Abstract
Transcription of the tumor necrosis factor (TNF) gene is rapidly and transiently induced by lipopolysaccharide in cells of monocyte/macrophage lineage. Previous studies have suggested that in the mouse, multiple NF-κB/Rel-binding sites contribute to the TNF transcriptional response to LPS. But the role of these regulatory elements in transcriptional activation of the TNF-α gene in human monocytes remains unclear. Previously, a transcription factor, termed lipopolysaccharide-induced TNF-α factor (LITAF), was found to regulate TNF-α gene expression. However, the specific protein domain(s) of human (h)LITAF that interact with the hTNF-α promoter had not been identified. In this study, we identify by footprinting a sequence motif, CTCCC (−515 to −511), within the TNF-α promoter that binds to hLITAF. We also identify the region of hLITAF (amino acids 165–180) that was named peptide B and specifically mediates binding to the hTNF-α promoter. When THP-1 cells were stimulated with this peptide B, it was sufficient to induce TNF-α secretion. Induction of TNF-α transcription by LPS or peptide B depended on the presence of the −515 to −511 promoter region, which was found to be essential for hLITAF binding. Together, these findings help to clarify the mechanism of hLITAF/hTNF-α interaction and the manner by which hLITAF contributes to hTNF-α regulation in an attempt to design new pharmacological interventions to address TNF-related diseases.
Keywords: lipopolysaccharide-induced tumor necrosis factor-α factor‖electrophoretic mobility-shift assay‖DNA-binding site
Tumor necrosis factor (TNF)-α is a pleiotropic cytokine that is mainly produced by cells of the monocyte/macrophage lineage. TNF-α was originally identified as an endogenous factor, induced in response to inflammatory stimuli. Many studies have revealed that TNF-α exhibits both beneficial and pathologic effects, a feature that requires rigorous control of its expression (1–5) and that highlights its importance. The regulation of TNF-α gene expression in cells of the monocytic lineage is stimulus-dependent and quite complex, involving controls at both transcriptional (6–8) and posttranscriptional levels (9). Many studies of the transcriptional regulation of TNF-α have focused on the investigation of transcription factors that bind to the responsive element sites within the TNF-α promoter, such as NF-κB (10), Ets (11), NF-AT (12), activating protein 1 (AP-1; ref. 13), cAMP response element-binding protein (14), signal transducers and activators of transcription (STAT1, ref. 15), and lipopolysaccharide (LPS)-induced TNF-α factor (LITAF, ref. 16). However, the relative contributions of these various regulatory elements in transcriptional activation of the TNF-α gene in human monocytes are poorly understood.
LPS, extracted from the outer membrane of Gram-negative bacteria, has been identified as a principal endotoxic component (17). LPS is a potent stimulator of monocytes and macrophages, inducing production and secretion of TNF-α and other inflammatory mediators (18). The effects of LPS on transcription factor activity and expression have been widely investigated. Previous studies suggested that in vivo, LPS up-regulates the DNA-binding activity of inducible transcription factors NF-κB, AP-1, and cAMP response element-binding protein in a time-dependent manner, but it down-regulates the DNA-binding activity of constitutive transcription factors specificity protein 1 (SP-1) and AP-2 (19, 20). NF-κB was found to play a major role in the up-regulation of TNF-α transcription in response to LPS in human monocytic THP-1 leukemia cells (21), because mutation or deletion of the NF-κB binding motif within the hTNF-α promoter abolished the response to LPS in reporter gene-transfected cells (22). However, inhibition of NF-κB binding did not completely abolish LPS-induced TNF-α expression (23). Thus, the role of NF-κB in the transcriptional activation of the human (h)TNF-α gene in human monocytes in response to LPS has remained controversial. It was then hypothesized that there may be another transcription factor, acting either independently or in concert with NF-κB, in the activation of hTNF-α transcription (24, 25).
We recently identified such a factor, termed hLITAF (16, 26), which mediates hTNF-α transcription. Inhibition of hLITAF mRNA expression in THP-1 cells resulted in a reduction of hTNF-α transcripts. We also found that high levels of hLITAF mRNA are expressed predominantly in the placenta, peripheral blood leukocytes, lymph nodes, and spleen. These findings suggested that both NF-κB and LITAF contribute to the activation of the hTNF-α gene in response to LPS. In the present study, we have identified the site within the hTNF-α promoter that is essential for hLITAF binding. Using DNase I footprint analysis and electrophoretic mobility-shift assay (EMSA), we have also identified the region within hLITAF that specifically mediates DNA binding. Furthermore, a synthetic peptide corresponding to amino acids 165–180 of hLITAF was found to be sufficient to bind to and activate the TNF-α promoter. Together, these findings help to clarify the role of hLITAF in the regulation of hTNF-α transcription.
Materials and Methods
Bacterial Strains.
All cloning constructions including mutagenesis were performed using Escherichia coli strain DH5α (Invitrogen). All clones involved in purification of GST fusion protein were performed in strain BL21 (Amersham Biosciences).
Cell Culture.
THP-1 monocytic cells were grown in RPMI medium 1640 with 10% FCS and maintained in an atmosphere of 5% CO2 at 37°C.
Plasmid Constructs.
The series of hLITAF DNA fragments was generated by PCR with the following primer pairs and subcloned into the pGEX4T-1 vector (Amersham Biosciences) (Fig. 1a): GST-hLITAF amino acids 1–75, 5′-CGGGATCCATGTCGGTTCCAGGACCT-3′ and 5′-cggaattcggtaattggattgttatt-3′; GST-hLITAF amino acids 1–151, 5′-CGGGATCCATGTCGTTCCAGGACCT-3′ and 5′-cggaattccagttgggacagtaatgg-3′; GST-hLITAF amino acids 76–151, 5′-CGGGATCCGTGCAGACGGTCTACGTG-3′ and 5′-cggaattccagttgggacagtaatgg-3′; GST-hLITAF amino acids 1–228, 5′-CGGGATCCATGTCGGTTCCAGGACCT-3′ and 5′-cgggatcctcagggtctcagggaggc-3′; GST-hLITAF amino acids 76–228, 5′-CGGGATCCGTGCAGACGGTCTACGTG-3′ and 5′-cgggatcctcagggtctcagggaggc-3′; GST-hLITAF amino acids 152–228, 5′-CGGGATCCCAGAGCTCTCCTGGGCAC-3′ and 5′-cgggatcctcagggtctcagggaggc-3′. In GST-hLITAF amino acids 152–228Δ180–195, the first in-frame mutant hLITAF DNA fragment was generated by PCR with primer pair 5′-CGGGATCCCAGAGCTCTCCTGGGCAC-3′ (coordinates 687–704 bp with BamHI) and 5′-gtggaaaggacttcctgc-3′ (coordinates 770–753 bp). The second hLITAF DNA fragment was generated by PCR with primer pair 5′-GCAGGAAGTCCTTTCCACCCTCTCCAGGGGGCCCAC-3′ (coordinates 753–836 bp) and 5′-cggaattctcagggtctcagggaggc-3′ (coordinates 983–966 bp with EcoRI). Both the first and second DNA fragments were purified and diluted as template to 1 ng per reaction and amplified by PCR with primer pair 5′-CGGGATCCCAGAGCTCTCCTGGGCAC-3′ (coordinates 687–704 bp with BamHI) and 5′-cggaattctcagggtctcagggaggc-3′ (coordinates 983–966 bp with EcoRI). Finally, the in-frame hLITAF mutant DNA fragment was inserted into the pGEX4T-1 vector. For GST-hLITAF amino acids 152–228Δ165–180, the first in-frame mutant hLITAF DNA fragment was generated by PCR with primer pair 5′-CGGGATCCCAGAGCTCTCCTGGGCAC-3′ (coordinates 687–704 bp with BamHI) and 5′-tccaccaggcgtgaagctggatgagagtcctacaaacgcttg-3′ (coordinates 797–708 bp). The second hLITAF DNA fragment was generated by PCR with primer pair 5′-TTCACGCCTGGTGGAGGT-3′ (coordinates 783–800 bp) and 5′-cggaattctcagggtctcagggaggc-3′ (coordinates 983–966 bp with EcoRI). Both first and second DNA fragments above were purified and diluted as template to 1 ng per reaction and amplified by PCR with 5′ and 3′ primers, 5′-CGGGATCCCAGAGCTCTCCTGGGCAC-3′ (coordinates 687–704 bp with BamHI) and 5′-cggaattctcagggtctcagggaggc-3′ (coordinates 983–966 bp with EcoRI). Finally, the in-frame mutant DNA fragment was inserted into the vector.
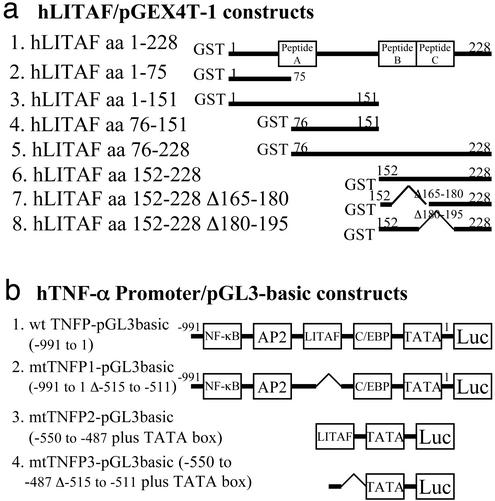
Figure 1.
Diagram of plasmid constructs used in this study for production of GST fusion protein (a) and for luciferase reporter assay (b). Three peptides, A, B, and C, were synthesized and indicated, respectively, by a box at the region of hLITAF (a). The major potential binding site for transcription factors (33) is indicated, respectively, by a box on the hTNF-α promoter DNA (b).
The following series of hTNF-α promoter DNA fragments was subcloned (Fig. 1b) into the pGL3-Basic vector (Amersham Biosciences), a promoterless and enhancerless luciferase reporter gene. The wild-type TNF promoter (wt TNFP; −991 to 1) was generated by PCR with primer pair 5′-AGCTCCTGGGAGATATGGCCAC-3′ and 5′-gggtgtgccaacaactgccttt-3′. The mutant TNFP1 (mtTNFP1; −991 to 1Δ−515 to −511), the first in-frame mutant hTNF-α promoter, was generated by PCR with primer pair 5′-AGCTCCTGGGAGATATGGCCAC-3′ and 5′-tgcgaaggagctgggggctt. The second mutant DNA was generated by PCR with primer pair 5′-CCTTCGCAGGGACCCAAACACAGGCCTCA-3′ and 5′-gggtgtgccaacaactgccttt-3′. Both first and second DNA fragments above were purified and diluted as template to 1 ng per reaction and finally amplified by PCR with primer pair 5′-AGCTCCTGGGAGATATGGCCAC-3′ and 5′-gggtgtgccaacaactgccttt-3′. mtTNFP2 (−550 to −487 plus TATA box) was generated by annealing with primer pair 5′-AGGCCTCAAGCCTGCCACCAAGCCCCCAGCTCCTTCTCCCCGCAGGGACCCAAACACAGGCCTCATATAAAGGCAGTTGTTGGCACACCC-3′ and 5′-gggtgtgccaacaactgcctttatatgaggcctgtgtttgggtccctgcggggagaaggagctgggggcttggtggcaggc ttgaggcct-3′. mtTNFP3 (−550 to −487Δ−515 to −511 plus TATA box) was generated by annealing with primer pair 5′-AGGCCTCAAGCCTGCCACCAAGCCCCCAGCTCCTTCGCAGGGACCCAAACACAGGCCTCATATAAAGGCAGTTGTTGGCACACCC-3′ and 5′-gggtgtgccaacaactgcctttatatgaggcctgtgtttgggtccctgcgaaggagctgggggcttggtggcaggcttgagg cct-3′.
Purification of GST-hLITAF Fusion Protein.
GST-hLITAF recombinant plasmids were transformed into competent BL21 cells. LBA medium (Lennox broth + ampicillin; 2 ml) was inoculated with a single colony of the appropriate transformant for culture at 37°C overnight. This 2-ml culture was then transferred to 100 ml of 2× YT broth plus ampicillin (16 g of tryptone + 10 g of yeast extract + 5 g of NaCl/liter; 100 μg/ml) and grown at 30°C with shaking until the absorbance at 600 nm reached 0.6, at which time isopropyl β-d-thiogalactoside (IPTG) was added to a final concentration of 0.1 mM. The culture was incubated for an additional 2–6 h, then subjected to centrifugation at 3,000 × g for 10 min at 4°C. The cells were washed with PBS and completely suspended in 2 ml of ice-cold PBS, then lysed by brief sonication for 10 s (output 20, Branson Sonifier 450), then centrifuged twice at 5,000 × g for 10 min at 4°C. The supernatant was transferred to a fresh container, to which was added 100 μl of glutathione Sepharose 4B beads (Amersham Biosciences), and the mixture was rocked for 30 min at 4°C, then washed three times with PBS. Protein samples were run in SDS/PAGE (10% gel).
DNase I Footprinting.
The protein-DNA binding site was analyzed by the DNase I footprinting method (27) with some modifications. Two oligonucleotides were synthesized. The first one was designed as a template, with a HindIII site at the 5′ end. For the reverse orientation, nucleotides from −487 to −550 bp in the hTNF-α promoter were represented (5′-TGAGGCCTGTGTTTGGGTCCCTGCGGGGAGAAGGAGCTGGGGGCTTGGTGGCAGGCTTGAGGCCT-3′). The second one was designed as a primer from −550 to −535 bp in the hTNF-α promoter, 5′-aggcctcaagcctgcc-3′. Template (0.5 μg) and 0.1 μg of primer were mixed and incubated at 37°C for 1 h, then 2 μl of 2.5 mM 4dNTP mix, 5 μl of 10× Klenow fragment buffer, 5 units of Klenow fragment (Invitrogen), and water to 50 μl were added and incubated at 37°C for 30 min. The DNA was purified, then precipitated with ethanol. After centrifugation, the DNA pellet was suspended in 10 μl of TE (10 mM Tris/1 mM EDTA, pH 7.5) buffer. DNA (0.5 μg) was labeled with [γ-32P]ATP by using T4 polynucleotide kinase (Promega) and then digested by HindIII as described (28). Labeled DNA was purified by using a Sephadex G-25 column (Roche Diagnostics) and precipitated with ethanol. After centrifugation, the DNA pellet was suspended in 10 μl of water. The [γ-32P]ATP-labeled DNA was then mixed with 25 μl of binding buffer (Promega), 0.1 μg of GST-hLITAF fusion protein (GST fusion protein alone as control), and nuclease-free water (Promega) to 50 μl and incubated on ice for 30 min, to which 50 μl of prewarmed Ca2/Mg2 solution at room temperature was added and incubated for 1 min; 3 μl of DNase I (Promega) was then added, mixed gently, incubated for an additional 5 min, and followed by reaction termination. The reaction mixture was treated with phenol and precipitated with ethanol. After centrifugation, the DNA pellet was suspended in 5 μl of TE buffer. The sample was applied to a 6% polyacrylamide sequencing gel (Invitrogen).
EMSA.
A reaction mixture containing 0.1 μg of GST-hLITAF fusion protein, 1 μl of radiolabeled (1 × 105 cpm/μl) double-stranded DNA oligonucleotide (2 pmol), 3 μg of poly(deoxyinosinic acid⋅deoxycytidylic acid) [poly(dI⋅dC), Sigma], 5 μg of BSA, 4 μl of gel shift binding 5× buffer (Promega), and nuclease-free water to 20 μl was incubated at room temperature for 30 min before electrophoresis on nondenaturing 6% polyacrylamide gels in Tris-borate-EDTA buffer (90 mM Tris-borate/2 mM EDTA Hepes, pH 8.0).
Peptides.
The synthetic peptides that follow were supplied by Lofstrand Laboratories (Gaithersburg, MD). Peptide A consisted of the sequence SYYTQPAPIPNNNPITVQTVY from the hLITAF amino acids 60–80, peptide B consisted of the sequence SQTWREPGAAGSPFHL from amino acids 165–180, and peptide C consisted of the sequence LSSSFTPGGGSALVVS from amino acids 180–195 (Figs. 1, 4, and 5). Hemagglutinin (HA) antigenic peptide served as control peptide and consisted of the sequence YPYDVPDYASL (Alpha Diagnostic, San Antonio, TX). All peptides were solubilized in DMSO and delivered into THP-1 cells by Chariot kit (Active Motif, Carlsbad, CA) for reporter assays as described in refs. 29 and 30.
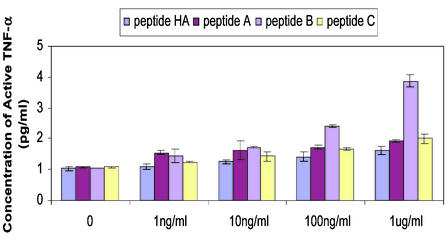
Figure 4.
TNF-α secretion upon stimulation. Production of TNF-α in THP-1 cells after transfection of peptide A, B, C, or HA with Chariot was as described (29, 30) for 24 h. The concentration of active TNF-α was induced by various concentrations of peptide, measured by duplicate ELISAs at the same condition, and graphed.
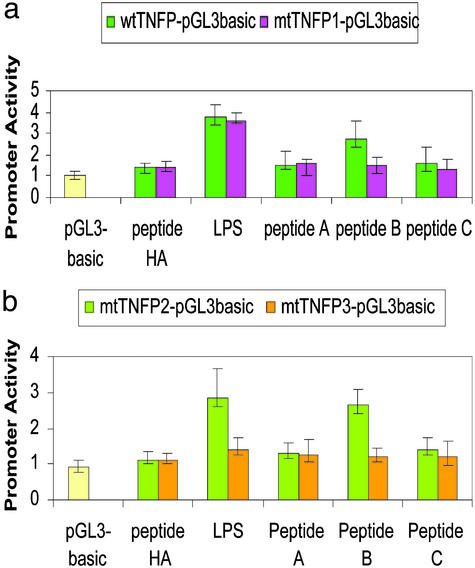
Figure 5.
Transcriptional activity of a series of deletion constructs of TNF-α promoter DNA. (a) The activity was measured due to the promoter wtTNFP (−991 to 1) or mtTNFP1 (−991 to 1Δ−515 to −511). pGL3-basic transfected cell was used as unstimulated control. After transfection of DNAs for 3 h, the cells were washed with PBS twice and stimulated with 100 ng/ml LPS (E. coli) or transfected with Chariot/peptide complex of 1 μg/ml peptide A, B, C, or HA. Triplicate assays were performed. Values are normalized by β-galactosidase production and graphed. (b) The activity was measured due to the promoter mtTNFP2 (−550 to −487 plus TATA box) or mtTNFP3 (−550 to −487Δ−515 to −511 plus TATA box). LPS or peptides as stimuli and controls were used at the same condition as described above.
ELISA.
THP-1 cells were induced to maturation by addition of 200 mM phorbol 12-myristate 13-acetate (Sigma) and incubated at 37°C, 5% CO2, for 20 h, then washed with PBS twice and stimulated (delivered) with Chariot/peptide complex of various concentrations of peptide A, B, C, or HA in a 96-well plate at 2 × 104 cells per well as indicated in the text. After 24 h of incubation at 37°C, 5% CO2, culture supernatants were harvested and centrifuged at 1,500 × g to remove cell debris, then TNF-α was measured by ELISA (Abraxis, Warminster, PA) and quantified on a model 680 Microplate Reader (Bio-Rad).
Transient Transfection and Luciferase Assay.
THP-1 cells (5 × 106 per well) were induced to maturation by addition of 200 mM phorbol 12-myristate 13-acetate (Sigma) and incubated at 37°C, 5% CO2, for 20 h, washed with PBS twice, cotransfected with 1 μg of DNA by using FuGENE 6 (Roche Molecular Biochemicals) for 3 h, washed with PBS, then individually transfected with Chariot/peptide complex of 1 μg/ml peptide A, B, C, or HA or stimulated with 100 ng/ml LPS (E. coli) for 3 h, washed with PBS, and incubated at 37°C, 5% CO2, overnight. The β-galactosidase gene was included in all transfections. The cells were harvested and lysed ≈12 h after transfection. Luciferase activity in the lysates was measured by using a commercial kit (Luciferase Reporter Assay System, Promega) and normalized by β-galactosidase assay in the same lysates as described (31).
Results
Determination of hLITAF-Binding Site CTCCC in TNF-α Promoter by Footprint.
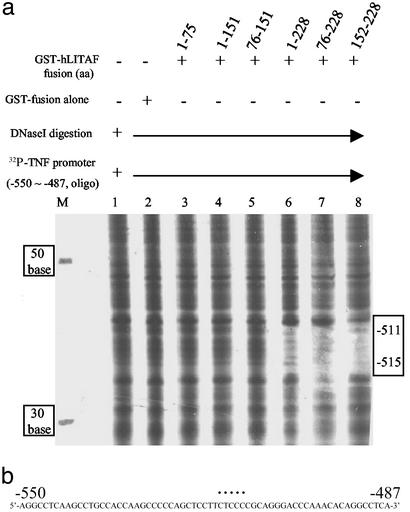
The site within the hTNF-α promoter that binds LITAF was determined by DNase I footprinting. In this experiment, six wild-type or mutant GST-hLITAF fusion proteins were used, as described in Fig. 1a. The [32P]ATP-labeled hTNF-α promoter DNA fragment (−550 to −487) was used as the probe. To label this probe only at one end from its 5′ flank, the DNA was particularly designed to contain a HindIII site at its 3′ flank. HindIII digestion thus removed the [32P]ATP label at the 3′ end, leaving a probe labeled only at its 5′ end. The DNA was then degraded base by base from its 3′ end by DNase I digestion but protected from degradation by its protein–DNA interaction (gap), and the surviving fragments were detected by electrophoresis and autoradiography. As described in Fig. 1a, the clone GST-hLITAF amino acids 1–228 expressed a full-length protein, whereas other clones expressed various deletion mutants. It is clear that the probed DNA was fully degraded by DNase I in the absence of any protection from the protein amino acids 1–75, 76–151, or 1–151 (Fig. 2a, lanes 3–5) but was partially protected by the protein amino acids 1–228, 76–228, or 152–228 (Fig. 2a, lanes 6–8). The region corresponding to amino acids 152–228 (lane 8) protected the probe DNA more strongly than did others, indicating that the protein amino acids 1–151 did not contain the site of protein–DNA binding. Furthermore, this protected region, ≈5 bases in length, seemed to contain a CTCCC motif, which seemed to act as a specific binding site for hLITAF (Fig. 2b).
Figure 2.
Detection of hLITAF/hTNF-α DNA interaction by using footprinting. (a) [32P]ATP-labeled TNF-α promoter DNA (−550 to −487) as probe was added to each tube of reaction buffer (lanes 1–8) but mixed with GST-hLITAF fusion protein in lanes 3–8. Nonprotein (lane 1), 0.1 μg of GST-fusion protein alone (lane 2), and 0.1 μg of GST-hLITAF fusion protein amino acids 1–75 (lane 3), 1–151 (lane 4), 76–151 (lane 5), 1–228 (lane 6), 76–228 (lane 7), and 152–228 (lane 8) were mixed with probe. The mixtures were digested with DNase I (Promega) for 5 min, then that reaction was terminated by adding stop solution. Samples were then run in 6% polyacrylamide sequencing gels. [32P]ATP-labeled DNAs (30, 50, and 70 bp) were used as markers on the left side of the gel, and those marker molecular weight values were indicated as shown (lane M). The protected, undigested DNA is in the gap, indicated by a box on the right side of the gel. The DNase I-degraded DNA was measured base by base in comparison with markers. (b) The sequence of TNF-α promoter DNA from −550 to −487 is shown. The specific site identified in response to hLITAF binding is indicated by a dotted line on the sequences.
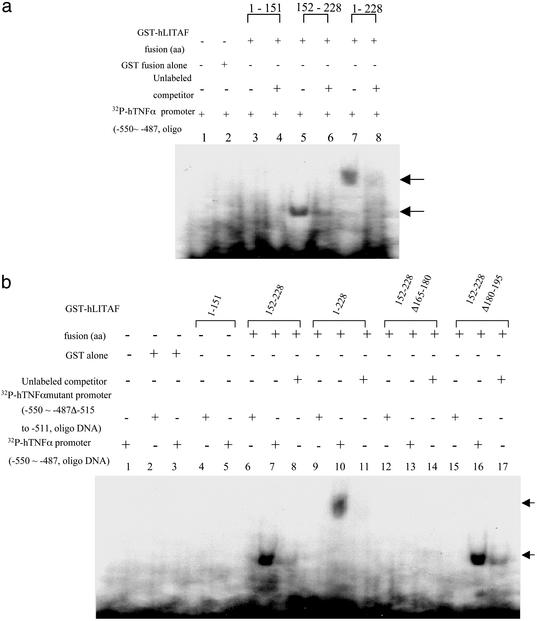
Identification of Binding Activity of the Short Sequence CTCCC in the hTNF-α Promoter.
From the results shown above, the short sequence CTCCC (−515 to −511) within the hTNF-α promoter seemed to be the specific binding site for hLITAF. Three GST-hLITAF fusion proteins, amino acids 1–151, 152–228, 1–228, or GST alone as control were used to test binding to the [32P]ATP-labeled hTNF-α promoter DNA containing the CTCCC sequence by using EMSA. It was clear that the CTCCC motif bound to the hLITAF amino acids 152–228, because both the fusion protein amino acids 152–228 and 1–228 shifted the DNA band, as indicated by arrows (Fig. 3a, lanes 5 and 7). In contrast, no shifted band was observed in lane 2 for GST alone or lane 3 for amino acids 1–151 (Fig. 3a). To directly prove binding activity by the short sequence CTCCC in the hTNF-α promoter, we constructed two mutants in which the CTCCC region was deleted (see Materials and Methods and Fig. 1b). In identical EMSA experiments, the proteins representing amino acids 1–151, 152–228, and 1–228 failed to bind to the mutant DNA probes (Fig. 3b, lanes 4, 6, and 9). As a positive control, the protein amino acids 152–228 or 1–228 were found to shift the wild-type probe containing the CTCCC motif (Fig. 3b, lanes 7 and 10). Together, these results indicate that the CTCCC motif is the specific site for hLITAF/hTNF-α binding. We subsequently identified the specific region within the hLITAF protein amino acids 152–228 binding to CTCCC. GST-hLITAF fusion proteins containing internal deletions (amino acids 152–228Δ180–195 and 152–228Δ165–180) were created, as shown in Fig. 1a. Their analysis by EMSA is shown in Fig. 3b, where it is evident that the protein lacking amino acids 165–180 did not shift the DNA (lane 13), whereas it did if amino acids 165–180 were present (lanes 7, 10, and 16). These findings suggest that the region of amino acids 165–180 participates in hLITAF/hTNF-α binding.
Figure 3.
EMSA of the protein–DNA interaction. (a) A probe consisting of [32P]ATP-labeled hTNF-α promoter DNA from −550 to −487 was added to each tube of reaction buffer. Probe alone (lane 1) and mixed with a 50-fold excess of unlabeled competitor (lanes 4, 6, and 8), 0.1 μg of GST-fusion protein alone (lane 2), and 0.1 μg of GST-hLITAF amino acids 1–151 (lanes 3 and 4), 152–228 (lanes 5 and 6), and 1–228 (lanes 7 and 8) are shown. The only shifted DNA band is indicated by an arrow. (b) [32P]ATP-labeled TNF-α promoter DNA from −550 to −487 (lanes 1, 3, 5, 7, 10, 13, and 16) or mutant DNA from −550 to −487 Δ−515 to −511 as probe was added to each tube of reaction buffer (lanes 2, 4, 6, 9, 12, and 15). The probed DNAs were mixed with a 50-fold excess of unlabeled competitor (lanes 8, 11, 14, and 17). The probe was also individually mixed with 0.1 μg of GST-fusion protein alone (lanes 2 and 3), 0.1 μg of GST-hLITAF fusion protein amino acids 1–151 (lanes 4 and 5), 152–228 (lanes 6–8), 1–228 (lanes 9–11), 152–228 Δ165–180 (lanes 12–14), and 152–228 Δ180–195 (lanes 15–17) and then incubated on ice for 30 min before electrophoresis on nondenaturing 6% polyacrylamide gel. The only shifted DNA band is indicated by an arrow.
hTNF-α ELISA After Peptide Stimulation.
The data reported above suggest that the hLITAF protein amino acids 165–180 might be sufficient to induce TNF-α expression in monocytic cells. Three peptides, referred to as A, B, and C, were synthesized based on the amino acid sequence of hLITAF. The peptide HA as negative control was used in this study (see Materials and Methods). Peptides were introduced into THP-1 cells by Chariot, a commercial kit for protein transduction. After pretreatment with phorbol 12-myristate 13-acetate, THP-1 cells were then stimulated by addition of 1, 10, or 100 ng/ml or 1 μg/ml peptide A, B, C, or HA for 24 h. Culture supernatants were harvested, and TNF-α was quantified by ELISA. As shown in Fig. 4, treatment with peptide A or C did not induce any significant measurable TNF-α secretion within 24 h, nor did treatment with peptide HA. In contrast, peptide B could increase in TNF-α secretion by as much as 2.4-fold over unstimulated levels (Fig. 4).
Analysis of Promoter Activity After Stimulation by LPS or Peptide A, B, or C.
To determine whether the short sequence CTCCC (−515 to −511) in the hTNF-α promoter is responsible for hLITAF binding activity, a series of hTNF-α promoter/reporter constructs was cloned (Fig. 1b) and individually transiently transfected into THP-1 cells. Cells were then stimulated by LPS or transfected by Chariot-compounded peptide A, B, C, or HA. TNF-α promoter activity was finally analyzed by using the luciferase assay. As shown in Fig. 5a, LPS similarly activated both the wild-type full-length and mutant TNF-α promoters. In contrast, treatment with peptide A, C, or HA did not result in any significant increase in hTNF-α promoter activity. However, peptide B caused an ≈2-fold increase in luciferase expression induced by the wild-type promoter, compared with HA-stimulated cells. The mutant TNF-α promoter, which lacks the CTCCC motif, was not induced by peptide B, nor by peptide A, C, or HA (Fig. 5a). Subsequent studies were performed using luciferase reporter plasmids under the control of a small fragment of the hTNF-α promoter that contains or lacks the LITAF binding site (Fig. 1b). THP-1 cells were transiently transfected with these shorter reporter plasmids and then stimulated with LPS or peptide A, B, C, or HA (Fig. 5b). These studies showed that activation of these shorter reporter plasmids by LPS strongly depended on the presence of the LITAF binding site. Peptides A and C were unable to activate these shorter reporter plasmids, whereas peptide B could induce 2.3-fold more luciferase expression in comparison with HA control. This activation was not observed in reporter plasmids lacking the LITAF binding site, consistent with data obtained by using the full-length hTNF-α promoter (Fig. 5a). Together, these studies support a role for LITAF in activation of the hTNF-α promoter by LPS. Furthermore, a specific peptide, corresponding to a portion of the DNA binding domain of LITAF, possesses the ability to activate the hTNF-α promoter when the LITAF binding site is present.
Discussion
A number of studies (1–5) have shown that TNF-α and TNF-α-induced factors contribute to the pathogenesis of inflammatory disorders. Hence, it is important for TNF-α gene expression to be kept under rigid control. The TNF-α promoter is very complex and contains specific consensus DNA sequences recognized by several transcription factors (6–14, 16). Gram-negative bacterial LPS, a potent inducer of TNF-α gene expression in monocytes and macrophages, activates TNF-α transcription via a mechanism that is highly dependent on NF-κB and other specific sites in the promoter (10, 32). Our previous studies identified an LPS-induced TNF-α factor, termed LITAF. At that time, we suggested that LITAF might be important for the control of TNF-α gene expression.
In this report, we have defined a regulatory element, located within the hTNF-α promoter, that mediates LPS-induced TNF-α gene expression in THP-1 cells. DNase I footprinting demonstrated that hTNF-α promoter DNA sequences located between −515 and −511 were protected by hLITAF protein. Sequence analysis identified the protected bases as CTCCC (Fig. 2b). After comparison of CTCCC with all other known regulatory elements by database analysis, it seems that this DNA is a unique regulatory element. Deletion of neighboring DNA adjacent to the CTCCC site did not particularly affect hLITAF binding (data not shown). hTNF-α promoter DNA could not be protected from DNase I digestion by the hLITAF amino acids 1–75, 1–151, and 76–151 (Fig. 2a, lanes 3–5), but DNA was totally protected by the protein amino acids 1–228, 76–228, and 152–228 (Fig. 2a, lanes 6–8). Deletion of this DNA sequence obliterated hLITAF/hTNF-α binding (Fig. 3b, lanes 6 and 9). We also have defined an hLITAF peptide (amino acids 165–180) that plays an important role for binding activity. This peptide can physically interact with CTCCC in vitro (Fig. 3a, lanes 5 and 7). Interestingly, the DNA–protein complex containing the protein amino acids 1–228 migrated more slowly in the nondenaturing gel (lane 7) compared with the much smaller peptide amino acids 152–228 (lane 5). Furthermore, deletion of the amino acid 165–180 region abolished hLITAF binding to DNA (Fig. 3b, lane 13). The findings suggest that hLITAF amino acids 165–180 are an important contributor to its DNA binding activity.
We had previously shown that hLITAF inhibition of LITAF mRNA levels in THP-1 cells resulted in a reduction of TNF-α transcripts, suggesting a role for LITAF in TNF-α gene regulation (16). The mechanism by which TNF-α gene expression might be regulated by LITAF remained unclear. We hypothesized that the DNA binding region of hLITAF, amino acids 165–180, might also function in living cells to control TNF-α gene expression. Hence, the peptides A, B, and C, each containing portions of the sequence of hLITAF, were synthesized. These peptides were also used as soluble stimuli for TNF-α induction in THP-1 cells. Surprisingly, peptide B significantly increased the production of TNF-α by 1.7- to 2.4-fold following the addition of 100–1,000 ng/ml for 24 h (Fig. 4).
We also investigated whether the stimulatory activity of hLITAF was solely dependent on the CTCCC motif within the hTNF-α promoter. As shown in Fig. 5a, THP-1 cells transfected with both the wild-type and mutant (mtTNFP1, 991 to 1Δ−515 to −511) TNF-α promoter/reporter plasmids were similarly activated by LPS. With regard to the reason why the promoter lacking the CTCCC motif was still activated by LPS, one explanation might be that the region −991 to +1 in the TNF-α promoter contains not only the LITAF binding site but also sites for NF-κB, activating protein-1, and others (see Fig. 1b) (10, 33). These transcription factors, in contrast to hLITAF, can also be induced by LPS, after which they bind to the promoter and regulate the target gene expression. In this situation, however, the endogenous hLITAF cannot work in the absence of CTCCC. Thus, we subsequently investigated the function of the CTCCC-containing region of the TNF-α promoter in the absence of other TNF-α promoter elements. The mtTNFP2 (−550 to −487 plus TATA box) and mtTNFP3 (−550 to −487Δ−515 to −511 plus TATA box) were constructed (Fig. 1b), as this region, −550 to −487, contains the only binding site for hLITAF. By using this short reporter construct, a clear dependence of the CTCCC-containing region on promoter activation by LPS and peptide B was observed (Fig. 5b). Clearly, either LPS-induced endogenous hLITAF or peptide B activates the mtTNFP2 reporter constructs, whereas no activation takes place for mtTNFP3 reporter constructs, presumably because mtTNFP2 contains CTCCC, whereas mtTNFP3 lacks this site. These findings suggest that the 165–180 amino acid region of hLITAF is an independent domain capable of activating TNF-α gene expression. Together, these findings help to clarify the mechanism of hLITAF/hTNF-α interaction and the manner by which hLITAF contributes to hTNF-α regulation. The elucidation of these mechanisms should help the design of new pharmacological approaches aimed at addressing TNF-related diseases.
Acknowledgments
This work was supported by National Institute of Craniofacial and Dental Research Grant DE14079 (to S.A.).
Abbreviations
- TNF
tumor necrosis factor
- h
human
- LPS
lipopolysaccharide
- EMSA
electrophoretic mobility-shift assay
- HA
hemagglutinin
- LITAF
LPS-induced TNF-α factor
- TNFP
TNF promoter
- mtTNFP
mutant TNFP
References
- 1.Alexander H R, Sheppard B C, Jensen J C, Langstein H N, Buresh C M, Venzon D, Walker E C, Fraker D L, Stovroff M C, Norton J A. J Clin Invest. 1991;88:34–39. doi: 10.1172/JCI115298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sugarman B J, Aggarwal B B, Hass P E, Figari I S, Palladino M A, Jr, Shepard H M. Science. 1985;22:943–945. doi: 10.1126/science.3933111. [DOI] [PubMed] [Google Scholar]
- 3.Beutler B, Cerami A. Annu Rev Immunol. 1989;7:625–655. doi: 10.1146/annurev.iy.07.040189.003205. [DOI] [PubMed] [Google Scholar]
- 4.Talmadge J E, Phillips H, Schneider M, Rowe T, Pennington R, Bowersox O, Lenz B. Cancer Res. 1988;48:544–550. [PubMed] [Google Scholar]
- 5.Uglialoro A M, Turbay D, Pesavento P A, Delgado J C, McKenzie F E, Gribben J G, Hartl D, Yunis E J, Goldfeld A E. Tissue Antigens. 1998;52:359–367. doi: 10.1111/j.1399-0039.1998.tb03056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards C K, III, Zhou T, Zhang J, Baker T J, De M, Long R E, Borcherding D R, Bowlin T L, Bluethmann H, Mountz J D. J Immunol. 1996;157:1758–1772. [PubMed] [Google Scholar]
- 7.Jue D M, Jeon K I, Jeong J Y. J Korean Med Sci. 1999;14:231–238. doi: 10.3346/jkms.1999.14.3.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conn D L. Arthritis Rheum. 2001;45:462–467. doi: 10.1002/1529-0131(200110)45:5<462::aid-art366>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 9.Anderson P. Ann Rheum Dis. 2000;59:13–15. doi: 10.1136/ard.59.suppl_1.i3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collart M A, Baeuerle P, Vassalli P. Mol Cell Biol. 1990;10:1498–1506. doi: 10.1128/mcb.10.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kramer B, Wiegmann K, Kronke M. J Biol Chem. 1995;270:6577–6583. doi: 10.1074/jbc.270.12.6577. [DOI] [PubMed] [Google Scholar]
- 12.Csonga R, Prieschl E E, Jaksche D, Novotny V, Baumruker T. J Immunol. 1998;160:273–283. [PubMed] [Google Scholar]
- 13.Rhoades K L, Golub S H, Economou J S. J Biol Chem. 1992;267:22102–22107. [PubMed] [Google Scholar]
- 14.Pope R, Mungre S, Liu H, Thimmapaya B. Cytokine. 2000;12:1171–1181. doi: 10.1006/cyto.2000.0691. [DOI] [PubMed] [Google Scholar]
- 15.Bouma G, Crusius J B, Oudkerk P M, Kolkman J J, von Blomberg B M, Kostense P J, Giphart M J, Schreuder G M, Meuwissen S G, Pena A S. Scand J Immunol. 1996;43:456–463. doi: 10.1046/j.1365-3083.1996.d01-65.x. [DOI] [PubMed] [Google Scholar]
- 16.Myokai F, Takashiba S, Lebo R, Amar S. Proc Natl Acad Sci USA. 1999;96:4518–4523. doi: 10.1073/pnas.96.8.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chappell V L, Le L X, LaGrone L, Mileski W J. Shock. 2000;14:400–403. doi: 10.1097/00024382-200014030-00027. [DOI] [PubMed] [Google Scholar]
- 18.Marrack P, Kappler J. Nature. 1988;332:840–843. doi: 10.1038/332840a0. [DOI] [PubMed] [Google Scholar]
- 19.Ye X, Liu S F. Biochem Biophys Res Commun. 2001;288:927–932. doi: 10.1006/bbrc.2001.5883. [DOI] [PubMed] [Google Scholar]
- 20.Shapira L, Takashiba S, Champagne C, Amar S, Van Dyke T E. J Immunol. 1994;153:1818–1824. [PubMed] [Google Scholar]
- 21.Yao J, Mackman N, Edgington T S, Fan S T. J Biol Chem. 1997;272:17795–17801. doi: 10.1074/jbc.272.28.17795. [DOI] [PubMed] [Google Scholar]
- 22.Kuprash D V, Udalova I A, Turetskaya R L, Kwiatkowski D, Rice N R, Nedospasov S A. J Immunol. 1999;162:4045–4052. [PubMed] [Google Scholar]
- 23.Goldfeld A E, Doyle C, Maniatis T. Proc Natl Acad Sci USA. 1990;87:9769–9773. doi: 10.1073/pnas.87.24.9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foxwell B, Browne K, Bondeson J, Clarke C, de Martin R, Brennan F, Feldmann M. Proc Natl Acad Sci USA. 1998;95:8211–8215. doi: 10.1073/pnas.95.14.8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai E Y, Falvo J V, Tsytsykova A V, Barczak A K, Reimold A M, Glimcher L H, Fenton M J, Gordon D C, Dunn I F, Goldfeld A E. Mol Cell Biol. 2000;20:6084–6094. doi: 10.1128/mcb.20.16.6084-6094.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takashiba S, Van Dyke T E, Shapira L, Amar S. Infect Immun. 1995;63:1529–1534. doi: 10.1128/iai.63.4.1529-1534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galas D J, Schmitz A. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donis-Keller H. Nucleic Acids Res. 1980;8:3133. doi: 10.1093/nar/8.14.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horng T, Barton G M, Medzhitov R. Nat Immunol. 2001;2:835–841. doi: 10.1038/ni0901-835. [DOI] [PubMed] [Google Scholar]
- 30.Morris M C, Depollier J, Mery J, Heitz F, Divita G. Nat Biotechnol. 2001;19:1173–1176. doi: 10.1038/nbt1201-1173. [DOI] [PubMed] [Google Scholar]
- 31.McClane S J, Hamilton T E, Burke C V, Raper S E. Hum Gene Ther. 1997;10:739–746. doi: 10.1089/hum.1997.8.6-739. [DOI] [PubMed] [Google Scholar]
- 32.Johnson P F, McKnight S L. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- 33.Amar S, Han X. Appl Genom Proteom. 2002;1:31–44. [Google Scholar]