Abstract
The salt tolerance locus SOS1 from Arabidopsis has been shown to encode a putative plasma membrane Na+/H+ antiporter. In this study, we examined the tissue-specific pattern of gene expression as well as the Na+ transport activity and subcellular localization of SOS1. When expressed in a yeast mutant deficient in endogenous Na+ transporters, SOS1 was able to reduce Na+ accumulation and improve salt tolerance of the mutant cells. Confocal imaging of a SOS1–green fluorescent protein fusion protein in transgenic Arabidopsis plants indicated that SOS1 is localized in the plasma membrane. Analysis of SOS1 promoter–β-glucuronidase transgenic Arabidopsis plants revealed preferential expression of SOS1 in epidermal cells at the root tip and in parenchyma cells at the xylem/symplast boundary of roots, stems, and leaves. Under mild salt stress (25 mM NaCl), sos1 mutant shoot accumulated less Na+ than did the wild-type shoot. However, under severe salt stress (100 mM NaCl), sos1 mutant plants accumulated more Na+ than did the wild type. There also was greater Na+ content in the xylem sap of sos1 mutant plants exposed to 100 mM NaCl. These results suggest that SOS1 is critical for controlling long-distance Na+ transport from root to shoot. We present a model in which SOS1 functions in retrieving Na+ from the xylem stream under severe salt stress, whereas under mild salt stress it may function in loading Na+ into the xylem.
INTRODUCTION
Plant growth depends on mineral nutrients absorbed from the soil by roots. Although Na+ is a major cation present in soil solutions, Na+ is not considered an essential mineral for most plants. In saline soils, high concentrations of Na+ disrupt K+ and other mineral nutrition, create hyperosmotic stress, and cause secondary problems such as oxidative stress (Zhu, 2001). These adverse effects contribute to plant growth inhibition and even plant death.
Many cytosolic enzymes are activated by K+ and inhibited by Na+ (Flowers et al., 1977). Three mechanisms are available to plant cells to prevent excessive accumulation of Na+ in the cytosol (Niu et al., 1995; Blumwald et al., 2000; Zhu, 2001). First, Na+ entry to plant cells may be restricted by selective ion uptake. Nonselective cation channels have been proposed to mediate substantial Na+ entry into plant roots, but genes encoding these channels have yet to be identified (Amtmann and Sanders, 1999; Tyerman and Skerrett, 1999). The cloned transporters HKT1 and LCT1 have Na+ permeability when expressed in yeast or oocytes, suggesting that they also may be candidate Na+ influx transporters (Rubio et al., 1995; Schachtman et al., 1997). Recently, studies in yeast demonstrated that the magnitude of the membrane potential affects net Na+ influx into cells. Mutations in the yeast PMP3 gene lead to membrane hyperpolarization, increased Na+ influx, and salt sensitivity (Navarre and Goffeau, 2000).
Second, internalized Na+ can be stored in vacuoles. Vacuolar compartmentation is an efficient strategy for plant cells to deal with salt stress because the stored Na+ contributes to osmotic adjustment (Flowers et al., 1977). Halophytes, plants that are adapted naturally to high salinity, are known to accumulate large amounts of Na+ in the vacuole (Flowers et al., 1977). Vacuolar Na+/H+ antiporters have been cloned from glycophytes (i.e., salt-sensitive plants) (Apse et al., 1999; Fukuda et al., 1999; Gaxiola et al., 1999; Darley et al., 2000; Quintero et al., 2000). Increased expression of a vacuolar Na+/H+ antiporter, AtNHX1, in transgenic Arabidopsis plants has been shown to improve plant salt tolerance (Apse et al., 1999).
Third, Na+ in the cytosol may be exported back to the growth medium or to apoplastic spaces. Na+/H+ antiporters on the plasma membrane are expected to fulfill this function (Blumwald et al., 2000). Na+ extrusion through these Na+/H+ antiporters is driven by an inwardly directed proton gradient created by H+-ATPases. Recently, we identified a putative plasma membrane Na+/H+ antiporter encoded by the Arabidopsis SOS1 gene (Shi et al., 2000). SOS1 was identified initially as a genetic locus required for salt tolerance in Arabidopsis (Wu et al., 1996). Loss-of-function mutations in SOS1 render plants extremely sensitive to Na+ inhibition. The transcript level of SOS1 is upregulated by NaCl stress but not by drought, cold, or abscisic acid (Shi et al., 2000). Salt upregulation of SOS1 expression is partially under the control of the SOS2/SOS3 regulatory pathway (Halfter et al., 2000; Liu et al., 2000; Shi et al., 2000; Zhu, 2000). Phylogenetic analysis of the SOS1 amino acid sequence suggests that it may be a plasma membrane Na+/H+ antiporter. However, neither its function as a Na+ transporter nor its subcellular localization has been demonstrated experimentally.
An important issue in salt stress physiology is Na+ translocation from the root to the shoot (Flowers et al., 1977; Epstein, 1998). Physiological evidence suggests that halophytes and salt-resistant glycophytes actively transport Na+ from the root to the shoot, whereas salt-sensitive glycophytes appear to limit Na+ entry into the transpirational stream to prevent Na+ accumulation in the shoot (Flowers et al., 1977; Lauchli, 1984). The transporter(s) responsible for Na+ transport to and from the xylem vessels are not known, although plasma membrane Na+/H+ antiporters have been proposed to fulfill this role (Lacan and Durand, 1996; Hasegawa et al., 2000). It also is not known which cell layer(s) might be critical for controlling the extent of Na+ entry or exit from the xylem. Theoretically, both endodermal and pericycle cell layers could be crucial in the root (Epstein, 1998).
Here we provide experimental evidence that SOS1 is a plasma membrane Na+ transporter essential for controlling long-distance Na+ movement in plants. The Na+ efflux capacity of SOS1 is demonstrated by its complementation of a yeast mutant defective in Na+ extrusion. Confocal imaging analysis with transgenic plants expressing a SOS1–green fluorescent protein (GFP) fusion protein shows that SOS1 is localized to the plasma membrane. Callus tissue from sos1 mutant plants accumulates more Na+ than does wild-type callus, suggesting that SOS1 is important for Na+ efflux in plant cells. We show that SOS1 is expressed preferentially in cells bordering the xylem vessels and strongly affects long-distance Na+ transport in plants. We suggest that SOS1 functions in Na+ retrieval from the xylem sap under severe salt stress conditions, whereas it may load Na+ into the root xylem when mild salt stress is applied.
RESULTS
SOS1 Complementation of Yeast Na+/H+ Antiporter Mutants
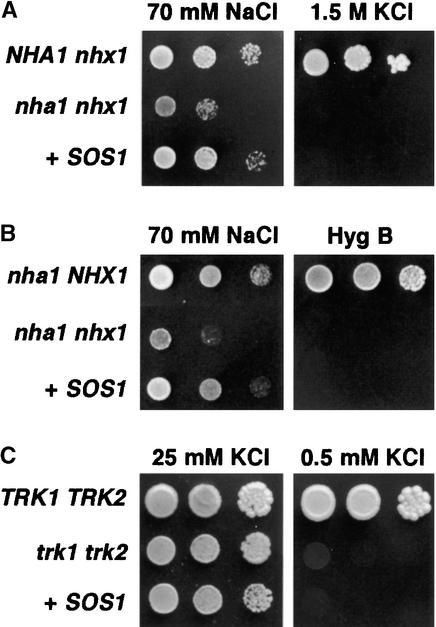
Saccharomyces cerevisiae cells contain two types of Na+/H+ antiporters with different localization and function. NHX1 is essential for the accumulation of Na+ into vacuoles and for resistance to various toxic cations, including hygromycin B (Nass and Rao, 1998; Gaxiola et al., 1999; Quintero et al., 2000). NHA1 functions in the plasma membrane and mediates Na+ and K+ efflux (Bañuelos et al., 1998). The Arabidopsis SOS1 protein shares significant sequence similarity with both NHA1 and NHX1 proteins. To determine the capacity of SOS1 to substitute for the yeast NHA1 or NHX1 antiporters, mutant complementation tests were performed. Because the Na+ tolerance of yeast is dominated by the activity of ENA1 to ENA4 Na+-ATPases mediating Na+ extrusion from the cell, a Δena1-ena4 mutant, in which the tandem array of ENA genes had been deleted, was used as the background for SOS1-dependent Na+ tolerance tests. In the Δena1-ena4 background, one or both of the Na+/H+ antiporter genes were deleted to create strains of genotype NHA1 nhx1, nha1 NHX1, and nha1 nhx1. The nha1 nhx1 double mutant is more sensitive to 70 mM NaCl (1 mM KCl) than is either of the single antiporter mutants (Figures 1A and 1B). Expression of SOS1 in the nha1 nhx1 mutant restored growth on 70 mM NaCl to the same level as that in endogenous NHA1 or NHX1 antiporters (Figures 1A and 1B). However, SOS1 failed to recover the sensitivity of a nha1 mutant to high external KCl (Bañuelos et al., 1998) and the distinctive sensitivity of a nhx1 mutant to hygromycin B (Gaxiola et al., 1999), indicating that SOS1 imparts specific tolerance to Na+ but does not fulfill all of the functions of these fungal antiporters (Figures 1A and 1B).
Figure 1.
Complementation of Na+ and K+ Transport Mutants of Yeast by SOS1.
S. cerevisiae strains AXT3 (nha1 nhx1) and WΔ3 (trk1 trk2) were transformed with plasmid containing SOS1 (+SOS1), whereas strains GX1 (NHA1 nhx1), ANT3 (nha1 NHX1), and W303 (TRK1 TRK2) were transformed with the empty vector.
(A) and (B) Transformants were grown overnight in liquid arginine-phosphate medium (AP medium; 10 mM l-arginine, 8 mM PO4H3, 2 mM MgSO4, 0.2 mM CaCl2, 2% glucose + vitamins and trace elements, pH 6.5) with 1 mM KCl. Five microliters of serial decimal dilutions were spotted onto plates of the same medium supplemented with 70 mM NaCl and in YPD medium containing either 1.5 M KCl (A) or 50 μg/mL hygromycin B (Hyg B) (B). Plates were incubated at 28°C and photographed after 2 days (hygromycin B) or 4 days (NaCl and KCl).
(C) Cells were grown in AP medium with 25 mM KCl, collected, and washed several times with distilled water until complete removal of KCl. Decimal dilutions were spotted onto AP plates containing either 25 mM KCl, to support the growth of trk1 trk2 mutants, or 0.5 mM KCl, a low concentration that prevents the growth of trk1 trk2 cells.
K+ uptake at low external concentration is impaired in sos1 mutant plants (Wu et al., 1996), raising the question of whether K+ ions function as a substrate for the SOS1 transporter. To test this possibility, the K+ tolerance of nha1 mutant cells expressing or not expressing SOS1 was examined at various KCl concentrations, but no growth differences indicative of K+ efflux capacity were found (data not shown). More importantly, SOS1 failed to restore K+ acquisition to a yeast trk1 trk2 double mutant deficient in K+ uptake (Figure 1C) (Ramos et al., 1994). The KCl concentration used (0.5 mM) is in the upper limit of K+ availability that still restricts the growth of trk1 trk2 mutants, so even a modest enhancement of K+ uptake would have promoted cell growth. Moreover, K+ acquisition was not stimulated by supplemental Na+, excluding the possibility that SOS1 is a Na+/K+ transporter (data not shown).
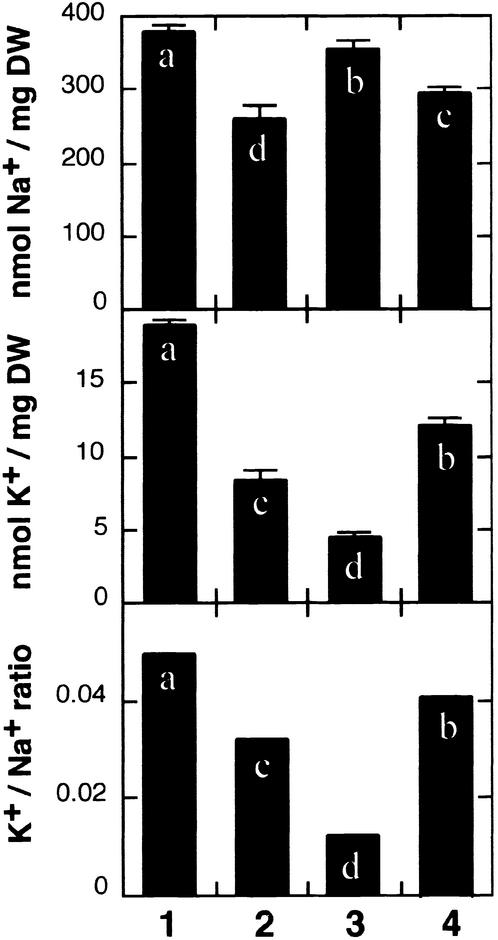
To determine the mechanism by which SOS1 conferred Na+ tolerance to yeast, Na+ and K+ contents were measured in cells transformed with SOS1. When grown in the presence of 70 mM NaCl and 1 mM KCl, SOS1-expressing cells maintained lower Na+ and higher K+ levels than did control cells without antiporter activity (nha1 nhx1) (Figure 2). NHA1 function in the plasma membrane produced similar results, reducing the cellular Na+ content and improving the K+ balance. On the other hand, the activity of the endomembrane protein NHX1 resulted in the accumulation of compartmentalized Na+ and K+ (Figure 2). The finding that Na+ tolerance conferred by SOS1 correlated with lower Na+ content, mimicking NHA1 activity, suggests that SOS1 directs Na+ efflux.
Figure 2.
Expression of SOS1 in Yeast Na+/H+ Antiporter Mutants Reduces Na+ Accumulation.
Cells of strains ANT3 (nha1 NHX1) (lane 1), GX1 (NHA1 nhx1) (lane 2), AXT3 (nha1 nhx1) (lane 3), and AXT3 transformed with plasmid containing SOS1 (lane 4) were grown in liquid AP medium supplemented with 1 mM KCl and 70 mM NaCl until midexponential growth phase and collected by filtration, and their Na+ and K+ contents were determined. Units are nanomoles of ion per milligram dry weight (DW) of cell samples. Data shown are averages and se values of ion contents of three independent cultures of each strain. Columns with different letters indicate significant difference at P < 0.05 (Fisher's protected LSD test).
SOS1 Is Localized in the Plasma Membrane of Plant Cells
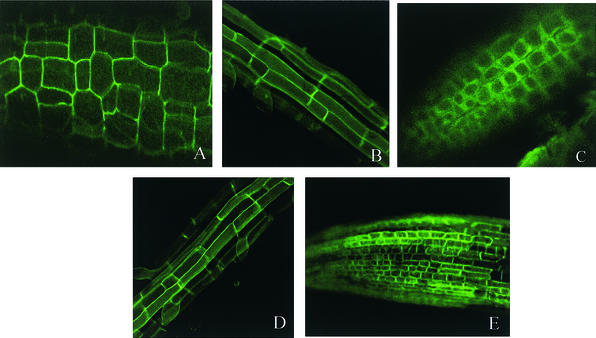
Sequence similarities and phylogenetic analysis suggested that SOS1 may be a plasma membrane Na+/H+ antiporter (Shi et al., 2000). Because our repeated attempts to obtain anti-SOS1 antibodies for immunolocalization were not successful, we sought to verify the subcellular localization of SOS1 by fusing it with the GFP. The SOS1-GFP fusion protein was expressed under the 35S promoter of Cauliflower mosaic virus in transgenic Arabidopsis plants. Subcellular localization of the fusion protein was visualized by confocal imaging of green fluorescence in the cells of transgenic seedlings. As shown in Figures 3A and 3B, green fluorescence was found on the surface of cells in hypocotyls and roots of SOS1-GFP transgenic plants. Similarly, GFP expression also was detected in undifferentiated cells of the root tip (Figure 3C). Because these root tip cells do not contain large central vacuoles, the cell surface green fluorescence reflects GFP expression on the plasma membrane or cell wall but not on the tonoplast or cytoplasm. To exclude the possibility of cell wall association of SOS1-GFP, the root tip cells were plasmolyzed by mannitol treatment. As shown in Figure 3C, the fluorescence in the plasmolyzed root tip cells was detached slightly from the cell wall, indicating a plasma membrane rather than a cell wall localization of the SOS1-GFP fusion protein. Fluorescence characteristics of SOS1-GFP are very similar to those exhibited by GFP fusion to PIP2a, a plasma membrane water channel protein (Cutler et al., 2000). Figures 3D and 3E show the cell surface green fluorescence in the young root and root tip cells of transgenic plants expressing the PIP2a-GFP fusion protein (Cutler et al., 2000). Together, these results strongly suggest that SOS1 is a plasma membrane protein.
Figure 3.
Subcellular Localization of the SOS1-GFP Fusion Protein in Transgenic Arabidopsis Plants.
(A) Localization of SOS1-GFP fluorescence in hypocotyl cells.
(B) Localization of SOS1-GFP fluorescence in young root cells.
(C) Localization of SOS1-GFP fluorescence in root tip cells plasmolyzed with 0.5 M mannitol.
(D) Localization of PIP2a-GFP in young root cells.
(E) Localization of PIP2a-GFP in root tip cells.
Function of SOS1 in Callus Tissues
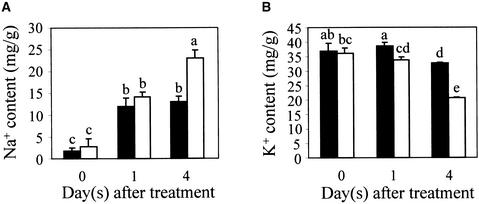
The results described above support the notion that SOS1 is a plasma membrane Na+ transporter. We next attempted to determine the function of SOS1 in callus cultures to avoid the complexities of long-distance transport in a whole plant system. Our previous data showed that sos1 mutant callus is more sensitive to NaCl inhibition than is wild-type callus, indicating that SOS1 is functional in the callus cells (Wu et al., 1996). If SOS1 functions in the plasma membrane to export Na+, we would expect sos1 knockout mutant cells to have a higher Na+ content than wild-type cells when treated with NaCl. As shown in Figure 4, the Na+ contents increased in both sos1 and wild-type calli, but after 4 days of NaCl treatment the Na+ content in sos1 callus was nearly twofold that of wild-type callus (Figure 4A). In contrast, the K+ content in sos1 was approximately two-thirds of the wild-type value (Figure 4B). These results suggest that SOS1 functions in Na+ efflux in the callus cells, which is consistent with the plasma membrane localization of SOS1. The cause of the reduced accumulation of K+ in sos1 mutant cells is unclear, but the result could be an indirect consequence of the higher Na+ content in these cells (i.e., a high concentration of internal Na+ may inhibit K+ uptake).
Figure 4.
Callus Cultures from sos1 Mutant Plants Accumulate More Na+ and Less K+ Than Do Wild-Type Calli.
(A) Na+ contents in wild-type and sos1 mutant calli.
(B) K+ contents in wild-type and sos1 mutant calli.
Closed bars, wild type; open bars, sos1-1 mutant. Data shown are averages and ±sd values of three independent experiments. Columns with different letters indicate significant difference at P < 0.05 (Fisher's protected LSD test).
Preferential Expression of SOS1 in Parenchyma Cells at the Xylem/Symplast Boundary
RNA gel blot experiments detected SOS1 mRNA in both the root and the shoot of Arabidopsis seedlings (Shi et al., 2000). In situ hybridization experiments performed to locate the expression of SOS1 failed to detect the transcript, probably as a result of the low level of expression of the SOS1 gene. A similar difficulty in detecting the transcript expression of the K+ channel SKOR by in situ hybridization has been reported (Gaymard et al., 1998). Therefore, we examined the expression pattern of SOS1 using transgenic plants carrying the Escherichia coli β-glucuronidase (GUS) gene under the control of the SOS1 promoter region. GUS activity was detected throughout the plant, mainly in the inner tissues surrounding the vasculature (Figure 5A). The vascular pattern of GUS staining was found in roots, hypocotyls, inflorescence stems, and leaves (Figures 5B to 5E). SOS1 promoter–driven GUS expression could not be detected in flowers and siliques (data not shown). In the root, cross-sectional analysis revealed that GUS activity was localized primarily in the pericycle and in other parenchyma cells bordering xylem vessels (Figure 5F). In stem and petiole cross-sections, GUS activity also was restricted to parenchyma cells at the xylem/symplast boundary (Figures 5G to 5I). The preferential expression of SOS1 at the xylem/symplast boundary, as indicated by the GUS reporter gene, suggests an important role of SOS1 in regulating long-distance Na+ transport by controlling xylem loading or unloading.
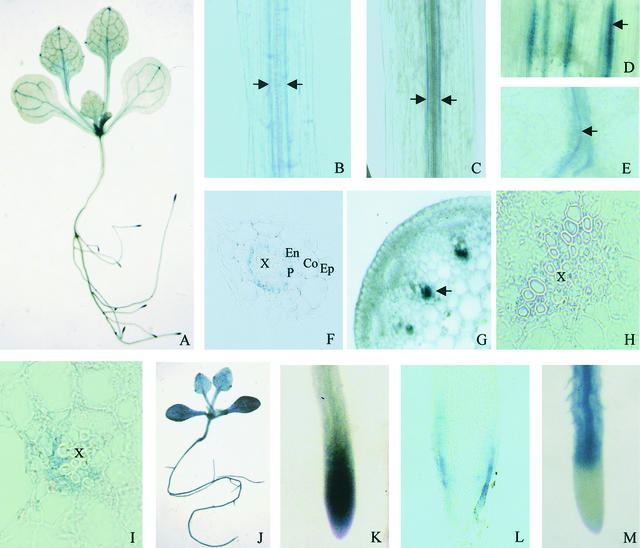
Figure 5.
SOS1 Promoter–GUS Expression Pattern in Transgenic Arabidopsis Plants, and Comparison with the Expression Pattern of AtNHX1 Promoter–GUS.
Except for panels (J) and (M), all panels show SOS1::GUS expression. (J) and (M) show AtNHX1::GUS expression.
(A) GUS staining in a SOS1::GUS seedling. Note the preferential GUS expression in the root tip and vasculature.
(B) GUS expression in cells surrounding the vascular system in a young root in a SOS1::GUS plant.
(C) GUS expression in cells surrounding the vascular system in the hypocotyl in a SOS1::GUS plant.
(D) GUS expression in cells associated with vascular strands in an inflorescence stem in a SOS1::GUS plant.
(E) GUS expression in cells surrounding leaf veins in a SOS1::GUS plant.
(F) Cross-section of a root in a SOS1::GUS plant, showing GUS expression in the pericycle.
(G) Hand section of an inflorescence stem in a SOS1::GUS plant, showing GUS expression associated with vascular strands.
(H) Cross-section of an inflorescence stem, showing an enlarged image of a vascular strand. Note the GUS expression in xylem parenchyma cells.
(I) Cross-section of a petiole in a SOS1::GUS plant, showing GUS expression in xylem parenchyma cells.
(J) GUS expression in a AtNHX1::GUS seedling. Note the strong and virtually ubiquitous GUS expression.
(K) GUS expression in a root tip in a SOS1::GUS plant.
(L) Longitudinal section of the root tip shown in (K), showing GUS expression in epidermal cells.
(M) GUS expression pattern in a root in a AtNHX1::GUS plant, showing that AtNHX1 is not expressed at the root tip.
Co, cortex; En, endodermis; Ep, epidermis; P, pericycle; X, xylem vessel. Arrows indicate GUS expression associated with the vasculature in SOS1::GUS plants.
Compared with SOS1, the expression pattern of the vacuolar Na+/H+ antiporter gene AtNHX1 was different. GUS activity was present in all cells of transgenic seedlings expressing the GUS gene under the control of the AtNHX1 promoter, although the expression was stronger in vascular tissues (Figure 5J). The cotyledons had higher GUS activity than did true leaves (Figure 5J). Moreover, GUS expression driven by the AtNHX1 promoter was much stronger than that driven by the SOS1 promoter. Visible GUS staining in SOS1 promoter–GUS transgenic plants could be seen only after overnight color reaction. However, strong GUS staining was obtained in AtNHX1 promoter–GUS transgenic plants after just 2 hr of color reaction. The lower expression level of SOS1 compared with AtNHX1 was confirmed by RNA gel blot experiments (data not shown).
SOS1 Expression in the Root Tip
Besides expression associated with the vasculature, SOS1 promoter–GUS expression was observed consistently at the root tip (Figure 5K). Cross-sectional analysis revealed that the root tip GUS activity was localized in the epidermal cells (Figure 5L). Because meristematic cells at the root tip lack large vacuoles for significant Na+ compartmentation, SOS1 might be critical to prevent Na+ from reaching these cells by actively extruding Na+ back to the root growth medium. Consistent with this notion, promoter-GUS analysis showed that the AtNHX1 gene encoding a vacuolar Na+/H+ antiporter was not expressed at the root tip (Figure 5M).
Role of SOS1 in Controlling Long-Distance Na+ Transport
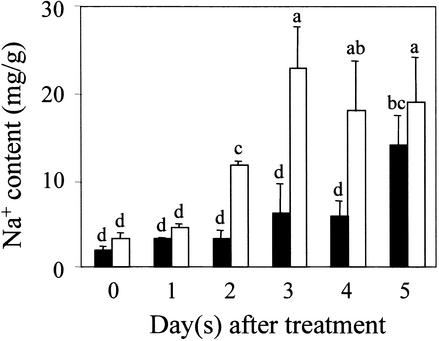
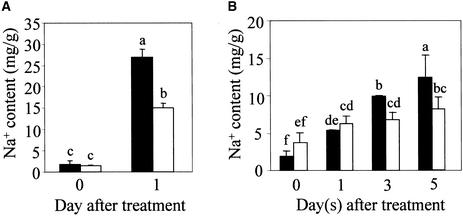
The preferential expression of SOS1 in cells surrounding the vasculature suggests a role of this transporter in long-distance Na+ movement in plants. To investigate this possibility, we compared Na+ accumulation in the root and shoot of sos1 mutant and wild-type plants. Because plant ion relations are influenced greatly by environmental conditions, especially by the root growth medium, we determined Na+ contents in plants grown in various soil and culture media. When grown in regular soil mix irrigated with 1/20 Murashige and Skoog (1962) (MS) nutrient solution supplemented with 100 mM NaCl, the aerial parts of sos1 mutant plants accumulated more Na+ than did wild-type plants, reaching a peak level of Na+ after 3 days of NaCl treatment (Figure 6). In comparison, Na+ accumulation was slower and less in the wild-type shoot (Figure 6). This faster and greater Na+ accumulation in the sos1 mutant shoot was observed consistently in five independent experiments.
Figure 6.
Soil-Grown sos1 Mutant Plants Accumulate More Na+ in the Shoot Than Do Wild-Type Plants in Response to 100 mM NaCl Treatment.
Five independent experiments were performed, and similar patterns of ion accumulation were obtained. Results from one typical experiment are shown. Closed bars, wild type; open bars, sos1-1. Error bars represent ±sd (n = 3). Columns with different letters indicate significant difference at P < 0.05 (Fisher's protected LSD test).
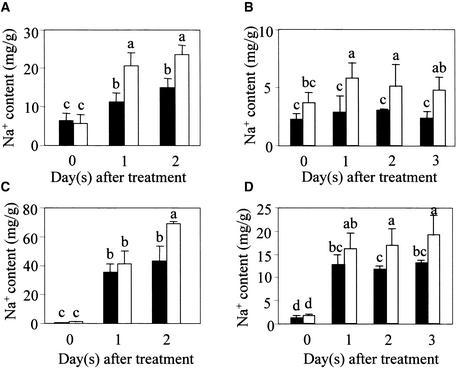
To determine the Na+ content in roots, plants were grown hydroponically or in Turface “hydroponic soil” to facilitate root harvest. As shown in Figure 7, the roots of sos1 mutant plants accumulated more Na+ than did wild-type plants in response to 100 mM NaCl treatment in hydroponic culture and in Turface soil. Under both culture conditions, the aerial parts of sos1 mutant plants also accumulated more Na+ than did the wild type (Figures 7C and 7D). The Na+ contents in sos1 and wild-type plants from the hydroponic culture were considerably higher than those from soil conditions, presumably reflecting the much faster exposure of plant roots to added NaCl in the hydroponic system.
Figure 7.
Increased Na+ Accumulation in Both the Root and Shoot of sos1 Mutant Plants Grown in Hydroponic Culture or in Turface Soil.
(A) Na+ contents in roots of hydroponically cultured plants.
(B) Na+ contents in roots of plants grown in Turface soil.
(C) Na+ contents in shoots of hydroponically cultured plants.
(D) Na+ contents in shoots of plants grown in Turface soil.
Closed bars, wild type; open bars, sos1-1. Error bars represent ±sd (n = 3). Columns with different letters indicate significant difference at P < 0.05 (Fisher's protected LSD test).
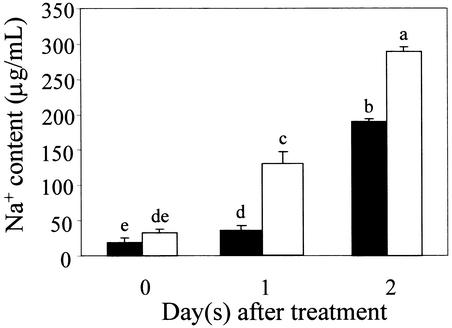
Na+ is transported from the root to the shoot via the xylem. To examine more directly the role of SOS1 in controlling Na+ transport from the root to the shoot, we measured the Na+ content in the xylem sap from sos1 mutant and wild-type plants. The method of xylem sap collection was as described by Gaymard et al. (1998) and Nublat et al. (2001). The Na+ concentration in the xylem sap of both sos1 mutant and wild-type plants increased substantially over time in 100 mM NaCl, but the Na+ content was always higher for sos1 (Figure 8). These results clearly reveal a role of wild-type SOS1 in retrieving Na+ from the xylem to prevent overaccumulation in the xylem transpirational stream under the treatment conditions used.
Figure 8.
Soil-Grown sos1 Mutant Plants Accumulate More Na+ in the Xylem Sap in Response to 100 mM NaCl Treatment.
Plants were grown in general soil mix in pots. After bolting, the plants were subjected to salt treatment by immersing the bottoms of the pots in a solution containing one-twentieth strength MS salts and 100 mM NaCl for the times indicated. Xylem sap was collected as described in Methods. Three independent experiments gave consistent results. Results from one typical experiment are shown. Closed bars, wild type; open bars, sos1-1. Error bars represent ±sd (n = 3). Columns with different letters indicate significant difference at P < 0.05 (Fisher's protected LSD test).
These results would seem to contradict our previous findings that sos1 mutant plants accumulated less Na+ than did the wild type in response to low levels of salt stress (e.g., 25 mM NaCl) in liquid culture (Ding and Zhu, 1997). To determine whether this apparent discrepancy was caused by different growth conditions and saline regimens, we grew plants in liquid culture for 8 days and then treated the seedlings with 25 mM NaCl for 1 day (Ding and Zhu, 1997). Consistent with our previous report, sos1 mutant seedlings had lower Na+ content than did the wild type under these conditions (Figure 9A). Interestingly, even when plants were grown in soil but were kept under 100% relative humidity (RH) to reduce transpiration, sos1 mutants also accumulated less Na+ than did the wild type after 3 or 5 days of treatment with 25 mM NaCl (Figure 9B). These findings suggest that SOS1 might function to load Na+ into the xylem under conditions of mild Na+ stress and low or no transpiration.
Figure 9.
sos1 Mutant Plants Accumulate Less Na+ in Response to Mild NaCl Stress.
(A) Na+ contents in wild-type and sos1 seedlings cultured in liquid medium. The medium consisted of half-strength MS salts and 2% sucrose, pH 5.6. Salt treatment was performed by adding NaCl to the medium to a final concentration of 25 mM.
(B) Na+ content in aerial parts of wild-type and sos1 mutant plants grown in general soil mix. After 3 weeks of growth, the plants were treated by immersing the bottoms of the pots in a solution containing one-twentieth strength MS salts supplemented with 25 mM NaCl for the times indicated. The treated plants were kept in a chamber with near 100% RH. Two independent experiments were performed, and consistent results were obtained. Results from one typical experiment are shown. Closed bars, wild type; open bars, sos1-1. Error bars represent ±sd (n = 3). Columns with different letters indicate significant difference at P < 0.05 (Fisher's protected LSD test).
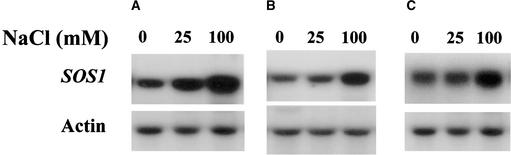
RNA gel blot analysis was performed to determine the levels of SOS1 transcript under the various treatment regimens (Figure 10). In wild-type plants grown in regular soil under low humidity, 25 mM NaCl treatment for 2 days caused a slight increase in SOS1 expression in the shoot, whereas 100 mM NaCl treatment for 2 days resulted in a more substantial increase in SOS1 expression in the shoot (Figure 10A). In young seedlings grown in liquid culture, 25 mM NaCl treatment did not increase SOS1 expression, whereas 100 mM NaCl treatment did (Figure 10B). In plants grown in regular soil under high humidity, 100 but not 25 mM NaCl treatment caused an obvious increase in SOS1 expression in the shoot (Figure 10C). These results indicate that SOS1 transcript levels increase when the Na+ load in the plant is high.
Figure 10.
SOS1 Transcript Expression under Various Salt Treatment Regimens.
(A) Shoot tissues from plants grown in regular soil with 2 days of salt treatment.
(B) Whole seedlings grown in liquid culture with 1 day of salt treatment.
(C) Shoot tissues from plants grown in regular soil under high humidity with 2 days of salt treatment.
Actin probe was used as a control for RNA loading.
DISCUSSION
Yeast cells devoid of endogenous Na+ transporters (i.e., Na+-ATPases and plasma membrane and vacuolar Na+/H+ antiporters) are very sensitive to NaCl stress. Expression of the Arabidopsis SOS1 gene improved salt tolerance to an extent that was comparable to that conferred by the yeast endogenous Na+/H+ antiporters NHA1 and NHX1 (Figure 1). Both yeast antiporters have broad substrate specificity, imparting tolerance to Na+, Li+, and K+ (Bañuelos et al., 1998; Quintero et al., 2000) (results not shown). In contrast, SOS1 activity was specific for Na+ because the plant protein was inefficient for K+ efflux or uptake in vivo (Figure 1). A similar reduction of Na+ content in SOS1- and NHA1-expressing yeast indicated that SOS1 mediated Na+ extrusion (Figure 2). A role of SOS1 in Na+ efflux in plant cells also was supported by ion content analysis of callus cultures generated from sos1 mutant plants (Figure 4). Consistent with such a role, confocal imaging of the SOS1-GFP fusion protein indicated a plasma membrane localization of SOS1 (Figure 3).
SOS1 was expressed preferentially in epidermal cells at the root tip. Meristematic cells are known to lack large vacuoles for Na+ compartmentation; thus, Na+ extrusion to the external medium by epidermal cells might be crucial to protect the root meristem. Moreover, we found that AtNHX1 was not expressed at the root tip (Figure 5M), suggesting a reliance on Na+ efflux by SOS1. Whether SOS1 is distributed equally on the plasma membrane of the epidermal cells or is directed asymmetrically to the external side of epidermal cells for maximal Na+ extrusion to the root, growth medium is unknown at present.
Another interesting expression pattern of SOS1 was revealed by the preferential activity of SOS1 promoter–GUS in parenchyma cells surrounding the xylem (Figures 5B to 5I). This is in sharp contrast to the more abundantly expressed vacuolar Na+/H+ antiporter gene AtNHX1, which is present almost ubiquitously in Arabidopsis tissues (Figure 5J). High-level expression of a Na+ efflux carrier in all cells may not be beneficial to plants because Na+ extruded by one cell would become toxic to its neighboring cells. On the other hand, a basal level of expression of a Na+ efflux carrier in all cells may prevent the rapid accumulation of Na+ in the cytosol, keeping Na+ entry commensurate with Na+ compartmentation in the vacuole.
The expression of SOS1 in cells surrounding the stele indicates a role of this transporter in controlling the Na+ load of the vascular system. We reported previously that sos1 mutant seedlings grown in liquid culture accumulated less Na+ than did the wild type when challenged with a moderate salt stress (e.g., 25 mM NaCl) (Ding and Zhu, 1997). These observations were confirmed and extended in this study (Figure 9A). In plants growing in soil that were treated with a mild NaCl stress, the sos1 mutation appeared to decrease Na+ accumulation in the shoot (Figure 9B). Therefore, under moderate salinity, SOS1 might function in loading Na+ into the xylem for transport to the shoot and subsequent compartmentation. Active Na+ loading in exchange for xylematic H+ would be favored energetically by the acidic pH of the xylem sap (Fisher, 2000). Fine-tuning of root ion uptake, xylem transport, and vacuolar compartmentation would facilitate osmotic adjustment and plant growth. However, under severe salt stress (e.g., 100 mM NaCl) for Arabidopsis plants, SOS1 appears to function in limiting Na+ loading of the xylem sap because sos1 mutant plants accumulated Na+ to higher levels in the xylem as well as the shoot (Figures 6 and 8). This finding could be attributable to defective Na+ extrusion at the root epidermal cells in the sos1 mutant, because the sos1 root also had more Na+ than did the wild-type root (Figures 7A and 7B). Excessive Na+ entry into the root might lead to unrestricted Na+ loading of the xylem and translocation to aerial parts of the plant.
Notwithstanding this simple model, Lacan and Durand (1996) have shown that excised soybean roots treated with NaCl reabsorbed Na+ from the xylem vessels in exchange for K+. Their evidence indicated that the net Na+/K+ exchange was achieved by coupling Na+/H+ and K+/H+ antiport activities at the xylem/symplast boundary. These two parallel transport processes were strongly linked because Na+ in the xylem sap enhanced K+ release, whereas increased xylematic K+ prompted Na+ reabsorption. Na+ removed from the xylem sap was excreted subsequently to the external medium. Net Na+/K+ exchange was energized by the proton pump and enhanced when the pH of the xylem sap was decreased. These authors suggested that reabsorption of Na+ from the acidic xylem sap by a Na+/H+ antiporter was enabled energetically through H+ cycling by K+/H+ antiport and H+/anion symport.
Considering this capacity for reversible Na+/K+ exchange at the root xylem/symplast boundary and the preferential expression of SOS1 in root pericycle cells, we propose a model in which SOS1 has a dual role in loading and reabsorption of Na+ from the xylem stream. When salinity is moderate, SOS1 functions to load Na+ into the xylem for controlled delivery to the shoot and storage in leaf mesophyll cells. Arabidopsis mesophyll cells may have sufficient capacity to compartmentalize a small amount of Na+ into their vacuoles, presumably through AtNHX1 activity. With more severe salt stress, Na+ might quickly accumulate in the shoot, exceeding the capacity of the Arabidopsis mesophyll cells to compartmentalize ions. Excessive Na+ accumulation in the shoot can be exacerbated by rapid leaf transpiration associated with low humidity. Under these conditions of severe salt stress, SOS1 would function to restrict net Na+ uptake at the root tip and to retrieve Na+ from the xylem in the mature root, and both of these processes would help to limit the rapid accumulation of Na+ in the shoot. It is important to note that this is only a model to explain our observations; as yet, there is no direct experimental proof for it.
If SOS1 catalyzes an electroneutral Na+/H+ exchange, the free energy of the process would be ΔG = −R · T · ln · [Na+]sap · [H+]cyt/[Na+]cyt · [H+]sap. At equilibrium (ΔG = 0), [Na+]sap · [H+]cyt = [Na+]cyt · [H+]sap. Thus, for a pH gradient of 1 unit between xylem sap (pHsap ∼ 6) and the xylem parenchyma cell (pHcyt ∼ 7), the activity of SOS1 could be reversed by a 10-fold difference in Na+ content between the xylem sap and the cytosol of the xylem parenchyma cell. Increases >10-fold in the Na+ concentration in the xylem sap of salinized plants can be expected (Flowers and Lauchli, 1983). For instance, Na+ concentration in the xylem sap of wild-type Arabidopsis was increased from ∼2 mM at the beginning of the salt treatment to ∼20 mM after 2 days (Figure 8). This dramatic increase would be sufficient to reverse the activity of SOS1 from Na+ efflux in low salt to Na+ reabsorption by xylem parenchyma cells under high salt treatment, provided that cytosolic Na+ in xylem parenchyma cells is kept in the low millimolar range. As proposed by Lacan and Durand (1996), the pH in the xylem sap would be buffered by the linked activity of a putative K+/H+ antiporter, allowing the reentry of proton and producing the net Na+/K+ exchange observed in planta. The proposed coupling between Na+ and K+ exchange at the xylem/symplast boundary also could provide an explanation for why sos1 mutant plants are not capable of growing on low K+ culture medium. Perhaps under low K+ availability, a functional SOS1 is required to enable K+ loading to the xylem.
The present study raises several interesting questions that need to be addressed in the future. For example, is the SOS1 protein localized asymmetrically on the plasma membrane of the xylem parenchyma and the root epidermal cells for vectorial Na+ transport? The animal plasma membrane Na+/H+ antiporter NHE3 is capable of interacting with the cytoskeleton through its C-terminal cytoplasmic tail (Kurashima et al., 1999). SOS1 is predicted to have a very long cytosolic tail at the C terminus (Shi et al., 2000), suggesting the possibility of interaction with many proteins, including cytoskeletal components. An interaction with the cytoskeleton would permit SOS1 to be localized asymmetrically in the cell.
Our model of Na+ loading into or retrieval from the xylem implies that SOS1 could function in either Na+ influx or efflux. Reversible bidirectional activity of Na+/H+ antiporters has been demonstrated for the SOD2 protein from fission yeast (Hahnenberger et al., 1996). When expressed in budding yeast, the SOD2 protein from fission yeast functions in Na+ influx when the external Na+ concentration is high. However, when internal Na+ concentration is higher, the antiporter functions in Na+ efflux (Hahnenberger et al., 1996). Whether SOS1 serves to mediate Na+ influx or efflux as a Na+/H+ antiporter may depend on the cellular and extracellular Na+ and H+ environments of the SOS1-expressing cells. Although without precedent, it remains a possibility that SOS1 may operate as an antiporter in the xylem-loading mode and as an ion channel or a Na+/H+ symporter in the xylem-unloading mode.
METHODS
Expression of SOS1 in Saccharomyces cerevisiae
S. cerevisiae strains ANT3 (Δena1::HIS3::ena4, Δnha1::LEU2), GX1 (Δena1::HIS3::ena4, Δnhx1::TRP1), AXT3 (Δena1::HIS3::ena4, Δnha1:: LEU2, Δnhx1::TRP1), and WΔ3 (Δtrk1::LEU2, Δtrk2::HIS3) are derivatives of W303 and have been described elsewhere (Madrid et al., 1998; Quintero et al., 2000). The entire SOS1 open reading frame was cloned into the yeast expression vector pYPGE15 under the control of the constitutive gene promoter PGK1 (Brunelli and Pall, 1993). K+ acquisition was determined in arginine-phosphate medium (AP medium; 10 mM l-arginine, 8 mM PO4H3, 2 mM MgSO4, 0.2 mM CaCl2, 2% glucose + vitamins and trace elements, pH 6.5), which is essentially free of alkali cations, supplemented with various concentrations of KCl as noted (Ramos et al., 1994). Na+ tolerance tests were performed in AP growth medium supplemented with 1 mM KCl and 70 mM NaCl (Quintero et al., 2000). For ion content measurements, yeast cells were collected during exponential growth in the same liquid AP medium, and their Na+ and K+ contents were determined by atomic emission spectrometry after acidic extraction (Quintero et al., 2000). Tolerance to KCl and hygromycin B was determined in regular YPD medium (1% yeast extract, 2% peptone, and 2% glucose).
Localization of the SOS1-GFP Fusion Protein
The green fluorescent protein (GFP) gene was fused in frame to SOS1 at its C terminus. The chimeric fragment replaced the β-glucuronidase (GUS) gene of plasmid pBI121, and the resulting construct was introduced into Agrobacterium tumefaciens strain GV3101. Arabidopsis thaliana (ecotype Columbia) wild-type plants were transformed using the vacuum infiltration method (Bechtold et al., 1993). Fifty independent transgenic lines were used for localization of the fusion protein. Roots of transgenic plants were treated with 0.5 M mannitol for 20 min for cell plasmolyzation. GFP was visualized using a Bio-Rad 1024 confocal scanning head attached to a Nikon (Tokyo, Japan) Optiphot 2 microscope.
SOS1 Promoter–GUS Analysis
An ∼1.9-kb SOS1 promoter region upstream of the ATG start codon was isolated by polymerase chain reaction amplification and cloned into pBI101 in front of the GUS reporter gene. The AtNHX1 promoter (∼1.9 kb upstream of the ATG start codon) was cloned into the pCAMBIA1391Z vector. The promoter-GUS constructs were introduced into transgenic Arabidopsis plants. For each promoter, at least 40 independent transgenic lines in the T2 generation were grown on Murashige and Skoog (1962) (MS) medium containing appropriate antibiotics (kanamycin for pBI101 and hygromycin for pCAMBIA1391Z) and subjected to GUS activity assays (Jefferson, 1987). For GUS staining of inflorescent tissues, antibiotic-resistant transgenic plants were transferred to soil. GUS-stained tissues were fixed in 4% paraformaldehyde in PBS, dehydrated, cleared, and embedded in Paraplast X-tra. Tissue sections were dewaxed and mounted directly in Permount (Fisher Scientific) mounting medium.
Plant Growth, NaCl Treatments, and Ion Determination
For collection of plant shoot or xylem sap, plants were grown in general soil (Scotts-Sierra Horticultural Products Co., Maysville, OH) for 3 weeks and then watered with one-twentieth strength MS salts plus 100 mM NaCl. For collection of plant shoot and root separately, plants were grown for 3 weeks either hydroponically in one-tenth strength MS nutrient solution or in Turface soil (Profile Products LLC, Buffalo Grove, IL) fertilized with Hoagland solution. Plants also were cultured in liquid medium containing one-half strength MS salts and 2% sucrose, pH 5.6, with shaking for 8 days for whole seedling collection. Plants in soil or hydroponic culture were grown in a growth chamber with a 16-hr-light (22°C)/8-hr-dark (18°C) cycle at 85% relative humidity (RH). Calli were induced from wild-type and sos1-1 mutant seed as described (Wu et al., 1996). The calli were transferred to the same callus induction medium (Wu et al., 1996) plus 100 mM NaCl for various times. Plant materials were collected and dried at 80°C for at least 2 days and weighed. The samples were digested with HNO3, and Na+ and K+ concentrations were determined with an atomic absorption spectrometer (model 3100; Perkin-Elmer, Norwalk, CT). For xylem sap collection, rosette leaves and inflorescence stems were cut at the base. The plants were kept in a chamber with 100% RH, and water drops that emerged on the cut surface of the inflorescence stem were collected (Gaymard et al., 1998). The xylem sap was diluted directly in HNO3 solution for ion content measurement. Plant ion content data are presented as averages with standard deviations. Statistical analysis for Fisher's protected LSD test was performed using SAS general linear models (SAS Institute, Inc., Cary, NC).
RNA Gel Blot Analysis
RNA extraction and RNA gel blot analysis were performed as described (Shi et al., 2000).
Acknowledgments
We thank Drs. S.R. Cutler, D.W. Ehrhardt, and C.R. Somerville for the generous gift of PIP2a-GFP transgenic Arabidopsis. This work was supported by National Institutes of Health Grant No. R01GM59138 to J.-K.Z. and by Grant No. BIO2000-0938 from the Spanish Comisión de Investigaciones Científicas y Técnica to J.M.P. and F.J.Q.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010371.
References
- Amtmann, A., and Sanders, D. (1999). Mechanisms of Na+ uptake by plant cells. Adv. Bot. Res. 29, 75–112. [Google Scholar]
- Apse, M.P., Aharon, G.S., Snedden, W.A., and Blumwald, E. (1999). Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285, 1256–1258. [DOI] [PubMed] [Google Scholar]
- Bañuelos, M.A., Sychrová, H., Bleykasten-Grosshans, C., Souciet, J.L., and Potier, S. (1998). The Nha1 antiporter of Saccharomyces cerevisiae mediates sodium and potassium efflux. Microbiology 144, 2749–2758. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316, 1194–1199. [Google Scholar]
- Blumwald, E., Aharon, G.S., and Apse, M.P. (2000). Sodium transport in plant cells. Biochim. Biophys. Acta 1465, 140–151. [DOI] [PubMed] [Google Scholar]
- Brunelli, J.P., and Pall, M.L. (1993). A series of yeast/Escherichia coli lambda expression vectors designed for directional cloning of cDNAs and cre/lox-mediated plasmid excision. Yeast 9, 1309–1318. [DOI] [PubMed] [Google Scholar]
- Cutler, S.R., Ehrhardt, D.W., Griffitts, J.S., and Somerville, C.R. (2000). Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97, 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darley, C.P., van Wuytswinkel, O.C., van der Woude, K., Mager, W.H., and de Boer, A.H. (2000). Arabidopsis thaliana and Saccharomyces cerevisiae NHX1 genes encode amiloride sensitive electroneutral Na+/H+ exchangers. Biochem. J. 351, 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, L., and Zhu, J.-K. (1997). Reduced Na+ uptake in the NaCl-hypersensitive sos1 mutant of Arabidopsis thaliana. Plant Physiol. 113, 795–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein, E. (1998). How calcium enhances plant salt tolerance. Science 280, 1906–1907. [DOI] [PubMed] [Google Scholar]
- Fisher, D.B. (2000). Long-distance transport. In Biochemistry and Molecular Biology of Plants, B.B. Buchanan, W. Gruissem, and R.L. Jones, eds (Rockville, MD: American Society of Plant Physiologists), pp. 730–784.
- Flowers, T.J., and Lauchli, A. (1983). Inorganic plant nutrition. V 3. Sodium versus potassium: Substitution and compartmentation. Encycl. Plant Physiol. 15B, 651–681. [Google Scholar]
- Flowers, T.J., Troke, P.F., and Yeo, A.R. (1977). The mechanism of salt tolerance in halophytes. Annu. Rev. Plant Physiol. 28, 89–121. [Google Scholar]
- Fukuda, A., Nakamura, A., and Tanaka, Y. (1999). Molecular cloning and expression of the Na+/H+ exchanger gene in Oryza sativa. Biochim. Biophys. Acta 1446, 149–155. [DOI] [PubMed] [Google Scholar]
- Gaxiola, R.A., Rao, R., Sherman, A., Grisafi, P., Alper, S.L., and Fink, G.R. (1999). The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc. Natl. Acad. Sci. USA 96, 1480–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaymard, F., Pilot, G., Lacombe, B., Bouchez, D., Bruneau, D., Boucherez, J., Michaux-Ferriere, N., Thibaud, J.B., and Sentenac, H. (1998). Identification and disruption of a plant shaker-like outward channel involved in K+ release into the xylem sap. Cell 94, 647–655. [DOI] [PubMed] [Google Scholar]
- Hahnenberger, K.M., Jia, Z., and Young, P.G. (1996). Functional expression of the Schizosaccharomyces pombe Na+/H+ antiporter gene, sod2, in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93, 5031–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter, U., Ishitani, M., and Zhu, J.K. (2000). The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 97, 3735–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa, P.M., Bressan, R.A., and Pardo, J.M. (2000). The dawn of plant salt tolerance genetics. Trends Plant Sci. 5, 317–319. [DOI] [PubMed] [Google Scholar]
- Jefferson, R. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405. [Google Scholar]
- Kurashima, K., D'Souza, S., Szászi, K., Ramjeesingh, R., Orlowski, J., and Grinstein, S. (1999). The apical Na+/H+ exchanger isoform NHE3 is regulated by the actin cytoskeleton. J. Biol. Chem. 274, 29843–29849. [DOI] [PubMed] [Google Scholar]
- Lacan, D., and Durand, M. (1996). Na+-K+ exchange at the xylem/symplast boundary: Its significance in the salt sensitivity of soybean. Plant Physiol. 110, 705–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauchli, A. (1984). Salt exclusion: An adaptation of legumes for crops and pastures under saline condition. In Salinity Tolerance in Plants: Strategies for Crop Improvement, R.C. Staples and G.H. Toennissen, eds (New York: Wiley), pp. 171–187.
- Liu, J., Ishitani, M., Halfter, U., Kim, C., and Zhu, J.-K. (2000). The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. USA 97, 3730–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid, R., Gómez, M.J., Ramos, J., and Rodríguez-Navarro, A. (1998). Ectopic potassium uptake in trk1 trk2 mutants of Saccharomyces cerevisiae correlates with a highly hyperpolarized membrane potential. J. Biol. Chem. 273, 14838–14844. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nass, R., and Rao, R. (1998). Novel localization of a Na+/H+ exchanger in a late endosomal compartment of yeast: Implications for vacuole biogenesis. J. Biol. Chem. 273, 21054–21060. [DOI] [PubMed] [Google Scholar]
- Navarre, C., and Goffeau, A. (2000). Membrane hyperpolarization and salt sensitivity induced by deletion of PMP3, a highly conserved small protein of yeast plasma membrane. EMBO J. 19, 2515–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, X., Bressan, R.A., Hasegawa, P.M., and Pardo, J.M. (1995). Ion homeostasis in NaCl stress environments. Plant Physiol. 109, 735–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nublat, A., Desplans, J., Casse, F., and Berthomieu, P. (2001). sas1, an Arabidopsis mutant overaccumulating sodium in the shoot, shows deficiency in the control of the root radial transport of sodium. Plant Cell 13, 125–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero, F.J., Blatt, M.R., and Pardo, J.M. (2000). Functional conservation between yeast and plant endosomal Na+/H+ antiporters. FEBS Lett. 471, 224–228. [DOI] [PubMed] [Google Scholar]
- Ramos, J., Alijo, R., Haro, R., and Rodriguez-Navarro, A. (1994). TRK2 is not a low-affinity potassium transporter in S. cerevisiae. J. Bacteriol. 176, 249–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio, F., Gassman, W., and Schroeder, J.I. (1995). Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270, 1660–1663. [DOI] [PubMed] [Google Scholar]
- Schachtman, D.P., Kumar, R., Schroeder, J.I., and Marsh, E.L. (1997). The structure and function of a novel cation transporter (LCT1) in higher plants. Proc. Natl. Acad. Sci. USA 94, 11079–11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H., Ishitani, M., Kim, C., and Zhu, J.-K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 97, 6896–6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyerman, S.D., and Skerrett, I.M. (1999). Root ion channels and salinity. Sci. Hortic. 78, 175–235. [Google Scholar]
- Wu, S.-J., Lei, D., and Zhu, J.-K. (1996). SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8, 617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.-K. (2000). Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol. 124, 941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.-K. (2001). Plant salt tolerance. Trends Plant Sci. 6, 66–71. [DOI] [PubMed] [Google Scholar]