β subunit (residues 252–277). However, the acidic crystal (∼pH 5.0), prepared by soaking the original native crystal, revealed unit cell shrinkage (see Materials and methods) and gave rise to a clear electron density of the α helix at the C-terminus of the β subunit (Figure 1A). The structure model was rebuilt and further refined against the diffraction data from the acidic crystal up to 2.1 Å resolution. The final model contains residues 7–281 (or 5–277 for the second molecule within the asymmetric unit) among the 286 residues for the α subunit, and residues 2–271 among the 277 residues for the β subunit, with an R-factor of 22.2% (Rfree = 26.2%) (Table I).
Table I. Data collection, phasing and refinement statistics.
| Data collection |
Native 1 |
Native 2 |
Gold derivative |
Platinum derivative |
||||
|---|---|---|---|---|---|---|---|---|
| Beamline |
SPring-8/BL45PX |
|
SPring-8/BL45PX |
SPring-8/BL44B2 |
||||
| Dataset | Remote | Peak | Edge | Remote | Peak | Edge | ||
| Wavelength (Å) | 1.0200 | 1.0200 | 1.0200 | 1.0395 | 1.0399 | 1.0688 | 1.0728 | 1.0731 |
| Resolution (Å) | 2.2 | 2.1 | 3.3 | 3.3 | 3.3 | 4.0 | 4.0 | 3.9 |
| Observed reflection | 210 797 | 228 951 | 119 941 | 119 763 | 116 669 | 73 776 | 73 476 | 80 138 |
| Unique reflection | 56 343 | 61 088 | 18 443 | 18 416 | 18 330 | 9849 | 9831 | 10 697 |
| Completeness (%)a | 99.8 (99.7) | 100.0 (100.0) | 99.8 (99.6) | 99.8 (99.6) | 99.8 (99.6) | 99.8 (100.0) | 99.8 (100.0) | 99.8 (100.0) |
| Rsyma,b | 0.061 (0.340) | 0.051 (0.265) | 0.080 (0.349) | 0.079 (0.331) | 0.076 (0.357) | 0.092 (0.383) | 0.095 (0.362) | 0.083 (0.365) |
|
I/σ(I) |
13.6 |
26.0 |
19.7 |
20.3 |
19.2 |
11.9 |
12.1 |
12.8 |
| Phasing | ||||||||
| Phasing power (cent./acent./anom.)c | na/na/2.02 | 2.40/3.16/1.88 | 1.85/2.69/1.78 | na/na/3.59 | 1.29/2.38/3.80 | 1.06/1.62/3.12 | ||
| Rcullis (cent./acent./anom.)d | na/na/0.77 | 0.52/0.49/0.80 | 0.55/0.53/0.83 | na/na/0.61 | 0.56/0.52/0.57 | 0.67/0.63/0.68 | ||
| Mean figure of merit | 0.542 | 0.611 | ||||||
| Refinement against Native 2 data | ||||||||
| Resolution (Å) | 50.0–2.1 | |||||||
| Reflections | 60 190 | |||||||
| Re | 0.222 | |||||||
| Rfreee | 0.262 | |||||||
| No. of atoms | ||||||||
| Protein | 8743 | |||||||
| Water | 513 | |||||||
| Nitrate ion | 16 | |||||||
| Average B (Å2) | 43.7 | |||||||
| Ramachandran plot | ||||||||
| Most favored (%) | 90.3 | |||||||
| Allowed (%) | 9.7 | |||||||
| R.m.s. deviations | ||||||||
| Bond length (Å) | 0.007 | |||||||
| Bond angle (°) | 1.378 | |||||||
| Luzzati error (Å) | 0.28 | |||||||
aValues in parentheses refer to data in the highest resolution shell.
bRsym = Σ|I – <I>|/ΣI, where I is the observed intensity and <I> is the average intensity from multiple observations of the symmetry-related reflections.
cPhasing power = <|FH(calc)|/E>, where E is the phase-integrated lack of closure. The three values for each wavelength are for centric (cent.), acentric isomorphous (acent.) and acentric anomalous (anom.) contributions.
dRcullis = <E>/<|FPH – FP|>.
eR = Σ||Fobs| – |Fcalc||/Σ|Fobs|. Rfree is the R-value for a subset of 5% of the reflection data, which were not included in the crystallographic refinement.
Fig. 1. Structure of the chicken CapZ α1β1 heterodimer. (A) The sigmaA-weighted Fo – Fc omit map with the refined structure of the C-terminal helix of the β subunit. The electron density map is contoured at 2.4 σ, and residues 252–271 are omitted for the phase calculation. (B and C) Stereo view of the ribbon representation of the CapZ crystal structure. The α and β subunits are shown in yellow and green, respectively. In (C), the molecule is viewed from the top side of that in (B), and residue 252 of the β subunit, the first residue of the disordered region in the Native 1 crystal, is indicated in red. (D) Sequence alignment of the proteins in the capping protein family. The sequences of chicken CapZ α1 (SWISS-PROT entry name CAZ1_CHICK) and β1 (CAPB_CHICK) are aligned based on their structures; those of chicken α2 (CAZ2_CHICK), mouse α3 (CAZ3_MOUSE), fruit fly α (CAPA_DROME) and yeast α (CAPA_YEAST) with chicken α1, and those of chicken β2 (alternative splice isoform of CAPB_CHICK, shown as CAPB_CHICK2 in the alignment), bovine β3 (CAPB_BOVINE), fruit fly β (CAPB_DROME) and yeast β (CAPB_YEAST) with chicken β1 are aligned with CLUSTAL W (Thompson et al., 1994). The residues conserved among ≥80% of the proteins of the same subunit, α or β, are indicated in yellow or green, and the strictly conserved residues are indicated in red or purple, respectively. The identical residues between chicken α1 and β1 are highlighted with boxes, and the important residues for stabilizing the structure described in the text are labeled with triangles. The secondary structures of chicken α1 and β1 are indicated above and below the sequences, respectively. The residue numbers for these two subunits are also shown.
Overall structure
CapZ has an elongated structure, with overall dimensions of ∼90 × 50 × 55 Å, in which the α and β subunits assemble tightly, constituting a single heterodimeric molecule (Figure 1B and C). The two subunits are strikingly similar to each other, so that the entire molecule has a pseudo 2-fold rotational symmetry around the axis perpendicular to the long axis of the molecule. Each subunit can be divided into three domains and an additional C-terminal extension. The N-terminal domain consists of a bundle of three antiparallel helices in an up–down–up arrangement (helices 1–3). The middle domain is composed of four β strands (strands 1–4 for the α subunit; three β strands 1–3 for the β subunit), making two reverse turns packed in a characteristic manner. The C-terminal domain comprises an anti parallel β sheet formed by five consecutive β strands (strands 5–9), flanked on one side by a shorter N-terminal helix (helix 4) and a long C-terminal helix (helix 5). The β strands of each subunit form a single 10-stranded antiparallel β sheet in the center of the molecule. The long α helix of each subunit is roughly aligned with the shorter α helix of the other subunit, being juxtaposed on the β sheet in an antiparallel manner. The C-terminal domain has an extension at the C-terminus, consisting of a long loop followed by a helix (helix 6). Helix 6 of the β subunit, which is longer than the counterpart of the α subunit, protrudes from the core structure. A DALI database search (Holm and Sander, 1996) did not reveal any known protein structures significantly similar to the CapZ structure.
Similarity between the subunits
No sequence similarity between the α and β subunits has been detected thus far. However, when the sequences are compared based on the present crystal structure (Figure 1D), the residues crucial for maintaining the overall architecture, such as Tyr131(α)/Tyr107(β), Tyr144(α)/Tyr120(β), Glu236(α)/Glu221(β), Asn237(α)/Asn222(β) and Arg259(α)/Arg244(β), are obviously conserved between the two subunits. Many hydrophobic residues are also conserved or equivalently substituted, which must substantially contribute to the stabilization of the individual subunit structure as well as that of the dimer association (see below). These residues are also conserved or equivalently substituted among the same subunits of different isoforms or species. The structures of the α and β subunits superimpose well, with a root mean square (r.m.s.) deviation of 3.43 Å over 183 equivalent Cα positions, except for the N-terminal domains and the C-terminal extensions, which are tilted differently. Despite the poor sequence similarity, the high degree of structural conservation strongly suggests that the α and β subunits of CapZ might have arisen from gene duplication and thus belong to the same (and possibly a new) protein superfamily.
Subunit interaction
The structure shows tight interactions between the α and β subunits, indicating that CapZ exists and functions as a stable heterodimer. The interactions are dominated by those between the C-terminal domains, but those between the N-terminal domains are also important. Of the total accessible surface area of each subunit (α, 17 200 Å2; β, 17 000 Å2), ∼23% (3900 Å2) is in the buried region for the inter-subunit contact. Of this buried area, 2900 Å (∼74% of 3900 Å2) is buried between the C-terminal domains, whereas 760 Å2 (∼19% of 3900 Å2) is buried between the N-terminal domains.
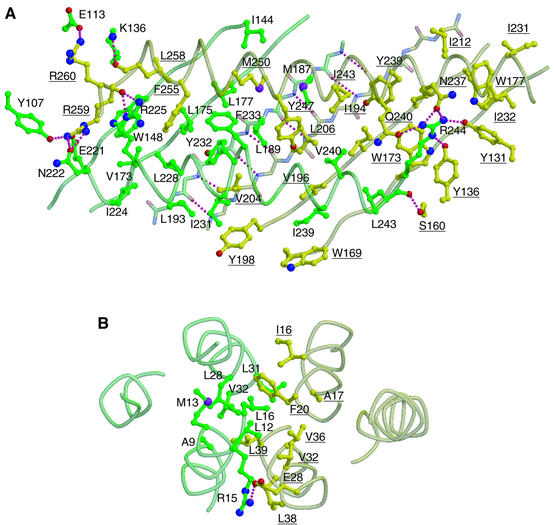
Between the C-terminal domains (Figure 2A), strand 9 of each subunit at the dimer interface, including residues 201–209 of the α subunit and 186–194 of the β subunit, is involved in interstrand main-chain hydrogen bonding and constitutes a single antiparallel β sheet together with the other strands in the domain. Between the β sheet and the overlying α helices, remarkable hydrophobic interactions are observed with the residues indicated in Figure 2A. The two helices are also tightly packed together, so that no groove is left between them, forming another inter-subunit interface. These hydrophobic interactions are further reinforced by the interactions involving Arg259 of the α subunit and Arg244 of the β subunit, two strictly conserved residues located at symmetrically related positions in the molecule at the C-terminal end of each helix 5. These residues form side-chain hydrogen bonds with the surrounding residues of the other subunit, such as Arg259(α) with Tyr107, Glu221 and Asn222 of the β subunit, and Arg244(β) with Tyr131, Tyr136, Asn237 and Gln240 of the α subunit (Figure 2A), which are also conserved across isoforms and species (Figure 1D). The main-chain atoms adjacent to the two arginines also contribute to the interactions, as do those of the carbonyl oxygen atoms of Leu258(α) and Arg259(α) with the side-chains of Lys136(β) and Arg225(β), respectively, and the main-chain carbonyl oxygen of Leu243(β) with the side-chain of Ser160(α). In addition to these inter-subunit interactions, these two arginine residues seem to play important roles in actin binding (see below).
Fig. 2. Residues contributing to the interactions between the two subunits. (A) Interface between the C-terminal domains and (B) interface between the N-terminal domains. In (A) and (B), the molecule is viewed from almost the same direction as in Figure 1C. The α and β subunits are shown in yellow and green, respectively. In the ball-and-stick models, the oxygen, nitrogen and sulfur atoms are shown in red, blue and purple, respectively. The pink dotted lines indicate inter-subunit hydrogen bonds. Residue numbers of the α subunit are underlined.
The dimer interactions between the N-terminal domains (Figure 2B) take place through the formation of a four-helix bundle, with two helices (helices 1 and 2) from each subunit. These interactions are also mainly hydrophobic, formed by the residues as indicated in Figure 2B. The interactions are further enforced by an inter-subunit salt bridge between Glu28(α) and Arg15(β).
The chicken CapZ deletion mutant consisting solely of the C-terminal domains retains the heterodimer assembly in vitro (Casella and Torres, 1994), indicating that the interaction between the C-terminal domains is robust and important for heterodimer assembly. On the other hand, the deletion of the N-terminal domains of yeast capping protein led to the loss of the protein in vivo (Sizonenko et al., 1996). Since in yeast null mutants of one subunit lead to the loss of the other subunit in vivo (Amatruda et al., 1992), capping protein probably only exists stably when the proper heterodimer is formed. Therefore, although the results of chicken CapZ and yeast capping protein appear to be inconsistent with each other with regard to the necessity of the N-terminal domain for heterodimer stability, the most straightforward interpretation of the yeast result would be that, in the in vivo expressed protein, the N-terminal domain is also required for heterodimer stability.
The properties of the dimer interfaces explain why αβ heterodimers, not αα or ββ homodimers, are exclusively formed. The surface topologies of the C-terminal domains of each subunit are complementary, so that the depressions or protrusions of each subunit interface make van der Waals contacts properly only when the heterodimer is formed. The different tilt of the N-terminal domains described above might also contribute to preventing homodimer assembly, since in the case of the homodimer the four-helix bundle formation at the interface might not occur and it would bring about a dramatic decrease in inter-subunit interactions.
Actin-binding sites
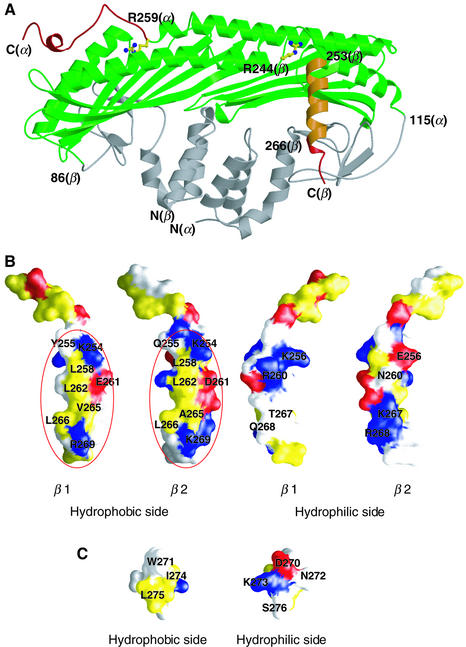
The actin-binding sites in CapZ are identified on two distinct regions (Figure 3A), based on the crystal structure in conjunction with previously reported deletion mutant analyses. The mutant analyses indicated that the C-terminal end of each subunit, especially residues 259–286 of the α subunit (based on the result from the yeast capping protein) and 266–277 of the β subunit, are crucially required for actin binding (Hug et al., 1992; Casella and Torres, 1994; Sizonenko et al., 1996). This interpretation is justified by the present crystal structure which probably excludes the possibility that the deletions cause destabilization of folding of each subunit and the heterodimer assembly. Since one CapZ binds to one barbed end (Caldwell et al., 1989; Schafer et al., 1993), consisting of two actin monomers, and since the two C-terminal extensions are rather far apart, it is highly likely that each of them interacts with each of the two actin monomers at the filament end. This two-to-two binding scheme nicely explains why CapZ specifically binds to an actin filament and not to an actin monomer. The 2-fold symmetrical architecture of CapZ is advantageous to provision of this two-to-two interaction.
Fig. 3. Actin-binding extensions of CapZ. (A) The regions crucial for actin binding [residues 259-C-terminus(α) and 266-C-terminus(β)], the segment which exhibits actin-binding ability [residues 253-C-terminus(β)] and the regions that are not required for high-affinity actin binding [residues N-terminus-115(α) and N-terminus-86(β)] are mapped in red, orange and gray, respectively, on the CapZ structure based on the previous deletion mutant experiments (Hug et al., 1992; Casella and Torres, 1994; Sizonenko et al., 1996). The two conserved arginine residues, Arg259(α) and Arg244(β), are indicated with ball-and-stick models. (B) Surface representations of the C-terminal extension (residues 246–271, the sole variant region between the isoforms) of two isoforms of β, β1 (the present structure) and β2 (a homology model). The left two panels show the hydrophobic sides of the helices, and the right two panels show the hydrophilic sides. Hydrophobic, acidic and basic residues are colored in yellow, red and blue, respectively. Possible actin-binding sites are highlighted with red circles. (C) Surface representations of helix 6 (residues 270–276) of the α subunit. The color coding is the same as in (B).
Apart from the C-terminal regions of the α and β subunits, there could be extra actin-binding sites on CapZ, although no evidence is available at present. The additional binding sites must reside within the C-terminal domain, since the N-terminal and middle domains are not required for the high-affinity binding (Casella and Torres, 1994). Thus the other regions in the C-terminal domains, such as helix 5 or the β-turn regions, could also contribute to actin binding. The other results of previous deletion experiments will not be discussed here, since, in the light of the present crystal structure, the deletions probably caused deterioration of the proper folding of the protein.
Multiple binding sites on the C-terminal helix of the β subunit
The C-terminal helix of the β subunit (helix 6) is amphiphilic: one side is hydrophobic and the other is hydrophilic (Figure 3B). Interestingly, although the common function of actin binding is attributed to this region, there is no apparent sequence homology among the isoforms or species in this region (Figure 1D). The puzzle is particularly represented by the two different isoforms β1 and β2, which are identical in the first 246 amino acids and differ exclusively in the C-terminal extension. Neverthe less, they exhibit almost the same actin-binding activity (Schafer et al., 1996). A closer inspection of the structure reveals that the residues on the hydrophobic side, such as Leu258, Glu261, Leu262, Val265, Leu266 and Arg269, are either almost conserved or equivalently substituted (Figures 1D and 3B). Indeed, mutational experiments suggested that some of the above residues (Leu262 and Leu266) are important for actin binding (Barron-Casella et al., 1995). These results indicate that CapZ binds to actin via the hydrophobic side of the C-terminal helix. In the C-terminal region of the α subunit, the other possible actin-binding region, the helix is also amphiphilic, with a hydrophobic surface composed of Trp271, Ile274 and Leu275 (Figure 3C), which are conserved or equivalently substituted.
On the other hand, the hydrophilic side of the helix of the β subunit has an isoform-characteristic charge distribution pattern (Figure 3B). Comparison of the β1 and β2 isoforms reveals differences at residues 256 (β1:Lys/β2:Glu), 260 (Arg/Asn), 267 (Thr/Lys) and 268 (Gln/Arg). The two isoforms display tissue-specific expression and distinct subcellular localizations, as β1 is localized at the Z-lines of sarcomeres whereas β2 is generally localized at the cell–cell junctions and the plasma membrane in striated muscle cells (Schafer et al., 1994b), and thus they are considered to have distinct functions in vivo (Hart and Cooper, 1999). Therefore the isoform-specific hydrophilic side might be recognized by isoform-specific target proteins, resulting in targeting of CapZ and thus of the actin filament ends in the cell. [However, at present it is not possible to exclude the possibility that the five-residue extension of the β1 C-terminus (Figure 1D) also contributes to the recognition by the targets.] The amphiphilic helix might provide two interaction sites, one on the hydrophobic side and the other on the hydrophilic side, as in the α helix of a mitochondrial protein presequence that accomodates multiple signals for protein import receptors (Abe et al., 2000). The helix of the CapZ β subunit has probably evolved with one side conserved for hydrophobic interactions with actin, while the other side diversified for binding to a specific target protein. Apart from the hydrophilic side of helix 6 of the β subunit, the insertions specifically found in the β3 isoform and in yeast protein could also contribute to interactions with target proteins and determination of the localization (Figure 1D).
Possible CapZ binding site on actin
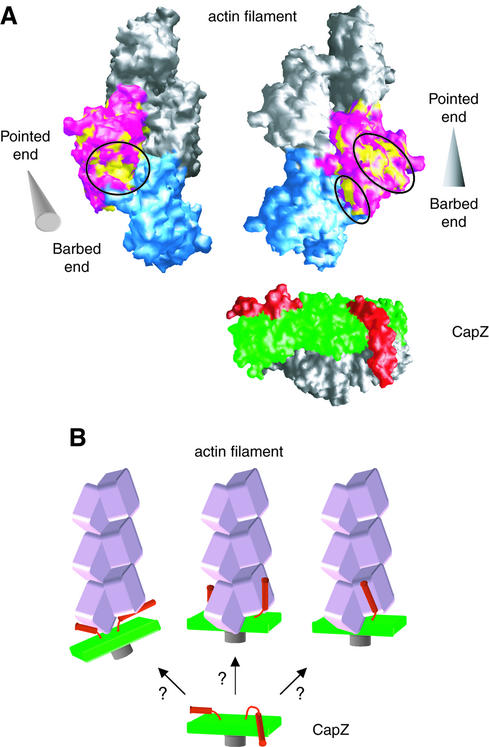
The hydrophobic nature of the actin-binding sites of CapZ suggests that the putative CapZ-binding sites on actin must also be hydrophobic. One obvious hydrophobic region is at the cleft between actin subdomains 1 and 3 (Figure 4A, left panel). Several actin-binding proteins, such as gelsolin (McLaughlin et al., 1993; Robinson et al., 1999) and the vitamin-D-binding protein (Otterbein et al., 2002), bind to this region with the hydrophobic surfaces of α helices. Other candidates are at the surface of subdomain 3 and the neighboring regions, or at the subunit interface of subdomain 3, at the cleft in the center of the actin filament (Figure 4A, right panel). At present, no experimental evidence is available to distinguish between these possible sites.
Fig. 4. Actin–CapZ interaction. (A) Surface representations of an actin filament model (Lorenz et al., 1993), viewed from two different directions. In the left panel, the filament axis runs across and oblique to the plane of the page with the barbed end in the foreground, while in the right panel, the axis runs on the plane. The cones at the sides of the filaments indicate the filament orientations with the apex and the base indicating the pointed and barbed ends, respectively. One of the two actin molecules at the barbed end is colored in pink, and the hydrophobic residues on its surface are in yellow. The hydrophobic patches are highlighted with circles. The other actin molecule at the end is colored in cyan. The molecular surface of CapZ is also shown on the same scale, with the pair of C-terminal extensions [residues α(260-C-terminus) and β(245-C-terminus)] in red, and the N-terminal domains (residues α(N-terminus-115) and β(N-terminus-86)] in gray. (B) Schematic drawings of the possible binding modes of capping protein to the actin filament (speculative models).
CapZ–actin filament interaction
With regard to the docking of CapZ to the actin filament, the symmetry mismatch is remarkable. The CapZ structure has a pseudo 2-fold rotational symmetry, while the two actin monomers at the filament end are related to each other by a pseudo 21 helical symmetry, i.e. by a rotation of 166° with a translation of 27.5 Å along the filament axis.
The present crystal structure of CapZ suggests that the actin-binding C-terminal region of each subunit is essentially mobile and flexible, thus reconciling the symmetry mismatch. First, the C-terminus of the β subunit has no interaction at all with the rest of the molecule. The region was originally disordered in the crystal at neutral pH, and became fixed due to the molecular packing arising from the unit cell shrinkage in the acidic crystal. The high average B-factor of helix 6 (53.7 Å2 for the main-chain atoms) also reflects the mobility of this region. The C-terminus of the α subunit is likely to be mobile and flexible as well, because it shares a similar architecture and might also have become fixed in the crystal as a result of the molecular packing, although it currently sits between helices 4 and 5 of the β subunit in the crystal structure. This extension also has significantly higher B-factors (an average of 36.1 Å2 for the main-chain atoms in helix 6) than the adjacent regions in the molecule, such as helix 4 (18.3 Å2) and helix 5a (23.3 Å2) of the β subunit. Secondly, each mobile extension is well defined by a highly con served arginine residue, either Arg259(α) or Arg244(β), which makes tight interactions with the surrounding residues as described above (Figures 2A and 3A). The arginine residue must anchor the extension to the rest of the molecule, forming a pivot on which the extension moves. The mutation of Arg259(α) together with Arg260(α) caused a loss of function in the yeast capping protein (Sizonenko et al., 1996), indicating the functional significance of this residue. Thirdly, the C-terminal regions of the α and β subunits in the present structure cannot interact simultaneously with the two actin molecules at the filament end without moving from the rest of the molecule. This is because the C-terminal extensions themselves and the hydrophobic sides of the amphiphilic helices 6 at their present positions in the crystal seem to be incorrectly oriented for binding to the filament end. Therefore, owing to their flexible and mobile nature, each C-terminal extension probably moves out from the ‘body’ to bind an actin filament, like a pair of ‘tentacles’, enabling the binding of CapZ to two actin molecules at the filament end (Figure 4B).
Another factor facilitating docking, despite the symmetry mismatch, is the fact that CapZ is a heterodimer rather than a homodimer. Despite the overall similarity of the two subunits, they differ in their finer details. In particular, the two C-terminal ‘tentacles’ of the α and β subunits actually have different loop lengths, helix lengths and residue compositions. These differences might have been acquired through the independent evolution of the two subunits from a common origin.
Dynamic capping by CapZ
The ‘tentacular’ binding mechanism with the two mobile extensions could account for the dynamic aspect of actin filament capping by CapZ at the Z-line; exogenous actin can be incorporated at the barbed end of thin filaments in striated muscle in vivo (Littlefield et al., 2001). The flexibility of the CapZ extensions probably enables temporal uncapping of the actin filament without disrupting the sarcomeric structure, while CapZ is kept anchored to the Z-line through the interaction with the target proteins as described previously (Papa et al., 1999). Alternatively or additionally, the double linkers might serve for actin monomer exchange at the end without complete detachment of CapZ from the actin filament by retaining one linker attached. The mobility of the ‘tentacles’ is also consistent with the ability of this molecule to nucleate polymerization of the actin filament, which is observed in vitro; the two mobile ‘tentacles’ may bind actin monomers in solution, facilitating and stabilizing the stochastic formation of the critical nucleus for filament growth.
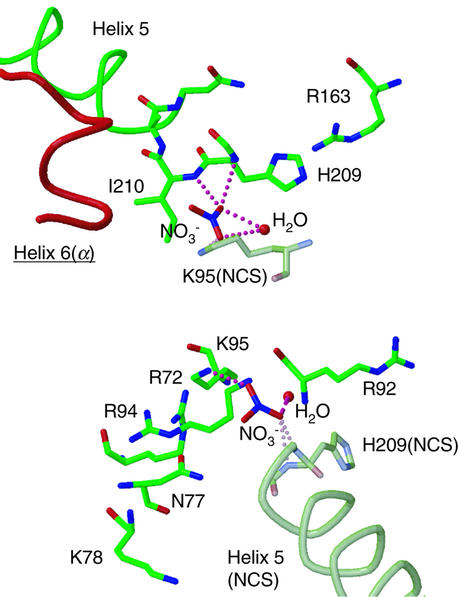
Nitrate ions and implication for PIP2 binding
The present crystal structure contains four nitrate ions, which were added for crystallization as magnesium nitrate, providing a clue to the possible binding sites of PIP2. The electron densities of two nitrate ions were found at the interface between the two molecules related by non-crystallographic symmetry (NCS), bound to the β subunit. One nitrate interacts with Lys95 while the other interacts with the helix dipole of helix 5 (Figure 5), and these binding characteristics were conserved between the two NCS molecules. In some phosphatase and kinase structures, nitrate ions have been found at the phosphate binding sites as mimics of the metaphosphate transition states (Fauman et al., 1996; Poland et al., 1996; Zhou et al., 1998; Cook et al., 2002). The specific nitrate binding observed in the CapZ structure suggests that the nitrate binding sites are the candidates for the phosphate binding sites of PIP2, although CapZ, unlike phosphatases, has no catalytic activity with phosphate groups. Several other basic residues are also found around these nitrate-binding sites, and thus PIP2 binding to these sites might be plausible. Since these binding sites are located near the actin-binding C-terminal extension of the α subunit (Figure 5), the PIP2 binding could modulate the binding of CapZ to the actin filament. Both sites must be solvent accessible under a physiological state, namely either in solution or probably even when CapZ binds to the actin filament in the manner proposed above, although these sites reside at the interface between two molecules so that these are not accessible in the crystal. Our attempts to bind PIP2 (with short alkyl chains or just using polar head groups as inositol-1,4,5-trisphosphate) to the CapZ molecule by soaking the crystals have not been successful; no electron density assignable to PIP2 molecules around the CapZ molecule has been revealed (data not shown).
Fig. 5. Two nitrate ions bound to the β subunits at the interface between two NCS molecules. Helix 6 of the actin-binding C-terminal extension of the α subunit is also shown in red. The pink dotted lines indicate hydrogen bonds. The β subunit of the NCS-related molecule is shown in shaded colors.
The other two nitrates were found at the NCS-equivalent positions in either of the two molecules in the asymmetric unit, which are in the cavities of the middle domain of the β subunit, surrounded by Gln45, Asp63 and Tyr71. This site is also solvent accessible and could be a candidate for PIP2 binding site. However, this is unlikely. Native 1 data (from the original crystal at neutral pH) showed no electron density of the nitrate there (data not shown), indicating that this nitrate binding is less specific than the two described above.
Conclusion
The crystal structure of CapZ provides a structural basis for understanding how capping protein caps the actin filament and how it localizes the filament to a target through interactions with other components. The symmetrical but heterodimeric structure, with a pair of mobile functional extensions, accounts for these mechanisms well. For further clarification of the mechanisms, more extensive work is required to elucidate the interactions of capping protein with other components, such as the actin filament and other target proteins. It is especially challenging and important to understand how CapZ binds to the barbed end of the actin filament. In order to address the question, structural studies using electron microscopy are currently in progress in our laboratory.
Materials and methods
Protein preparation and crystallization
The chicken CapZα1 and β1 subunits were co-expressed in Escherichia coli using pET-3d-βI′/βII, an expression vector generously provided by Professor T.Obinata of Chiba University, and were purified as described (Soeno et al., 1998) with some modifications. Briefly, cells expressing the CapZ α1 and β1 subunits were lysed by sonication, and the supernatant was fractionated with ammonium sulfate (50–70% saturation). The CapZ α1β1 heterodimer was purified using a Macro-Prep ceramic hydroxyapatite (type I) column (Bio-Rad), followed by chromatography on DEAE cellulose (DE52, Whatman) and Uno-S columns (Bio-Rad). Throughout the purification procedures, the two subunits formed a stable heterodimer. The dynamic light scattering and the small-angle X-ray scattering analyses confirmed that the purified CapZ sample existed as a stable 1:1 heterodimer molecule, without any dissociation of the subunits or formation of higher aggregates (data not shown). Crystals of CapZ were obtained at 20°C by the hanging-drop vapor diffusion method from the protein sample (10 mg/ml, in 5 mM Tris–HCl pH 8.0, 1 mM dithiothreitol and 5% Jeffamine M-600) combined in a 1:1 ratio with a reservoir solution consisting of 18% PEG3350, 0.2 M Mg(NO3)2 and 0.1 M MES–NaOH pH 6.0. The crystals were transferred to a stock solution of 20% PEG3350, 0.2 M Mg(NO3)2, 20% ethylene glycol, 5% Jeffamine M-600 and 50 mM MES–NaOH pH 6.0 (the final pH of the mixed solution was ∼6.5, and the crystals under this condition are denoted as ‘crystals at neutral pH’), and were flash frozen in a cold nitrogen flow. The gold derivative was obtained by soaking the crystal in the stock solution (without Jeffamine M-600) containing 3 mM KAu(CN)2 for 2 days, and the platinum derivative was obtained by soaking in the solution containing 1 mM K2Pt(SCN)6 for 2 days. The native crystal under acidic conditions (the acidic crystal) was prepared by soaking the crystal in a solution of 20% PEG3350, 0.2 M Mg(NO3)2, 20% ethylene glycol, 5% Jeffamine M-600 and 50 mM sodium acetate buffer pH 4.0 (the final pH of the mixed solution was ∼5.0). The CapZ crystals belong to the space group P21, with the following unit cell dimensions. Crystal at neutral pH (Native 1): a = 100.7 Å, b = 59.4 Å, c = 102.2 Å and β = 113.6°. Acidic crystal (Native 2): a = 99.2 Å, b = 57.2 Å, c = 99.2 Å and β = 113.0°. Gold derivative: a = 100.9 Å, b = 59.6 Å, c = 103.8 Å and β = 114.0°. Platinum derivative: a = 100.3 Å, b = 60.1 Å, c = 105.1 Å and β = 114.1°. Each asymmetric unit contains two heterodimer molecules and 38–45% solvent.
Data collection and structure determination
Diffraction data were collected at 90 K with an R-AXIS V imaging plate detector at beamline BL45XU-PX or with a MARCCD165 detector at beamline BL44B2, SPring-8. The data were processed using DENZO and SCALEPACK (Otwinowski and Minor, 1997). The positions of the six gold atoms were determined using SOLVE (Terwilliger and Berendzen, 1999), and the refinement and phase calculations were carried out using SHARP (de La Fortelle and Bricogne, 1997). The electron density map obtained at 3.3 Å enabled five α helices (including helix 5 of the α and β subunits) of each heterodimer molecule to be traced. The NCS operator and the molecular masks were determined using these helices. Density modification, including NCS averaging, solvent flattening and histogram matching, were performed with DM (Cowtan, 1994). The Au-MAD phases were also further improved by combination of the Pt-MAD phases (calculated with SHARP) using SIGMAA (Read, 1986) (resultant mean figure of merit, 0.600) followed by DM, and the density map derived was used to assist chain tracing in some ambiguous parts. Model building was carried out with TURBO-FRODO (Roussel and Cambillau, 1996), and the phases derived from a partial model were combined with the Au-MAD phases using SIGMAA for further improvement. At first, the structure model was refined against the Native 1 dataset (crystal at neutral pH) using CNS (Brünger et al., 1998), including rigid body, simulated annealing, minimize and individual B-factor refinement, followed by manual rebuilding, in several cycles. However, this dataset did not yield a clear electron density at the C-terminal part of the β subunit (residues 252–277). We then switched to the Native 2 dataset (acidic crystal) for refinement, which enabled model-building of the missing C-terminus of the β subunit. The model was further refined against dataset 2 with CNS, as described above. No NCS restraints were applied at the final stage of the refinement. The structures of the two heterodimer molecules in an asymmetric unit have no major differences, and one of the heterodimer structures is indicated as a representative in the figures. Graphical presentations were prepared using the programs TURBO-FRODO (Roussel and Cambillau, 1996), MOLSCRIPT (Kraulis, 1991), RASTER3D (Meritt and Murphy, 1994), GRASP (Nicholls et al., 1991) and ALSCRIPT (Barton, 1993).
Structure data
The atomic coordinates and the structure factors of CapZ have been deposited in the Protein Data Bank (PDB accession code 1IZN).
Acknowledgments
Acknowledgements
We thank T.Obibata for the expression vector, K.-I.Sano for making baculovirus expression constructs in early crystallization stages, Y.Kawano, T.Hikima and S.Takeda for help with X-ray data collection, Y.Nishikawa and T.Fujisawa for small-angle X-ray scattering analysis, R.C.Robinson and R.Dominguez for providing the Arp2/3 complex and the vitamin-D-binding protein–actin complex coordinates and discussions, J.A.Cooper, D.Sept, K.Mizuguchi and A.G.Murzin for discussions, and D.Popp for critical reading of the manuscript. Part of this study was supported by the Special Coordination Funds for Promoting Science and Technology from the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government.
References
- Abe Y., Shodai,T., Muto,T., Mihara,K., Torii,H., Nishikawa,S., Endo,T. and Kohda,D. (2000) Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell, 100, 551–560. [DOI] [PubMed] [Google Scholar]
- Amatruda J.F. and Cooper,J.A. (1992) Purification, characterization and immunofluorescence localization of Saccharomyces cerecisiae capping protein. J. Cell Biol., 117, 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amatruda J.F., Gattermeier,D.J., Karpova,T.S. and Cooper,J.A. (1992) Effects of null mutations and overexpression of capping protein on morphogenesis, actin distribution and polarized secretion in yeast. J. Cell Biol., 119, 1151–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron-Casella E.A., Torres,M.A., Scherer,S.W., Heng,H.H.Q., Tsui,L.-C. and Casella,J.F. (1995) Sequence analysis and chromosomal localization of human CapZ. Conserved residues within the actin-binding domain may link CapZ to gelsolin/severin and profilin protein families. J. Biol. Chem., 270, 21472–21479. [DOI] [PubMed] [Google Scholar]
- Barton G.J. (1993) ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng., 6, 37–40. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for maclomolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Caldwell J.E., Heiss,S.G., Mermall,V. and Cooper,J.A. (1989) Effects of CapZ, an actin capping protein of muscle, on the polymerization of actin. Biochemistry, 28, 8506–8514. [DOI] [PubMed] [Google Scholar]
- Casella J.F. and Torres,M.A. (1994) Interaction of CapZ with actin. The NH2-terminal domains of the α1 and β1 subunits are not required for actin capping, and α1β and α2β heterodimers bind differentially to actin. J. Biol. Chem., 269, 6992–6998. [PubMed] [Google Scholar]
- Casella J.F., Craig,S.W., Maack,D.J. and Brown,A.E. (1987) CapZ(36/32), a barbed end actin-capping protein, is a component of the Z-line of skeletal muscle. J. Cell Biol., 105, 371–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook A., Lowe,E.D., Chrysina,E.D., Skamnaki,V.T., Oikonomakos,N.G. and Johnson,L.N. (2002) Structural studies on phospho-CDK2/cyclin A bound to nitrate, a transition state analogue: implications for the protein kinase mechanism. Biochemistry, 41, 7301–7311. [DOI] [PubMed] [Google Scholar]
- Cowtan K. (1994) DM: an automated procedure for phase improvement by density modification. Joint CCP4 and ESF–EACBM Newsletter on Protein Crystallography, 31, 34–38. [Google Scholar]
- Czech M.P. (2000) PIP2 and PIP3: complex roles at the cell surface. Cell, 100, 603–606. [DOI] [PubMed] [Google Scholar]
- de La Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Fauman E.B., Yuvaniyama,C., Schubert,H.L., Stuckey,J.A. and Saper,M.A. (1996) The X-ray crystal structures of Yersinia tyrosine phosphatase with bound tungstate and nitrate. J. Biol. Chem., 271, 18780–18788. [DOI] [PubMed] [Google Scholar]
- Hart M.C. and Cooper,J.A. (1999) Vertebrate isoforms of actin capping protein β have distinct functions in vivo. J. Cell Biol., 147, 1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1996) Mapping the protein universe. Science, 273, 595–602. [DOI] [PubMed] [Google Scholar]
- Hug C., Miller,T.M., Torres,M.A., Casella,J.F. and Cooper,J.A. (1992) Identification and characterization of an actin-binding site of CapZ. J. Cell Biol., 116, 923–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenberg G., Aebi,U. and Pollard,T.D. (1980) An actin-binding protein from Acanthamoeba regulates actin filament polymerization and interactions. Nature, 288, 455–459. [DOI] [PubMed] [Google Scholar]
- Jung G., Remmert,K., Wu,X., Volosky,J.M. and Hammer,J.A.,III (2001) The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. J. Cell Biol., 153, 1479–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots for protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Littlefield R., Almenar-Queralt,A. and Fowler,V.M. (2001) Actin dynamics at pointed ends regulates thin filament length in striated muscle. Nat. Cell Biol., 3, 544–551. [DOI] [PubMed] [Google Scholar]
- Loisel T.P., Boujemaa,R., Pantaloni,D. and Carlier,M.-F. (1999) Reconstruction of actin-based motility of Listeria and Shigella using pure proteins. Nature, 401, 613–616. [DOI] [PubMed] [Google Scholar]
- Lorenz M., Popp,D. and Holmes,K.C. (1993) Refinement of the F-actin model against X-ray fiber diffraction data by the use of a directed mutation algorithm. J. Mol. Biol., 234, 826–836. [DOI] [PubMed] [Google Scholar]
- McLaughlin P.J., Gooch,J.T., Mannherz,H.-G. and Weeds,A.G. (1993) Structure of gelsolin segment 1–actin complex and the mechanism of filament severing. Nature, 364, 685–692. [DOI] [PubMed] [Google Scholar]
- Maruyama K., Kurokawa,H., Oosawa,M., Shimaoka,S., Yamamoto,H., Ito,M. and Maruyama,K. (1990) β-actinin is equivalent to Cap Z protein. J. Biol. Chem., 265, 8712–8715. [PubMed] [Google Scholar]
- Meritt E.A. and Murphy,M.E.P. (1994) Raster3D Version 2.0—a program for photorealistic molecular graphics. Acta Crystallogr. D, 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Genet., 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Otterbein L.R., Cosio,C., Graceffa,P. and Dominguez,R. (2002) Crystal structure of the vitamin D-binding protein and its complex with actin: structural basis of the actin-scavenger system. Proc. Natl Acad. Sci. USA, 99, 8003–8008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Palmgren S., Ojala,P.J., Wear,M.A., Cooper,J.A. and Lappalainen,P. (2001) Interactions with PIP2, ADP-actin monomers and capping protein regulate the activity and localization of yeast twinfilin. J. Cell Biol., 155, 251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaloni D., Le Clainche,C. and Carlier,M.-F. (2001) Mechanism of actin-based motility. Science, 292, 1502–1506. [DOI] [PubMed] [Google Scholar]
- Papa I., Astier,C., Kwiatek,O., Raynaud,F., Bonnal,C., Lebart,M.-C., Roustan,C. and Benyamin,Y. (1999) Alpha actinin-CapZ, an anchoring complex for thin filaments in Z-line. J. Muscle Res. Cell Motil., 20, 187–197. [DOI] [PubMed] [Google Scholar]
- Poland B.W., Fromm,H.J. and Honzatko,R.B. (1996) Crystal structure of adenylosuccinate synthetase from Escherichia coli complexed with GDP, IMP, hedaacidin, NO3– and Mg2+. J. Mol. Biol., 264, 1013–1027. [DOI] [PubMed] [Google Scholar]
- Read R.J. (1986) Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr., A, 42, 140–149. [Google Scholar]
- Robinson R.C., Mejillano,M., Le,V.P., Burtnick,L.D., Yin,H.L. and Choe,S. (1999) Domain movement in gelsolin: a calcium-activated switch. Science, 286, 1939–1942. [DOI] [PubMed] [Google Scholar]
- Roussel A. and Cambillau,C. (1996) TURBO-FRODO Manual. AFMB-CNRS, Marseille, France.
- Schafer D.A. and Cooper,J.A. (1995) Control of actin assembly at filament ends. Annu. Rev. Cell. Dev. Biol., 11, 497–518. [DOI] [PubMed] [Google Scholar]
- Schafer D.A., Mooseker,M.S. and Cooper,J.A. (1992) Localization of capping protein in chicken epithelial cells by immunofluorescence and biochemical fractionation. J. Cell Biol., 118, 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer D.A., Waddle,J.A. and Cooper,J.A. (1993) Localization of CapZ during myofibrillogenesis in cultured chicken muscle. Cell Motil. Cytoskeleton, 25, 317–335. [DOI] [PubMed] [Google Scholar]
- Schafer D.A., Gill,S.R., Cooper,J.A., Heuser,J.E. and Schroer,T.A. (1994a) Ultrastructural analysis of the dynactin complex: an actin-related protein is a component of a filament that resembles F-actin. J. Cell Biol., 126, 403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer D.A., Korshunova,Y.O., Schroer,T.A. and Cooper,J.A. (1994b) Differential localization and sequence analysis of capping protein β-subunit isoforms of vertebrates. J. Cell Biol., 127, 453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer D.A., Jennings,P.B. and Cooper,J.A. (1996) Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J. Cell Biol., 135, 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizonenko G.I., Karpova,T.S., Gattermeir,D.J. and Cooper,J.A. (1996) Mutational analysis of capping protein function in Saccharomyces cerevisiae. Mol. Biol. Cell, 7, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeno Y., Abe,H., Kimura,S., Maruyama,K. and Obinata,T. (1998) Generation of functional β-actinin (CapZ) in an E.coli expression system. J. Muscle Res. Cell Motil., 19, 639–646. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressivemultiple sequence alignment through sequence weighting,position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G., Somasundaram,T., Blanc,E., Parthasarathy,G., Ellington,W.R. and Chapman,M.S. (1998) Transition state structure of arginine kinase: implications for catalysis of bimolecular reactions. Proc. Natl Acad. Sci. USA, 95, 8449–8454. [DOI] [PMC free article] [PubMed] [Google Scholar]