Abstract
In Arabidopsis, SHY2 encodes IAA3, a member of the auxin-induced Aux/IAA family. Gain-of-function mutations in SHY2/IAA3 cause enlarged cotyledons, short hypocotyls, and altered auxin-regulated root development. Here we show that the gain-of-function mutation shy2-2 decreases both the induction and repression of auxin-regulated genes, suggesting that SHY2/IAA3 acts as a negative regulator in auxin signaling. shy2-2 affects auxin induction of many previously characterized primary response genes, implying that it might repress primary auxin responses. In addition, shy2-2 also affects expression of multiple auxin-nonresponsive genes. Light regulates expression of SHY2/IAA3, suggesting a possible link between light and auxin response pathways.
INTRODUCTION
Auxins regulate numerous cellular and developmental responses in plants, including cell division, expansion, and differentiation; patterning of embryos, vasculature, and other tissues; and distribution of growth between primary and lateral root and shoot meristems (Thimann, 1977; Sachs, 1991). This multiplicity of regulatory activities has spurred considerable interest in mechanisms of auxin signaling and response. Although auxin may regulate some cellular responses (such as expansion or polarity) through direct effects on membrane or cytoskeletal functions, auxin also regulates expression of numerous genes whose products probably perform most developmental responses (Guilfoyle, 1999).
Three major classes of early auxin response genes have been identified from various plant species: Aux/IAA, SAUR (small auxin upregulated), and GH3 (Guilfoyle, 1999). Some ACS genes, encoding 1-aminocyclopropane-1-carboxylate synthase, which is required for ethylene biosynthesis, also can be induced by auxin (Abel et al., 1995a). Auxin induces many of these genes rapidly, specifically, and in the absence of de novo protein synthesis, and these genes are considered primary response genes. The protein synthesis inhibitor cycloheximide can induce some Aux/IAA, SAUR, GH3, and ACS genes, indicating that a short-lived protein normally represses these transcripts (Franco et al., 1990; Abel et al., 1995b; Roux and Perrot-Rechenmann, 1997; Hsieh et al., 2000).
Aux/IAA genes are the best characterized auxin-responsive genes, and there are at least 24 such genes in Arabidopsis (Liscum and Reed, 2001). Analysis of IAA1 to IAA14 transcripts revealed different spatial expression patterns and varied profiles of induction by auxin (Abel et al., 1995b). Some, such as IAA3 and IAA6, were induced within minutes and returned to baseline levels after 2 hr. These genes, as well as genes whose induced expression persisted for several hours, behaved as primary auxin response genes. Others, such as AXR2/IAA7 and IAA8, were induced much more slowly, and this induction depended on protein synthesis. Therefore, they may be secondary response genes.
Functional auxin response elements (AuxREs) containing the consensus core sequence 5′-TGTCTC-3′ have been identified in promoters of primary response genes (Guilfoyle, 1999; Hagen and Guilfoyle, 2001). ARF (auxin response factor) proteins can bind to these elements, and there are 23 ARF genes in Arabidopsis. ARF proteins have an N-terminal DNA binding domain, and most ARFs have domains III and IV, which are conserved among both ARFs and Aux/IAA proteins, at their C termini. Between the N-terminal domain and the C-terminal domain is a middle region that is quite divergent among different ARFs. Transient assays using carrot protoplasts showed that the ARF proteins can regulate expression of promoters having AuxREs (Ulmasov et al., 1997). Some of the ARF proteins (ARF5, ARF6, ARF7, and ARF8) with Q-rich middle domains will activate transcription, whereas others (ARF1) may repress transcription (Ulmasov et al., 1999).
Aux/IAA genes are targets of auxin regulation, and they encode proteins likely to regulate auxin-responsive gene expression. Semidominant mutations in several Aux/IAA genes, such as SHY2/IAA3, AXR3/IAA17, AXR2/IAA7, MSG2/ IAA19, IAR2/IAA28, and SLR/IAA14, have been isolated, and each one causes pleiotropic auxin-related phenotypes, suggesting that Aux/IAA genes play a central role in auxin signaling (Reed, 2001). Aux/IAA genes encode short-lived nuclear proteins that contain four highly conserved motifs called domains I, II, III, and IV. Aux/IAA proteins that have been tested have half-lives as short as 6 to 8 min (Abel et al., 1994). All of these semidominant mutations cause amino acid changes in a very conserved amino acid sequence (VGWPPV) in domain II, and they probably increase protein function by stabilizing the corresponding proteins, indicating that domain II serves as a degradation signal (Worley et al., 2000; Ouellet et al., 2001; Ramos et al., 2001). Domain III is a dimerization domain that mediates homodimerization and heterodimerization among Aux/IAA and ARF proteins (Kim et al., 1997; Ulmasov et al., 1997; Morgan et al., 1999; Ouellet et al., 2001). Transient expression studies showed that Aux/IAA proteins can repress the auxin-dependent activation of AuxRE-mediated reporter genes (Ulmasov et al., 1997). Because there is no evidence that Aux/IAA proteins bind directly to DNA, it is likely that the interactions among Aux/IAA proteins and ARF proteins regulate gene expression. Some of the aux/iaa mutants show altered expression of auxin-inducible genes or reporter constructs, consistent with Aux/IAA proteins regulating auxin-induced transcription (Timpte et al., 1994; Abel et al., 1995b; Leyser et al., 1996; Rogg et al., 2001).
Semidominant shy2 (short hypocotyl 2) mutations were identified in screens for suppressors of the long hypocotyl phenotype of phytochrome-deficient hy2 or phyB mutants (Kim et al., 1996; Reed et al., 1998). SHY2 encodes IAA3, one of the Aux/IAA genes (Soh et al., 1999; Tian and Reed, 1999). shy2-1, shy2-2, and shy2-3 have missense mutations in conserved domain II, and shy2-2 plants have higher steady state levels of SHY2/IAA3 protein than wild-type plants (Colón-Carmona et al., 2000). shy2-2 seedlings have short hypocotyls and enlarged cotyledons both in the dark and in the light, have upcurled leaves in the light, and flower early (Kim et al., 1996, 1998; Reed et al., 1998; Tian and Reed, 1999). shy2-2 seedlings also make leaves in the dark. Consistent with SHY2/IAA3 having a role in auxin responses in roots, shy2-2 seedlings also have fewer lateral roots, less wavy root growth on tilted hard agar plates, and slower root gravitropic response than wild-type seedlings (Tian and Reed, 1999). shy2-1 mutants, which have the same mutation as shy2-2, have similar shoot phenotypes and express CAB and PSBA genes when grown in the dark (Kim et al., 1998). shy2-3 plants have similar but less severe phenotypes; therefore, shy2-3 is a weaker allele. The dark phenotypes of these mutants suggest that SHY2/IAA3 may regulate light responses. We deduced that shy2-2 is a gain-of-function mutation because it is semidominant, a shy2-2 mutant transgene confers mutant phenotypes in wild-type plants, and it can be suppressed by intragenic loss-of-function mutations (Tian and Reed, 1999).
The shy2-24 mutation is an intragenic suppressor of shy2-2, and it introduces a stop codon just upstream of the shy2-2 mutation. Therefore, shy2-24 is a putative null mutant. In contrast to shy2-2, shy2-24 mutants have no drastic leaf or hypocotyl phenotype. However, they have root phenotypes, such as increased number of lateral roots, enhanced wavy root growth on tilted hard agar plates, and faster root gravitropic response.
The Aux/IAA protein biochemistry results discussed above suggested that shy2 mutant phenotypes might arise from altered auxin-regulated gene expression. To determine whether this is true, and to identify potential regulatory targets of SHY2/IAA3, we used gene chips, RNA gel blot hybridizations, and promoter::reporter constructs to compare gene expression in the wild type, shy2-2, and shy2-24 mutants with or without auxin treatment. We found that shy2-2 affects the expression of many of the previously identified auxin-inducible genes, such as Aux/IAA, SAUR, GH3, and ACS genes, as well as numerous previously unknown auxin-responsive genes. shy2-2 also negatively regulates itself. In addition, the shy2-2 mutation affects many genes not regulated by auxin, which might be responsible for developmental phenotypes. Light also regulates the expression of SHY2/IAA3, suggesting that light and auxin signaling pathways interact.
RESULTS
Autoregulation of SHY2/IAA3
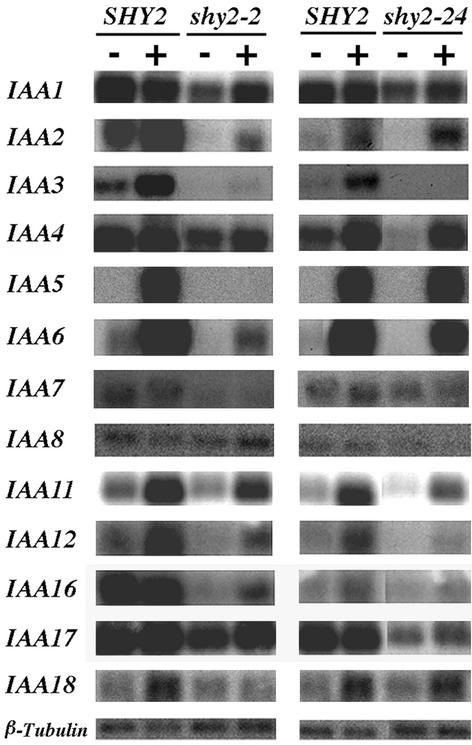
Six-day-old light-grown wild-type, shy2-2, and shy2-24 seedlings were either mock treated or treated with 20 μM indoleacetic acid (IAA) for 2 hr, and the overall expression levels of SHY2/IAA3 were examined by RNA gel blot hybridization. As shown in Figure 1, wild-type seedlings had a low steady state level of SHY2/IAA3 expression, and upon auxin treatment, the transcript level was increased approximately threefold. In gain-of-function shy2-2 seedlings, both the steady state level and the induction of SHY2/IAA3 by auxin were reduced severely compared with wild-type seedlings, suggesting that shy2-2 negatively autoregulates its own gene. In loss-of-function shy2-24 seedlings, the transcript level also was lower than in wild type at both steady state and induced levels. The shy2-24 mutation causes a premature stop codon, and the decreased transcript level in this case might arise from transcript instability.
Figure 1.
RNA Gel Blot Analysis of Expression of Aux/IAA Genes in SHY2 (Wild Type) and shy2-2 and shy2-24 Mutants.
Poly(A)+ RNA (5 μg) was isolated from 6-day-old SHY2, shy2-2, and shy2-24 light-grown seedlings that had been either mock treated (−) or treated with 20 μM IAA for 2 hr (+). RNA was separated in 1% agarose, transferred to a nylon membrane, and hybridized with 32P-labeled Aux/IAA probes or with a 32P-labeled β-tubulin probe. shy2-2 and shy2-24 were assayed in different experiments, and the wild-type controls for each experiment are shown. Exposure times were 72 hr for IAA1, IAA2, IAA4, IAA11, IAA12, IAA16, and IAA17 and 288 hr for IAA3, IAA5, IAA6, IAA7, IAA8, and IAA18.
Previous RNA gel blot hybridization data indicated that SHY2/IAA3 mRNA is most abundant in etiolated seedlings and in leaves, stems, and flowers of light-grown plants; and that it is present in roots as well (Abel et al., 1995b). To determine more precisely where SHY2/IAA3 is expressed and how shy2 mutations affect the expression, we transformed wild-type plants with a reporter gene consisting of 2.2 kb of DNA upstream of the SHY2/IAA3 start codon fused to the Escherichia coli β-glucuronidase gene (GUS). This same upstream fragment, when fused to shy2-2 and transformed into wild-type plants, conferred shy2-2–like phenotypes, suggesting that it is a functional promoter (Tian and Reed, 1999). Six independent transgenic lines each with an insertion at a single locus were generated and analyzed by histochemical staining for GUS activity. All six lines showed similar 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (X-gluc) staining patterns. We treated seedlings of one line with auxin and found that the expression of GUS mRNA was induced approximately twofold, as expected. This line then was crossed to gain-of-function and loss-of-function shy2 mutants.
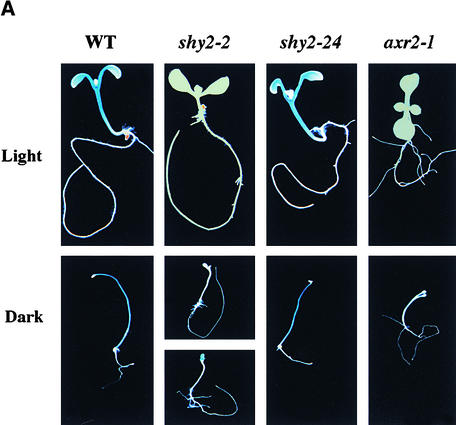
Figure 2A shows the X-gluc staining of 9-day-old wild-type, shy2-2, and shy2-24 seedlings carrying the PSHY2/IAA3::GUS transgene. Expression in light-grown wild-type seedlings was prominent in hypocotyls and cotyledons. Newly formed leaf primordia did not show any staining. This expression pattern varied over time. For example, after 14 days of growth, the first pair of leaves had detectable staining, as did hypocotyls and cotyledons, but newly formed leaves did not stain. After 21 days of growth, staining disappeared in hypocotyls and persisted in older leaves (data not shown). Dark-grown wild-type seedlings stained in the hypocotyls but not in the cotyledons, and this pattern persisted over time. Overall, X-gluc staining was stronger in dark-grown seedlings than in light-grown seedlings. There was no staining in the roots in either light- or dark-grown seedlings.
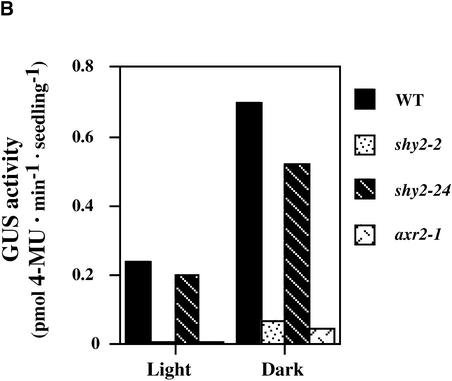
Figure 2.
Histochemical Analysis of Expression of PSHY2/IAA3::GUS.
(A) X-gluc staining of 9-day-old wild type (WT), shy2-2, shy2-24, and axr2-1 PSHY2/IAA3::GUS seedlings grown in the light or in the dark.
(B) 4-Methylumbelliferyl β-d-glucuronide (4-MU) assays of light-grown and dark-grown wild-type (WT), shy2-2, shy2-24, and axr2-1 PSHY2/IAA3::GUS seedlings.
Consistent with our mRNA expression data (Figure 1), we observed less X-gluc staining in shy2-2 PSHY2/IAA3::GUS seedlings than in wild-type seedlings in both light and dark (Figure 2A). Light-grown shy2-2 PSHY2/IAA3::GUS seedlings had almost no staining, and dark-grown shy2-2 PSHY2/IAA3::GUS seedlings either had no staining or had staining only in cotyledons but not in the hypocotyl. In shy2-24 PSHY2/IAA3::GUS seedlings, X-gluc staining was identical to that in the wild type, which is consistent with our suggestion that the decreased SHY2/IAA3 transcript level in shy2-24 reflects transcript instability rather than decreased promoter activity. Quantitative GUS activity assays (Figure 2B) supported these results.
Light Regulates SHY2/IAA3 Expression
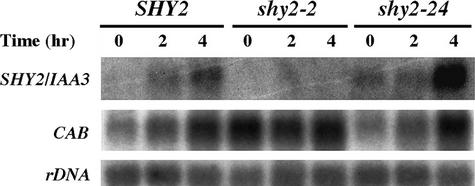
The expression of PSHY2/IAA3::GUS was greater in dark-grown than in light-grown seedlings, implying that light might repress SHY2/IAA3 gene expression. To test this idea, dark-grown wild-type seedlings were either given a pulse of red light and then returned to darkness or shifted to continuous white light. After 4 hr, RNA from the light-treated seedlings was isolated, and the expression of SHY2/IAA3 was compared with that in dark-grown seedlings by RNA gel blot hybridization. As shown in Figure 3, consistent with the X-gluc staining data, light inhibited SHY2/IAA3 expression. In the same experiment, both light treatments induced the expression of CAB, as expected.
Figure 3.
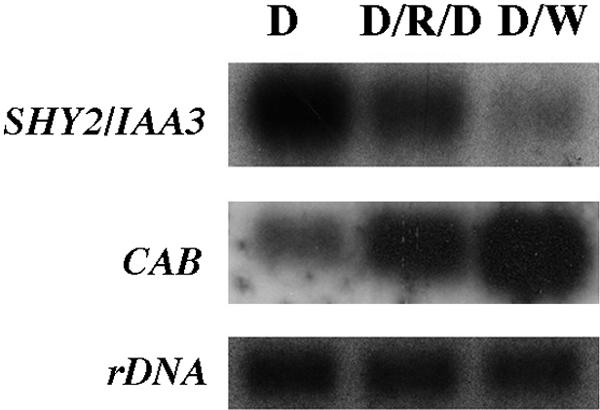
Inhibition of SHY2/IAA3 Expression by Light in the Presence of Sucrose.
Six-day-old dark-grown wild-type seedlings were either given a pulse of red light (R) and then returned to darkness (D) or shifted to continuous white light (W). mRNA was isolated after 4 hr, and RNA gel blots were hybridized with SHY2/IAA3 cDNA, CAB, or rDNA probes. Exposure times were 1 hr (CAB and rDNA) and 72 hr (SHY2/IAA3).
We found that this light regulation of SHY2/IAA3 depended on growth conditions. The experiments showing light repression of SHY2/IAA3 expression were all performed in medium containing sucrose. In contrast to these results, in the absence of sucrose, light induced SHY2/IAA3 expression. Seedlings were grown in the dark for 6 days in the absence of sucrose and were given a pulse of red light and returned to darkness for 2 or 4 hr. As shown in Figure 4, wild-type seedlings had less basal expression of SHY2/IAA3 than did wild-type seedlings grown in the presence of sucrose (Figure 3), and red light induced SHY2/IAA3 expression in both wild-type and shy2-24 seedlings, despite the deduced instability of the shy2-24 transcript. Red light did not induce expression in shy2-2 seedlings, presumably because of the negative autoregulation by shy2-2. As expected, the expression of CAB was induced by red light in wild-type seedlings. Previous studies showed that shy2-1 seedlings grown in the presence of sucrose had increased expression of CAB in the dark (Kim et al., 1998). As shown in Figure 4, in the absence of sucrose, shy2-2 seedlings also had increased expression of CAB compared with that of the wild type, and the induction of CAB in shy2-2 seedlings was abolished. We discuss a simple model to explain these contrasting effects of light under different growth conditions below.
Figure 4.
Induction of SHY2/IAA3 Expression by Light in the Absence of Sucrose.
Six-day-old dark-grown seedlings were given a pulse of red light and returned to darkness for 0, 2, or 4 hr. RNA was isolated as described in Methods, and RNA gel blots were hybridized with SHY2/IAA3, CAB, and rDNA probes. Exposure times were 1 hr (CAB and rDNA) and 360 hr (SHY2/IAA3).
shy2-2 Impedes Auxin-Regulated Expression of Multiple Genes
To determine how SHY2/IAA3 affects expression of other genes, we performed a gene chip experiment. Wild-type, shy2-2, and shy2-24 seedlings were grown in the light for 6 days, mock treated or treated with 20 μM IAA for 2 hr, and their RNA was isolated. Probes made from a mixture of RNA isolated from three independent experiments were hybridized to an Affymetrix gene chip having ∼8000 Arabidopsis genes. Approximately two-thirds of the genes (5500 genes) had detectable signals in our experiment. As listed in Tables 1 and 2, by using a cutoff of ≥2.5-fold auxin induction or repression in at least two genotypes, we identified 74 auxin-induced genes and 26 auxin-repressed genes. Twenty of the 74 auxin-induced genes were previously identified genes of the Aux/IAA, SAUR, GH3, and ACS gene families.
Table 1.
Auxin-Induced Genes
| Auxin Induction (fold)c
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Category | Coded Protein/Other Featuresa | GenBank Number |
Affymetrix Number |
Number of AuxREsb |
SHY2 | shy2-2 | shy2-24 | |
| Previously identified | ||||||||
| auxin-regulated genes | ||||||||
| IAA1 | AAA16569 | 13289 | 7 | ∼12.4 | ∼4.6 | ∼9.7 | ||
| IAA2 | AAB97164 | 13297 | 3 | 3.8 | 3.5 | d | 7.5 | |
| IAA5 | AAC49046 | 13660 | 3 | ∼77.2 | ∼4.4 | ∼69.0 | ||
| IAA6 | AAC49047 | 13661 | 4 | ∼4.0 | ∼1.2 | ∼5.5 | ||
| IAA13 | AAB80649 | 13293 | 2 | 4.3 | 3.2 | d | 4.3 | |
| IAA19 | AAB84356 | 13296 | 7 | 17.7 | 5 | 13.6 | ||
| IAA20 | AAB84357 | 13298 | 4 | ∼4.5 | ∼−1.2 | ∼4.7 | ||
| SAUR-1e | CAB45438 | 20488 | 1 | 2.7 | −1.9 | 3.1 | ||
| SAUR-9 | CAA18505 | 12947 | NDf | 3 | 1.8 | 2.5 | ||
| SAUR-10 | AAD20125 | 13781 | 3 | 5.5 | −1.3 | 4.6 | ||
| SAUR-13 | CAB38618 | 19695 | 3 | 2.6 | −1 | 2.4 | ||
| SAUR-15 | AAB30527 | 12608 | 2 | ∼15.5 | ∼1.1 | ∼17.9 | ||
| SAUR-25 | CAB36843 | 13395 | 3 | ∼4.3 | ∼1.6 | ∼5.4 | ||
| GH3-1e | AAC61292 | 12553 | 2 | 14.4 | 6.9 | 25.7 | ||
| GH3-2 | CAB38206 | 13565 | ND | 17.9 | 12.1 | 16.8 | ||
| GH3-3 | AAB87114 | 16995 | 6 | 74.5 | ∼137.4 | d | 131.2 | |
| GH3-5 | CAA19720 | 16989 | 4 | 8.4 | 5.4 | 11.1 | ||
| ACS4 | AAA85039 | 17107 | 3 | 3.3 | −1.2 | 4.1 | ||
| ACS6 | AAC63850 | 12891 | 5 | 3.2 | 2.5 | d | 3.1 | |
| ACS8 | CAB38925 | 16387 | 6 | ∼20.6 | ∼4.0 | ∼20.7 | ||
| Transcription related | ||||||||
| HAT2 | AAA56901 | 18950 | 4 | 9.3 | 4.5 | 8.9 | ||
| Zinc finger protein | AAA87298 | 16581 | 7 | ∼14.4 | ∼4.5 | ∼11.9 | ||
| ARF19 (IAA22)e | AAB91321 | 13300 | ND | 2.9 | 2.1 | 2.8 | ||
| Putative heat shock transcription factor | AAB84350 | 18765 | ND | 3 | ∼2.8 | ∼5.3 | ||
| Contains similarity to GATA-type zinc fingers | AAB61058 | 17180 | ND | ∼8.0 | 3.4 | d | ∼6.7 | |
| Signal transduction | ||||||||
| Similar to protein kinases | AAD30630 | 16878 | 5 | 5.6 | 2.7 | 5.5 | ||
| Putative protein kinase | AAC31848 | 15005 | 2 | ∼7.4 | ∼2.6 | ∼5.6 | ||
| Receptor protein kinase–like protein | CAA18216 | 19418 | 1 | 2.8 | 1.9 | d | 3.3 | |
| Putative protein kinase | AAC34357 | 19025 | ND | 4.2 | 1.9 | d | 4.1 | |
| Putative receptor-like protein kinase | AAC02766 | 14112 | ND | 3.6 | 2.5 | 4.4 | ||
| Putative protein kinase | AAC79621 | 12388 | ND | 2.6 | ∼4.9 | 1.9 | ||
| Putative protein kinase | AAC14522 | 12357 | ND | 2.5 | 2.1 | 2.7 | ||
| Transport | ||||||||
| Similar to nitrate and oligopeptide transporters | AAD39317 | 19835 | 3 | 3.6 | 1.4 | 3.8 | ||
| AUX1-like amino acid permease | AAD29811 | 20612 | ND | 2.7 | 2 | 2.7 | ||
| Cell wall establishment | ||||||||
| Putative expansin | AAB87577 | 19660 | 3 | 2.5 | 1.6 | 3 | ||
| Similar to xyloglucan endotransglycosylase– related protein XTR4 | AAD39577 | 19490 | 4 | 2.6 | −1.1 | 3.2 | ||
| Contains similarity to expansins | AAC72858 | 19346 | 2 | 3.4 | 1.7 | d | 5.4 | |
| Similar to putative glucan synthase | AAF24822 | 18515 | ND | 3.6 | ∼2.7 | −1.2 | ||
| Putative glucosyl transferase | AAD20156 | 15496 | ND | 3.4 | 3.2 | 2.7 | ||
| Metabolic enzymes | ||||||||
| Putative cytochrome P450 | AAC34228 | 19045 | 6 | 8.8 | 2.7 | 2.3 | ||
| Cytochrome P450–like protein | CAB38204 | 14032 | 4 | 3.9 | 4.8d | 3.7 | ||
| Similar to mitochondrial protein and AAA-type ATPase | CAB46004 | 15424 | 1 | ∼6.2 | ∼2.0d | ∼6.9 | ||
| Peroxidase ATP24a | CAA72484 | 18946 | 3 | 2.9 | 1.5 | 4.2 | ||
| Lupeol synthase | AAD05032 | 15653 | 4 | ∼3.0 | ∼−1.5d | ∼5.5 | ||
| Similar to mandelonitrile lyase | AAD39305 | 18869 | 8 | 2.8 | ∼2.1 | 3 | ||
| Spermine synthase | AAF01311 | 17999 | 6 | 5.2 | 2.1d | 5.9 | ||
| Glutamate decarboxylase | AAA33709 | 18508 | 6 | 3.3 | 1.9d | 2.7 | ||
| Similar to glutaredoxin | AAB07873 | 14766 | 3 | 5.9 | 2.9 | ∼6.4 | ||
| Short-chain alcohol dehydrogenase–like protein | CAB41928 | 20685 | ND | 3.2 | 3.1 | 4.1 | ||
| 12-oxophytodienoate reductase OPR1 | AAC78440 | 18253 | ND | 2.6 | 3.4 | 2.6 | ||
| Putative glutathione S-transferase | AAC95196 | 18966 | 4 | 2.6 | 4.7 | 3.6 | ||
| Putative alanine acetyl transferase | AAD15402 | 14918 | ND | ∼4.8 | ∼3.2 | ∼3.3 | ||
| Putative laccase (diphenol oxidase) | AAD20177 | 17287 | ND | ∼4.5 | ∼2.5 | 2.3 | ||
| Monooxygenase | CAA07574 | 17877 | 5 | 3.5 | 4.1 | 3.5 | ||
| Putative peroxidase | AAD22357 | 20328 | ND | ∼4.6 | ∼2.8 | ∼5.0 | ||
| Similar to indole-3-acetate β-glucosyltransferase |
AAD30627 | 20297 | ND | 3.3 | 2.5 | 3.3 | ||
| Putative pectinesterase | AAC62855 | 17338 | ND | 2.5 | 2.1 | 2.9 | ||
| Others | ||||||||
| Similar to TOLB (colicin tolerance) protein precursor | AAC80599 | 20519 | ND | 4.8 | 3.1 | 3.5 | ||
| GASA5 | AAA98520 | 19565 | ND | 3.6 | 2.6 | 3.4 | ||
| Dehydrin | AAB00374 | 19186 | ND | 3.3 | 3.4 | 5.5 | ||
| ATAF2 | CAA52772 | 18591 | ND | 2.5 | 2.1 | 3.1 | ||
| Similar to hookless1 (HLS1) | AAC17822 | 20122 | 2 | ∼13.7 | ∼10.0 | ∼10.7 | ||
| Translation factor EF-1 α–like protein | CAA16563 | 13439 | ND | 4 | 2.1 | 2.5 | ||
| Unknown | ||||||||
| Hypothetical protein | CAB10516 | 13016 | 2 | 13.7 | 5.8 | 15.8 | ||
| Hypothetical protein | AAD15394 | 12095 | 3 | 7 | 2.9 | 3.5 | ||
| Putative protein | CAB39613 | 14951 | 3 | 3.9 | 2.2 | 4.1 | ||
| Unknown protein | AAD21443 | 18885 | 2 | 3.4 | 1.8 | 3.5 | ||
| Hypothetical protein | AAD23726 | 16180 | 6 | ∼5.7 | ∼2.5d | ∼4.2 | ||
| Unknown protein | AAC27828 | 12090 | 4 | ∼21.6 | 3.7 | 14.5 | ||
| Hypothetical protein | AAD23725 | 15288 | 1 | ∼14.6 | 2.8 | 6.2 | ||
| Predicted protein of unknown function | AAD22649 | 13656 | 3 | 4.7 | 4.1 | 4.5 | ||
| Unknown protein | AAB82622 | 16247 | ND | 4.3 | 3.2 | 4.1 | ||
| Unknown protein | AAC63640 | 20550 | ND | 3.2 | 1.9 | 3.3 | ||
| Hypothetical protein | AAC24177 | 17154 | ND | ∼3.8 | 2d | ∼4.0 | ||
Boldface indicates that shy2-2 causes less auxin induction compared with the wild type. Roman indicates that the auxin induction is not affected by shy2-2.
Number of TGTC_C and G_GACA sequences in the 2-kb fragments upstream of the start codon.
Fold difference of expression levels after auxin treatment relative to without auxin treatment. ∼ indicates that one of the expression levels for comparison is below the detection limit; therefore, the auxin induction fold is approximate.
shy2-2 had both reduced basal and reduced auxin-induced expression levels.
Gene numbers for ARF, SAUR, and GH3 genes are from Hagen and Guilfoyle (2001).
ND, not determined.
Table 2.
Auxin-Repressed Genes
| Auxin Induction (fold)c
|
|||||||
|---|---|---|---|---|---|---|---|
| Category | Coded Protein/Other Featuresa | GenBank Number | Affymetrix Number | Number of AuxREsb | SHY2 | shy2-2 | shy2-24 |
| Transcription related | |||||||
| Zinc finger protein | AAA87299 | 17071 | 4 | ∼−3.9 | 1 | ∼−2.9 | |
| RING-H2 finger protein RHA4a | AAC69852 | 20040 | NDd | −4 | −1.7 | −3.5 | |
| TINY | AAC29139 | 19722 | 5 | −2.6 | −1.5 | −3 | |
| ATK1 | CAA57121 | 12928 | 2 | ∼−3.4 | 1.5 | ∼−2.9 | |
| Putative transcription factor | AAD53097 | 17610 | ND | −2.7 | −1.2 | ∼−2.9 | |
| Putative transcription factor | AAC83598 | 18790 | ND | −3.3 | −2 | ∼−3.0 | |
| Putative WRKY DNA-binding protein | AAD17441 | 14679 | ND | ∼−3.9 | ∼−5.1 | ∼7.1 | |
| R2R3-MYB transcription factor | CAB09190 | 18747 | ND | ∼−2.8 | ∼−2.3 | ∼−2.6 | |
| Signal transduction | |||||||
| Putative histidine kinase | AAD03576 | 13362 | ND | −2.6 | −1.7 | −2.5 | |
| Receptor protein kinase–like protein | AAC14033 | 16356 | ND | −4.3 | −4.2 | ∼−4.2 | |
| Transport | |||||||
| Nodulin-26–like protein | CAA16760 | 19847 | 4 | −10.7 | −2.4 | ∼−9.9 | |
| Similar to Arabidopsis Fe(II) transport protein | AAB71447 | 19718 | ND | ∼−3.6 | ∼−3.7 | −3 | |
| Metabolic enzymes | |||||||
| Putative cytochrome P450 protein | AAD10659 | 14366 | 2 | −5.8 | −3.4 | −6.5 | |
| Peroxidase | CAA66963 | 16971 | 4 | ∼−18.0 | −1.7 | ∼−13.5 | |
| Putative peroxidase | AAC79614 | 18150 | 3 | −3.1 | −1.1 | −2.9 | |
| Putative endochitinase | AAB64046 | 12542 | 4 | ∼−4.1 | −1.1 | ∼−3.0 | |
| UDP-glucose 4-epimerase–like protein | CAB45812 | 17748 | 4 | ∼−4.1 | 1.6 | −3.5 | |
| CER1-like protein | AAC23640 | 19237 | 3 | −3.1 | −1.5 | −3.6 | |
| Putative cytochrome P450 | AAD22364 | 19549 | ND | −3.8 | −3.6 | −2.2 | |
| Putative pectinesterase | AAC17097 | 12352 | ND | −3.6 | −1.8 | −3 | |
| Others | |||||||
| HSR201–like protein | CAB10318 | 16045 | 7 | −4.1 | −1.5 | −5.5 | |
| Putative RNA binding protein | AAC23648 | 18817 | 4 | ∼23.5 | −1.6 | ∼2.9 | |
| pEARLI 1–like protein | CAB41722 | 18983 | ND | −4.3 | −3.3 | −4.5 | |
| Similar to NPH-3 (nonphototropic hypocotyl-3) | CAA16538 | 16213 | ND | ∼−3.8 | ∼−3.5 | −1.6 | |
| Unknown | |||||||
| Putative protein | CAA22575 | 16510 | 5 | −4.7 | 1 | −3.7 | |
| Chromosome I bacterial artificial chromosome F3F20 genomic sequence | AAD30618 | 16832 | ND | −2.7 | −2.8 | −2 | |
Boldface indicates that shy2-2 causes less auxin repression compared with the wild type. Roman indicates that the auxin repression is not affected by shy2-2.
Number of TGTC_C and G_GACA sequences in the 2-kb fragments upstream of the start codon.
Fold difference of expression levels after auxin treatment relative to without auxin treatment. ∼ indicates that one of the expression levels for comparison is below the detection limit; therefore, the auxin repression fold is approximate.
ND, not determined.
We compared the expression of these genes among the wild-type and shy2 mutants. Consistent with the subtle phenotypes of shy2-24 plants, expression levels of auxin-responsive genes in shy2-24 generally paralleled those in the wild type (except for SHY2/IAA3 itself). shy2-24 plants had reduced expression of just four genes, and none of these four genes was auxin responsive. Thus, the differences between wild-type and shy2-24 plants are slight, and in practice, the shy2-24 mutant data served as a quasi-replicate of wild-type data.
In contrast to shy2-24, many genes had altered expression in shy2-2 seedlings. Forty-two of the 74 auxin-induced genes were less induced in shy2-2 seedlings than in wild-type seedlings. Seventeen of these 42 genes were previously identified auxin-inducible Aux/IAA, SAUR, GH3, and ACS genes. Fourteen of the 26 auxin-repressed genes had decreased repression in shy2-2 seedlings. In addition, the chip has 19 more genes of the Aux/IAA, SAUR, GH3, and ACS gene families that were either induced (16 genes) or repressed (three genes) by auxin <2.5-fold. On the basis of previous RNA gel blot hybridization data, we also consider these genes to be auxin-responsive genes (Abel et al., 1995b). Six of these genes also were less induced in shy2-2 seedlings (Figure 5).
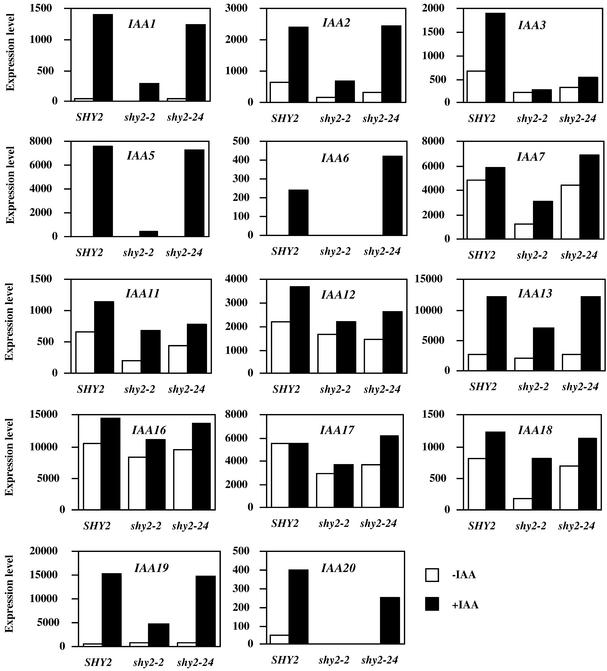
Figure 5.
Analysis of Aux/IAA Gene Expression in SHY2 (Wild-Type), shy2-2, and shy2-24 Seedlings.
Six-day-old light-grown seedlings were either mock-treated (open bars) or treated with 20 μM IAA (closed bars) for 2 hr. Total RNA was isolated, and the gene chip experiment was performed as described in Methods. The expression levels (y axis) were computed using Affymetrix software and represent differential hybridizations between perfectly matched and mismatched oligodeoxyribonucleotides.
Aux/IAA Genes
In addition to the gene chip experiment, we also examined expression of selected Aux/IAA genes by RNA gel blot hybridization (Figure 1). In total, we assessed expression of 22 Aux/IAA genes, 12 of them by both methods. In most cases for which we obtained data from both experiments, these were qualitatively consistent. The collected data revealed that other than SHY2/IAA3, expression of 11 Aux/IAA genes was lower in shy2-2 seedlings than in wild-type seedlings. As shown in Figures 1 and 5, for IAA2, AXR2/IAA7, IAA11, AXR3/IAA17, and IAA18, both steady state and auxin-induced levels were lower in shy2-2 than in the wild type. For IAA1, IAA5, IAA6, IAA13, MSG2/IAA19, and IAA20, steady state levels were similar but auxin-induced levels were less in shy2-2 seedlings than in the wild type. For all of these genes, expression levels in shy2-24 generally paralleled those in the wild type. The data also revealed 10 Aux/IAA genes (IAA4, IAA8, IAA9, IAA10, IAA12, SLR/IAA14, IAA16, IAA26, IAA27, and IAR2/IAA28) whose expression was not affected by shy2 mutations either at the steady state level or at the auxin-induced level (Figures 1 and 5 and data not shown). Although RNA gel blot hybridization showed that the auxin induction level of IAA12 was reduced in shy2-24 seedlings, quantitative data from the gene chip experiment indicated that the difference was less than twofold. Therefore, we considered the expression of IAA12 in wild-type and shy2-24 seedlings to be similar.
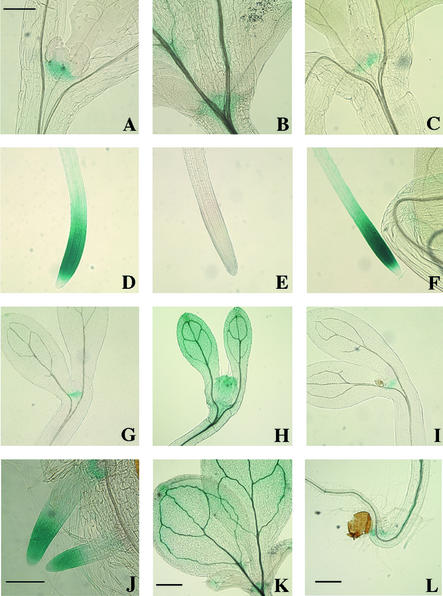
Among the Aux/IAA genes characterized previously, AXR2/ IAA7 and IAA8 are suggested to be secondary response genes. Gene chip and RNA gel blot hybridization data showed that shy2-2 seedlings had reduced levels of AXR2/IAA7. To provide additional insight into how shy2 mutations affect AXR2/IAA7 expression, we fused 2.3 kb of DNA upstream of AXR2/IAA7 to the GUS gene and introduced this construct into wild-type plants. The same upstream fragment, when fused to axr2-1 (which has a gain-of-function mutation in AXR2/IAA7) and transformed into wild-type plants, conferred axr2-1–like phenotypes, suggesting that it is a functional promoter (P. Nagpal and J.W. Reed, unpublished results). We generated seven independent lines, each with an insertion at a single locus, and analyzed them for histochemical staining of GUS activity. As shown in Figure 6, PAXR2/IAA7::GUS expression was detected in shoot and root meristems in light-grown wild-type seedlings (Figures 6A and 6D) and only in shoot meristems, but not primary root meristems, in dark-grown seedlings (Figure 6G and data not shown). In addition, we observed X-gluc staining in the vasculature of hypocotyl/root junctions and occasionally in lateral root tips in dark-grown wild-type seedlings (Figure 6L and data not shown).
Figure 6.
Histochemical Analysis of PAXR2/IAA7::GUS Expression.
X-gluc staining of 9-day-old light-grown ([A] to [F], [J], and [K]) and dark-grown ([G] to [I] and [L]) wild-type ([A], [D], [G], [J], and [L]), shy2-2 ([B], [E], [H], and [K]), and shy2-24 ([C], [F], and [I]) PAXR2/IAA7::GUS seedlings.
(A) to (C) and (G) to (I) Shoot meristems.
(D) to (F) Primary root tips.
(J) Lateral root tips.
(K) Cotyledons and leaves.
(L) Hypocotyl/root junction.
Bar in (A) = 0.25 mm for (A) to (I); bar in (J) = 0.15 mm; and bars in (K) and (L) = 0.5 mm.
We crossed this construct into shy2 mutant seedlings. Compared with the wild type, both light- and dark-grown shy2-2 seedlings had ectopic PAXR2/IAA7::GUS expression in cotyledons, leaf primordia, and vasculature of hypocotyls in addition to expression in shoot meristems (Figures 6B, 6H, and 6K). They did not have any PAXR2/IAA7::GUS expression in root meristems or in other parts of the roots (Figure 6E and data not shown). These data suggest that although the total AXR2/IAA7 expression level was reduced in shy2-2 seedlings, shy2-2 had contrasting effects in different tissues. SHY2/IAA3 may regulate AXR2/IAA7 expression positively in aerial parts of the seedlings but negatively in the roots. The X-gluc staining in light-grown shy2-24 PAXR2/IAA7::GUS mutant seedlings was similar to that of wild-type seedlings (Figures 6C and 6F). In dark-grown shy2-24 seedlings, X-gluc staining also was detected in the shoot meristems (Figure 6I) and the vasculature of hypocotyl/root junctions, as in wild-type seedlings. However, compared with wild-type seedlings, dark-grown shy2-24 seedlings had stronger X-gluc staining in lateral root tips, and they also had X-gluc staining in primary root tips (data not shown).
axr2-1 is a gain-of-function mutation in AXR2/IAA7, and axr2-1 seedlings have similar shoot phenotypes to shy2-2 seedlings, such as short hypocotyls and upcurled leaves (Wilson et al., 1990; Timpte et al., 1992, 1994; Nagpal et al., 2000). They also have agravitropic shoot and root growth. However, axr2-1 seedlings have more lateral roots, whereas shy2-2 seedlings have fewer lateral roots than does the wild type. We introduced PSHY2/IAA3::GUS and PAXR2/IAA7::GUS into axr2-1 mutants to examine how axr2-1 affected the expression of these two genes. Consistent with previous mRNA data (Abel et al., 1995b; Nagpal et al., 2000), axr2-1 PAXR2/IAA7::GUS seedlings had almost no staining when grown either in light or in darkness, indicating that AXR2/IAA7 also negatively regulates itself (data not shown). Similarly, axr2-1 PSHY2/IAA3::GUS seedlings had almost no staining when grown either in light or in darkness, suggesting that AXR2/IAA7 has a negative effect on SHY2/IAA3 expression (Figure 2A). Because AXR2/IAA7 is considered a secondary response gene, these results indicate that products of both primary (SHY2/IAA3) and secondary (AXR2/IAA7) auxin-responsive genes can feed back to repress SHY2/IAA3 expression.
To determine how shy2 mutations affect IAA2 expression, we transformed the previously described PIAA2::GUS construct (Luschnig et al., 1998) into wild-type plants and crossed a transgenic line with an insertion at a single locus into shy2 mutants. Light-grown wild-type PIAA2::GUS seedlings had prominent X-gluc staining in petioles and root tips. They also had staining in cotyledons, leaves, and mature tissues of roots. Compared with wild type, shy2-2 PIAA2::GUS seedlings had much less staining in petioles, consistent with the decreased IAA2 expression in shy2-2 seedlings in the chip experiment, but they had similar staining in other tissues. shy2-24 PIAA2::GUS seedlings had similar X-gluc staining to wild type (data not shown).
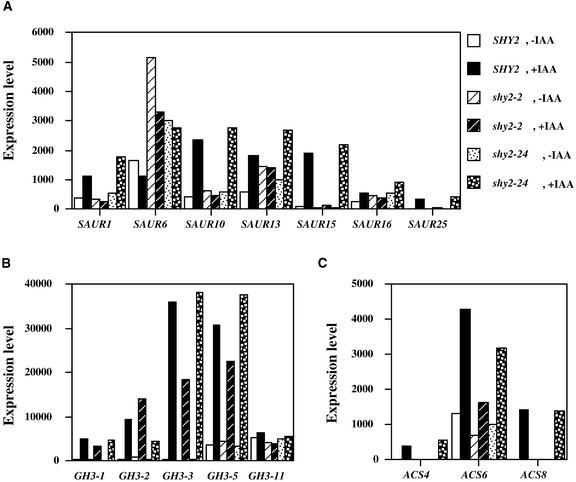
SAUR Genes
There are 16 SAUR genes on the chip, and 11 of them had detectable signals. Among these, auxin induced 10 SAUR genes and repressed one, SAUR6. As shown in Figure 7A, auxin inductions of SAUR1, SAUR10, SAUR13, SAUR15, SAUR16, and SAUR25 were abolished almost completely in shy2-2 seedlings. Expression of SAUR9, SAUR12, SAUR32, and SAUR36 were not changed by shy2 mutations (data not shown). Compared with the wild type, shy2-2 seedlings had similar repression of the SAUR6 gene by auxin. However, both the baseline and the auxin-repressed levels of SAUR6 were higher in shy2-2 seedlings than in wild-type seedlings.
Figure 7.
Analysis of SAUR (A), GH3 (B), and ACS (C) Gene Expression in SHY2 (Wild-Type), shy2-2, and shy2-24 Seedlings.
Six-day-old light-grown seedlings were either mock treated (−IAA) or treated with 20 μM IAA (+IAA) for 2 hr. Total RNA was isolated, and the gene chip experiment was performed as described in Methods. The expression levels (y axis) were computed using Affymetrix software and represent differential hybridizations between perfectly matched and mismatched oligodeoxyribonuleotides.
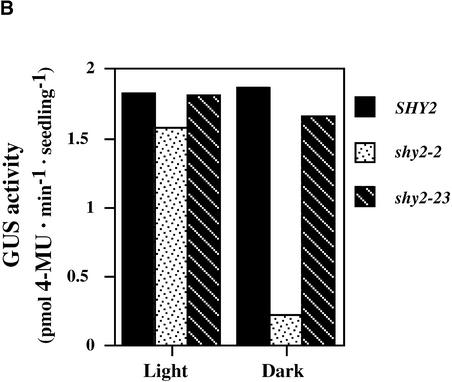
We examined the effects of shy2 mutations on SAUR15 (previously named SAUR-AC1) spatial expression using the PSAUR-AC1::GUS fusion (Gil and Green, 1997). Figure 8A shows X-gluc staining of 9-day-old wild-type, shy2-2, and shy2-23 (which has the same mutation as shy2-24) seedlings. In the wild type, PSAUR-AC1::GUS was expressed in the elongating region of the hypocotyls of light- and dark-grown seedlings and in the cotyledons and primary leaves of light-grown seedlings. Occasionally, we observed moderate expression in the roots. Compared with the wild type, shy2-2 seedlings had much less GUS activity, and X-gluc staining was most abundant in cotyledons and shoot meristems rather than in the hypocotyls, in both light- and dark-grown seedlings. The difference of PSAUR-AC1::GUS expression between shy2-2 and wild-type seedlings was more obvious in dark-grown seedlings. Staining in shy2-23 seedlings was similar to that in the wild type. Quantitative GUS activity assays supported these results (Figure 8B).
Figure 8.
Histochemical Analysis of PSAUR-AC1::GUS Expression.
(A) X-gluc staining of 9-day-old light- and dark-grown SHY2 (wild-type), shy2-2, and shy2-23 PSAUR-AC1::GUS seedlings.
(B) 4-Methylumbelliferyl β-d-glucuronide (4-MU) assay of 9-day-old dark-grown SHY2 (wild-type), shy2-2, and shy2-23 PSAUR-AC1::GUS seedlings.
GH3 Genes
The shy2-2 mutation had less profound effects on GH3 genes. There are seven GH3 genes on the chip, and five of them had detectable signals. As shown in Figure 7B, among these GH3 genes, expression of GH3-2 and GH3-11 was almost normal in shy2-2. The auxin induction, but not the baseline levels, of GH3-3 and GH3-5 was reduced by 20 to 50% in shy2-2 mutants, and the auxin induction of GH3-1 was reduced only slightly in shy2-2.
ACS Genes
There are three ACS genes on the chip (ACS4, ACS6, and ACS8). As shown in Figure 7C, shy2-2 reduced either auxin induction or baseline expression levels of all of these genes.
Other Auxin-Induced Genes
In addition to the 25 previously identified auxin-responsive genes described above, there are 38 more auxin-regulated genes that were affected by the shy2-2 mutation. Twenty-four of these genes were auxin-inducible genes, and shy2-2 seedlings had reduced baseline expression levels or induction levels (Table 1), suggesting that, as for the Aux/IAA, SAUR, GH3, and ACS genes, SHY2/IAA3 also is a repressor of transcription of these genes. Fourteen of these 38 genes were auxin-repressed genes (Table 2). However, the repression of these genes by auxin was reduced in shy2-2 seedlings, and some of them were induced by auxin in shy2-2, indicating that shy2-2 might inhibit transcription factors that normally repress their transcription.
Auxin-Nonresponsive Genes
We searched the chip data for all of the auxin-nonresponsive genes expressed differently between shy2 mutants and the wild type either with or without auxin treatment. As listed in Table 3, there were 81 such genes in several categories affected by the shy2-2 mutation. These genes may be responsible for establishing cellular consequences visible as shy2-2 mutant phenotypes. We discuss the possible physiological roles of some of these genes below.
Table 3.
Auxin Nonresponsive Genes Affected by shy2-2
| Category | Coded Protein/Other Featuresa | GenBank Number | Affymetrix Number |
shy2-2/SHY2, −IAAb(fold difference) | shy2-2/SHY2, +IAAc(fold difference) |
|---|---|---|---|---|---|
| Transcription related | |||||
| Ethylene-responsive element binding factor 1 | BAA32418 | 16063 | ∼223.5 | −7.1 | |
| NAM (no apical meristem)–like protein | AAB81668 | 13806 | ∼25.1 | −1.3 | |
| Similar to gb|X92204NAM gene product from Petunia hybrida | AAD39614 | 12643 | ∼23.1 | −2.9 | |
| Putative c2h2 zinc finger transcription factor | AAB80922 | 15665 | ∼24.9 | −1.6 | |
| RING-H2 finger protein RHA1b | AAC68670 | 16553 | −3.3 | −2.5 | |
| Signal transduction | |||||
| Receptor-like serine/threonine protein kinase ARK3 | CAA20203 | 16360 | ∼−6.2 | −3.8 | |
| Putative receptor-like protein kinase | AAD32284 | 12497 | −3 | −2.7 | |
| Serine/threonine kinase–like protein | CAA18465 | 13550 | −3.5 | −3.2 | |
| Wall-associated kinase 1 | CAA08794 | 15616 | −3.4 | −2.6 | |
| Putative serine/threonine protein kinase | CAB38914 | 18459 | −3.4 | −5.5 | |
| Calmodulin-like protein | CAB42906 | 13217 | −9.5 | −6.8 | |
| Putative calmodulin binding protein | CAA18193 | 19857 | −6.8 | −2.9 | |
| Light related | |||||
| Chlorophyll a/b binding protein | CAB41095 | 16004 | 3 | 2.8 | |
| Lhcb2 protein | AAD28772 | 15153 | 5.3 | 4.6 | |
| Putative photosystem I reaction center subunit II | AAD15351 | 18086 | 2.7 | 3 | |
| PII protein | AAC78333 | 17505 | 3.7 | 4.7 | |
| Transport | |||||
| Putative ligand-gated ion channel protein | AAC33239 | 18844 | ∼−7.9 | −3.3 | |
| Contains similarity to sugar transporters | AAC26243 | 14116 | −3.2 | −2.9 | |
| Contains similarity to Medicago sativa corC (magnesium and cobalt efflux protein) | AAC19312 | 20468 | 4.8 | 4.1 | |
| Sulfate transporter protein | AAC78252 | 17042 | 4.3 | 4.5 | |
| Cell wall establishment | |||||
| Similar to the family of glycosyl hydrolases | AAC13634 | 12336d | −7.4 | −1 | |
| Xyloglucan endotransglycosylase related protein | AAC05572 | 16620 | −2.8 | −2.6 | |
| Putative polygalacturonase | CAA23048 | 13628 | −3.2 | −2.8 | |
| Putative β-glucosidase | CAB43970 | 18917 | ∼−7.6 | −3.8 | |
| Putative xyloglucan endo-1,4-β-D-glucanase | CAB39603 | 16630 | −4.4 | −3.7 | |
| Endo-xyloglucan transferase | AAC39467 | 16927 | −3.5 | −2.4 | |
| β-1,3-Glucanase 2 | CAB68132 | 13212 | −9.7 | −8.1 | |
| β-1,3-Glucanase | AAA32756 | 12364 | −17.9 | −12.1 | |
| Xyloglucan endotransglycosylase–like protein | CAA22967 | 19294 | −3.6 | −5.9 | |
| Contains similarity to Nicotiana alata pistil extensin-like protein | AAC28181 | 15049 | −3.8 | −3.9 | |
| Strong similarity to extensin-like protein gb|Z34465from Zea mays | AAC17609 | 19826 | ∼−10.8 | ∼−7.5 | |
| Contains similarity to Zea mays embryogenesis transmembrane protein | AAC33958 | 17185 | ∼−13.4 | −14.1 | |
| Xyloglucan endotransglycosylase–related protein | AAB18367 | 17533 | 6.3 | 16 | |
| Thioglucosidase | CAA55787 | 12740 | ∼6.0 | ∼5.5 | |
| Metabolic enzymes | |||||
| Cytochrome P450–like protein | CAB10312 | 18951 | ∼−8.5 | ∼−6.5 | |
| Putative cytochrome P450 | AAC26690 | 12989 | −8.5 | −3.5 | |
| Peroxidase ATP19a | CAB51413 | 13610 | −2.7 | −2.7 | |
| Peroxidase | CAA67092 | 12400 | −2.8 | −2.7 | |
| Strong similarity to Arabidopsis peroxidase ATP11A | AAB71454 | 17932 | −4.6 | −6.8 | |
| Peroxidase | CAA66967 | 20296 | −5.2 | −4.4 | |
| Putative peroxidase | CAB39666 | 15562 | ∼−14.3 | −7.2 | |
| Putative oxidoreductase | AAC79100 | 14338 | ∼−38.4 | −8.5 | |
| Caffeoyl-CoA O-methyltransferase–like protein | CAB38951 | 19348d | −4 | −1.2 | |
| Hydroxymethylglutaryl CoA reductase | AAA32814 | 12920d | −1.2 | ∼2208.3 | |
| Putative tropinone reductase | AAC95203 | 20491 | −3.1 | −6.1 | |
| Putative anthocyanin 5-aromatic acyltransferase | AAB95283 | 18876 | −4.3 | −4.8 | |
| Putative tyrosine aminotransferase | AAD23027 | 17008 | −7.7 | −5.7 | |
| Chitinase-like protein | CAA19692 | 12514 | ∼−10.9 | ∼−10.0 | |
| Putative trehalose-6-phosphate synthase | AAD08939 | 13706 | ∼−4.4 | −2.3 | |
| Putative glutathione S-transferase | AAC32912 | 12764 | −2.5 | −2.4 | |
| Squalene epoxidase homolog | CAA06769 | 19704 | ∼24.8 | ∼−5.5 | |
| Similar to PHZF, catalyzing the hydroxylation of phenazine-1-carboxylic acid to 2-hydroxy- phenazine-1-carboxylic acid | AAD15344 | 14497 | 6 | ∼5.1 | |
| Cytochrome P450 monooxygenase-like protein | CAB38208 | 14117 | 3.7 | 4.9 | |
| Pyrophosphate-dependent phosphofructo-1-kinase | CAB38956 | 19741d | −1.6 | 10.9 | |
| Drought-inducible cysteine proteinase RD21A precursor-like protein | CAB51416 | 12746 | 6.5 | 8 | |
| Carbonic anhydrase | AAA50156 | 15144 | 3.3 | 3.1 | |
| Disease responses | |||||
| Putative disease resistance protein | AAC12833 | 12251 | ∼−5.2 | −2.7 | |
| Pathogenesis-related protein 1 | CAA65420 | 12933d | −3.3 | −1.3 | |
| Downy mildew resistance protein RPP5 | AAF08790 | 20208 | ∼−5.0 | ∼−5.5 | |
| Putative disease resistance protein | CAA18120 | 19443 | ∼−3.8 | −5.1 | |
| Putative disease resistance response protein | CAB43056 | 14013 | ∼11.3 | ∼8.6 | |
| Resistence protein-like | CAA16927 | 17294 | ∼5.3 | ∼5.0 | |
| Contains similarity to pathogenesis-related protein 1 precursors and SCP-like extracellular proteins | AAD17355 | 20395 | ∼4.9 | ∼4.4 | |
| Others | |||||
| Contains similarity to WB domains, G-β repeats | AAC35546 | 19589d | −1.1 | ∼−30.4 | |
| Putative surface protein | AAD25652 | 14076d | −1 | −3.8 | |
| Thioredoxin h | AAC49356 | 13187 | −3 | −3 | |
| Thioredoxin h | AAC49356 | 13189 | −4.6 | −4.1 | |
| Peroxiredoxin TPx2 | AAD28243 | 15116 | −3.8 | −3.9 | |
| Pre-hevein–like protein | AAA20642 | 15162 | −3.4 | −2.6 | |
| ORF1 | CAA50905 | 16439 | −3.3 | −3.7 | |
| Putative protein | CAA19766 | 13937 | ∼−6.4 | ∼−7.0 | |
| Unknown protein | AAD15461 | 15846 | −7.7 | −7.1 | |
| Unknown protein | AAD15461 | 14704 | −26.3 | −13.4 | |
| Selenium binding protein–like | CAB46000 | 18215 | 4.1 | 5.3 | |
| Ubiquitin fusion degradation protein 1 homolog | CAB10321 | 19879 | ∼5.6 | 2.9 | |
| T15B16.1 gene product | AAC72861 | 13935 | 13.2 | 20 | |
| Hypothetical protein | AAC78273 | 18761 | ∼16.7 | ∼24.0 | |
| Putative protein | CAB45069 | 15531 | ∼13.4 | ∼23.5 | |
| Putative protein | CAB43702 | 12660 | 5.9 | 8 | |
| Putative protein | CAB41004 | 18648 | ∼4.3 | ∼6.8 | |
| Putative protein | CAA19878 | 12169 | 4.6 | ∼23.7 |
Boldface indicates that gene expression is decreased in shy2-2. Roman indicates that gene expression is increased in shy2-2 compared with the wild type.
Fold difference of expression levels in shy2-2 relative to the wild type without auxin treatment. ∼indicates that one of the expression levels for comparison is below the detection limit; therefore, the fold difference is approximate.
Fold difference of expression levels in shy2-2 relative to the wild type after auxin treatment. ∼indicates that one of the expression levels for comparison is below the detection limit; therefore, the fold difference is approximate.
shy2-2 causes reduced basal expression level or auxin-induced level but not both.
DISCUSSION
SHY2/IAA3 Affects Gene Expression
shy2-2 seedlings have short hypocotyls, slightly auxin-resistant hypocotyl and root growth, reduced gravitropic root response, and decreased lateral root number (Soh et al., 1999; Tian and Reed, 1999). We have interpreted these phenotypes as indicating that shy2-2 seedlings have reduced auxin responses. The experiments described in this article confirm at a molecular level that shy2-2 seedlings have decreased auxin responses. Of the 74 genes that were induced >2.5-fold by auxin in wild-type seedlings, 42 genes were not induced or were induced much less in shy2-2 seedlings. Similarly, of the 26 genes that were repressed >2.5-fold by auxin in wild-type seedlings, 14 genes were less repressed in shy2-2 seedlings. These results indicate that SHY2/IAA3 can decrease both induction and repression of a substantial fraction of auxin-regulated genes.
Genetic studies have shown that shy2-2 causes a gain of function, and shy2-2 seedlings have higher SHY2/IAA3 protein levels than do wild-type seedlings (Tian and Reed, 1999; Colón-Carmona et al., 2000). Therefore, increased SHY2/IAA3 protein activity is responsible for shy2-2 phenotypes and for the changes in gene expression profiles. The protein synthesis inhibitor cycloheximide can superinduce several primary auxin response genes (Franco et al., 1990; Abel et al., 1995b), implying that a short-lived protein normally represses expression of auxin-induced genes. That stabilization of SHY2/IAA3 by the shy2-2 mutation decreases auxin regulation of gene expression suggests that SHY2/IAA3 may be such a repressor. Although there is no evidence that the SHY2/IAA3 protein can bind to DNA, it might dimerize with ARF proteins that have either activator or repressor middle domains, thereby impeding either activation or repression of gene expression by ARFs. Recent evidence indicates that auxin can regulate stability of other Aux/IAA proteins, suggesting that regulated turnover of SHY2/IAA3 may mediate auxin gene expression responses (Gray et al., 2001; Zenser et al., 2001).
Auxin rapidly induces expression of SHY2/IAA3, and the shy2-2 mutation almost completely abolished both the basal expression level and the auxin induction of SHY2/IAA3. Auxin induction of SHY2/IAA3, therefore, potentiates a negative feedback on expression of SHY2/IAA3 and other auxin-induced genes. Such negative feedback might allow precise quantitative control of auxin responses.
Genes Regulated by shy2-2
Most Aux/IAA, SAUR, GH3, and ACS genes affected by shy2-2 are primary auxin response genes, indicating that shy2-2 inhibits primary auxin responses. shy2-2 also affected expression of one previously characterized secondary response gene, AXR2/IAA7. shy2-2 might regulate AXR2/IAA7 and other secondary or late response genes directly or indirectly through primary response genes. shy2-2 does not affect expression of some other known auxin-regulated genes, indicating that SHY2/IAA3 has some specificity in its regulatory targets. This specificity might arise from functional specialization of the protein or from tissue-specific SHY2/IAA3 expression.
Some of the semidominant mutants in other Aux/IAA genes also have altered expression of primary auxin response genes. For example, axr2-1 caused reduced expression and auxin induction of a number of Aux/IAA genes, especially SHY2/IAA3, IAA5, and itself, and it also caused decreased induction of SAUR-AC1 expression (Timpte et al., 1994; Abel et al., 1995b; Nagpal et al., 2000). iaa28-1 caused reduced expression and auxin induction of the synthetic BA::GUS auxin-responsive promoter::reporter fusion (Rogg et al., 2001). In contrast, axr3-1 caused ectopic SAUR-AC1 expression in the root vasculature (Leyser et al., 1996). These results suggest that to mediate auxin responses precisely, these Aux/IAA genes might form a tightly regulated network in which some Aux/IAA proteins (such as IAA28, AXR2/IAA7, and SHY2/IAA3) act as repressors and some Aux/IAA proteins (such as AXR3/IAA17) act as activators. They may have overlapping and/or distinct downstream targets, again depending on the specificity of the proteins and their spatial expression patterns.
Other than the Aux/IAA, SAUR, GH3, and ACS genes, shy2-2 affects expression of a number of auxin-respon-sive and auxin-nonresponsive genes. Some of the auxin-responsive genes have multiple AuxREs in their promoters (Table 1) and may be primary or secondary auxin response genes. shy2-2 may regulate expression of the auxin-nonresponsive genes indirectly.
The genes that are misregulated in shy2-2 presumably cause the mutant phenotypes; therefore, they may provide clues about cellular or signaling functions that are altered in the mutant. For example, several genes encoding protein kinases were less induced in shy2-2 than in wild-type seedlings, and these might be components of signal transduction pathways that regulate SHY2/IAA3-mediated inhibition of auxin-regulated gene expression. Several metabolic enzymes that are misregulated, such as cytochrome P450s and a putative tyrosine aminotransferase homologous to the ROOTY protein, might affect biosynthesis or breakdown of auxin or other signaling molecules (Barlier et al., 2000; Hull et al., 2000; Bak et al., 2001). Genes encoding enzymes that might synthesize or modify the cell wall may affect cell enlargement, leading to the short hypocotyls and upcurled leaves of shy2-2 mutants. Further molecular and genetic studies will help reveal how much each of these genes contributes to shy2 mutant phenotypes.
Novel Auxin-Regulated Genes
A number of other genes were induced or repressed by auxin but unaffected by the shy2-2 mutation, and these may reveal aspects of auxin response that are independent of SHY2/IAA3 action. Expression of an AUX1-like permease is induced by auxin, possibly to allow auxin influx into cells to activate auxin responses. Expression of a gene encoding an IAA β-glucosyltransferase, which is involved in IAA conjugation (Normanly and Bartel, 1999), is induced by auxin, indicating that negative feedback controls on auxin responses might act to increase auxin conjugation and affect auxin signaling. HOOKLESS1 encodes a putative acetyltransferase that affects differential cell growth during apical hook formation (Lehman et al., 1996), and expression of an HLS1 homolog is also induced significantly (>10-fold) by auxin, consistent with the idea that these acetyltransferases function in auxin responses.
The genes listed in Tables 1 and 2 are those that are either induced or repressed by >2.5-fold after 2 hr of auxin treatment. Previous studies showed that some primary response genes had transient responses to auxin and some were induced <2.5-fold (Abel et al., 1995b). Moreover, we assayed only approximately one-fourth of the genes in Arabidopsis. Therefore, our results include only a subset of auxin-responsive genes in the genome. Other experiments exploring the kinetics of global gene regulation response to auxin might reveal additional aspects of auxin responses.
SHY2/IAA3 Acts in Hypocotyls and Cotyledons
The PSHY2/IAA3::GUS fusion was expressed in the hypocotyls of dark-grown and young light-grown wild-type seedlings, in cotyledons of light-grown seedlings, and in leaves of older seedlings. shy2-2 mutants have short hypocotyls, enlarged cotyledons, and upcurled leaves, and PSHY2/IAA3::GUS expression in these organs suggests that SHY2/IAA3 regulates hypocotyl, cotyledon, and leaf growth cell autonomously. Consistent with this idea, several auxin-regulated promoter::GUS fusions were misexpressed in hypocotyls, cotyledons, and leaves of shy2-2 seedlings. For example, compared with wild-type seedlings, light-grown shy2-2 seedlings had less expression of PSAUR-AC1::GUS and PSHY2/IAA3:: GUS in hypocotyls and less expression of PIAA2::GUS in the petioles of the cotyledons. They also had more expression of PAXR2/IAA7::GUS in the cotyledons. Dark-grown shy2-2 seedlings also had almost no PSHY2/IAA3::GUS expression in hypocotyls but more expression in the cotyledons than wild-type seedlings. Similarly, dark-grown shy2-2 seedlings had more expression of PAXR2/IAA7::GUS and PSAUR-AC1::GUS in cotyledons than wild-type seedlings. Because shy2-2 mutants have enlarged cotyledons in the dark, these effects on gene expression in cotyledons might be an indirect effect of cotyledon growth in shy2-2 seedlings.
Both shy2-2 and shy2-24 mutants have altered root gravitropism and lateral root numbers. However, we observed no X-gluc staining in roots of plants carrying the PSHY2/IAA3:: GUS transgene. A previous report showed that SHY2/IAA3 was expressed at a very low level in roots (Abel et al., 1995b), and our reverse transcriptase–polymerase chain reaction from root RNA also showed expression in the root (data not shown). These results suggest that this transgene does not reflect normal expression precisely, that the expression in the roots was too low to be detected by X-gluc staining, or that the mRNA experiments suffered from nonspecific cross-hybridization. If it is true that SHY2/IAA3 is not expressed in the roots, then it is possible that it regulates root growth through some indirect mechanism, for example, by affecting polar auxin transport from shoot to root. We did find that shy2-2 caused reduced PAXR2/IAA7::GUS expression in roots. shy2-3 also caused ectopic PAXR3/IAA17:: GUS expression in root tips (O. Leyser, personal communication). All of these results suggest a role of SHY2/IAA3 in roots.
In contrast to the dramatic phenotypes of shy2-2 gain-of-function seedlings, shy2-24 mutants do not have strong hypocotyl, cotyledon, or leaf phenotypes, most likely because of genetic redundancy. IAA4 is phylogenetically related most closely to SHY2/IAA3, and previous work showed that IAA4 is expressed throughout the seedlings, including the cotyledons, hypocotyls, and roots (Abel et al., 1995b). Therefore, IAA4, or other Aux/IAA proteins with similar expression patterns, could act redundantly with SHY2/IAA3 to regulate hypocotyl or cotyledon growth.
Role of SHY2/IAA3 in Light Responses
The shy2-2 mutation can bypass the function of phyB, and shy2-2 seedlings have short hypocotyls and expanded cotyledons and make leaves in the dark. Therefore, shy2-2 might cause activation of light responses and photomorphogenesis. Consistent with this idea, in both dark and light, several light-regulated genes, such as CAB, and some photosynthesis factors are upregulated in shy2-2 (Kim et al., 1998) (Table 3). Recently, auxin was shown to downregulate expression of some light-induced genes (Gil et al., 2001). Therefore, the overexpression of light-induced genes in shy2-2 might arise from the reduced auxin responses.
Our data that light regulated SHY2/IAA3 expression suggest further links between light and SHY2/IAA3 activity. Overall, PSHY2/IAA3::GUS expression was lower in light than in darkness, and RNA gel blot hybridizations also showed that light regulates SHY2/IAA3 expression in dark-grown wild-type seedlings. In the presence of sucrose, a pulse of red light or a shift to continuous white light inhibited SHY2/IAA3 expression, whereas in the absence of sucrose, a red light pulse induced SHY2/IAA3 expression. These results suggest that both light and sucrose signaling pathways modulate SHY2/IAA3 gene expression and/or protein activity.
Considering that SHY2/IAA3 can inhibit expression of its own gene, our data could be explained by light regulation of SHY2/IAA3 protein activity. In the dark in the absence of sucrose, there may be little SHY2/IAA3 protein present, and light can induce the gene by an unknown mechanism. Sucrose increases SHY2/IAA3 expression in the dark, similarly to its effect on other light-regulated genes such as CAB (Brusslan and Tobin, 1992), so there may be more SHY2/IAA3 protein present. In this situation, light might activate the autoinhibitory activity of SHY2/IAA3, and this apparently overcomes the light activation of gene expression seen in the absence of sucrose. Light did not activate SHY2/IAA3 expression in the shy2-2 mutant, suggesting that light normally regulates SHY2/IAA3 protein activity. Consistent with the possibility that photoreceptors regulate SHY2/IAA3 protein activity, oat phyA can phosphorylate SHY2/IAA3 in vitro (Colón-Carmona et al., 2000) and SHY2/IAA3 also interacts with Arabidopsis phyB (Q. Tian, L. Krall, and J.W. Reed, unpublished results).
METHODS
Growth Conditions and Tissue Treatment
Arabidopsis thaliana seedlings were surface-sterilized, plated on Murashige and Skoog (1962) (MS)/agar plates (1 × MS salts [Gibco BRL], 0.8% [w/v] phytagar [Gibco BRL], and 1 × Gamborg's B5 vitamin mix [Sigma]) with or without 2% (w/v) sucrose, and stored at 4°C for 40 hr before being moved to the appropriate light conditions. For dark growth, seed were treated with constant white light for 6 hr to induce germination and then wrapped in three layers of aluminum foil to create a dark condition. For the red light induction experiments, we used light-emitting diode red light sources, which emit light with a peak at 670 nm and a half-bandwidth of 25 nm (Quantum Devices, Inc., Barneveld, WI). Seedlings were first grown in the dark for 6 days, and then they were given a pulse of red light with an intensity of 460 μmol·m−2·sec−1 for 5 min and returned to darkness for the indicated period of time. For the white light induction experiment, seedlings were treated with constant white light with an intensity of 45 μmol·m−2·sec−1 for 4 hr after 6 days of growth in the dark. For all experiments, the growth temperature was 21°C. For the auxin induction experiments, seedlings were grown in MS/sucrose liquid medium for 6 days with moderate shaking at 100 rpm. The growth temperature was 23°C, and the white light intensity was 6 to 10 μmol·m−2·sec−1. After 6 days of growth, seedlings were either mock treated with 0.1% ethanol or treated with 20 μM indoleacetic acid (IAA) dissolved in ethanol for 2 hr.
RNA Gel Blot Analysis
Seedlings were taken carefully from the plates, or drained with 3MM Whatman paper if they were grown in liquid medium, and frozen with liquid nitrogen. The frozen tissues were homogenized with mortars and pestles in liquid nitrogen. For the light induction experiments, total RNA was isolated using the acid guanidinium thiocyanate–phenol extraction method as described previously (Chomczynski and Sacchi, 1987). For other experiments, total RNA was extracted using Trizol reagent (Gibco BRL). Poly(A)+ RNA was extracted using oligo(dT)25 Dynabeads according to the manufacturer's instructions (Dynal, Lake Success, NY). mRNA isolated from 50 μg of total RNA was loaded on formaldehyde gels and transferred onto nylon membranes (Hybond N+; Amersham, Piscataway, NJ) by capillary blotting.
Blots were washed for 5 sec with 2 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate), baked at 80°C for 1 hr, and hybridized as described (Church and Gilbert, 1984). All Aux/IAA cDNA probes were kindly provided by A. Theologis (Plant Gene Expression Center, Albany, CA). We either amplified the cDNA fragments by polymerase chain reaction (PCR) using M13F1 and M13R3 vector primers or degenerate primers recognizing Aux/IAA genes, or we cut the fragments from the cDNA clones using restriction endonucleases and used those fragments as probes. The 32P probes were prepared by labeling the cDNA fragments (Aux/IAA) or the whole plasmids (CAB) using a random priming kit (Boehringer Mannheim, Indianapolis, IN). The blots were hybridized with the probes in Church buffer (250 mM NaPO4, pH 7.4, 7% SDS, 1 mM EDTA, and 1% BSA) at 65°C overnight and then washed in 2 × SSC once for 15 min followed by 0.2 × SSC and 0.1% SDS twice for 15 min each time at 65°C. The blots then were exposed to x-ray films (Eastman Kodak, Rochester, NY).
Microarray Method
Total RNA was extracted using Trizol reagent (Gibco BRL). Seven micrograms of total RNA was used to synthesize cDNA. A custom cDNA kit (Gibco BRL) was used with a T7-(dT)24 primer for this reaction. Biotinylated cRNA then was generated from the cDNA reaction using the Enzo BioArray High Yield RNA Transcript Kit (Affymetrix, Sunnyvale, CA). The cRNA then was fragmented in fragmentation buffer (1 × fragmentation buffer is 40 mM Tris acetate, pH 8.1, 100 mM KOAc, and 30 mM MgOAc) at 94°C for 35 min before chip hybridization. Fifteen micrograms of fragmented cRNA then was added to a hybridization cocktail [0.05 μg/μL fragmented cRNA, 50 pM control oligonucleotide B2, BioB, BioC, BioD, and cre hybridization controls, 0.1 mg/mL herring sperm DNA, 0.5 mg/mL acetylated BSA, 100 mM 2-(N-morpholino)-ethanesulfonic acid, pH 6.7, 1 M NaCl, 20 mM EDTA, and 0.01% Tween 20]. Arrays were hybridized for 16 hr in the GeneChip Fluidics Station 400 and were washed and scanned with the Hewlett-Packard GeneArray Scanner (Boise, ID). Affymetrix GeneChip Microarray Suite 4.0 software was used for washing, scanning, and basic analysis. Sample quality was assessed by examination of 3′ to 5′ intensity ratios of certain genes.
Construction of Promoter::Reporter Fusions
The 2.2-kb SHY2/IAA3 promoter was amplified by PCR using primers 5′-AACTGCAGTGCTATAATCAACCAGCG-3′ and 5′-CGGGAT CCTGAGGTTAACAAACTCATCC-3′ (underlined are PstI and BamHI sites, respectively). The 2.3-kb AXR2/IAA7 promoter was amplified by PCR using primers 5′-AACTGCAGCAATATGTGTATGTGCACG-3′ and 5′-CGGGATCCGTTGGCCGATCATGTTACTTG-3′ (underlined are PstI and BamHI sites, respectively). These PCR products were cloned into pPZP211 (Hajdukiewicz et al., 1994) with PstI–BamHI sites. The 2.0-kb β-glucuronidase gene (GUS) was cut from pBI101.1 (Clontech, Palo Alto, CA) and placed at the 3′ ends of the SHY2/IAA3 and AXR2/IAA7 promoters with BamHI–EcoRI sites.
PSHY2/IAA3::GUS was transformed into Landsberg erecta (Ler) plants by vacuum infiltration (Bechtold et al., 1993). We obtained 35 independent T1 plants and screened T2 progeny of these plants for 3:1 segregation for kanamycin resistance, indicating a single locus insertion. Six individual lines segregated 3:1, and each of them showed a similar 5-bromo-4-chloro-3-indolyl glucuronide (X-gluc) staining pattern. We crossed one of these lines into shy2 and axr2-1 mutants. For each cross, we screened for homozygous lines either by phenotype (shy2-2 and axr2-1) or by cleaved amplified polymorphic sequence polymorphism (shy2-24) (Tian and Reed, 1999). PAXR2/IAA7::GUS was transformed into Wassilewskija (Ws) and Ler. We obtained 13 T1 lines in Ws and three T1 lines in Ler, and among them were seven individual lines in Ws and one line in Ler with an insertion at a single locus. Each of the lines showed a similar staining pattern, and we used one line in Ws for the crossing. A previously described PIAA2::GUS construct (Luschnig et al., 1998) was transformed into Ler. We obtained 22 T1 lines, and seven of them had insertions at a single locus. Each of the seven lines showed similar X-gluc staining, and we crossed one of them into shy2 mutants. Previously described PSAUR-AC1::GUS (in Columbia background) (Gil and Green, 1997) was crossed directly into shy2 mutants.
GUS Staining
Seedlings were taken carefully from the MS/sucrose plates and placed directly in 50 mM NaHPO4 buffer, pH 7.0, containing 2 mM X-gluc. The 0.2 M X-gluc stock in N,N-dimethylformaldehyde was prepared fresh every time before use and diluted into 50 mM NaHPO4 buffer. The seedlings then were vacuum infiltrated for 5 min and subsequently incubated at 37°C for 16 hr or until sufficient staining developed. After clearing in 70% ethanol overnight, the seedlings were photographed through a dissecting microscope or a compound microscope with differential interference contrast optics.
GUS Activity Assay
Seedlings were grown on MS/sucrose plates as described above. After 6 days, 20 seedlings were taken carefully from the plates and homogenized with blue pestles in 100 μL of GUS extraction buffer (50 mM NaHPO4, pH 7.0, 10 mM EDTA, 0.1% Triton X-100, 0.1% N-lauroyl sarcosine, and 10 mM 2-mercaptoethanol) until no obvious debris was visible. The insoluble debris was spun out in a microcentrifuge. The supernatants were used directly for the assay or, if necessary, kept at −80°C until processing. For the GUS assay, 50 μL of the extracts was mixed with 150 μL of 1 mM 4-methylumbelliferyl β-d-glucuronide in GUS extraction buffer prewarmed to 37°C. The mixture was incubated at 37°C for 5, 30, 60, 120, and 240 min. At each time point, 40 μL of the mixture was removed and mixed with 360 μL of stop buffer (0.2 M Na2CO3). The resulting fluorescence was measured in a luminescence spectrometer (LS-5B; Perkin-Elmer), which was adjusted to read 900 units for 9 μM 4-methylumbelliferone. We read the fluorescence of each sample three times, plotted the average, and determined the GUS activity from the slope of this line.
Accession Numbers
Accession numbers for the genes described in this article are listed in the tables.
Acknowledgments
We thank Punita Nagpal for her generous help with RNA gel blot analysis and photography using the compound microscope, helpful discussion, and her comments on the manuscript; Jeff Dangl and Sarah Grant for use of their compound microscope; and Mike Vernon and Brian Popko for performing the microarray hybridizations. This work was supported by National Institutes of Health Grant No. R29-GM52456 to J.W.R.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010283.
References
- Abel, S., Oeller, P.W., and Theologis, A. (1994). Early auxin-induced genes encode short-lived nuclear proteins. Proc. Natl. Acad. Sci. USA 91, 326–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel, S., Nguyen, M.D., Chow, W., and Theologis, A. (1995. a). ACS4, a primary indoleacetic acid-responsive gene encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis thal-iana. J. Biol. Chem. 270, 19093–19099. [DOI] [PubMed] [Google Scholar]
- Abel, S., Nguyen, M.D., and Theologis, A. (1995. b). The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J. Mol. Biol. 251, 533–549. [DOI] [PubMed] [Google Scholar]
- Bak, S., Tax, F.E., Feldmann, K.A., Galbraith, D.W., and Feyereisen, R. (2001). CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell 13, 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlier, I., Kowalczyk, M., Marchant, A., Ljung, K., Bhalerao, R., Bennett, M., Sandberg, G., and Bellini, C. (2000). The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc. Natl. Acad. Sci. USA 97, 14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316, 1194–1199. [Google Scholar]
- Brusslan, J., and Tobin, E. (1992). Light-independent developmental regulation of cab gene expression in Arabidopsis thaliana seedlings. Proc. Natl. Acad. Sci. USA 89, 7791–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski, P., and Sacchi, N. (1987). Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform-extraction. Anal. Biochem. 162, 156–159. [DOI] [PubMed] [Google Scholar]
- Church, G.M., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Carmona, A., Chen, D.L., Yeh, K.-C., and Abel, S. (2000). Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol. 124, 1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco, A.R., Gee, M.A., and Guilfoyle, T.J. (1990). Induction and superinduction of auxin-responsive mRNAs with auxin and protein synthesis inhibitors. J. Biol. Chem. 265, 15845–15849. [PubMed] [Google Scholar]
- Gil, P., and Green, P.J. (1997). Regulatory activity exerted by the SAUR-AC1 promoter region in transgenic plants. Plant Mol. Biol. 34, 803–808. [DOI] [PubMed] [Google Scholar]
- Gil, P., Dewey, E., Friml, J., Zhao, Y., Snowden, K.C., Putterill, J., Palme, K., Estelle, M., and Chory, J. (2001). BIG, a calossin-like protein required for polar auxin transport in Arabidopsis. Genes Dev. 15, 1985–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCFTIR1-dependent degradation of Aux/IAA proteins. Nature 414, 271–276. [DOI] [PubMed] [Google Scholar]
- Guilfoyle, T.J. (1999). Auxin-regulated genes and promoters. In Biochemistry and Molecular Biology of Plant Hormones, P.J.J. Hooykaas, M. Hall, and K.L. Libbenga, eds (Leiden, the Netherlands: Elsevier), pp. 423-459.
- Hagen, G., and Guilfoyle, T. (2001). Auxin-responsive gene expression: Genes, promoters, and regulatory factors. Plant Mol. Biol. 15, 1985–1997. [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Hsieh, H.L., Okamoto, H., Wang, M., Ang, L.H., Matsui, M., Goodman, H., and Deng, X.W. (2000). FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev. 14, 1958–1970. [PMC free article] [PubMed] [Google Scholar]
- Hull, A.K., Vij, R., and Celenza, J.L. (2000). Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. USA 97, 2379–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, B.C., Soh, M.S., Kang, B.J., Furuya, M., and Nam, H.G. (1996). Two dominant photomorphogenic mutations of Arabidopsis thaliana identified as suppressor mutations of hy2. Plant J. 9, 441–456. [DOI] [PubMed] [Google Scholar]
- Kim, B.C., Soh, M.S., Hong, S.H., Furuya, M., and Nam, H.G. (1998). Photomorphogenic development of the Arabidopsis shy2-1D mutation and its interaction with phytochromes in darkness. Plant J. 15, 61–68. [DOI] [PubMed] [Google Scholar]
- Kim, J., Harter, K., and Theologis, A. (1997). Protein-protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 94, 11786–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman, A., Black, R., and Ecker, J.R. (1996). HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell 85, 183–194. [DOI] [PubMed] [Google Scholar]
- Leyser, H.M.O., Pickett, F.B., Dharmasiri, S., and Estelle, M. (1996). Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 10, 403–413. [DOI] [PubMed] [Google Scholar]
- Liscum, E., and Reed, J.W. (2001). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol., in press. [PubMed]
- Luschnig, C., Gaxiola, R.A., Grisafi, P., and Fink, G.R. (1998). EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 12, 2175–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, K.E., Zarembinski, T.I., Theologis, A., and Abel, S. (1999). Biochemical characterization of recombinant polypeptides corresponding to the predicted beta-alpha-alpha fold in Aux/IAA proteins. FEBS Lett. 454, 283–287. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nagpal, P., Walker, L., Young, J., Sonawala, A., Timpte, C., Estelle, M., and Reed, J.W. (2000). AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 123, 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly, J., and Bartel, B. (1999). Redundancy as a way of life: IAA metabolism. Curr. Opin. Plant Biol. 2, 207–213. [DOI] [PubMed] [Google Scholar]
- Ouellet, F., Overvoorde, P.J., and Theologis, A. (2001). IAA17/AXR3: Biochemical insight into an auxin mutant phenotype. Plant Cell 13, 829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos, J.A., Zenser, N., Leyser, O., and Callis, J. (2001). Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13, 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J.W. (2001). Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 6, 420–425. [DOI] [PubMed] [Google Scholar]
- Reed, J.W., Elumalai, R.P., and Chory, J. (1998). Suppressors of an Arabidopsis thaliana phyB mutation identify genes that control light signalling and hypocotyl elongation. Genetics 148, 1295–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogg, L.E., Lasswell, J., and Bartel, B. (2001). A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13, 465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux, C., and Perrot-Rechenmann, C. (1997). Isolation by differential display and characterization of a tobacco auxin-responsive cDNA Nt-gh3, related to GH3. FEBS Lett. 419, 131–136. [DOI] [PubMed] [Google Scholar]
- Sachs, T. (1991). Pattern Formation in Plant Tissues. (Cambridge, UK: Cambridge University Press).
- Soh, M.S., Hong, S.H., Kim, B.C., Vizir, I., Park, D.H., Choi, G., Hong, M.Y., Chung, Y.-Y., Furuya, M., and Nam, H.G. (1999). Regulation of both light- and auxin-mediated development by the Arabidopsis IAA3/SHY2 gene. J. Plant Biol. 42, 239–246. [Google Scholar]
- Thimann, K.V. (1977). Hormone Action in the Whole Life of Plants. (Amherst, MA: University of Massachusetts Press).
- Tian, Q., and Reed, J.W. (1999). Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126, 711–721. [DOI] [PubMed] [Google Scholar]
- Timpte, C.S., Wilson, A.K., and Estelle, M. (1992). Effects of the axr2 mutation of Arabidopsis on cell shape in hypocotyl and inflorescence. Planta 188, 271–278. [DOI] [PubMed] [Google Scholar]
- Timpte, C., Wilson, A., and Estelle, M. (1994). The axr2-1 mutation of Arabidopsis thaliana is a gain-of-function mutation that disrupts an early step in auxin response. Genetics 138, 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999). Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 96, 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, A.K., Pickett, F.B., Turner, J.C., and Estelle, M. (1990). A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol. Gen. Genet. 222, 377–383. [DOI] [PubMed] [Google Scholar]
- Worley, C.K., Zenser, N., Ramos, J., Rouse, D., Leyser, O., Theologis, A., and Callis, J. (2000). Degradation of Aux/IAA proteins is essential for normal auxin signaling. Plant J. 21, 553–562. [DOI] [PubMed] [Google Scholar]
- Zenser, N., Ellsmore, A., Leasure, C., and Callis, J. (2001). Auxin modulates the degradation rate of Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 20, 11795–11800. [DOI] [PMC free article] [PubMed] [Google Scholar]