Abstract
Ribonucleotide reductase consists of two nonidentical proteins, R1 and R2, and catalyzes the rate-limiting step in DNA precursor synthesis: the reduction of ribonucleotides to deoxyribonucleotides. A strictly balanced supply of deoxyribonucleotides is essential for both accurate DNA replication and repair. Therefore, ribonucleotide reductase activity is under exquisite control both transcriptionally and posttranscriptionally. In proliferating mammalian cells, enzyme activity is regulated by control of R2 protein stability. This control, which responds to DNA damage, is effective until cells pass into mitosis. We demonstrate that the mitotic degradation and hence the overall periodicity of R2 protein levels depends on a KEN box sequence, recognized by the Cdh1–anaphase-promoting complex. The mouse R2 protein specifically binds Cdh1 and is polyubiquitinated in an in vitro ubiquitin assay system. Mutating the KEN signal stabilizes the R2 protein during mitosis/G1 in R2 protein-overexpressing cells. The degradation process, which blocks deoxyribonucleotide production during G1, may be an important mechanism protecting the cell against unscheduled DNA synthesis. The newly discovered p53-induced p53R2 protein that lacks a KEN box may supply deoxyribonucleotides for DNA repair during G0/G1.
Ribonucleotide reductase is essential for de novo synthesis of deoxyribonucleotides required for DNA replication and repair (1). In mammalian cells, the enzyme consists of two nonidentical homodimeric subunits, proteins R1 and R2, which are both required for activity. Transcription of the R1 and R2 genes is cell cycle-regulated with low or undetectable levels of transcripts in G0/G1 phase and maximal levels during S phase (2). The levels of the R1 protein are in excess and almost constant during the cell cycle in proliferating cells due to a long half-life (3, 4). Therefore, overall enzyme activity is controlled by the levels of the limiting R2 protein that are undetectable in G1 and increase in S phase (3, 5). In proliferating cells, the transcription of the R2 gene, once activated at the beginning of S phase, reaches its maximum about 7 h later and then declines. This R2 promoter activity is neither increased nor prolonged by DNA damage or replication blocks. Instead, the cell cycle activity of the mammalian ribonucleotide reductase is controlled by an S-phase/DNA damage-specific delayed degradation of the R2 protein, which is effective until cells pass into mitosis. This mechanism ensures an adequate supply of dNTPs for replication and repair in S phase and during G2 (6).
The question of how human or mouse G1 cells, which lack the R2 protein, obtain dNTPs for DNA repair was recently answered by the discovery of a new, p53-induced R2 protein called p53R2 (7, 8). The p53R2 gene is localized to a different chromosome from the R2 gene. In nonproliferating cells, DNA damage induces expression of the p53R2 protein and also the R1 protein. Together they can form an active ribonucleotide reductase complex supplying resting cells with dNTPs for DNA repair (9). These results demonstrate that mammalian RNR genes, like the Mec1/Rad 53 pathway-regulated yeast RNR genes (10, 11), are subject to transcriptional regulation after DNA damage. In human cells, the Mec1 homolog ATR and the Rad 53 homolog CHK2 function upstream of p53, which is the central player in DNA damage response in mammalian cells and is mutated in >50% of human cancers (reviewed in ref. 12).
In this work, we were interested in identifying the system responsible for R2 protein degradation during mitosis and to understand why cells express two different but very homologous forms of the R2 protein. Inhibition by N-acetyl-Leu-Leu-norleucinal indicated that the proteolysis of the R2 protein during mitosis was proteasome-dependent but the signal for the regulated proteolysis was not determined (6). There are two ubiquitin protein ligases that have central roles in cell cycle regulation: the Skip1/Cullin/F-box (SCF) complex and the anaphase-promoting complex (APC) (13–15). Recruitment to the SCF complex occurs via the F-box subunit and requires substrate phosphorylation. The R2 protein is indeed uniquely phosphorylated on Ser-20 by cell cycle-dependent kinases both in vivo and in vitro but the function of this phosphorylation has not been determined (6, 16, 17). On the other hand, the mouse R2 protein contains an N-terminally located sequence RVPL similar to the APC-dependent destruction box RXXL (18) where the two invariant residues are conserved also in the human, hamster, and guinea pig R2 proteins. Yet, a truncated mouse R2 protein lacking the first 20-aa residues including this putative destruction box showed the same degradation pattern as the wild-type R2 protein (6).
However, there are two different activators of the APC, Cdc20 and Cdh1. Both activators were shown to specifically associate with substrates via their N termini and therefore act as substrate recognition and activating modules for APC (19). Cdc20–APC recognizes destruction box-containing proteins and is active during mitosis where one of the targets is securin (reviewed in ref. 20). Cdh1–APC recognizes either a destruction box or a newly discovered recognition signal, the KEN box, and it is active in late mitosis and during G1/Go (19, 21, 22). In S phase, cell cycle-dependent phosphorylation of Cdh1 causes it to dissociate from the APC, which is then kept inactive during S phase and G2 until the following mitosis (23).
In this article, we show that the mouse R2 protein contains an N-terminally located conserved KEN box and is a new substrate for Cdh1–APC. The wild-type R2 protein binds specifically to the APC activating protein Cdhl in pull-down assays and it is polyubiquitinated in an in vitro Cdh1–APC ubiquitination assay system. Mutating the KEN box to AAN stabilizes the protein against degradation during mitosis/G1 in stably transformed mouse fibroblast cells overexpressing R2 protein. Interestingly, the p53R2 protein lacks a KEN box and therefore should evade cell cycle-specific proteolysis mediated by Cdh1–APC.
Materials and Methods
Cell Culture.
Mouse Balb/3T3 (ATCC no. CCL-163) cells were cultivated in DMEM (Sigma) supplemented with 10% heat-inactivated horse serum. Flow cytometry was carried out as described (6). Transfection of cells by electroporation with HindIII-linearized vectors and selection of stably transformed clones were performed as described (24).
Vector Construction.
A vector construct containing the genomic R2 gene in pUC18 was obtained by partial digestion of the pM2Hind13 plasmid with ClaI (at nucleotide −1006) and NdeI (25). The long DNA fragment containing the entire R2 genomic gene including 1,000 bp of promoter was isolated, blunted with the Klenow fragment of DNA polymerase, and religated (D construct). The R2 gene expressing the AAN-mutated protein was created by overlap extension PCR of an NgoMI–NgoMI fragment (nucleotides −169 to +181), where the required mutation was introduced. The fragment containing the AAN mutation (AAGGAGAAC replaced by GCGGCGAAC) was then ligated into the D construct instead of the original fragment to give the mutated KEN (MK) construct. For in vitro translation of the mouse R2 protein, the T7 RNA polymerase responsive vector pETM2 was used (26). The AAN, S20A, and S20D mutations were introduced by overlap extension PCR of an XbaI (cuts between the T7 promoter and the AGGA sequence) to NcoI (cuts at nucleotide 503 downstream from the ATG in the R2 cDNA) fragment where the required mutations were introduced. The fragments containing the mutations (GCTGCGAAC replacing the original AAGGAGAAC and GCG or GAT replacing the original TCG) were then ligated into the R2 cDNA replacing the original fragment. All constructs were verified by restriction analysis and sequencing of PCR-amplified regions.
In Vitro Ubiquitination Assays.
These assays were performed as described (22) by using 35S-labeled in vitro-translated R2 proteins (Promega TNT kit). Instead of UBCx alone as a ubiquitin-conjugating enzyme (E2), the assays were done with a combination of UBCx and bacterially expressed UBC4.
Pull-Down Assays Using in Vitro-Translated Myc-Tagged Proteins.
Cold in vitro-translated myc-tagged Cdh1 or Cdc20 were bound to 9E10 anti-myc-coupled beads (Santa Cruz Biotechnology) and after the prebinding, 35S-labeled R2 protein was added as described (19).
Immunoblotting.
Cells were rinsed twice with ice-cold PBS (pH 7.4) and then lysed by the addition of 200 μl per 5-cm dish of PBS containing 0.5% Nonidet P-40 and protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 2 mM pepstatin, 0.6 mM leupeptin, and 2 mM benzamidine, prepared as a 100× stock solution in 100% ethanol). The lysates were collected with a rubber policeman, and debris was removed by centrifugation in an Eppendorf centrifuge for 3 min at 7,900 × g in the cold room (4°C). From the supernatants, 150 μl were transferred to new tubes and immediately frozen in liquid nitrogen and stored at −70°C. Equal amounts of protein from the lysates were electrophoresed, and then transferred to Hybond-C extra membranes as described (6). After transfer, the membranes were blocked in 100 mM Tris-Cl (pH 8), 1.5 M NaCl, and 0.5% Tween 20 containing 2% nonfat dry milk for 30 min. Then JC4 anti-R2 rat mAbs were added followed by a horseradish peroxidase-conjugated secondary Ab, and finally detection was done by using the ECL Plus system as described (6).
Results
A Putative Cdh1–APC Recognition Signal Is Located in the N-Terminal Part of the Mouse Ribonucleotide Reductase R2 Protein.
The identification of a new APC recognition signal, composed of the amino acid sequence KEN (22), prompted us to look for this sequence in the mouse R2 protein. Indeed, a KEN sequence is present at position 30–32 from the N-terminal and is completely conserved, both in position and amino acid residues, in the human, mouse, hamster, and guinea pig R2 proteins (Fig. 1). An N-terminally located KEN sequence can also be found in the R2 protein homologues present in Caenorhabditis elegans and Drosophila but not in budding or fission yeast.
Figure 1.
The KEN sequence is conserved in human, mouse, hamster, and guinea pig (GenBank accession no. AY209181) R2 proteins. A comparison of the N-terminal amino acid sequence in the human, mouse, hamster, and guinea pig R2 proteins is shown. The conserved KEN sequence and the uniquely phosphorylated Ser-20 are indicated in bold.
The APC Substrate-Specific Adaptor Cdh1 but Not Cdc20 Specifically Binds the KEN-Containing Mouse R2 Protein.
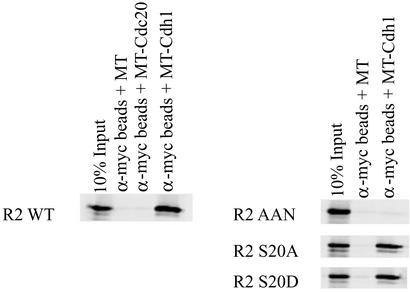
To investigate the interaction between APC and the mouse R2 protein, we incubated in vitro-translated myc-tagged (MT) Cdc20 or Cdh1 bound to α-myc-coated beads with 35S-labeled in vitro-translated wild-type R2 protein or R2 protein where the KEN box was mutated to AAN (Fig. 2). Cdh1 binds the wild-type R2 protein to at least 10% of the input signal whereas Cdc20 and the myc-tag alone do not bind. No Cdh1 binding was observed to the AAN mutant form of the R2 protein.
Figure 2.
Wild-type mouse R2 protein but not the AAN mutant R2 protein specifically binds to Cdh1. In vitro-translated 35S-labeled wild-type R2 protein (R2 WT), the AAN mutant R2 protein (R2 AAN), the S20A mutant R2 protein (R2 S20A), or the S20D mutant R2 protein (R2 S20D) was incubated in the presence of cold in vitro-translated myc-tagged Cdh1 (MT-Cdh1) or Cdc20 (MT-Cdc20) bound to α-myc beads or in the presence of myc tag (MT) alone bound to α-myc beads. The left-most lane in each image indicates 10% of the in vitro translation material supplied in the binding assay.
We and others have observed that the S20 residue of the mouse R2 protein is uniquely phosphorylated both in vivo and in vitro by cell cycle-dependent kinases. To study whether this phosphorylation affected Cdh1 binding, we compared the binding to wild-type R2 protein with the binding to R2 proteins where the Ser-20 residue was mutated to alanine or aspartic acid. As seen in Fig. 2, neither mutation affected binding to Cdh1.
In Vitro Polyubiquitination of the Mouse R2 Protein by Cdh1–APC Requires an Intact KEN Box.
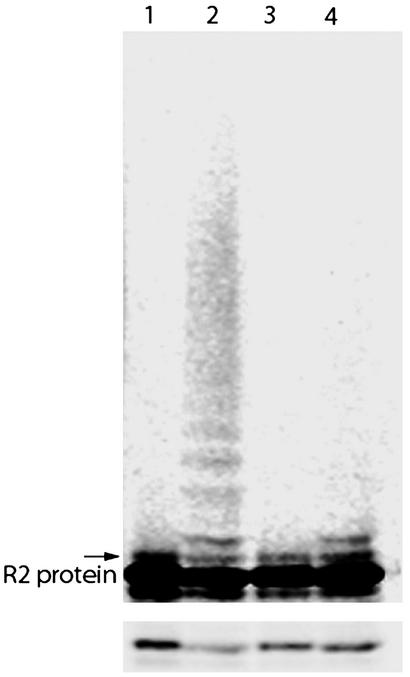
Knowing that the mouse R2 protein binds specifically to Cdh1 in vitro, we tested whether the R2 protein is also a Cdh1–APC substrate by using a purified in vitro ubiquitination system. In this assay, 35S-labeled, in vitro-translated mouse R2 protein was incubated with immunopurified Xenopus APC, baculovirus-expressed and purified Cdh1, E1, UBC4/UBCx (=E2), ubiquitin, and an energy-regenerating system. For wild-type R2 protein, both monoubiquitinated species and the light-smearing characteristic of polyubiquitinated higher molecular weight conjugates can be seen (Fig. 3). For the AAN mutant, no conjugation at all or sometimes one or two monoubiquitinated species were observed. The overall amount of conjugation is best seen in the lighter exposure in Fig. 3 Lower showing only the region of full-length R2 protein. There is a clear decrease in the signal from the wild-type protein after ubiquitination whereas the AAN mutant signal remains the same. In the initial experiments where only UBCx was used as the ubiquitin conjugating enzyme, no clear polyubiquitination was observed. However, a significant burst in conjugation was observed after the addition of UBC4 to the assay indicating different specificities.
Figure 3.
Wild-type mouse R2 protein but not the AAN mutant R2 protein is recognized as a substrate by Cdh1–APC. Incubation of 35S-labeled in vitro-translated R2 protein in an in vitro ubiquitination assay containing immunopurified Xenopus APC (E3); and baculovirus-expressed and purified Cdh1, E1, UBC4/UBCx (E2), ubiquitin, and an energy-regenerating system. Lane 1, wild-type R2 protein input; lane 2, wild-type R2 protein after a 1-h incubation; lane 3, AAN mutant R2 protein input; lane 4, AAN mutant R2 protein after a 1-h incubation. Lower shows a lighter exposure where the quantitative loss of the unmodified protein can be seen in lane 2 but not in lane 4. The arrow indicates the position of monoubiquitinated R2 protein.
A Mouse R2 Protein with Mutated KEN Box Is Not Degraded in Late Mitosis.
To test whether a mutation in the R2 protein KEN box affects protein stability in vivo, we transfected mouse Balb/3T3 fibroblasts with plasmid constructs containing the entire R2 gene including the promoter region. One plasmid encoded the wild-type R2 protein while another plasmid encoded R2 protein where the KEN box was mutated to AAN. Immunoblotting confirmed that the levels of wild-type or mutated R2 protein in the stably transformed cells were similar and about 10 times higher than in the nontransformed cells (data not shown). Both R2 proteins were active in vivo, because the transformed cells showed increased resistance to hydroxyurea with IC50 values of 0.2–0.4 mM as compared with 0.04 mM for nontransformed cells (27).
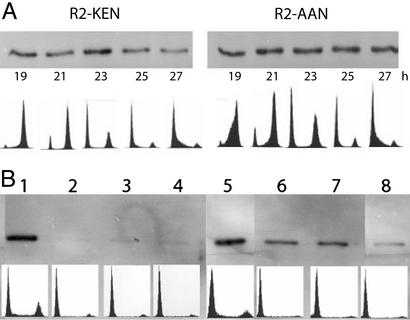
Cells stably transformed with the wild-type R2 gene construct or the AAN mutant R2 gene construct were synchronized by serum starvation. At different time points after serum readdition, cells were harvested for flow cytometry or analysis of R2 protein levels by immunoblotting (Fig. 4A). In the cells overexpressing wild-type R2 protein, maximal levels of R2 protein were observed in cells harvested 23 h after serum readdition. These cells were in mitosis/early G1. With more cells entering G1, the levels of R2 protein decreased (25- and 27-h time points) similar to published observations (6). In contrast to this pattern, no decrease in R2 protein levels was observed in cells overexpressing the AAN mutant R2 protein.
Figure 4.
Mutating the KEN signal stabilizes the R2 protein during mitosis and G1. (A) Stably transformed mouse Balb/3T3 fibroblasts overexpressing wild-type R2 protein (clone D2) or the AAN mutant R2 protein (clone MK6) were explanted on 5-cm dishes (2 × 105 cells per dish) and allowed to grow for 24 h in DMEM containing 10% horse serum. Then the cells were synchronized in DMEM containing 1% horse serum for 45 h and finally released by the addition of DMEM containing 20% horse serum. At different time points after serum readdition (19–27 h), cells overexpressing wild-type R2 protein (lanes 1–5) or the AAN mutant R2 protein (lanes 6–10) were harvested for flow cytometry and immunoblotting. (B) Stably transformed cells overexpressing native or AAN-mutated R2 protein (clones D4 and MK 2, respectively) were explanted on 5-cm dishes and allowed to grow for 24 h in DMEM containing 10% horse serum. After harvesting four dishes for flow cytometry and immunoblotting, the medium was changed to DMEM containing 1% serum and cells were harvested as before after 24 and 36 h. Finally, the cells were released from starvation with DMEM containing 20% serum and harvested 4 h after serum readdition. To make sure that the same amount of protein (0.3 μg) was loaded in each lane, the protein concentration of each cell extract was determined by the Bio-Rad protein assay before loading. Lane 1, logarithmically growing, wild-type R2 protein-overexpressing cells; lane 2, the same after 24 h of serum starvation; lane 3, the same after 36 h of serum starvation; lane 4, the same 4 h after serum readdition; lane 5, logarithmically growing, AAN mutant R2 protein-overexpressing cells; lane 6, the same after 24 h of serum starvation; lane 7, the same after 36 h of serum starvation; lane 8, the same 4 h after serum readdition. In both A and B, Lower shows the flow cytometry profile corresponding to each time point.
Cells synchronized in G1 by serum starvation tend to lose synchronization late in the cell cycle and therefore the degradation of R2 protein during mitosis can be difficult to monitor. With this in mind, we designed an experiment where the degradation of the R2 protein was monitored while the cells accumulated in G0/G1 during serum starvation. Wild-type and AAN mutant R2 protein overexpressing stably transformed cells were explanted and allowed to grow for 24 h in a complete medium, and then four dishes were harvested for flow cytometry and R2 protein immunoblotting (logarithmically growing cells). The remaining cells were synchronized by serum starvation, and additional dishes were harvested for flow cytometry and immunoblotting after 24 and 36 h of starvation. Finally, the last dishes were harvested and analyzed as before 4 h after serum readdition (Fig. 4B). As seen by flow cytometry, most of the cells were in G0/G1 already after 24 h of serum starvation and no significant change in cell cycle distribution was observed for the 36- and 4-h time points. The immunoblotting analyses showed a clear difference in R2 protein stability between cells overexpressing the wild-type R2 protein and cells overexpressing the AAN mutant protein. No R2 protein could be detected in serum-starved cells stably transformed with the wild-type R2 construct although a significant level of R2 protein remained in the cells transformed with the AAN mutant construct (Fig. 4B, compare lanes 2–4 to 6–8).
Discussion
The N terminus of the mouse R2 protein up to amino acid residue 65 is disordered and not visible in the crystal structure (28). Furthermore, in NMR studies, a number of resolved resonances not belonging to the highly mobile C terminus may be due to flexible N-terminal residues (29). Recombinant mouse R2 protein lacking the N-terminal 61 residues is fully active in a ribonucleotide reductase assay together with the R1 protein, showing that this part of the polypeptide chain is dispensable for catalytic activity (L.T., unpublished data). This N-terminal part is also missing in the homologous R2 proteins of Escherichia coli or herpes simplex virus, indicating a function not related to enzyme activity. The identification of the KEN box responsible for Cdh1–APC-mediated proteolysis in residues 30–32 suggests a function for the N-terminal part of the mouse R2 protein.
The N terminus also contains a serine residue uniquely phosphorylated by cell cycle-dependent kinases and located 10 residues upstream from the KEN box (6, 17). Phosphorylation of Ser-20 does not affect the catalytic activity of the R2 protein. It was reported that negative charge close to the KEN box could inhibit ubiquitination by APC, which is quite the opposite of the Skip1/Cullin/F-box complex where phosphorylation is a positive regulator of ubiquitination (22). Our data with the S20A R2 protein mutant clearly showed that phosphorylation was not required for Cdh1 binding and no difference was seen in the ubiquitination assay between the S20A and wild-type R2 proteins (data not shown). Also, the S20D R2 protein, where the serine is changed to an aspartic acid to mimic a phosphoserine, showed the same binding and ubiquitination as the wild-type protein. However, it is more difficult to state conclusively from these in vitro results that phosphorylation does not inhibit ubiquitination in vivo because aspartic acid is not the same as phosphoserine even though this seems to be a good approximation in many cases. The results from the Cdh1 binding experiments are in full agreement with in vivo data where the same stability of the R2 protein during the cell cycle was observed for the wild-type protein, which is phosphorylated immediately after synthesis, as for the S20A mutant, the S20D mutant, and a Δ 20 mutant, which lacks amino acid residues 2–20 (6).
The mouse p53R2 protein shows 81% amino acid sequence identity compared with the mouse R2 protein and all of the iron ligands, the tyrosyl free radical, the amino acid residues involved in long-range radical transfer, and the C-terminal heptapeptide, essential for binding to the R1 protein, are conserved (9). The major difference is that the p53R2 protein has a 33-aa residue truncation in the N terminus compared with the R2 protein and therefore lacks the KEN box. This difference may explain why there are two different R2 proteins in a cell. The normal R2 protein would not be stable in G0/G1 cells, which contain an active Cdh1–APC. The p53R2 protein lacking a KEN box should be stable during G0/G1 and, together with the R1 protein, it could supply dNTPs required for DNA repair.
Why is it important for the cell to degrade the R2 protein in late mitosis and thereby stop deoxyribonucleotide production? We believe that this degradation could be one mechanism to inhibit unscheduled DNA replication because the availability of the R1 protein all through the cell cycle in proliferating cells would otherwise lead to S-phase levels of dNTPs during G1. Inhibition of APC in cultured cells with a Cdh1 Ab led to early entry into S phase (30). The inability to degrade the R2 protein in late mitosis in these cells may be one factor contributing to overreplication. A deregulated R2 protein might be a tumor progressor determinant because constitutively expressed R2 protein in Balb/3T3 cells stably transformed with a retroviral expression vector containing mouse R2 cDNA led to a greatly increased frequency of focus formation in cooperation with Ha-ras transformation (31).
Deoxyribonucleotides are required for recombination, replication, and repair of DNA both in chromosomes and mitochondria, and the importance of correct dNTP pools in a cell is reflected in the many different ways that are used to control ribonucleotide reductase. In addition to the complicated allosteric regulation by nucleoside triphosphate effectors (32), ribonucleotide reductase is controlled at the level of transcription (2) and by the regulated proteolysis of the limiting R2 protein by the Cdh1–APC both in the normal cell cycle and after DNA damage in proliferating cells. The normal R2 gene is not induced by DNA damage but instead DNA damage leads to the accumulation of p53, which induces the p53R2 gene (7, 8). This induction would be most important in resting cells and cells in G1 phase, which lack the R2 protein and have very low levels of dNTPs. Finally, a fourth level of control of ribonucleotide reductase was recently discovered. A previously uncharacterized type of cytoplasmic poly(A) polymerase, Cid13, specifically targeting the mRNA encoding the small subunit of ribonucleotide reductase, was identified in fission yeast (33). After DNA damage or initiation of DNA synthesis when there is a rapid consumption of dNTPs, Cid13 may polyadenylate R2 mRNA leading to higher R2 protein levels and increased dNTP synthesis.
Acknowledgments
This work was supported by the Swedish Research Council, the Swedish Foundation for Strategic Research, the Kempe Foundation, the Medical Faculty of Umeå University, and the U.S. National Institute of General Medical Sciences.
Abbreviation
- APC
anaphase-promoting complex
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY209181).
References
- 1.Reichard P. Science. 1993;260:1773–1777. doi: 10.1126/science.8511586. [DOI] [PubMed] [Google Scholar]
- 2.Björklund S, Skog S, Tribukait B, Thelander L. Biochemistry. 1990;29:5452–5458. doi: 10.1021/bi00475a007. [DOI] [PubMed] [Google Scholar]
- 3.Engström Y, Eriksson S, Jildevik I, Skog S, Thelander L, Tribukait B. J Biol Chem. 1985;260:9114–9116. [PubMed] [Google Scholar]
- 4.Mann G J, Musgrove E A, Fox R M, Thelander L. Cancer Res. 1988;48:5151–5156. [PubMed] [Google Scholar]
- 5.Eriksson S, Gräslund A, Skog S, Thelander L, Tribukait B. J Biol Chem. 1984;259:11695–11700. [PubMed] [Google Scholar]
- 6.Chabes A, Thelander L. J Biol Chem. 2000;275:17747–17753. doi: 10.1074/jbc.M000799200. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 8.Nakano K, Bálint É, Ashcroft M, Vousden K H. Oncogene. 2000;19:4283–4289. doi: 10.1038/sj.onc.1203774. [DOI] [PubMed] [Google Scholar]
- 9.Guittet O, Håkansson P, Voevodskaya N, Fridd S, Gräslund A, Arakawa H, Nakamura Y, Thelander L. J Biol Chem. 2001;276:40647–40651. doi: 10.1074/jbc.M106088200. [DOI] [PubMed] [Google Scholar]
- 10.Huang M, Elledge S J. Mol Cell Biol. 1997;17:6105–6113. doi: 10.1128/mcb.17.10.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang M, Zhou Z, Elledge S J. Cell. 1998;94:595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- 12.Zhou B B, Elledge S J. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 13.Peters J-M. Curr Opin Cell Biol. 1998;10:759–768. doi: 10.1016/s0955-0674(98)80119-1. [DOI] [PubMed] [Google Scholar]
- 14.Zachariae W, Nasmyth K. Genes Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- 15.Jackson P K, Eldrige A G, Freed E, Furstenthal L, Hsu J Y, Kaiser B K, Reimann J D. Trends Cell Biol. 2000;10:429–439. doi: 10.1016/s0962-8924(00)01834-1. [DOI] [PubMed] [Google Scholar]
- 16.Chan A K, Litchfield D W, Wright J A. Biochemistry. 1993;32:12835–12840. doi: 10.1021/bi00210a036. [DOI] [PubMed] [Google Scholar]
- 17.Chan A K, Persad S, Litchfield D W, Wright J A. Biochim Biophys Acta. 1999;1448:363–371. doi: 10.1016/s0167-4889(98)00115-3. [DOI] [PubMed] [Google Scholar]
- 18.Glotzer M, Murray A W, Kirschner M W. Nature. 1991;349:132–137. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 19.Pfleger C M, Lee E, Kirschner M W. Genes Dev. 2001;15:2396–2407. doi: 10.1101/gad.918201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nasmyth K. Annu Rev Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- 21.Gieffers C, Peters B H, Kramer E R, Dotti C G, Peters J M. Proc Natl Acad Sci USA. 1999;96:11317–11322. doi: 10.1073/pnas.96.20.11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfleger C M, Kirschner M W. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- 23.Jaspersen S L, Charles J F, Morgan D O. Curr Biol. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- 24.Björklund S, Skogman E, Thelander L. EMBO J. 1992;11:4953–4959. doi: 10.1002/j.1460-2075.1992.tb05602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thelander M, Thelander L. EMBO J. 1989;8:2475–2479. doi: 10.1002/j.1460-2075.1989.tb08383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mann G J, Gräslund A, Ochiai E-I, Ingemarson R, Thelander L. Biochemistry. 1991;30:1939–1947. doi: 10.1021/bi00221a030. [DOI] [PubMed] [Google Scholar]
- 27.Åkerblom L, Ehrenberg A, Gräslund A, Lankinen H, Reichard P, Thelander L. Proc Natl Acad Sci USA. 1891;78:2159–2163. doi: 10.1073/pnas.78.4.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kauppi B, Nielsen B B, Ramaswamy S, Kjoller Larsen I, Thelander M, Thelander L, Eklund H. J Mol Biol. 1996;262:706–720. doi: 10.1006/jmbi.1996.0546. [DOI] [PubMed] [Google Scholar]
- 29.Lycksell P-O, Ingemarson R, Davis R, Gräslund A, Thelander L. Biochemistry. 1994;33:2838–2842. doi: 10.1021/bi00176a013. [DOI] [PubMed] [Google Scholar]
- 30.Sørensen C S, Lukas C, Kramer E R, Peters J-M, Bartek J, Lukas J. Mol Cell Biol. 2000;20:7613–7623. doi: 10.1128/mcb.20.20.7613-7623.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan H, Villegas C, Wright J A. Proc Natl Acad Sci USA. 1996;93:14036–14040. doi: 10.1073/pnas.93.24.14036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reichard P, Eliasson R, Ingemarson R, Thelander L. J Biol Chem. 2000;275:33021–33026. doi: 10.1074/jbc.M005337200. [DOI] [PubMed] [Google Scholar]
- 33.Saitoh S, Chabes A, McDonald W H, Thelander L, Yates J R, III, Russell P. Cell. 2002;109:563–573. doi: 10.1016/s0092-8674(02)00753-5. [DOI] [PubMed] [Google Scholar]