Abstract
Gene duplication creates evolutionary novelties by using older tools in new ways. We have identified evidence that the genes for enamel matrix proteins (EMPs), milk caseins, and salivary proteins comprise a family descended from a common ancestor by tandem gene duplication. These genes remain linked, except for one EMP gene, amelogenin. These genes show common structural features and are expressed in ontogenetically similar tissues. Many of these genes encode secretory Ca-binding phosphoproteins, which regulate the Ca-phosphate concentration of the extracellular environment. By exploiting this fundamental property, these genes have subsequently diversified to serve specialized adaptive functions. Casein makes milk supersaturated with Ca-phosphate, which was critical to the successive mammalian divergence. The innovation of enamel led to mineralized feeding apparatus, which enabled active predation of early vertebrates. The EMP genes comprise a subfamily not identified previously. A set of genes for dentine and bone extracellular matrix proteins constitutes an additional cluster distal to the EMP gene cluster, with similar structural features to EMP genes. The duplication and diversification of the primordial genes for enamel/dentine/bone extracellular matrix may have been important in core vertebrate feeding adaptations, the mineralized skeleton, the evolution of saliva, and, eventually, lactation. The order of duplication events may help delineate early events in mineralized skeletal formation, which is a major characteristic of vertebrates.
New biological function arises from new genes produced by gene duplication (1). Even highly diverged proteins today have been generated from older tools. Enamel matrix proteins (EMPs), caseins, and some salivary proteins are secretory Ca-binding phosphoproteins (SCPPs), secreted from the epithelium-derived tissues formed by epithelium-mesenchymal interactions (2, 3). If duplicated genes such as SCPP genes retain common functional and sequence features even after extensive divergence, we infer that the genes were generated from a common ancestor with a related function.
EMPs are responsible for organizing hydroxyapatite crystallization in the enamel organ and are principally coded by three genes: amelogenin (AMEL), ameloblastin (AMBN), and enamelin (ENAM). The evolutionary relationship among the EMP genes was unknown previously; no paralogous genes for any one of these genes were identified (4).
Milk caseins provide mammalian infants with Ca-phosphate to help bone and tooth development as well as to provide required amino acids. Bovine caseins are coded by four distinct genes: αS1 (CSN1S1), αS2 (CSN1S2), β (CSN2), and κ (CSN10) (5). The α- and β-caseins are termed Ca-sensitive, because they precipitate in the presence of Ca ions but are stabilized in colloidal suspension by their interaction with the Ca-insensitive κ-casein (6). Ca-sensitive casein genes (CSN) arose from a common origin by gene duplication (7, 8).
Statherin and histatins are salivary proteins that protect teeth by regulating the spontaneous precipitation of Ca-phosphate salts on enamel surface (9). Histatins also show antibacterial and antifungal properties (10). These proteins are coded by three distinct genes, STATH, HTN1, and HTN3, which originated from a common origin (11).
We have found that these SCPP genes including two EMPs (AMBN and ENAM), four caseins, and three salivary proteins as well as four other salivary proteins [three proline-rich proteins (PROL1, -3, and -5) and a mucin (MUC7)] all are clustered on human chromosome 4q13, whereas two distinct AMELs are located in nonpseudoautosomal regions of the X and Y chromosomes (12–16). We investigated the origin and functional divergence of these SCPP genes in vertebrate evolution.
Methods
Nucleotide Sequences.
The following nucleotide sequences were obtained from GenBank: human casein αS1 (NM001890), β (AF027807, M86237), κ (NM005212), STATH (NM003154), HTN1 (NM002159), HTN3 (NM000200), FLJ20513 (AK000520), NYD-SP26 (AF380838), PROL5 (D89501), PROL3 (NM006685), PROL1 (NM021225), MUC7 (L13283), AMBN (AF209780), ENAM (NM031889), AMEL (AF436849 and M86933), and SPARCL1 (secreted protein, acidic, cysteine-rich related, X86693); DSPP (dentin sialophosphoprotein, AF163151); DMP1 (dentine matrix acidic phosphoprotein 1, U89012); IBSP (integrin-binding sialoprotein, NM004967); MEPE (matrix, extracellular, phosphoglycoprotein, AJ276396); SPP1 (secreted phosphoprotein 1, BC022844); cow (M16644) and pig (X54975) CSN1S2; caiman (AY043290) AMBN; pig ENAM (U52196); alligator (AF095568) and African clawed toad (AF095569) AMEL; and human genome sequence for 4q13 (NT006216) and 4q21 (NT006204).
Computer Analysis.
We used DOTTER, a dot-matrix analysis program (17) to determine exon–intron boundaries by searching identical sequences between cDNA and genome sequences. Multiple sequence alignments were generated by using CLUSTALW (18) through the DNA Data Bank of Japan web site (www.ddbj.nig.ac.jp) and manually modified. Phylogenetic and molecular evolutionary analyses were conducted by using MEGA 2.1 (19). Both SIGNALP (www.cbs.dtu.dk) and PSORT II (http://psort.nibb.ac.jp) were used to predict cleavage sites and the tripartite structures within signal peptides (SPs) (20, 21).
Results
A Gene Cluster for EMP, Casein, and Salivary Proteins.
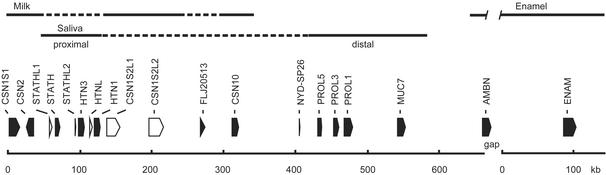
We localized 12 functional genes for the secretory proteins for milk, saliva (and/or tear), and enamel within a cluster spanning >776 kb on 4q13 in humans (Fig. 1). Five nonfunctional pseudogenes related to these proteins (CSN1S2L1, CSN1S2L2, STATHL1, STATHL2, and HTNL) and two genes with unknown functions (FLJ20513 and NYD-SP26) were also identified. CSN1S2L2 had a termination codon within exon 4 but retained all canonical splice sites, suggesting recent loss of function. Bovine CSNs are arranged in the same order and polarity as in humans, but CSN1S2 is functional (22). The proximal salivary-protein gene complex (STATH/HTN3/HTN1) is located within the Ca-sensitive CSN complex and is separated from the distal complex (PROL5, -3, -1, and MUC7). AMBN is located 109 kb distal to MUC7 but the 3′ half of this gene resides within a clone gap in the human draft genome sequence (23). The organization of this gene cluster is conserved in the mouse.
Figure 1.
Casein, salivary, and EMP gene cluster on human 4q13. Locations of the gene products (milk, saliva, and enamel) are shown on the top. Two salivary protein gene complexes, proximal and distal, are shown. Gene symbols, locations, and transcriptional polarities are indicated at the bottom. Filled and open boxes represent the locations of functional genes and pseudogenes, respectively. The scale represents sequence contigs. The 3′ half of AMBN is not included in the human draft genome sequence (gap). The length of AMBN was inferred from the mouse gene.
Structure of the EMP Genes.
The introns of the three EMP genes are all phase-0, that is, their boundaries do not disrupt codons (Fig. 2A). A recent study has revised the translation initiation site of mammalian AMBN (24). As a result, AMEL (both X and Y chromosomes) and AMBN show similar gene structure: both exon 1 and the 5′ end of exon 2 (12 or 15 bp) constitute 5′ UTR; exon 2 also codes 16 aa of SP and 2 aa of the N-terminal mature protein (25).
Figure 2.
Structures and intron phases. White, gray, and black regions represent UTR, SP, and the mature protein, respectively. (A) Exon–intron structures of EMP, casein, and salivary protein genes on 4q13 and AMEL on Xp22 are shown. The length of each exon (bp) is shown in the boxes. The phases of introns are indicated at the bottom of exon boundaries. CSN1S2L2 is a pseudogene, but the exon–intron boundaries were determined unambiguously by comparing bovine and porcine CSN1S2 sequences with the human genome sequence. (B) Exon–intron structures of SPARCL1 and dentine/bone ECM protein genes on 4q21 are shown. The structure of SPARC is the same as SPARCL1 except that exon 4 is missing in the 5′ region of SPARC. (C) The structure of the first four exons of the primordial EMP gene is shown. A protein kinase phosphorylates the Ser residue (P) in the SXE motif coded by the 3′ end of exon 3. The introns are exclusively phase-0.
The translation initiation site has not been determined experimentally for ENAM, but we found an authentic SP sequence of 16 aa consisting of a canonical tripartite structure: short, positively charged n-region, a central hydrophobic h-region, and a more polar c-region with a cleavage site (26, 27). Mouse Enam has an additional 5′-untranslated exon, which was detected within the human first intron but has not been identified in the transcripts (28). If these two exons corresponding to mouse exons 1 and 2 or exons 2 and 3 were inserted secondarily, the initial ENAM consisted of a single untranslated exon, and the entire 5′ structure was exactly the same as AMBN and AMEL (Fig. 2A).
Sequence Similarities Among EMPs.
Database searches and attempts by hybridization-based library screening failed to detect related sequences for any one of the three EMP genes (29). However, through analysis of the structure of these genes, we now can show that they all are in a single gene family. Large numbers of identical or similar residues in all these proteins were detected in the N-terminal sequences, at least up to the intermediate regions coded by exon 4 (see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). Seven residues completely conserved across known species were identified. Among them, the SXE motif (Ser-Xaa-Glu, where Xaa represents any residue) coded by the 3′ end of exon 3 is a putative phosphorylation site (refs. 30–32; Fig. 2C).
A phylogenetic tree was constructed (Fig. 3A). The highest sequence similarity was obtained between human AMBN and ENAM: 36.2% identity and 68.1% similarity for 47 aa. This suggests that the first gene duplication generated the AMEL and the AMBN/ENAM lineages. AMEL was translocated away at this time or later. We assume a primordial EMP gene that shows the following features: exon 1 and the 5′ end of exon 2 (12–15 bp) constitute 5′ UTR; exon 2 also codes SP (16 aa) and the N-terminal mature protein (2 aa); and exon 3 codes an SXE motif at the 3′ end. In addition, the introns are exclusively phase-0 (Fig. 2C).
Figure 3.
Phylogenetic trees for EMPs and casein-salivary proteins. MEGA II was used to construct gene trees based on the neighbor-joining method and to calculate substitution rates and bootstrap values. Both trees were drawn based on the timing of the inferred first gene duplication. (A) A phylogenetic tree for the three EMPs was constructed based on the amino acid sequences coded by exons 2–4, which are conserved among all EMP genes. The topology was the same when using sequences coded by exons 2 and 3. (B) A phylogenetic tree for Ca-sensitive CSN and three salivary protein genes was constructed based on the nucleotide sequences of the last exons (3′ UTR): exon 16 of CSN1S1, exon 18 of bovine CSN1S2 (human has two pseudogenes), exon 9 of CSN2, and exon 6 of STATH/HTNs. The same topology was obtained when using human CSN1S2L2. No ubiquitous repeat sequences were identified in these regions.
Structural Similarity Between EMP Genes and Other Genes in the Cluster.
In the casein and the salivary protein genes in the cluster, all but one intron in CSN1S1 are phase-0 (Fig. 2A). The last intron of CSN1S1 located within the termination codon is phase-2, which is conserved in cow (33). This phase-2 intron was probably generated by a mutation that abolished the termination codon and elongated the protein-coding region, because the last exon of CSN1S1 has sequence homology to the last exons of CSN1S2 and CSN2.
Each of CSN1S2, CSN2, STATH, and HTN1 has an SXE phosphorylation motif coded by the end of exon 3 (11, 34, 35). An SEE motif was found in CSN1S1 but is coded by the end of exon 4 (33). The phosphorylated residue in casein directly associates with a Ca ion and is responsible for the Ca-binding property (6). The SHE motif in HTN1 is shifted upstream by 1 aa, and the corresponding sequence has been substituted to SHA in HTN3 by a point mutation (36).
All genes for casein and salivary protein in the cluster show similar 5′ structure, although the C terminus of the SP spans to exon 3 in CSN10 and all salivary protein genes (Fig. 2A). CSN1S2 and CSN2 show especially high similarity to the primordial EMP gene: exon 1 and 5′-exon 2 (12 bp) constitute 5′ UTR, exon 2 encodes SP (15 aa) and the N-terminal mature protein (2 aa), and exon 3 codes an SXE motif at the 3′ end (8). In addition, their SPs show high sequence similarities to the EMPs in the n- and h-regions. Thus, we conclude that the ancestral Ca-sensitive CSN was derived from one of the three EMP genes, probably either from ENAM or AMBN inferred from their chromosomal locations.
Phylogenetic Analysis of Caseins, Statherin, and Histatins.
The Ca-sensitive casein genes and STATH/HTN share an additional feature; the last exon consists of only 3′ UTR except for CSN1S1 as described above (Fig. 2A). Comparison of the last exons revealed as high as 59.4% sequence homology for 251 bp between bovine CSN1S2 and human HTN1, although the protein-coding regions of these genes showed no sequence similarity except in the N-terminal half of the SPs. Lower sequence homology in coding regions rather than intergenic regions has been observed between STATH and HTN1 (11). A phylogenetic tree based on sequences of the last exon shows that STATH/HTN arose from CSN1S2 (Fig. 3B). This result is supported by a high bootstrap value and is consistent even if CSN sequences from other species were added. This is also supported by the order and polarity of these genes (Fig. 1). The close relationship between CSN1S2 and CSN2 corroborates the previous observation based on the 5′-flanking sequences (7).
Dentine and Bone Extracellular Matrix (ECM) Proteins.
In a 375-kb region on 4q21, ≈15 Mb distal to ENAM, there is the small integrin-binding ligand, n-linked glycoprotein (SIBLING) gene cluster for dentine and bone ECM proteins: DSPP, DMP1, IBSP, MEPE, and SPP1 (37). All these genes have the same transcriptional polarity and the structural features common to the primordial EMP genes (Fig. 2B). Their SPs show sequence similarity to the EMP genes. In addition, each of SPP1, DMP1, and IBSP codes an SXE motif and two following amino acids at the 3′ end of exon 3. Indeed, the same protein kinase phosphorylates Ser residues of casein and SPP1 (38). These data suggest that all these dentine/bone ECM protein genes and the primordial EMP gene arose from a common ancestor by gene duplication and comprise the SCPP gene family.
Discussion
Early in the vertebrate lineage, genes for transcription factors and signaling molecules were duplicated extensively (39). Epithelial–mesenchymal interactions facilitated these partly redundant tools for spatial and temporal regulations of organogenesis, which eventually produced various tissues such as teeth, fish scales, hair, feathers, mammary glands, and salivary glands (3, 40). Duplications of terminal differentiation genes also contributed to the generation of these tissues. By exploiting SP and SXE motifs, SCPPs regulate the Ca-phosphate concentration of the extracellular environment. Crystallization of the extracellular Ca-phosphate eventually created tooth, a mineralized hard tissue. By using common regulatory mechanisms, other SCPP genes could be expressed in ontogenetically similar but distinct tissues and contributed to the functions of mammary glands and salivary glands.
The introns for these SCPP genes are exclusively phase-0. Such nonrandom distribution of intron phases is mainly due to exon duplications (41). Exon duplication is common in Ca-sensitive CSN and contributed considerably to increasing the capacity of Ca-phosphate transport in milk (8). Our observations support this model; an ancient duplication of exon 3 coding an SXE motif generated the major phosphorylation sites. Exon duplications were also detected in exons 7–9 of human AMBN (42). Exon skipping is common in both casein and EMP genes (2, 43). This suggests that exon deletions also contributed to the current structure of these genes. Due to recurrent exon duplications and deletions, the number of exon is different among these genes (Fig. 2A).
CSN10 and four salivary genes in the distal complex show several similarities with the SCPP genes (Fig. 2A). Previously, amino acid sequence homology between CSN10 and fibrinogen γ-polypeptide (FGG) has been reported (44). However, the “homologous” regions are coded by six different exons in FGG, whereas these are solely coded by exon 4 in CSN10. We found that CSN10 has sequence similarity to the mouse mucin 10 gene (Muc10) in 5′ UTR (exon 1), SP, and portions of exon 3 (45). Muc10 is located in the distal salivary gene complex and has the same structural features with genes in the complex, whereas no MUC10 was identified in human. PROL1, -3, and -5 have sequence homologies to one another (46, 47) and show similar structure to MUC7 (Fig. 2A). We speculate that both Ca-sensitive and -insensitive CSNs and salivary protein genes in both proximal and distal complexes belong to the SCPP gene family.
Phase-0 introns are the most abundant in nature and account for 43% of all introns (41). However, genes exclusively consisting of phase-0 introns are uncommon. We searched intron phases of a number of other genes: whey proteins (α-lactalbumin, lysozyme, β-lactoglobulin, and lactotransferrin), salivary proteins (mucin 5B, proline-rich proteins located on 1q21 and 12p13, DMBT1, and cystatins), enamel proteins (tuftelin, matrix metalloproteinase 20, and kallikrein 4), two unknown genes within the same gene cluster, Ca-binding protein genes, and genes involved in bone formation, but none of these genes exclusively consisted of phase-0 introns. Furthermore, none of them showed the features common to the 5′ portion of the SCPP genes. Thus, no other SCPP genes were identified elsewhere in the genome outside of the cluster on chromosome 4 with the exception of AMELs.
It has been suggested that milk emerged as a cutaneous secretion that protects eggs from microorganisms and subsequently shifted to a directly nutritional product in mammal-like reptiles (48). Lactation enabled immature birth of altricial infants, and shortened the time during which mothers had to carry large fetuses. This led to the key characteristic adaptation of mammals (49). Casein made milk supersaturated with Ca-phosphate. Because available tools could produce carbohydrates, proteins, and lipids, the development of CSN may be critical to the mammalian adaptation. Indeed, CSN has been identified exclusively from mammals. The primordial Ca-sensitive CSN probably diverged either from ENAM or AMBN before the appearance of monotremes in the Jurassic (Fig. 4). Marsupials have two Ca-sensitive caseins, α and β (50, 51). Comparison of the 3′-UTR sequences revealed a close relationship between CSNα and CSN1S1, whereas CSNβ showed the highest homology to marsupial CSNα, not to eutherian CSNs. CSNβ may be specific to marsupials. Recurrent gene duplications of Ca-sensitive CSN imply the significance of the Ca-phosphate requirement for the postnatal growth in mammals.
Figure 4.
Divergence of the primordial EMP gene. The phylogeny of Ca-sensitive CSNs (milk), STATH/HTN (saliva), and EMP genes (enamel) was based on the two phylogenetic trees in Fig. 3. The primordial EMP gene appeared early in vertebrate evolution, perhaps in conodont (parenthesized). Immunohistochemical studies suggested the possibility that AMEL and ENAM emerged before the divergence of shark (parenthesized). AMBN diverged before caiman (reptiles). The primordial Ca-sensitive CSN diverged from one of the EMP genes, probably either from ENAM or AMBN before the emergence of monotremes. STATH then diverged from CSN1S2 before the divergence of rodents. Except for AMEL, genes are arranged in this order on chromosome 4.
As α-lactalbumin arose from lysozyme, gene duplications developed more adaptive milk (52). Likewise, STATH/HTN arose from CSN1S2 and developed more adaptive saliva. Statherin and histatin have been identified only from primates, suggesting the recent origin of these salivary proteins (11). We identified Stath but not Htn pseudogenes in the mouse genome at the locus corresponding to human. This indicates that STATH arose from CSN1S2 at least before the divergence of rodents 96 million years ago (53), and HTN subsequently descended (Fig. 4). A previous study estimated the STATH-HTN duplication date at 40–50 million years ago and the subsequent HTN1-HTN3 duplication date at 15–30 million years ago based on the nucleotide substitution rates (11). Rapid sequence divergence of HTN suggests the functional divergence from STATH. Indeed, HTN3 lost the SXE motif and cannot work as a Ca-binding phosphoprotein but developed novel antimicroorganism properties (10).
The transition from protochordates to vertebrates was associated with a shift from a passive to an active mode of predation (54). Mineralized feeding apparatus such as oropharyngeal denticles and teeth enabled food apprehension; hence this innovation was critical to the adaptive vertebrate lineage (55). The extant agnathans, hagfish and lamprey that diverged early from the stem craniate, have no mineralized exoskeleton. Conodonts (naked agnathan in the Late Cambrian; 510 million years ago) seem to have developed the earliest mineralized exoskeleton as an oral feeding apparatus. The conodont elements are almost entirely composed of enamel (lamellar crown), and dentine (basal body) underlays the crown in at least some species (56–58), whereas dentine is more common in dermal exoskeleton of ostracoderm (59). Thus, the earliest vertebrate history of the mineralized exoskeletal formation is controversial. We speculate that the primordial EMP is one of the most likely components used in the earliest enamel, conodont elements (Fig. 4). Replaceable sets of specialized teeth on the mandibular arch are confined to jawed vertebrates (60). Antibodies against mouse amelogenin and pig enamelin detected distinct shark (Heterodontus francisci) antigens (61). This suggests the possibility that AMEL and ENAM had diverged by the emergence of chondrichthyans in the Early Silurian (62). However, these EMP genes need to be isolated from chondrichthyans to identify this divergence (Fig. 4). The three EMP genes were fully established by the Carboniferous, as shown by the isolation of AMBN from the caiman (24).
The Ordovician agnathans such as Astraspidae developed dermal exoskeleton consisting of enamel, dentine, and bone (aspidin) (63). Today, these tissues interact in the development of tooth, each cooperatively leading to different tissue layers. Functionally and phylogenetically related genes need not retain close linkage alignment, but highly conserved linkage may be an indicator of current function or phylogenetic history. Novel functions can be developed relatively easily in ontogenetically similar tissues, because it requires small modifications to the original regulatory elements. Genes for enamel, dentine, and bone ECM protein genes arose from a common ancestor. We speculate that their tandem linkage arrangement facilitated a common spatial and temporal control of these paralogous genes in embryogenesis, that is, coordinately expressed in focal denticle germs (tubercles or scales on dermal exoskeleton). This supports a scenario in which bone initially appeared as attachment bone closely associated with dentine in scales, and later a continuous sheet such as a head shield emerged. The long-hypothesized relationship between teeth as feeding apparatus and ancestral exoskeletal scales is reinforced by the nature of these genes and their arrangement (55, 64, 65), and additional salivary function reinforces the idea of common usage of related genes for feeding function in vertebrate evolution. Lactation function came later but is evolutionarily related.
Interestingly, enamel and dentine are produced by different, interacting, and juxtaposed cell types (3). The colocalization of enamel and dentine ECM protein genes may also be related to the dental patterning mechanism by which dental epithelium and mesenchyme communicate, but nothing is as yet known about this aspect of the regulation of these closely syntenic genes expressed in a coordinated way in the two intercommunicating tissues. Similarly, it is not known whether variation in these genes is related to variation in dental or related patterning (L. J. Hlusko, J. Rogers, M. C. Mahaney, K.K., and K.M.W., unpublished data).
SPARC is located on 5q31.3-q32 and encodes a major noncollagenous protein of bone matrix (66, 67). We found that SPARCL1 (SPARC-related gene) is located 79 kb proximal to the dentine and bone ECM protein gene cluster (68). In addition, exons 1 and 2 of both SPARC and SPARCL1 show similar structural features to the other SCPP genes (Fig. 2B). Previously, nucleotide sequence homology was observed between exon 2 of AMEL and SPARC/SPARCL1 (29). However, this study did not consider the limits of sequence variation in the SP and a subsequent amino acid residue (26). Although SPARCL1 seems to be of recent origin, SPARC is identified in nematode and fruit fly (69). By exon shuffling, SPARC obtained a Follistatin-like domain (70), which is not seen in the other dental (enamel/dentine) and bone ECM protein genes in the cluster. SPARC also encodes a Ca-binding protein but does not have an SXE motif. Nevertheless, SPARC is the only gene that has been suggested as having a possible evolutionary relationship to the ECM protein genes in the cluster and that has been identified in protostome. The 5′ exons of SPARC may have originated the primordial dental/bone ECM protein gene.
We have hypothesized that the dental/bone ECM protein genes on human 4q13-q21 were created from a single ancestor by gene duplication. The emergence of enamel, dentine, and bone ECM protein genes would suggest when each hard tissue appeared. The sequence divergence of the duplicated genes may reflect complexity of the mineralized tissues and the complex nature of their appearance in various vertebrate lineages. Thus, analysis of the chromosomal loci containing these genes of agnathans and primitive gnathostomes may delineate early events in mineralized skeletal formation, which is a major characteristic of vertebrates.
Supplementary Material
Acknowledgments
We thank Drs. A. Walker, W. Miller, A. Buchanan, L. J. Hlusko, J. Rogers, and M. C. Mahaney and Mr. S. Sholtis for critical discussions and collaboration. This work is supported by National Science Foundation Grant SBER 9804907 and Pennsylvania State University.
Abbreviations
- EMP
enamel matrix protein
- SCPP
secretory Ca-binding phosphoproteins
- SP
signal peptide
- ECM
extracellular matrix
References
- 1.Ohno S. Evolution by Gene Duplication. New York: Springer; 1970. [Google Scholar]
- 2.Zeichner-David M, Diekwisch T, Fincham A, Lau E, MacDougall M, Moradian-Oldak J, Simmer J, Snead M, Slavkin H C. Int J Dev Biol. 1995;39:69–92. [PubMed] [Google Scholar]
- 3.Thesleff I, Sharpe P. Mech Dev. 1997;67:111–123. doi: 10.1016/s0925-4773(97)00115-9. [DOI] [PubMed] [Google Scholar]
- 4.Fincham A G, Moradian-Oldak J, Simmer J P. J Struct Biol. 1999;126:270–299. doi: 10.1006/jsbi.1999.4130. [DOI] [PubMed] [Google Scholar]
- 5.Ginger M R, Grigor M R. Comp Biochem Physiol B Biochem Mol Biol. 1999;124:133–145. doi: 10.1016/s0305-0491(99)00110-8. [DOI] [PubMed] [Google Scholar]
- 6.Farrell H M, Jr, Thompson M P. In: Calcium-Binding Proteins. Thompson M P, editor. II. Boca Raton, FL: CRC; 1988. pp. 117–137. [Google Scholar]
- 7.Yu-Lee L Y, Richter-Mann L, Couch C H, Stewart A F, Mackinlay A G, Rosen J M. Nucleic Acids Res. 1986;14:1883–1902. doi: 10.1093/nar/14.4.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosen J M. In: The Mammary Gland. Neville M C, Daniel C W, editors. New York: Plenum; 1987. pp. 301–322. [Google Scholar]
- 9.Hay D I, Moreno E C. In: Human Saliva: Clinical Chemistry and Microbiology. Tenovuo J O, editor. I. Boca Raton, FL: CRC; 1989. pp. 131–150. [Google Scholar]
- 10.Amerongen A V, Veerman E C. Oral Dis. 2002;8:12–22. doi: 10.1034/j.1601-0825.2002.1o816.x. [DOI] [PubMed] [Google Scholar]
- 11.Sabatini L M, Ota T, Azen E A. Mol Biol Evol. 1993;10:497–511. doi: 10.1093/oxfordjournals.molbev.a040022. [DOI] [PubMed] [Google Scholar]
- 12.Sabatini L M, Azen E A. Biochem Biophys Res Commun. 1989;160:495–502. doi: 10.1016/0006-291x(89)92460-1. [DOI] [PubMed] [Google Scholar]
- 13.vanderSpek J C, Wyandt H E, Skare J C, Milunsky A, Oppenheim F G, Troxler R F. Am J Hum Genet. 1989;45:381–387. [PMC free article] [PubMed] [Google Scholar]
- 14.Lau E C, Mohandas T K, Shapiro L J, Slavkin H C, Snead M L. Genomics. 1989;4:162–168. doi: 10.1016/0888-7543(89)90295-4. [DOI] [PubMed] [Google Scholar]
- 15.Rijnkels M, Meershoek E, de Boer H A, Pieper F R. Mamm Genome. 1997;8:285–286. doi: 10.1007/s003359900413. [DOI] [PubMed] [Google Scholar]
- 16.Dong J, Gu T T, Simmons D, MacDougall M. Eur J Oral Sci. 2000;108:353–358. doi: 10.1034/j.1600-0722.2000.108005353.x. [DOI] [PubMed] [Google Scholar]
- 17.Sonnhammer E L, Durbin R. Gene. 1995;167:GC1–GC10. doi: 10.1016/0378-1119(95)00714-8. [DOI] [PubMed] [Google Scholar]
- 18.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar S, Tamura K, Jakobsen I B, Nei M. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Int J Neural Syst. 1997;8:581–599. doi: 10.1142/s0129065797000537. [DOI] [PubMed] [Google Scholar]
- 21.Nakai K, Horton P. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 22.Rijnkels M, Kooiman P M, de Boer H A, Pieper F R. Mamm Genome. 1997;8:148–152. doi: 10.1007/s003359900377. [DOI] [PubMed] [Google Scholar]
- 23.Lander E S, Linton L M, Birren B, Nusbaum C, Zody M C, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 24.Shintani S, Kobata M, Toyosawa S, Fujiwara T, Sato A, Ooshima T. Gene. 2002;283:245–254. doi: 10.1016/s0378-1119(01)00848-4. [DOI] [PubMed] [Google Scholar]
- 25.Salido E C, Yen P H, Koprivnikar K, Yu L C, Shapiro L J. Am J Hum Genet. 1992;50:303–316. [PMC free article] [PubMed] [Google Scholar]
- 26.von Heijne G. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 27.Hu C C, Hart T C, Dupont B R, Chen J J, Sun X, Qian Q, Zhang C H, Jiang H, Mattern V L, Wright J T, Simmer J P. J Dent Res. 2000;79:912–919. doi: 10.1177/00220345000790040501. [DOI] [PubMed] [Google Scholar]
- 28.Hu J C, Zhang C H, Yang Y, Karrman-Mardh C, Forsman-Semb K, Simmer J P. J Dent Res. 2001;80:898–902. doi: 10.1177/00220345010800031001. [DOI] [PubMed] [Google Scholar]
- 29.Delgado S, Casane D, Bonnaud L, Laurin M, Sire J Y, Girondot M. Mol Biol Evol. 2001;18:2146–2153. doi: 10.1093/oxfordjournals.molbev.a003760. [DOI] [PubMed] [Google Scholar]
- 30.Fukae M, Tanabe T, Murakami C, Dohi N, Uchida T, Shimizu M. Adv Dent Res. 1996;10:111–118. doi: 10.1177/08959374960100020201. [DOI] [PubMed] [Google Scholar]
- 31.Salih E, Huang J C, Strawich E, Gouverneur M, Glimcher M J. Connect Tissue Res. 1998;38:225–235. doi: 10.3109/03008209809017041. [DOI] [PubMed] [Google Scholar]
- 32.Salih E, Huang J C, Strawich E, Gouverneur M, Glimcher M J. Connect Tissue Res. 1998;38:241–246. doi: 10.3109/03008209809017041. [DOI] [PubMed] [Google Scholar]
- 33.Koczan D, Hobom G, Seyfert H M. Nucleic Acids Res. 1991;19:5591–5596. doi: 10.1093/nar/19.20.5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones W K, Yu-Lee L Y, Clift S M, Brown T L, Rosen J M. J Biol Chem. 1985;260:7042–7050. [PubMed] [Google Scholar]
- 35.Groenen M A, Dijkhof R J, Verstege A J, van der Poel J J. Gene. 1993;123:187–193. doi: 10.1016/0378-1119(93)90123-k. [DOI] [PubMed] [Google Scholar]
- 36.Sabatini L M, Warner T F, Saitoh E, Azen E A. J Dent Res. 1989;68:1138–1145. doi: 10.1177/00220345890680070101. [DOI] [PubMed] [Google Scholar]
- 37.Fisher L W, Torchia D A, Fohr B, Young M F, Fedarko N S. Biochem Biophys Res Commun. 2001;280:460–465. doi: 10.1006/bbrc.2000.4146. [DOI] [PubMed] [Google Scholar]
- 38.Lasa M, Chang P L, Prince C W, Pinna L A. Biochem Biophys Res Commun. 1997;240:602–605. doi: 10.1006/bbrc.1997.7702. [DOI] [PubMed] [Google Scholar]
- 39.Holland P W. Semin Cell Dev Biol. 1999;10:541–547. doi: 10.1006/scdb.1999.0335. [DOI] [PubMed] [Google Scholar]
- 40.Krejsa R J. In: Hyman's Comparative Vertebrate Anatomy. Wake M H, editor. Chicago: Univ. of Chicago Press; 1979. pp. 112–191. [Google Scholar]
- 41.Fedorov A, Fedorova L, Starshenko V, Filatov V, Grigor'ev E. J Mol Evol. 1998;46:263–271. doi: 10.1007/pl00006302. [DOI] [PubMed] [Google Scholar]
- 42.Toyosawa S, Fujiwara T, Ooshima T, Shintani S, Sato A, Ogawa Y, Sobue S, Ijuhin N. Gene. 2000;256:1–11. doi: 10.1016/s0378-1119(00)00379-6. [DOI] [PubMed] [Google Scholar]
- 43.Menon R S, Chang Y F, Jeffers K F, Ham R G. Genomics. 1992;12:13–17. doi: 10.1016/0888-7543(92)90400-m. [DOI] [PubMed] [Google Scholar]
- 44.Jolles P, Loucheux-Lefebvre M H, Henschen A. J Mol Evol. 1978;11:271–277. doi: 10.1007/BF01733837. [DOI] [PubMed] [Google Scholar]
- 45.Denny P C, Mirels L, Denny P A. Glycobiology. 1996;6:43–50. doi: 10.1093/glycob/6.1.43. [DOI] [PubMed] [Google Scholar]
- 46.Isemura S, Saitoh E. J Biochem (Tokyo) 1994;115:1101–1106. doi: 10.1093/oxfordjournals.jbchem.a124464. [DOI] [PubMed] [Google Scholar]
- 47.Dickinson D P, Thiesse M. Curr Eye Res. 1996;15:377–386. doi: 10.3109/02713689608995828. [DOI] [PubMed] [Google Scholar]
- 48.Blackburn D G. Mamm Rev. 1989;19:1–26. [Google Scholar]
- 49.Pond C M. Evolution (Lawrence, Kans) 1976;31:177–199. [Google Scholar]
- 50.Collet C, Joseph R, Nicholas K. J Mol Endocrinol. 1992;8:13–20. doi: 10.1677/jme.0.0080013. [DOI] [PubMed] [Google Scholar]
- 51.Ginger M R, Piotte C P, Otter D E, Grigor M R. Biochim Biophys Acta. 1999;1427:92–104. doi: 10.1016/s0304-4165(99)00008-2. [DOI] [PubMed] [Google Scholar]
- 52.Brew K, Vanaman T C, Hill R L. J Biol Chem. 1967;242:3747–3749. [PubMed] [Google Scholar]
- 53.Nei M, Xu P, Glazko G. Proc Natl Acad Sci USA. 2001;98:2497–2502. doi: 10.1073/pnas.051611498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gans C, Northcutt R G. Science. 1983;220:268–274. doi: 10.1126/science.220.4594.268. [DOI] [PubMed] [Google Scholar]
- 55.Smith M M, Coates M I. In: Development, Function, and Evolution of Teeth. Teaford M F, Smith M M, Ferguson M W J, editors. Cambridge, U.K.: Cambridge Univ. Press; 2000. pp. 133–151. [Google Scholar]
- 56.Sansom I J. Zool J Linn Soc. 1996;118:47–57. [Google Scholar]
- 57.Smith M M, Sansom I J, Smith M P. Modern Geology. 1996;20:303–319. [Google Scholar]
- 58.Donoghue P C J, Aldridge R J. In: Major Events in Vertebrate Evolution. Ahlberg P E, editor. London: Taylor & Francis; 2001. pp. 85–105. [Google Scholar]
- 59.Forey P, Janvier P. Nature. 1993;361:129–134. [Google Scholar]
- 60.Smith M M, Coates M I. In: Major Events in Early Vertebrate Evolution. Ahlberg P E, editor. London: Taylor & Francis; 2001. pp. 223–240. [Google Scholar]
- 61.Satchell P G, Anderton X, Ryu O H, Luan X, Ortega A J, Opamen R, Berman B J, Witherspoon D E, Gutmann J L, Yamane A, et al. J Exp Zool. 2002;294:91–106. doi: 10.1002/jez.10148. [DOI] [PubMed] [Google Scholar]
- 62.Sansom I J, Smith M M, Smith M P. In: Major Events in Early Vertebrate Evolution. Ahlberg P E, editor. London: Taylor & Francis; 2001. pp. 156–171. [Google Scholar]
- 63.Smith M M, Hall B K. Biol Rev. 1990;65:277–373. doi: 10.1111/j.1469-185x.1990.tb01427.x. [DOI] [PubMed] [Google Scholar]
- 64.Ørvig T. In: Problems in Vertebrate Evolution. Andrews S M, Miles R S, Walker A D, editors. London: Academic; 1977. pp. 53–75. [Google Scholar]
- 65.Schaeffer B. In: Problems in Vertebrate Evolution. Andrews S M, Miles R S, Walker A D, editors. London: Academic; 1977. pp. 25–52. [Google Scholar]
- 66.Termine J D, Kleinman H K, Whitson S W, Conn K M, McGarvey M L, Martin G R. Cell. 1981;26:99–105. doi: 10.1016/0092-8674(81)90037-4. [DOI] [PubMed] [Google Scholar]
- 67.Yan Q, Sage E H. J Histochem Cytochem. 1999;47:1495–1506. doi: 10.1177/002215549904701201. [DOI] [PubMed] [Google Scholar]
- 68.Girard J P, Springer T A. Immunity. 1995;2:113–123. doi: 10.1016/1074-7613(95)90083-7. [DOI] [PubMed] [Google Scholar]
- 69.Martinek N, Zou R, Berg M, Sodek J, Ringuette M. Dev Genes Evol. 2002;212:124–133. doi: 10.1007/s00427-002-0220-9. [DOI] [PubMed] [Google Scholar]
- 70.Patty L. In: Human Genome Evolution. Jackson M, Strachan T, Dover G, editors. Oxford: BIOS Scientific; 1996. pp. 35–71. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.