Abstract
In vitro-selected RNA aptamers are potential inhibitors of disease-related proteins. Our laboratory previously isolated an RNA aptamer that binds with high affinity to human transcription factor NF-κB. This RNA aptamer competitively inhibits DNA binding by NF-κB in vitro and is recognized by its target protein in vivo in a yeast three-hybrid system. In the present study, yeast genetic selections were used to optimize the RNA aptamer for binding to NF-κB in the eukaryotic nucleus. Selection for improved binding to NF-κB from RNA libraries encoding (i) degenerate aptamer variants and (ii) sequences present at round 8 of 14 total rounds of in vitro selection yielded RNA aptamers with dramatically improved in vivo activity. Furthermore, we show that an in vivo-optimized RNA aptamer exhibits specific “decoy” activity, inhibiting transcriptional activation by its NF-κB target protein in a yeast one-hybrid assay. This decoy activity is enhanced by the expression of a bivalent aptamer. The combination of in vitro and in vivo genetic selections was crucial for obtaining RNA aptamers with in vivo decoy activity.
The artificial modulation of gene expression is a major goal of modern disease therapeutics. Recently, RNA aptamers have been investigated as inhibitors of disease-related proteins (reviewed in ref. 1). In vitro selections using large combinatorial RNA libraries have made trivial the identification of RNA sequences that bind with high affinities to target macromolecules. However, it is not intuitive that an RNA molecule selected for binding in vitro likewise will bind and inhibit the target protein within the complex environment of a living eukaryotic cell. Despite this caveat, some RNA aptamers identified through in vitro selection have been shown to perform the desired inhibitory function when expressed in cells. For example, Famulok and colleagues (2) used in vitro selection to identify an RNA aptamer that inhibits the intracellular domain of the β2 integrin LFA-1. A second example of an in vitro-selected RNA aptamer that subsequently was shown to function in vivo is an inhibitory aptamer identified by Lis and colleagues (3) against the Drosophila B52 splicing protein.
We have been intrigued by the possibility of identifying an RNA “decoy” for a model transcription factor, human NF-κB. Such a decoy molecule might fold so as to bind at or near the DNA-binding site of the transcription factor, sequestering NF-κB and reducing its ability to activate target genes. A member of the rel homology protooncogene family of proteins, NF-κB exists in cells primarily as a homodimer composed of two p50 subunits [(p50)2] or a heterodimer composed of one p50 and one p65 subunit (p50/p65). Both forms of NF-κB bind similar sites in duplex DNA [e.g., 5′-GGGACTTTCC (4)] but appear to activate transcription through distinct mechanisms because, unlike p65, p50 lacks a potent carboxyl-terminal transcription activation domain (5). Inhibition of NF-κB activity by the controlled expression of an RNA decoy potentially could have important therapeutic applications, because NF-κB is involved in disease processes such as inflammation, the prevention of apoptosis in tumor cells, and HIV-1 transcription (6). In fact, inhibition of NF-κB by other approaches has been shown to reduce myocardial damage after ischemia and reperfusion (7), potentiate tumor cell killing by tumor necrosis factor and radio- and chemotherapies (8), and cause spontaneous apoptosis in Epstein–Barr virus-transformed lymphoblastoid cells (9).
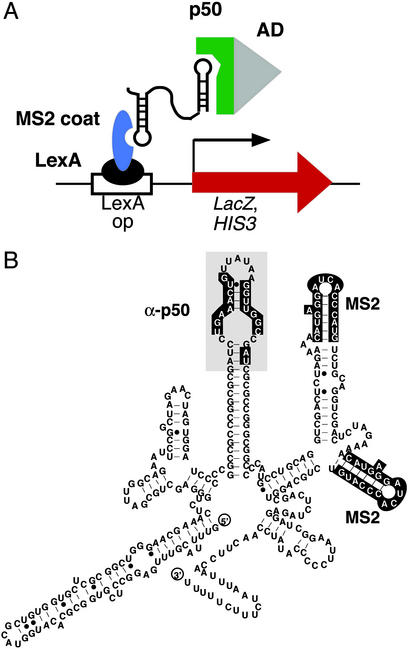
Our laboratory previously conducted an in vitro selection seeking RNA ligands for NF-κB, identifying a 31-nt RNA aptamer (α-p50) that binds with high affinity (Kd ≈ 1 nM) to the target protein (10). Although selected for binding to the p50 homodimer form of NF-κB, the α-p50 RNA aptamer also was shown to recognize the p50/p65 heterodimer (10). In addition, α-p50 functioned as an in vitro decoy for NF-κB, competing with radiolabeled κB DNA for transcription factor binding (10). Furthermore, the α-p50 RNA aptamer was shown to recognize its (p50)2 target within a eukaryotic nucleus (11) in a yeast three-hybrid assay (12). In this sensitive assay for detecting RNA–protein interactions, transcription of HIS3 and lacZ reporter genes depends on a trimolecular interaction within the yeast nucleus (Fig. 1A). Fig. 1B shows a predicted secondary structure (13–15) of the α-p50 aptamer engrafted into the MS2 hybrid RNA required for the yeast three-hybrid system (12).
Figure 1.
(A) The yeast three-hybrid system. Transcription of lacZ and HIS3 reporter genes (red) depends on a trimolecular interaction between hybrid protein 1 (LexA/MS2 coat protein fusion, black/blue), a hybrid RNA composed of the MS2 recognition sequence and the α-p50 aptamer sequence, and hybrid protein 2 (GAL4AD/p50, gray/green). (B) Predicted secondary structure of the initial α-p50/MS2 hybrid RNA. The in vitro-selected α-p50 RNA aptamer domain is shaded, with highlighting of nucleotides previously shown to be important for (p50)2 binding (10). MS2 coat protein recognition sequences are also highlighted.
In the present study, we have further used the yeast three-hybrid system to optimize the α-p50 RNA aptamer for binding to (p50)2 in the eukaryotic nucleus. Previous studies have investigated RNA aptamer modification for optimized in vivo expression, typically by appending rationally designed or in vitro selected sequences that increase aptamer expression, stability, or presentation in cells (2, 3, 16). In contrast, we now present yeast genetic selections to identify aptamer variants that show improved interaction with the target protein. This in vivo optimization yielded dramatic improvements in aptamer function, including subtle nucleotide changes that could not have been predicted to improve in vivo activity. We further show that our best optimized RNA aptamer serves as a specific RNA decoy for (p50)2 in a yeast one-hybrid system. These results demonstrate that RNA decoys can be used to inhibit the function of DNA-binding proteins in vivo and underscore the power of combining in vitro and in vivo genetic selections in RNA engineering.
Materials and Methods
Synthesis and Cloning of Degenerate α-p50/MS2 Hybrid RNA Library.
A degenerate α-p50 DNA oligonucleotide library was synthesized such that the average α-p50 variant had two to three mutations in the α-p50 DNA sequence 5′-GATC2TGA3CTGT2AT-A2G2T2G2C2GATC. The degenerate α-p50 DNA oligonucleotide library was PCR-amplified and cloned between XhoI and SphI sites of pJ714, a version of pIIIA/MS2–2 (12) encoding MS2 hybrid RNAs with a GC “clamp” flanking the inserted sequence.
Cloning of Early-Round in Vitro Selection RNA Library.
Sixty-nucleotide RNA sequences present at round 8 of the prior 14-round in vitro selection for (p50)2 RNA ligands (10) were reverse-transcribed, PCR-amplified, and cloned between XhoI and SphI sites of pJ713, a version of pIIIA/MS2–2 (12) without a GC clamp flanking the inserted sequence.
Hybrid Protein Constructs.
The yeast GAL4 transcriptional activation domain (GAL4AD)/p50 hybrid protein yeast expression plasmid used in yeast three-hybrid screens encodes human p50 amino acid residues 18–502 downstream of the yeast GAL4AD in the yeast shuttle vector pGAD424 (CLONTECH). For decoy assays, the alcohol dehydrogenase-1 promoter–GAL4AD-p50 cassette from the GAL4AD/p50 construct in pGAD424 was PCR-amplified, appending 5′ ApaI and 3′ AatII restriction sites for cloning into the ARS4/Cen6 vector pEXP-AD502 (CLONTECH). The GAL4AD/Sp1 hybrid protein yeast expression plasmid was generated in a similar fashion, by cloning an alcohol dehydrogenase-1-GAL4AD-Sp1 cassette from pGAD424 (CLONTECH) into pEXP-AD502 (CLONTECH). The resulting plasmid encodes the 167 carboxyl-terminal residues of human Sp1 downstream of the yeast GAL4AD.
Yeast Three-Hybrid Screens for Optimized α-p50 RNAs.
The host yeast strain L40-coat (12) expressing the GAL4AD/p50 hybrid protein was transformed with three-hybrid RNA plasmid libraries encoding (i) a degenerate α-p50 RNA library or (ii) an early-round in vitro selection RNA library. The transformation mixtures were plated on selective medium containing 3 mM 3-amino-1,2,4-triazole (3-AT). An estimated 3 × 105 plasmid sequences were screened in each case. Aptamer-encoding plasmids were isolated from HIS3- and lacZ-positive transformants as described (17) and retransformed into fresh L40-coat expressing GAL4AD/p50 to confirm phenotypes. True positive clones then were sequenced to analyze selected hybrid RNAs.
Reporter Gene Assays.
For the HIS3 reporter gene assay, yeast strains were grown to an optical density at 600 nm of ≈1.0 and diluted for plating of 1,000-cell spots on selective medium containing a 3-AT gradient over the concentration range specified in the figure legends. The lacZ reporter gene assay was conducted by using the substrate o-nitrophenyl β-d-galactopyranoside as described (18). Three to six independent transformants were assayed for each yeast strain.
Determination of in Vitro RNA Affinity and in Vivo RNA Accumulation.
RNA affinities for (p50)2 were determined by using a nitrocellulose filter-binding assay. RNA aptamers 1 (5′-GAUC2UGA3CUGU4A2G2U2G2C2GAUC) and 6 (5′-CAUACU2GA3CUGUA2G2U2G2CGUAUG) were purchased from Dharmacon Research (Lafayette, CO), deprotected according to the protocol suggested by the supplier, and purified by denaturing PAGE and elution. Purified RNAs were radiolabeled with [γ-32P]ATP by using T4 polynucleotide kinase (New England Biolabs). (p50)2 protein (amino acid residues 39–363) was expressed in Escherichia coli and purified as described (19).
Binding reactions consisted of a known concentration (0.85 nM) of the indicated radiolabeled RNA aptamer and increasing concentrations (50 pM to 5 μM) of recombinant, purified (p50)2. (p50)2 and labeled RNA 1 or 6 were incubated in binding buffer (10 mM Hepes, pH 7.5/100 mM NaCl/1 mM DTT) for 30 min at 25°C, filtered over nitrocellulose membranes, and quantitated by scintillation counting. Estimates of Kd were obtained by least-squares curve-fitting of binding isotherms (10).
To compare the accumulation in yeast of hybrid RNAs 1, 4, and 6, total RNA was extracted from yeast three-hybrid strains as described (20). Northern blot analysis was conducted as described (11), simultaneously hybridizing with radiolabeled oligonucleotide probes complementary to the RNase P RPR1 leader (5′-AGCAC2ACAGCGTAC2ATGT) and to the U6 small nuclear RNA (snRNA) (5′-TC2T2ATGCAG4A2CTGC). Radioactivity was detected and analyzed by storage phosphor technology.
RNA Decoy Assays.
For decoy assays, hybrid proteins were expressed from the ARS4/Cen6 vector pEXP-AD502 (CLONTECH) in a κB yeast one-hybrid dual reporter strain (11). RNA sequences were cloned between the XhoI and SphI restriction sites of pJ903, a version of pIIIA/MS2–2 (12) that carries a leu2-d marker in place of the URA3 marker, to express ≈300-nt hybrid RNA 6, identical to RNA 6 expressed in the three-hybrid system. Briefly, pJ903 was constructed by homologous in vivo recombination between pJ713 and a PCR product encoding the leu2-d sequence from pBS24-ΔAB6 (kindly provided by David Brow, University of Wisconsin, Madison) flanked on both sides by 40-nt regions of homology to 5′ and 3′ URA3 sequences.
Results
Degenerate α-p50/MS2 Hybrid RNA Library Screen.
Previously, we tested our in vitro selected α-p50 RNA aptamer in the yeast three-hybrid system and showed that the aptamer was recognized by its target protein, NF-κB [(p50)2], in a eukaryotic nucleus (11). We wanted to use yeast genetic selections to optimize the α-p50 RNA aptamer for (p50)2 recognition in yeast, with the ultimate goal of identifying an RNA decoy for (p50)2 that functions in vivo. As a starting point, we reasoned that by changing only one or a few nucleotides within the α-p50 RNA aptamer framework, we might achieve improved in vivo activity. To this end, a degenerate α-p50 DNA library was synthesized such that the average α-p50 variant carried two to three mutations. The resulting 31-bp, double-stranded, degenerate α-p50 DNA library was cloned into the three-hybrid system RNA expression vector and transformed into the yeast three-hybrid host strain L40-coat (12) expressing GAL4AD/p50. Degenerate α-p50 hybrid RNA sequences were screened for improved in vivo binding to (p50)2 by plating the transformation mixture on selective medium containing a higher 3-AT concentration than that tolerated by the yeast strain coexpressing GAL4AD/p50 with the original, in vitro-selected α-p50 RNA aptamer. This procedure seeks hybrid RNAs that induce higher levels of HIS3 reporter gene expression than previously achieved.
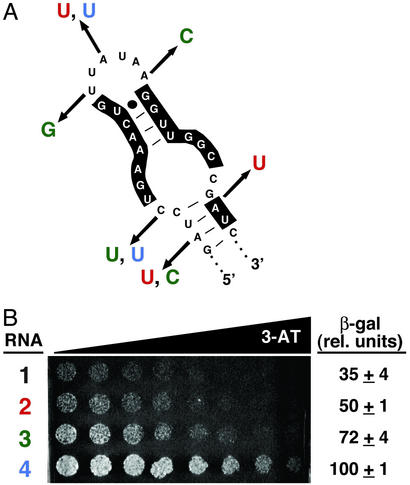
Reselection yielded improved aptamers. After confirming and sequencing, three α-p50 variants were identified that show improved (p50)2 binding in the yeast three-hybrid system (Fig. 2). Combinations of mutations found to improve the aptamer–(p50)2 interaction in yeast are indicated in Fig. 2A. With the exception of one A-to-U mutation in RNA 2 (Fig. 2, red), all mutations occurred outside of the region previously shown to be important for (p50)2 binding to the original in vitro-selected α-p50 aptamer (10). Interestingly, at three nucleotide positions, mutations occurred in two of the three selected sequences (Fig. 2A), suggesting that these might be important nucleotide changes for improving the aptamer–(p50)2 interaction in vivo. As shown in Fig. 2B, yeast three-hybrid strains expressing RNAs 2, 3, and 4 exhibited enhanced HIS3 and lacZ reporter gene expression, as compared with the original in vitro- selected α-p50 aptamer (RNA 1). In particular, mutating only two nucleotide positions within the α-p50 sequence (RNA 4, blue) resulted in significantly increased growth on a 3-AT gradient plate and ≈3-fold increased β-galactosidase levels, supporting the idea that subtle changes in α-p50 aptamer sequence can greatly improve in vivo binding activity.
Figure 2.
In vivo optimization of degenerate α-p50 aptamers. (A) Nucleotide changes selected for improved (p50)2 binding in yeast. Shown is the in vitro- selected α-p50 aptamer (1). Colored nucleotides indicate the combinations of mutations found to improve aptamer–(p50)2 interaction in yeast, yielding RNAs 2 (red), 3 (green), and 4 (blue). Nucleotides previously shown to be important for (p50)2 binding (10) are highlighted. (B) Yeast three-hybrid system HIS3 and lacZ reporter gene assays. Growth assay for HIS3 expression is on selective medium containing a gradient of up to 4 mM 3-AT. β-Galactosidase data reflect three independent transformants for each yeast strain.
Early-Round in Vitro Selection RNA Library Screen.
We further reasoned that sequences present midway through the in vitro selection for (p50)2 RNA aptamers (10) might actually show improved in vivo activity compared with the α-p50 aptamer that ultimately prevailed after 14 rounds of in vitro selection. Therefore, 60-nt RNA sequences present at round 8 of the 14-round in vitro selection for (p50)2 RNA ligands (10) were reverse-transcribed, PCR-amplified, and cloned into the yeast three-hybrid system RNA vector. This early-round in vitro selection library then was transformed into the yeast three-hybrid host strain L40-coat (12) expressing GAL4AD/p50. As before, to select for α-p50 hybrid RNA sequences with improved in vivo binding to (p50)2, the transformation mixture was plated on selective medium containing a higher 3-AT concentration than that tolerated by the yeast strain coexpressing GAL4AD/p50 with the original, in vitro selected α-p50 RNA aptamer.
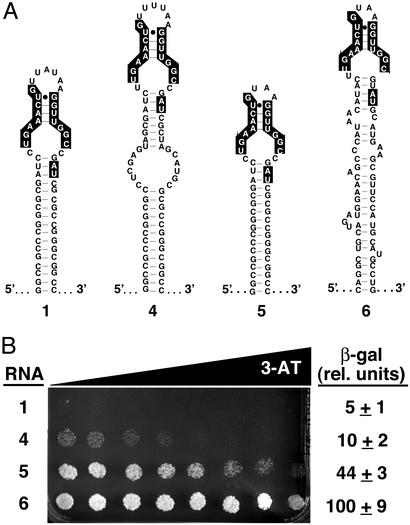
Analysis of selected transformants revealed a single RNA sequence (Fig. 3A, RNA 6). Interestingly, the sequence included the nucleotides previously shown to be important for (p50)2 binding to the original in vitro-selected α-p50 aptamer (10). However, the remainder of the sequence appears to have evolved independently from the α-p50 RNA aptamer that had prevailed originally in vitro. The most striking change is the presence of a 5′-GUAA tetraloop in place of the 7-nt terminal loop of α-p50 (Fig. 3A, compare RNAs 1 and 6). This tetraloop modification may enhance the proper in vivo folding of RNA 6. Other more subtle changes from the in vitro-selected α-p50 sequence are also evident, including a C-to-U change in the left side of the internal unpaired region of α-p50 that also was selected in the degenerate RNA library screen (Figs. 2A and 3A, RNA 4) and the deletion of a C residue from the right side of the internal unpaired region of α-p50. In addition, the RNA 6 sequence provided its own selected “clamp” in place of the engineered GC clamp found to aid the proper folding/presentation of RNA 1 (11).
Figure 3.
A yeast-optimized RNA aptamer selected from an early-round in vitro selection library exhibits drastically improved binding to NF-κB. (A) Predicted secondary structures of α-p50 RNA aptamer domains selected in vitro and in yeast: 1, in vitro-selected (α-p50 + GC clamp); 4, (α-p50 + GC clamp) variant 4 selected in yeast from degenerate library (Fig. 2); 5, (α-p50 + GC clamp + tetraloop); 6, α-p50 aptamer selected in yeast from a library encoding sequences present at round 8 of the 14-round in vitro selection. Nucleotides previously shown to be important for (p50)2 binding (10) are highlighted. (B) Yeast three-hybrid system HIS3 and lacZ reporter gene assays. Growth assay for HIS3 expression is on selective medium containing a gradient of up to 5 mM 3-AT. β-Galactosidase data reflect three independent transformants for each yeast strain.
As shown in Fig. 3B, the yeast three-hybrid strain expressing RNA 6 showed dramatically enhanced HIS3 and lacZ reporter gene expression, as evidenced by increased growth on a 3-AT gradient plate and β-galactosidase levels that were ≈20- and ≈10-fold higher, respectively, than for RNA 1 or degenerate RNA 4. We wondered whether the increased (p50)2-binding activity of RNA 6 was due solely to the presence of the GUAA tetraloop. To answer this question, a sequence was cloned into the hybrid RNA vector to express a version of RNA 1 with a tetraloop substituted in place of the terminal loop of α-p50 (Fig. 3A, RNA 5). When transformed into the yeast three-hybrid strain expressing GAL4AD/p50, hybrid RNA 5 shows increased (p50)2 binding in HIS3 and lacZ reporter assays compared with RNAs 1 and 4 (Fig. 3B), indicating that the tetraloop makes an important contribution to the improved in vivo binding of RNA 6 to (p50)2. However, the yeast three-hybrid strain expressing RNA 6 shows increased growth on a 3-AT gradient plate and ≈2-fold-higher β-galactosidase levels than the strain expressing RNA 5, suggesting that additional sequence features present in RNA 6 significantly enhance its in vivo activity.
Contribution of in Vivo-Selected RNA Clamp.
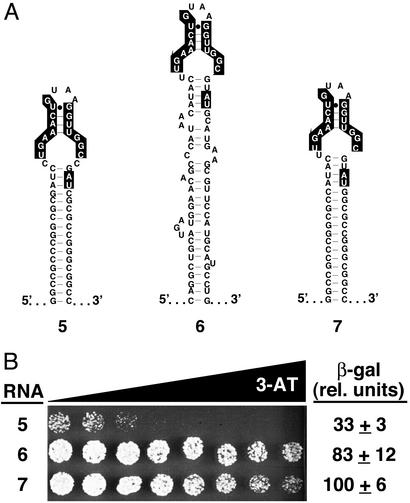
The predicted secondary structure of RNA 6, selected from an early-round in vitro selection RNA library, suggests the presence of a double-stranded clamp structure different from the GC clamp engineered into RNAs 1–5 (see Fig. 3A). Thus, we wondered whether this potential clamp contributed to the improved activity of RNA 6 in the yeast three-hybrid system. To answer this question, the selected clamp of RNA 6 was replaced with the GC clamp sequence of RNA 5, resulting in RNA 7 (Fig. 4A). This RNA then was compared with RNAs 5 and 6 for binding to (p50)2 in the yeast three-hybrid system. 3-AT gradient plate and β-galactosidase assays revealed that, within error, RNA 7 binds (p50)2 comparably to RNA 6 (Fig. 4B), suggesting the selected clamp of RNA 6 does not contribute to its improved (p50)2 binding. Intriguingly, although RNAs 5 and 7 differ at only four nucleotide positions, none of which are predicted to affect RNA folding, yeast three-hybrid strains expressing RNA 7 yield ≈3-fold-higher β-galactosidase reporter gene activity than strains expressing RNA 5 (Fig. 4B). These subtle changes could not have been rationally designed or predicted, highlighting the power of yeast genetic selections for RNA aptamer optimization.
Figure 4.
Yeast-selected RNA clamp is not essential. (A) Predicted secondary structures of α-p50 RNA aptamers 5, 6, and 7 (RNA 6 with GC clamp substituted for yeast three-hybrid-selected clamp). Nucleotides previously shown to be important for (p50)2 binding (10) are highlighted. (B) Yeast three-hybrid system HIS3 and lacZ reporter gene assays. Growth assay for HIS3 expression is on selective medium containing a gradient of up to 8 mM 3-AT. β-Galactosidase data reflect three independent transformants for each yeast strain.
In Vitro RNA Affinity and in Vivo RNA Accumulation.
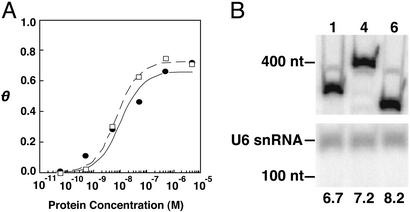
In an attempt to discover the basis for improved (p50)2 binding by RNA 6 in yeast three-hybrid assays, the affinities of RNAs 1 and 6 for (p50)2 were compared in vitro (Fig. 5A). Nitrocellulose filter experiments measuring the binding of radiolabeled RNA competitors 1 or 6 to increasing concentrations of recombinant, purified (p50)2 resulted in the binding curves shown in Fig. 5A. Curve-fitting of binding isotherms (10) yielded equilibrium dissociation constant (Kd) estimates for RNAs 1 and 6 binding to (p50)2 of 6.8 ± 4 nM and 5.4 ± 2 nM, respectively. Likewise, RNAs 1 and 6 were found to bind comparably to the p50/p65 form of NF-κB, with Kd values of 17.4 ± 6 nM and 22.1 ± 7 nM, respectively (data not shown). Within error, the in vitro affinities of RNAs 1 and 6 for (p50)2 were indistinguishable, suggesting that RNA 6 was not selected from the yeast three-hybrid screen on the basis of improved affinity for the (p50)2 target protein.
Figure 5.
Yeast-optimized α-p50 RNA aptamers do not exhibit increased (p50)2 affinity in vitro or improved accumulation in yeast. (A) Plot of θ [the fractional saturation of radiolabeled RNA 1 (●) or 6 (□)] vs. (p50)2 concentration (M) for increasing concentrations of (p50)2 protein, as determined by nitrocellulose filter-binding assays. Curve fit is to equation 1 in ref. 10. (B) Northern blot comparing accumulation of RNAs 1, 4, and 6 in yeast. Yeast total RNA was extracted and assayed by Northern blotting, probing with radiolabeled oligonucleotides complementary to the RNase P RPR1 leader and to the U6 snRNA. Mobilities of DNA size markers are indicated. At the bottom of each lane is shown the signal intensity of RNA 1, 4, or 6 relative to the U6 snRNA internal control.
To examine the accumulation of RNAs 1, 4, and 6 in yeast, a Northern blot analysis of total yeast RNA was conducted, probing simultaneously with radiolabeled oligonucleotides complementary to the RNase P RPR1 leader (present in all of the hybrid RNAs) and to the U6 snRNA as an internal control. As shown in Fig. 5B, although there is a slight increase in RNA levels with increasing yeast three-hybrid activity ([RNA] 1 < 4 < 6), the difference in RNA accumulation does not appear to be sufficient to explain the ≈20-fold increased activity of RNA 6 compared with RNA 1 (Fig. 3B). Thus, variable(s) other than increased (p50)2 affinity or RNA accumulation are implicated in the selection of RNA 6 from the early-round in vitro selection library. Other possible factors that may explain the improved in vivo (p50)2 binding by RNA 6 include improved folding, enhanced nuclear localization, or increased specificity for the target protein vs. nonspecific partners within the yeast cell.
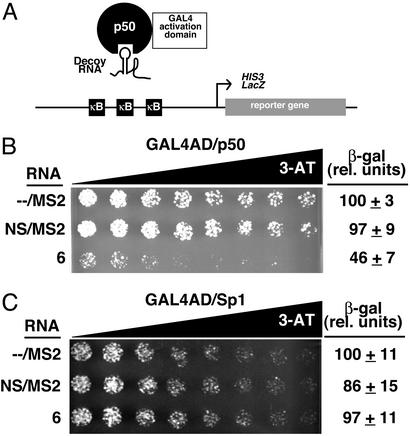
RNA Decoy Assays.
We wanted to determine whether optimized RNA 6 could function as a competitive RNA decoy for its transcription factor target in yeast. To this end, we generated a yeast one-hybrid decoy assay system (Fig. 6A). Binding of an RNA decoy to the DNA-binding site of (p50)2 should block access of the transcription factor to DNA, thereby reducing expression of HIS3 and lacZ reporter genes. When constructs encoding (i) only MS2-coat protein recognition sites (−/MS2), (ii) a nonspecific aptamer/MS2 hybrid RNA (NS/MS2), or (iii) RNA 6 were transformed into the yeast one-hybrid strain and assayed, only RNA 6 showed a decoy effect, as evidenced by decreased survival on 3-AT gradient plates and by ≈55% reduced β-galactosidase levels (Fig. 6B). To ensure that the decoy effect caused by RNA 6 was specific for (p50)2, a second yeast one-hybrid decoy strain was constructed with three copies of the Sp1-binding site upstream of HIS3 and lacZ reporter genes and expressing GAL4AD/Sp1. When this yeast one-hybrid strain was transformed with the same three-hybrid RNA constructs and assayed for HIS3 and lacZ reporter gene expression, none of these RNAs exhibited a decoy effect (Fig. 6C). This result is consistent with the notion that RNA 6 is a specific decoy for (p50)2.
Figure 6.
Yeast-optimized RNA 6 functions as a specific (p50)2 decoy in yeast. (A) Yeast one-hybrid decoy assay system. Binding of an RNA decoy to GAL4AD/p50 blocks binding of the fusion protein to κB sites on DNA, thus reducing transcription of HIS3 and lacZ reporter genes. (B) (p50)2 one-hybrid decoy system HIS3 and lacZ reporter gene assays. Growth assay for HIS3 expression is on selective medium containing a gradient of up to 0.5 mM 3-AT. β-Galactosidase data reflect six independent transformants for each yeast strain. (C) Negative control Sp1 one-hybrid decoy system HIS3 and lacZ reporter gene assays.
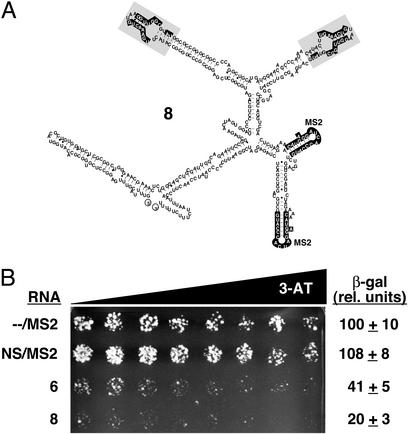
Effect of Increasing Intracellular Concentration of RNA 6.
To investigate the effect of increasing aptamer concentration upon RNA decoy activity, a “bivalent” aptamer (RNA 8; Fig. 7A) containing two copies of the optimized α-p50 domain was cloned. A GC clamp and the yeast three-hybrid-selected clamp of RNA 6 were used to aid in the folding and/or presentation of the aptamer domains within hybrid RNA 8. When RNA 8 was expressed in the yeast one-hybrid decoy assay system shown in Fig. 6A, this strain showed decreased survival on a 3-AT gradient plate and ≈2-fold-decreased β-galactosidase activity compared with RNA 6 (Fig. 7C). This result shows that the optimized α-p50 RNA aptamer exhibits a dose-dependent effect upon (p50)2 DNA binding and implies that by increasing the concentration of the optimized α-p50 aptamer, a greater decoy effect may be achieved.
Figure 7.
Effect of increasing intracellular concentration of RNA 6. (A) Predicted secondary structure of bivalent RNA 8. The in vitro-selected α-p50 RNA aptamer domain is shaded, with highlighting of nucleotides previously shown to be important for (p50)2 binding (10). MS2 coat protein-recognition sequences also are highlighted. (B) HIS3 and lacZ reporter assays showing that RNA 8 exhibits an ≈2-fold-enhanced decoy effect compared with RNA 6. Growth assay for HIS3 expression is on selective medium containing a gradient of up to 0.5 mM 3-AT. β-Galactosidase data reflect six independent transformants for each yeast strain.
Discussion
In Vivo RNA Aptamer Optimization.
After showing that an in vitro- selected RNA aptamer to human transcription factor NF-κB is recognized in the yeast nucleus by its target protein (11), we now have used yeast genetic selections to optimize this RNA–protein interaction in vivo. Screening a library of degenerate α-p50 hybrid RNAs for improved binding to (p50)2 in a yeast three-hybrid system resulted in the selection of three α-p50 variants with markedly improved in vivo activity (Fig. 2). This result demonstrated that even modest changes in nucleotide sequence [e.g., two nucleotide “mutations” in RNA 4 (Fig. 2)] can have a significant impact on in vivo binding of an RNA aptamer to its protein target.
In addition, we screened RNA sequences present at round 8 of the original 14-round in vitro selection for RNA ligands for (p50)2 (10), again seeking improved binding to the target protein in the yeast three-hybrid system. A selected sequence (RNA 6) retains a conserved region of the α-p50 aptamer but appears to have evolved independently from the α-p50 RNA aptamer that ultimately prevailed after 14 rounds of in vitro selection (Fig. 3). RNA 6 shows dramatically improved in vivo activity relative to both the original in vitro-selected α-p50 aptamer (RNA 1) and RNA 4 selected from the degenerate α-p50 hybrid RNA library (Fig. 3). Our results show that during the course of in vitro selection, sequences may become extinct that actually would function better within the much more complex environment of the eukaryotic cell. Thus, by complementing the massive screening capability of a first stage of in vitro selections with a second stage of yeast genetic selections, optimized RNA aptamers with improved in vivo activity may be identified.
Factors Driving Improved in Vivo RNA Aptamer Activity.
Factors that may influence the activity of RNA aptamers in vivo include proper RNA folding, intracellular accumulation, subcellular localization, and specificity for the target protein in the face of immense competition from other cellular factors. In our system, the single most important aptamer modification for improved (p50)2 binding was the substitution of a tetraloop in place of the 7-nt terminal loop of RNA 1 (Fig. 3). This modification resulted in >8-fold-increased β-galactosidase levels in a yeast three-hybrid system lacZ reporter gene assay (Fig. 3B). The tetraloop sequence, which is known to preferentially form U turns, may aid in the proper folding of the α-p50 aptamer sequence within the much larger (≈300-nt) hybrid RNA required for the yeast three-hybrid system. Interestingly, initial in vitro experiments assessing α-p50/MS2 hybrid RNA binding to (p50)2 suggested that the substitution of a tetraloop for the in vitro-selected α-p50 7-nt terminal loop adversely affected binding of the RNA aptamer to (p50)2 (11). Furthermore, when RNAs 1, 4, and 6 were transcribed in vitro, folded, radiolabeled, and electrophoresed under native conditions, each of the RNAs migrated as a single major species (data not shown), suggesting that these ≈300-nt hybrid RNAs are each similarly capable of adopting a single, preferred structure in vitro. These observations suggest that the tetraloop sequence may be particularly important for proper hybrid RNA folding in the context of the cell, again highlighting the differences in selective pressures encountered in vivo vs. in vitro.
Increased affinity for the (p50)2 target protein does not appear to have contributed significantly to enhanced aptamer activity, because the in vitro affinities of RNAs 1 and 6 for (p50)2 are indistinguishable (Fig. 5A). Increased intracellular RNA stability and/or accumulation also do not appear to be significant contributing factors, because the levels of RNAs 1, 4, and 6 within the yeast cell are comparable (Fig. 5B). Particularly enigmatic is the ≈3-fold-increased β-galactosidase levels in a yeast three-hybrid strain expressing RNA 7 vs. RNA 5, because both hybrid RNAs contain tetraloop and GC clamp sequences and differ only at four nucleotide positions. Perhaps these subtle nucleotide changes, one of which also was selected in the degenerate α-p50 library screen (Fig. 2A), are sufficient to allow discrimination against illegitimate protein partners that compete with the intended (p50)2 target.
Transcription Factor Decoy Activity of RNA 6.
The in vivo RNA optimizations described above were undertaken in an attempt to identify an RNA ligand that binds (p50)2 with sufficient in vivo affinity and specificity to function as a transcription factor decoy. In a yeast one-hybrid system, optimized RNA 6 was shown to exhibit decoy activity for the p50 homodimer form of NF-κB but not for a nonspecific transcription factor, Sp1 (Fig. 6). This decoy activity resulted in decreased expression of HIS3 and lacZ reporter genes (Fig. 6B). The α-p50 RNA aptamer apparently binds to (p50)2 with a 1:1 stoichiometry in solution (21), suggesting the aptamer targets the DNA-binding groove of the transcription factor. Binding of κB DNA and α-p50 RNA to overlapping sites would explain the observed in vitro (10, 11) and in vivo (Fig. 6B) competition of the RNA aptamer with κB DNA for (p50)2 binding. The decoy activities of RNAs 1, 4, and 6 in the yeast one-hybrid system correlated with the strengths of interaction with (p50)2 in the three-hybrid system (data not shown), indicating that yeast three-hybrid system aptamer optimization was key to achieving a strong RNA decoy effect.
Unlike the sensitive yeast three-hybrid system, the activity of α-p50 RNA aptamers in the yeast one-hybrid system appears to highly depend on RNA concentration. Initial decoy assays expressing hybrid RNA 6 from the 2-μm vector used in yeast three-hybrid selections [a version of pIIIA/MS2–2 (12)] resulted in sporadic decoy activity that depended on slight differences in RNA accumulation among individual yeast transformants (unpublished observations). By increasing the level of RNA expression ≈2-fold (unpublished observations) through the use of an RNA expression vector bearing a leu2-d marker (22), a reproducible decoy affect was achieved (Fig. 6B). By further increasing the concentration of RNA 6 through the expression of a bivalent aptamer (RNA 8; Fig. 7A), the (p50)2 decoy effect was enhanced ≈2-fold compared with the analogous “monovalent” aptamer, RNA 6 (Fig. 7B). This result highlights the potential to completely saturate the DNA-binding site of (p50)2 by further increasing the intracellular concentration of α-p50 RNA aptamers.
Implications.
The occurrence of apparent natural RNA decoys (23) suggests the possibility of expressing therapeutic decoy RNAs in cells to modulate gene expression (24). Such RNA decoys might offer certain advantages over RNA-based knockdown strategies that require many cellular components for RNA processing and activity [i.e., RNA interference technologies (25)]. In the case of RNA interference for cancer, tumor cell variants likely will overcome such therapies by mutation of any of a number of components in the nonessential RNA interference-processing pathway. Similarly, antiviral RNA interference therapies will be sensitive to point mutations in the target RNA sequence. It may be more difficult for diseased cells to evolve resistance to RNA decoy therapies. We have shown that genetic selections in yeast can dramatically improve in vivo RNA aptamer function relative to what was achieved after in vitro selection alone. It will be fascinating to determine whether optimization in one eukaryotic system will lead to improved aptamer performance in other eukaryotic cells.
Acknowledgments
This work is respectfully dedicated to the memory of Frank M. Rusnak. We thank Marvin Wickens, Stanley Fields, David Brow, Gourisanker Ghosh, and Claus Scheidereit for materials and advice and W. Scott Moye-Rowley, Joseph Gera, Wendy Olivas, Roy Parker, and Grazia Isaya for technical suggestions. We acknowledge the excellent technical assistance of Jeff Zimmerman. This work was supported by the Mayo Foundation and American Cancer Society Grant GMC-98585. L.A.C. is a Howard Hughes Medical Institute Predoctoral Fellow.
Abbreviations
- GAL4AD
yeast GAL4 transcriptional activation domain
- 3-AT
3-amino-1,2,4-triazole
- snRNA
small nuclear RNA
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Sullenger B A, Gilboa E. Nature. 2002;418:252–258. doi: 10.1038/418252a. [DOI] [PubMed] [Google Scholar]
- 2.Blind M, Kolanus W, Famulok M. Proc Natl Acad Sci USA. 1999;96:3606–3610. doi: 10.1073/pnas.96.7.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi H, Hoffman B E, Lis J T. Proc Natl Acad Sci USA. 1999;96:10033–10038. doi: 10.1073/pnas.96.18.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kunsch C, Ruben S M, Rosen C A. Mol Cell Biol. 1992;12:4412–4421. doi: 10.1128/mcb.12.10.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujita T, Nolan G P, Ghosh S, Baltimore D. Genes Dev. 1992;6:775–787. doi: 10.1101/gad.6.5.775. [DOI] [PubMed] [Google Scholar]
- 6.Baeuerle P A, Baltimore D. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 7.Morishita R, Sugimoto T, Aoki M, Kida I, Tomita N, Moriguchi A, Maeda K, Sawa Y, Kaneda Y, Higaki J, et al. Nat Med. 1997;3:894–899. doi: 10.1038/nm0897-894. [DOI] [PubMed] [Google Scholar]
- 8.Beg A A, Baltimore D. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 9.Cahir-McFarland E D, Davidson D M, Schauer S L, Duong J, Kieff E. Proc Natl Acad Sci USA. 2000;97:6055–6060. doi: 10.1073/pnas.100119497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lebruska L L, Maher L J., III Biochemistry. 1999;38:3168–3174. doi: 10.1021/bi982515x. [DOI] [PubMed] [Google Scholar]
- 11.Cassiday L A, Maher L J., III Biochemistry. 2001;40:2433–2438. doi: 10.1021/bi002376v. [DOI] [PubMed] [Google Scholar]
- 12.SenGupta D J, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. Proc Natl Acad Sci USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaeger J A, Turner D H, Zuker M. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaeger J A, Turner D H, Zuker M. Methods Enzymol. 1989;183:281–306. doi: 10.1016/0076-6879(90)83019-6. [DOI] [PubMed] [Google Scholar]
- 15.Zuker M. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 16.Martell R E, Nevins J R, Sullenger B A. Mol Ther. 2002;6:30–34. doi: 10.1006/mthe.2002.0624. [DOI] [PubMed] [Google Scholar]
- 17.CLONTECH. Yeast Protocols Handbook PT3024–1. Palo Alto, CA: CLONTECH; 1999. pp. 34–36. [Google Scholar]
- 18.Zhang X, Moye-Rowley W S. J Biol Chem. 2001;276:47844–47852. doi: 10.1074/jbc.M106285200. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh G, Duyne G V, Ghosh S, Sigler P. Nature. 1995;373:303–310. doi: 10.1038/373303a0. [DOI] [PubMed] [Google Scholar]
- 20.Caponigro G, Muhlrad D, Parker R. Mol Cell Biol. 1993;13:5141–5148. doi: 10.1128/mcb.13.9.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cassiday L A, Lebruska L L, Benson L M, Naylor S, Owen W G, Maher L J. Anal Biochem. 2002;306:290–297. doi: 10.1006/abio.2002.5710. [DOI] [PubMed] [Google Scholar]
- 22.Erhart E, Hollenberg C P. J Bacteriol. 1983;156:625–635. doi: 10.1128/jb.156.2.625-635.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cassiday L A, Maher L J., III Nucleic Acids Res. 2002;30:4118–4126. doi: 10.1093/nar/gkf512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishizaki J, Nevins J R, Sullenger B A. Nat Med. 1996;2:1386–1389. doi: 10.1038/nm1296-1386. [DOI] [PubMed] [Google Scholar]
- 25.Hannon G J. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]