Abstract
The equilibrium unfolding of the Formin binding protein 28 (FBP) WW domain, a stable three-stranded β-sheet protein, can be described as reversible apparent two-state folding. Kinetics studied by laser temperature jump reveal a third state at temperatures below the midpoint of unfolding. The FBP free-energy surface can be tuned between three-state and two-state kinetics by changing the temperature, by truncation of the C terminus, or by selected point mutations. FBP WW domain is the smallest three-state folder studied to date and the only one that can be freely tuned between three-state and apparent two-state folding by several methods (temperature, truncation, and mutation). Its small size (28–37 residues), the availability of a quantitative reaction coordinate (φT), the fast folding time scale (10s of μs), and the tunability of the folding routes by small temperature or sequence changes make this system the ideal prototype for studying more subtle features of the folding free-energy landscape by simulations or analytical theory.
Keywords: β-sheet‖β-strand‖protein folding mechanism
The WW-domain family (1, 2), named after the two conserved tryptophan residues, adopts an isolated antiparallel and highly twisted three-stranded β-sheet structure (Fig. 1). The small size (28–37 residues for the variants reported here) and ready availability from either recombinant or chemical synthesis (3–5), combined with their ability to fold into the desired structure after mutagenesis at virtually every position (ref. 3 and unpublished data) make them excellent model systems to provide further insight into β-sheet folding and stability.
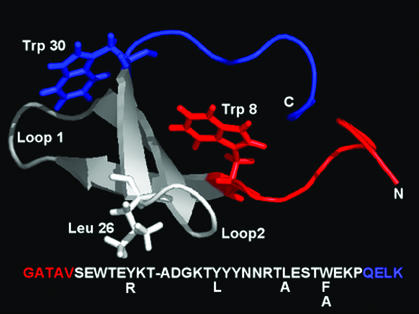
Figure 1.
Structure of the WT FBP/WW domain showing the main mutation sites and Trp-8. The ΔN/ΔC truncations are shown in red and blue, respectively. The sequence is summarized below the structure. This figure was created by using PYMOL (available at http://pymol.sourceforge.net/).
WW domains have helped to clarify the folding dynamics of β-sheet formation (3, 6–9). Hints of the complexity of the free-energy landscape associated with the folding of the hYap WW domain first emerged with reports that transition-state structure could be altered or tuned with changes in temperature (7). An extensive mutation study in the hPin1 WW-domain sequence, evaluated by discrete and continuous φ-value analysis, revealed residues important in defining transition-state structure (3). The loop connecting β-strand 1 and β-strand 2 (loop 1) is formed in the rate-limiting step for folding of hPin1 at physiological temperature. Although hPin1 was an apparent two-state folder under all conditions, these studies also suggested that it is possible to modify the folding free-energy surface through temperature and mutational tuning, such that different structural motifs could control the free energy of the transition state.
Here, we report kinetic data on another WW-domain family member, the domain derived from Formin binding protein 28 (FBP; ref. 10). The FBP WW domain (37 residues) is smaller than the hYap WW domain (57 residues), yet it is significantly more resistant toward thermal denaturation. Fersht and coworkers (8) used a continuous-flow mixing apparatus to study the folding mechanism of the FBP WW domain at ambient temperature, using chaotropes as the folding perturbant. Linear denaturant extrapolation was used to estimate the folding rate constant in aqueous solution. The value of 32,000 s−1 indicates faster folding than for the fastest hPin1 mutants (≈10,000 s−1).
Our direct measurements of FBP folding in denaturant-free aqueous buffer, using temperature denaturation and laser temperature-jump relaxation, reveal a richer kinetic picture. At temperatures below the midpoint of unfolding (TM), folding of the WT FBP WW domain is strongly biphasic. Trp-30 in particular reports on the slow phase. The folding mechanism can be tuned to an apparent two-state process by increasing the temperature, or by small truncations at the termini. Three-state folding in the truncated protein is partially restored by mutation in loop 2 (near the C terminus). These observations are consistent with our previous report of a broad and rough transition-state region describing WW-domain energetics between the folded and unfolded states (3, 7). The shape of the free-energy surface and associated folding channels can be sculpted easily by subtle changes in experimental conditions or by mutagenesis, and at least a two-dimensional free-energy surface is required to account for the observations in a single consistent picture. This responsive behavior from FBP is not specific only to WW-domain proteins, as other proteins have displayed similar trends (11). Karanicolas and Brooks (12) have recently carried out off-lattice folding kinetic and free-energy simulations that seem to be quite consistent with our data. These simulations provide detailed structural information into the folding process of this β-sheet that has been useful in our ongoing analysis of the folding mechanism.
Materials and Methods
Reagents.
The WT FBP28 WW-domain gene and its variants were amplified by using PCR and cloned into the prokaryotic expression vector pGEX-2T (Amersham Pharmacia). The WW domain was expressed as a GST-fusion protein and purified as described (3). Some variants were chemically synthesized by using solid-phase synthesis methods (13) employing fluorenylmethoxycarbonyl chemistry (Applied Biosystems). Protein mass and identity were confirmed by electrospray-ionization mass spectrometry and reversed-phase HPLC, respectively. Protein concentrations were determined spectrophotometrically by using calculated extinction coefficients (14). All variants reported are monomeric, as confirmed by analytical ultracentrifugation (ref. 15 and data not shown).
Equilibrium Unfolding Experiments.
Steady-state experiments were conducted in 10 mM sodium phosphate buffer (pH 7.0). Protein fluorescence was studied by excitation at 295 ± 0.5 nm and detected at 330 nm. Equilibrium thermal denaturation curves were recorded by far-UV CD (30 nm) and by fluorescence-emission intensity. Measurements were performed in 2-mm (far-UV CD) and 10-mm (fluorescence) Teflon-sealed quartz cuvettes. The temperature dependence was measured in 2°C increments from 2 to 108°C. Protein concentrations varied from 10 to 100 μM. The normalized unfolding transitions were superimposable within this concentration range. Equilibrium unfolding transitions were >95% reversible for final temperatures below 80°C. Exposure to higher temperatures resulted in partial irreversibility.
Thermodynamic parameters were determined as described (3, 7). Briefly, thermal unfolding transitions were fitted to a two-state unfolding model
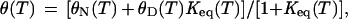 |
1 |
where θN(T) and θD(T) are linear pretransition and posttransition baselines, and Keq = exp(−ΔG(T)/RT) is the temperature-dependent equilibrium unfolding constant. ΔG(T) was approximated by a second-order Taylor series expansion about TM. Thermal denaturation curves were normalized to the fraction of unfolded protein (FU) to allow for comparison of different spectroscopic techniques.
Kinetic Experiments.
Relaxation kinetics of the FBP WW domain were measured by using a nanosecond resolution laser temperature-jump system. Temperature jumps between 8 and 13°C were used to perturb the N ⇔ D equilibrium. Protein concentrations ranged from 10 to 100 μM, and concentration dependences of rates were studied. Fluorescence decays (induced by a 292 ± 2 nm UV pulse) were collected every 14 ns for 500 μs after each temperature jump to follow the relaxation kinetics of the polypeptide chain to the new equilibrium value. The observed relaxation rates were calculated from the fluorescence decays by using our previously reported χ analysis, where the time-evolving shape f of each fluorescent decay is fitted to a linear combination of the denatured (f1) and final (f2) decay shapes (16, 17). The result is a curve of relaxation kinetics normalized to span the 1–0 range. This analysis yields a single exponential decay for apparent two-state reactions. Multistate processes cause a nonexponential trace. For internal consistency, all fits reported here were to double exponentials with f2 evaluated at 300 μs, which allowed straightforward distinction of single exponential from nonsingle exponential kinetics. The slowest processes could not be followed to the baseline, so their reported k2 is an upper bound.
When the folding reaction was apparently two-state, the unfolding equilibrium constant Keq = ku/kf and observed relaxation rate constant kobs = kf + ku were combined to extract folding rates kf and unfolding rates ku. For three-state reactions, the observed relaxation rates and amplitudes were used to characterize the kinetics.
Where kf could be evaluated, activation-free energies were calculated by simultaneously fitting the folding and unfolding rate constants to a Kramers model by using a temperature-dependent free energy of activation (18)
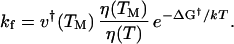 |
2 |
A viscosity-corrected frequency v† of 20 MHz, based on experimental observations of minimal chain-diffusion times (19, 20), describes the characteristic diffusional motion at the barrier, according to the high friction limit in the Kramers' theory. The folding rate is used to calculate the activation-free energy in the form of a second-order Taylor series polynomial, as described (3, 7). For consistency, the same quadratic functional form is used for ΔG† as for ΔG.
Results
Redesign of the FBP WW Domain.
The WT FBP WW domain folds into a twisted three-stranded antiparallel β-sheet tertiary structure, bracketed by two conserved Trp residues (Fig. 1; ref. 15). In an effort to reduce the size of the FBP WW domain, variants were constructed by shortening the N and/or C termini (Fig. 1). Inspection of the ensemble of 10 NMR-solution structures, selected on the basis of minimum total energy (15), indicates that the deleted N-terminal sequence (residues Gly-1–Val-5, Fig. 1) is conformationally flexible and does not interact with the remainder of the WW domain. The lower stability of ΔNΔC Y11R FBP (see next section) results principally from deletion of the C-terminal Leu-36 residue, part of a delocalized hydrophobic core, involving the side chains of Trp-8, Tyr-20, and Pro-33. This Trp-Tyr-Pro cluster is conserved among WW-domain sequences (15, 21–25). In the homologous hPin1 WW domain, even conservative sequence manipulation in this core region results in drastic thermodynamic destabilization as well as disruption of transition-state structure (3). Efforts to truncate the C terminus beyond the invariant Pro-33 residue proved unsuccessful: an ensemble of nonnative conformers with a high propensity to aggregate resulted (data not shown).
Several mutants were also investigated. Tyr-11 in strand 1 is part of a solvent-exposed hydrophobic cluster, including the side chains of Tyr-19 and Trp-30. A Tyr-11 → Arg (Tyr11Arg) mutation adds charge to the exterior of this potentially aggregation-prone hydrophobic cluster. In the homologous hPin1 WW domain, Tyr is replaced by an Arg residue (3). The Tyr11Arg mutation increases the stability of the WT FBP domain. The Tyr19Leu mutation examines the role of this position and residue identity in core integrity and folding stability. A Tyr or Phe residue is the preferred amino acid at this position in the WW-domain family, and a Tyr → Leu mutation in the context of the Pin WW domain is destabilizing, as it is for FBP. Leu-26 is a moderately large hydrophobic residue in loop 2, which may help register that loop during folding by interacting with other hydrophobic residues. It was mutated to Ala in the truncated mini protein. Finally, Trp-30 is located near the C terminus and reports on the dynamics of the 11/19/30 cluster and the C-terminal domain. It was replaced by Phe and Ala; the latter was also studied by Fersht and coworkers, who reported position 30 to be engaged in nonnative interactions in the transition state (φM = −0.4; ref. 9).
Thermal Denaturation Experiments.
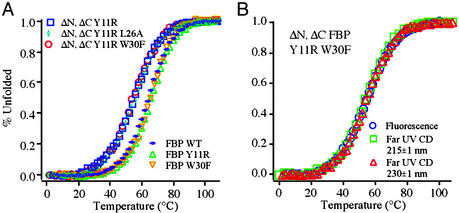
Thermal denaturation curves obtained for representative variants of the FBP WW domain are depicted in Fig. 2A and were independent of concentration in the 10–100 μM range. The ΔNΔC Y11R-truncated mini-variants are slightly less stable than the full-length mutants. All variants have simple sigmoidal unfolding transitions. They are slightly less cooperative than those obtained for the hPin1 WW domain (3, 4). Broad thermal denaturation curves are expected for proteins of the size of a WW domain, because of small heat capacity changes associated with the unfolding process (26). The far-UV CD spectra of the FBP WW domain and the ΔNΔC Y11R-truncated mini-versions are very similar (data not shown) and comparable to published spectra (15), suggesting that terminal truncation does not significantly perturb the WW-domain fold. Although atypical for β-sheet proteins, the maximum near 230 nm is characteristic of folded WW domains and most likely results from strong coupling of the aromatic side chains to the amide chromophore. Under strongly denaturing conditions, all FBP WW domains described display random-coil spectra, with a single minimum ≈200 nm. Thermal equilibrium denaturation curves can be fitted well with a two-state model (Eq. 1). Normalized transitions obtained by fluorescence and far-UV CD spectroscopy at different wavelengths are superimposable within experimental error, supporting the validity of a simple two-state model (Fig. 2B). A summary of the thermodynamic parameters is provided in Table 1.
Figure 2.
(A) Thermodynamic analysis based on CD (230 nm) of WT FBP WW domain, its mutants, and truncations. The truncated variants denature ≈10°C lower but have essentially the same cooperativity as WT and variants thereof. (B) Different spectroscopic probes yield superimposable transitions, the hallmark of thermodynamic two-state transitions. All other FBP variants tested behave similarly (data not shown).
Table 1.
Thermodynamic and kinetic parameters for FBP and its variants
| Name | TM, °C | ΔG0 | ΔG1 | ΔG2 | k1 (40 ± 2°C) | k2 (40 ± 2°C) | k (65 ± 2°C) |
|---|---|---|---|---|---|---|---|
| FBP WT | 64 | 0.13 (2) | 0.352 (2) | 0.00220 (20) | 0.030 (1) | <0.0011 | 0.071 (2) |
| FBP Y11R | 66 | −0.05 (3) | 0.384 (3) | 0.00172 (22) | 0.025 (4) | <0.0014 | 0.040 (1) |
| FBP Y19L | 55 | 0.76 (7) | 0.197 (4) | 0.00029 (10) | 0.035 (2) | <0.0021 | 0.073 (2) |
| FBP W30A | 57 | −0.33 (8) | 0.171 (5) | −0.00037 (17) | 0.027 (1) | — | — |
| FBP W30F | 66 | 0.05 (4) | 0.401 (5) | 0.00235 (32) | 0.054 (2) | — | 0.110 (6) |
| ΔN Y11R | 66 | −0.31 (2) | 0.402 (3) | 0.00186 (19) | 0.026 (3) | <0.0016 | 0.038 (1) |
| ΔNΔC Y11R | 55 | −0.10 (3) | 0.315 (3) | 0.00148 (16) | 0.050 (2) | — | 0.106 (4) |
| ΔNΔC Y11R/L26A | 56 | 0.02 (2) | 0.322 (2) | 0.00074 (14) | 0.044 (1) | <0.0020 | 0.097 (2) |
| ΔNΔC Y11R/W30F | 54 | −0.29 (4) | 0.322 (4) | 0.00080 (23) | 0.035 (1) | — | 0.088 (1) |
The free-energy parameters are defined by using a second-order Taylor series expansion about the melting temperature, TM. Uncertainties in parentheses are one SD. All ΔG in kJ⋅mol−1, all k in μs−1; the melting temperatures are nominal (±1°), so ΔG0 ≈ 0 was fitted also; k2 is the nominal slow biexponential component for f2 at 300 μs, which is a lower limit to the true slow-rate constant. —, These proteins exhibit single exponential kinetics and hence k2 is not relevant.
Tunable Multistate Kinetics of the FBP Domain.
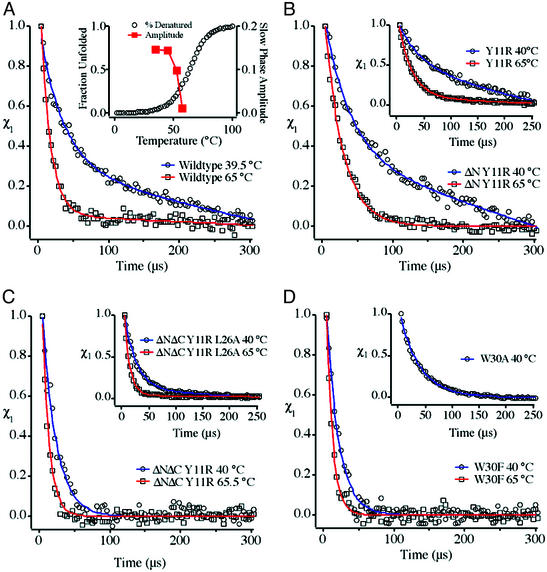
Kinetic experiments were carried out on the mutants of full-size and truncated FBP variants shown in Table 1. Fig. 3 summarizes the most important results at about 40 and 65°C (below and above the transition midpoint). Below the transition midpoint, WT FBP kinetics deviate strongly from a single-exponential decay, implying at least an apparent three-state folding scenario (Fig. 3A). Slow and fast phases are observed, with amplitudes essentially independent of protein concentration (10–250 μM, data not shown), so the observed kinetic heterogeneity cannot be attributed to intermolecular aggregation. The fast phase has a time constant of ≈30 μs, while the time constant of the slowest phase is >900 μs. As temperature is increased, the slow phase disappears completely near the midpoint of thermal unfolding (Fig. 3A Inset). Single exponential relaxation with a time constant of <15 μs is observed at 65°C. Temperature jumps after cooling the protein samples back to 35°C yielded the same biexponential kinetics again. To rule out the possibility that the slow FBP kinetic phase was a measurement artifact, we also interleaved measurements of WT hPin1 WW domain, previously shown to be a two-state folder (3), and WT FBP at different laser power levels. Individual relaxation measurements consistently showed either single exponential (hPin1) or nonsingle exponential kinetics (WT FBP). Thus, it seems that additional free-energy minima complicating the folding mechanism can be tuned out of free-energy range by raising the temperature.
Figure 3.
(A) Relaxation kinetics of WT FBP WW domain below and above the transition midpoint. (Inset) Increase in the slow-phase amplitude as temperature is lowered, overlaid on the fraction of unfolded protein from Fig. 2A. (B) A Tyr11Arg mutation, even combined with an N-terminal truncation, does not alleviate the double-exponential kinetics. (C) Further modification of the construct in B by truncation of the four C-terminal residues restores apparent two-state behavior at all temperatures, whereas a Leu26Ala mutation in loop 2, which may enhance misregistration there, partially restores three-state behavior at low temperatures in the ΔNΔC Y11R variant background. (D) Trp30Phe and Trp30Ala mutations of the full-length protein fold in a single phase, which may be caused by a reporter effect, or by involvement of other C-terminal residues in the formation of an intermediate with nonnative structure.
As shown in Fig. 3B and its Inset, the ΔN truncation and Tyr11Arg mutant have little effect on the biphasic kinetics at low temperature. For the N-terminal truncation, this finding is not surprising in light of the NMR structure, which shows little interaction with the rest of the protein. The insensitivity to Tyr11Arg, which participates in the Tyr-11/Tyr-19/Trp-30 crossstrand cluster, indicates that reduced hydrophobicity in that cluster does not destabilize the nonnative state(s). The Y19L core mutant similarly retains biexponential kinetics, providing further evidence against the cluster being associated with biphasic kinetics (data not shown).
However, temperature is not the only variable that simplifies the folding scenario for FBP WW domain. Fig. 3C shows the kinetics of the ΔNΔC Y11R doubly truncated version revealing that the additional C-terminal truncation completely restores apparent two-state folding over the entire temperature range we measured. The C terminus, including Leu-36, is, therefore, a candidate for involvement in deviations from two-state behavior. Concentration studies of the ΔNΔC Y11R truncated mini-protein revealed the presence of a slow phase at high concentration (>50 μM), consistent with aggregation inhibiting fast folding only at higher concentrations. This result is to be contrasted with full-length FBP, which showed no decrease in the slow phase when protein concentration was lowered from 150 to 10 μM.
Another structural region of the FBP WW domain, which could be involved in stabilizing nonnative states, is loop 2 (centered on Arg-24 and Thr-25). Previous studies of hPin1 domain indicated that loop 2 was close energetically to contributing to the folding mechanism (3). Because the strand-crossing 11/19/30 hydrophobic cluster is not a likely cause of the three-state scenario, we investigated loop 2 next. Fig. 3C Inset shows that a Leu26Ala mutation of the ΔNΔC Y11R-truncated FBP domain partially restores biexponential kinetics in the ΔN ΔC mini-domain that is normally a two-state folder. Thus, very small sequence truncations at the C terminus and point mutations can both modulate the two- and three-state behavior of FBP WW domain.
A final set of mutants investigated the effect of a Trp30Phe substitution. The folding kinetics of the ΔNΔC Y11R mini-variant were only slightly affected by this mutation and remained single exponential at all accessible temperatures with or without the tryptophan (data not shown). Interestingly, the full-length FBP WW domain with the C-terminal Trp30Phe mutation exhibits single-exponential kinetics under all conditions tested, even at temperatures below the midpoint of folding, where a slow phase is clearly evident in the WT construct (Fig. 3D). The relaxation time constant varies from 18 μs at 35°C to 9 μs at 56.1°C. This relaxation could be caused by a further C-terminal effect on intermediate state(s), which are tuned to high free energy by this fairly conservative mutation. It is more likely that a reporter effect is responsible, where the Trp30Phe mutation eliminates the indole probe that has the ability to detect the multistate nature of the transition: the Trp 30-containing ΔNΔC Y11R-truncated mini-protein certainly is single exponential (Fig. 3C), demonstrating that Trp-30 by itself cannot be responsible for the more complicated kinetics we observe in WT FBP.
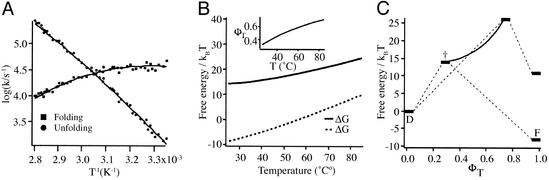
Although the Trp30Phe mutation of the ΔNΔC Y11R FBP is not the key to controlling three-state behavior, it is nonetheless of great utility. Unlike the Trp variant, which transiently aggregates at concentrations >50 μM, the Phe variant meets thermodynamic and kinetic two-state folding criteria at concentrations up to 300 μM. The relaxation time constants vary from 28 μs at 40°C to 11 μs at 65°C. The absence of a second phase at high concentrations makes ΔNΔC Y11R W30F an ideal system for studying two-state β-sheet folding. Folding and unfolding rate constants for this mutant, therefore, were obtained at many temperature points, and the free energies of folding and activation as a function of temperature were calculated according to Keq = e−ΔG(T)/RT and Eq. 2, as shown in Fig. 4 A and B. The entropic reaction coordinate
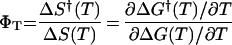 |
3 |
was used to quantify the progress toward the native state achieved at the transition state. φT measures the change in reaction entropy achieved in going from the denatured state to the transition state, relative to the full entropy of folding to the native state. The plot in Fig. 4C shows how the transition state moves up in free energy (manifested in slower folding rates) and toward the native state (a larger ΦT) as the native state is destabilized.
Figure 4.
(A) Folding and unfolding rate constants from temperature-jump experiments of ΔNΔC Y11R W30F. Excellent fits can be obtained to a quadratic expansion of the free energy with temperature; these fits are used to calculate free energies as a function of temperature. (B) Free energies derived from the rates in A and from the thermodynamic data in Fig. 2A. (C) Energetics of the ΔNΔC Y11R W30F WW domain, plotted as a function of φT from Eq. 3. The transition and native states are shown “unstressed” (40°C) and stressed (65°C).
Discussion
The Free-Energy Landscape of the FBP WW Domain.
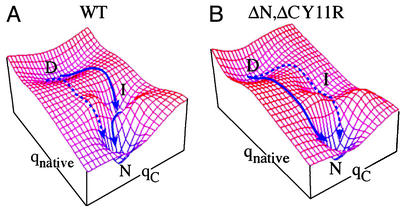
Proteins that explore multiple regions of the free-energy surface by redesign can provide particularly stringent tests for folding models (3, 27, 28). FBP WW domain is the smallest and most flexibly tunable of these proteins so far. Fig. 5 summarizes the folding process of the FBP WW domain in terms of a simplified, smoothed two-dimensional free-energy surface (29). Here, qnative measures the number of native contacts, whereas qC measures nonnative interactions in the C-terminal region. In addition to the native and denatured basins, there is a third basin corresponding to nonnative interactions in loop 2 and at the C terminus. This basin is clearly not obligatory for folding because the ΔNΔC-truncated variant folds in a single phase and faster than WT FBP, but with the same cooperativity (Fig. 2). Thus, we assign the slow phase of WT FBP to nonnative interactions stabilizing the intermediate basin, but only at low temperatures and in the presence of the C-terminal residues. Hence, the intermediate basin does not lie “between” the D and N basins in Fig. 5, but at larger values of qloop2,ΔC.
Figure 5.
Folding free-energy landscape of the FBP WW domain. The energy landscape is defined by two coordinates, the number of native contacts (qnative) and nonnative contact formation (qC) involving loop 2/C terminus. The two dominant minima represent the ensemble of denatured states (D) and the native state (N). The third local minimum accounts for the third species (I) manifested in the folding of the WT FBP WW domain under conditions that favor the native state. The experimental results are best rationalized in terms of such a two-dimensional surface (see text).
The experimental results thus can be rationalized as follows: in WT FBP, the energy for the three-state path is lowest, and an apparent three-state mechanism is seen, although some protein can simultaneously fold directly (dotted curve in Fig. 5A). As the temperature is raised, or if the C terminus is removed, the intermediate basin is destabilized, and the direct path is stabilized, yielding apparent two-state folding. A mutation in loop 2 such as Leu26Ala may again stabilize a misfolded intermediate, most likely by increasing the probability of misregistration in loop 2 during folding. The Trp30Phe mutation is not needed to lower the energy of the third state because the ΔNΔC-truncated protein with Trp-30 folds in a single phase (Fig. 3C). Although some studies have implicated Trp-30 in local nonnative contact formation (9), it clearly is by itself not responsible for the more complicated folding of WT FBP. Similarly, a Trp30Ala mutation decreases the rate and yields a single phase (Fig. 3D).
Karanicolas and Brooks have recently carried out extensive off-lattice simulations of FBP WW-domain folding (12). They conclude that the folding is biphasic, with the slow phase caused by states with nonnative (e.g., misregistered) structure in loop 2. This simulation agrees with our results, including the reemergence of some degree of three-state behavior when Leu-26 in loop 2 is mutated in the mini-protein. It will be very interesting to see whether further simulations of ΔC-truncated proteins reveal a connection with loop 2 during misfolding. Our results and the work by Karanicolas and Brooks show that modeling can provide microscopic insight into the folding mechanism, when subtle features of the experimental data are successfully reproduced by the model.
Energetics of the FBP and hPin1 WW Domains.
The free energy of activation for folding of the ΔNΔC Y11R W30F variant is about 3 kJ/mol lower than that for hPin1 (Fig. 4C and ref. 3). The folding free-energy curves for both proteins cross at about 70°C. This kind of behavior would be unusual if we were comparing a Pin variant with WT Pin. However, there are enough differences in sequence and structure between WT Pin and ΔNΔC Y11R W30F to justify such intersections in stability. For example, loop 1 in FBP is a (3:5) type-I β-bulge (15), which seems to be the preferred loop type in the WW-domain family at this position. The hPin1 loop, on the other hand, carries a one-residue insertion and adopts an unusual (4:6)-conformation with a type-II β-turn found within the larger (4:6) loop conformation (22). The hPin1 domain is more twisted than the FBP WW domain, which may be a direct consequence of the unusual loop 1 conformation in the former. Most importantly, the hydrophobic core of ΔNΔC FBP is partly truncated, owing mainly to the loss of Leu-36 upon domain minimization.
Fig. 4B depicts the transition-state energetics for the truncated protein plotted against the entropic reaction coordinate φT. As expected from Hammond's postulate, imposing thermodynamic stress against the native state by increasing the temperature shifts the transition-state ensemble both vertically (free energy) as well as horizontally (entropically) from an early (≈40% native-like) to a later, low-entropy ensemble (≈80% native-like). The findings are in excellent qualitative agreement with studies on the hPin1 domain (3). ΔNΔC FBP, however, has a larger φT value than hPin1 at low temperature [φT (Pin) ≈ 0.1; φT (FBP) ≈ 0.35]. This observation indicates that there is a higher entropic penalty in the ΔNΔC FBP domain for forming the crucial transition-state contacts that lead to productive downhill folding. One possibility is that partial core truncation associated with FBP domain shortening not only removes energetically favorable contacts in the native protein, but may also lead to an expansion of the denatured-state ensemble by weakening transient residual structure. Or, additional nonnative interactions, as those seen in the immunity protein Im7 (11), may play a dominant role in modifying the free-energy landscape on which the protein traverses. Irrespective of the exact source of the differences between the transition-state position characterizing the Pin and FBP WW domains, our quantitative φT-value approach does not support the substantially more compact transition state of WT FBP (≈70% native-like) at ambient temperature, inferred by continuous-flow experiments and linear extrapolation to aqueous buffer conditions (8).
Acknowledgments
We thank John Karanicolas and Dr. Charles Brooks III for many helpful discussions and for sharing the results of their simulations with us before publication. M.J. thanks the La Jolla Interfaces in Science program for fellowship support and enabling interactions with the Brooks group on aspects of theory and simulation. This work was supported by National Institutes of Health Grants GM 057175 (to M.G. and H.N.) and GM 051105, The Skaggs Institute of Chemical Biology, and the Lita Annenberg Hazen Foundation (to J.W.K., M.J., and A.M.). M.J. was also supported by a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft.
Abbreviation
- FBP
Formin binding protein 28
Footnotes
See commentary on page 3555.
References
- 1.Sudol M. Prog Biophys Mol Biol. 1996;65:113–132. doi: 10.1016/s0079-6107(96)00008-9. [DOI] [PubMed] [Google Scholar]
- 2.Sudol M, Hunter T. Cell. 2000;103:1001–1004. doi: 10.1016/s0092-8674(00)00203-8. [DOI] [PubMed] [Google Scholar]
- 3.Jäger M, Nguyen H, Crane J, Kelly J, Gruebele M. J Mol Biol. 2001;311:373–393. doi: 10.1006/jmbi.2001.4873. [DOI] [PubMed] [Google Scholar]
- 4.Kaul R, Angeles A, Jäger M, Powers E T, Kelly J W. J Am Chem Soc. 2001;123:5206–5212. doi: 10.1021/ja0102890. [DOI] [PubMed] [Google Scholar]
- 5.Toepert F, Pires J R, Landgraf C, Oschkinat H, Schneider-Mergener J. Angew Chem Int Ed Engl. 2001;40:897–900. doi: 10.1002/1521-3773(20010302)40:5<897::AID-ANIE897>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 6.Koepf E K, Petrassi H M, Sudol M, Kelly J W. Protein Sci. 1999;8:841–853. doi: 10.1110/ps.8.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crane J C, Koepf E K, Kelly J W, Gruebele M. J Mol Biol. 2000;298:283–292. doi: 10.1006/jmbi.2000.3665. [DOI] [PubMed] [Google Scholar]
- 8.Ferguson N, Johnson C M, Macias M, Oschkinat H, Fersht A. Proc Natl Acad Sci USA. 2001;98:13002–13007. doi: 10.1073/pnas.221467198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferguson N, Pires J R, Toepert F, Johnson C M, Pan Y P, Volkmer-Engert R, Schneider-Mergener J, Daggett V, Oschkinat H, Fersht A. Proc Natl Acad Sci USA. 2001;98:13008–13013. doi: 10.1073/pnas.221467398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan D C, Bedford M T, Leder P. EMBO J. 1996;15:1045–1054. [PMC free article] [PubMed] [Google Scholar]
- 11.Capaldi A P, Kleanthous C, Radford S E. Nat Struct Biol. 2002;9:209–216. doi: 10.1038/nsb757. [DOI] [PubMed] [Google Scholar]
- 12.Karanicolas J, Brooks C L., III Proc Natl Acad Sci USA. 2003;100:3954–3959. doi: 10.1073/pnas.0731771100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merrifield R B. J Am Chem Soc. 1963;85:2149–2154. [Google Scholar]
- 14.Gill S C, Hippel P H V. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 15.Macias M J, Gervais V, Civera C, Oschkinat H. Nat Struct Biol. 2000;7:375–379. doi: 10.1038/75144. [DOI] [PubMed] [Google Scholar]
- 16.Ervin J, Sabelko J, Gruebele M. J Photochem Photobiol B. 2000;54:1–15. doi: 10.1016/s1011-1344(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 17.Ballew R M, Sabelko J, Reiner C, Gruebele M. Rev Sci Instrum. 1996;67:3694–3699. [Google Scholar]
- 18.Kramers H A. Physica. 1940;7:284. [Google Scholar]
- 19.Lapidus L J, Eaton W A, Hofrichter J. Proc Natl Acad Sci USA. 2000;97:7220–7225. doi: 10.1073/pnas.97.13.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bieri O, Wirz J, Hellrung B, Schutkowski M, Drewello M, Kiefhaber T. Proc Natl Acad Sci USA. 1999;96:9597–9601. doi: 10.1073/pnas.96.17.9597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macias M J, Hyvonen M, Beraldi E, Schultz J, Sudol M, Saraste M, Oshkinat H. Nature. 1996;382:646–649. doi: 10.1038/382646a0. [DOI] [PubMed] [Google Scholar]
- 22.Ranganathan R, Lu K, Hunter T, Noel J P. Cell. 1997;89:875–886. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- 23.Huang X, Poy F, Zhang R, Joachimiak A, Sudol M, Eck M J. Nat Struct Biol. 2000;7:634–638. doi: 10.1038/77923. [DOI] [PubMed] [Google Scholar]
- 24.Kanelis V, Rotin D, Forman-Kay J D. Nat Struct Biol. 2001;8:407–412. doi: 10.1038/87562. [DOI] [PubMed] [Google Scholar]
- 25.Kowalski J A, Liu K, Kelly J W. Biopolymers. 2002;63:111–121. doi: 10.1002/bip.10020. [DOI] [PubMed] [Google Scholar]
- 26.Alexander P, Fahnestock S, Lee T, Orban J, Bryan P. Biochemistry. 1992;31:3597–3603. doi: 10.1021/bi00129a007. [DOI] [PubMed] [Google Scholar]
- 27.Bieri O, Wildegger G, Bachmann A, Wagner C, Kiefhaber T. Biochemistry. 1999;38:12460–12470. doi: 10.1021/bi9909703. [DOI] [PubMed] [Google Scholar]
- 28.Nauli S, Kuhlman B, Baker D. Nat Struct Biol. 2001;8:602–605. doi: 10.1038/89638. [DOI] [PubMed] [Google Scholar]
- 29.Gruebele M. Curr Opin Struct Biol. 2002;12:161–168. doi: 10.1016/s0959-440x(02)00304-4. [DOI] [PubMed] [Google Scholar]