Abstract
Somatic hypermutation (SHM), coupled to selection by antigen, generates high-affinity antibodies during germinal center (GC) B cell maturation. SHM is known to affect Bcl6, four additional oncogenes in diffuse large B cell lymphoma, and the CD95/Fas gene and is regarded as a major mechanism of B cell tumorigenesis. We find that mutations in the genes encoding the B cell receptor (BCR) accessory proteins B29 (Igβ, CD79b) and mb1 (Igα, CD79a) occur as often as Ig genes in a broad spectrum of GC- and post-GC-derived malignant B cell lines, as well as in normal peripheral B cells. These B29 and mb1 mutations are typical SHM consisting largely of single nucleotide substitutions targeted to hotspots. The B29 and mb1 mutations appear at frequencies similar to those of other non-Ig genes but lower than Ig genes. The distribution of mb1 mutations followed the characteristic pattern found in Ig and most non-Ig genes. In contrast, B29 mutations displayed a bimodal distribution resembling the CD95/Fas gene, in which promoter distal mutations conferred resistance to apoptosis. Distal B29 mutations in the cytoplasmic domain may contribute to B cell survival by limiting BCR signaling. B29 and mb1 are mutated in a much broader spectrum of GC-derived B cells than any other known somatically hypermutated non-Ig gene. This may be caused by the common cis-acting regulatory sequences that control the requisite coexpression of the B29, mb1, and Ig chains in the BCR.
B cell malignancies account for 90–95% of all adult leukemias and lymphomas (1). The majority of these malignant B cells carry somatic hypermutation (SHM) in their Ig variable (IgV) regions, identifying them as germinal center (GC) or post-GC in origin (1). Originally thought to be confined to IgV regions, genes translocated into the Ig locus, or transgenes under the control of Ig enhancers, SHM is now known to also mutate non-Ig genes. Mistargeted SHM was first identified in Bcl6 in approximately half of GC-derived malignancies (2) and a minor fraction of normal GC-derived B cells (3). The CD95/Fas gene was subsequently shown to be mutated in a limited set of normal and malignant post-GC B cells (4). Recently the Pax5, Pim1, RhoH/TTF, and germ-line c-myc oncogenes were reported to be hypermutated, but only in diffuse large B cell lymphoma (DLBCL) (5). With the exception of CD95/Fas, all known hypermutated non-Ig genes undergo translocations into the Ig locus. SHM of the four oncogenes in DLBCL is predicted to reflect selection (5), as shown for the CD95/Fas death domain mutations associated with resistance to apoptosis (6).
We (7) and others (8, 9) previously reported multiple B29 coding region mutations in chronic lymphocytic leukemia (CLL) cells with low or undetectable surface B cell receptor (BCR). We have also demonstrated that selected CLL-B29 mutations reproduced the low BCR surface expression and diminished BCR signaling that are the hallmarks of CLL (10). The features of these CLL-B29 mutations led us to predict that they were generated by SHM. Here we report that B29 is mutated as often as IgV in normal peripheral B cells as well as GC-derived Burkitt lymphoma (BL), DLBCL, primary effusion lymphoma (PEL), and myeloma cell lines. We have also determined that mb1, the gene encoding the second essential BCR accessory protein, is similarly mutated in the same spectrum of B cells. The mutation frequencies and characteristics of B29 and mb1 mutations closely resemble SHM of other non-Ig genes. However, B29 and mb1 are the first non-Ig genes shown to be targeted by SHM that are not protooncogenes or tumor suppressor genes. In addition, neither gene is known to undergo translocation or to be directly involved in tumorigenesis. The occurrence of B29 and mb1 mutations in all GC-derived B cells with somatically hypermutated IgV may be due to similarities in the transcription control regions of these coordinately expressed BCR components.
Materials and Methods
Cell Lines and Peripheral Blood Lymphocytes (PBLs).
A variety of B cells lines representing different stages of B cell development from pre-B to post-GC were selected for use in this study (see Tables 5–7, which are published as supporting information on the PNAS web site, www.pnas.org). BLIN-1 and Nalm6 cell lines were received from D. J. Rawlings (University of Washington); BCBL-1 and BJAB lines were from M. A. Teitell (University of California, Los Angeles; refs. 11–13) the CL-01 subclone of BL-16 was from A. Saxon (University of California, Los Angeles, ref. 14); NU-DHL1 and SU-DHL 5-9 were from Cancer Therapeutics (Los Angeles) with the permission of A. Epstein (University of Southern California Medical Center, Los Angeles, ref. 15); OCI Ly1–19 was from E. Davis (National Institutes of Health, Bethesda) with the permission of H. Messner (University of Ontario, refs. 16 and 17); and L428 was from Amgen (Thousand Oaks, CA). All other cell lines were purchased from American Type Culture Collection.
Blood samples were obtained with informed consent from randomly selected normal donors. PBLs were separated from whole blood by centrifugation on Ficoll-Paque PLUS (Amersham Pharmacia). CD19+ (bound) and CD19− (flow through) cells were isolated by incubation with CD19 magnetic beads. CD19+ PBLs were removed from the beads using CD19 DETACHaBEAD (both from Dynal, Oslo) following the manufacturer's directions.
Isolation of Cell Line Genomic DNA (gDNA), PCR Amplification, and Direct Sequencing.
gDNA from ANBL6, EJM, Karpas 620, KMS12, and L363 myeloma cell lines was a kind gift of W. M. Kuehl (National Cancer Institute, Bethesda, ref. 18). gDNA from 2 × 107 normal or cultured cells was prepared using the Wizard Genomic DNA Purification kit (Promega). The B29 and mb1 genes were amplified from 200 ng of gDNA with a Thermal AceDNA Polymerase kit (Invitrogen) and B29 or mb1 gene specific primers (see Table 8, which is published as supporting information on the PNAS web site). Genomic PCR was repeated for selected samples using PfuUltra DNA Polymerase (Stratagene) PCR error rate was 4 × 10−7 to 6 × 10−6 per bp, as reported by the respective manufacturers. PCR products were purified using a StrataPrep PCR purification kit (Stratagene) following the manufacturer's directions, and directly sequenced with PCR and internal primers (see Table 9, which is published as supporting information on the PNAS web site) and the ABI Prism Dye Terminator Cycle Sequencing Reaction kit (Applied Biosystems). Sequence analyses were performed using macvector software (Oxford Molecular Group, Campbell, CA). B29 genomic sequences were aligned to GenBank accession number L27587 (19), and mb1 genomic sequences were aligned to GenBank accession number L32754 (20). The Transfac database (21) was used to search promoters for transcription factor binding sites.
Single Cell PCR from PBLs.
Single CD19+ or CD19− (negative control) PBLs were isolated by limiting dilution, transferred into 0.2 ml PCR tubes and lysed as described (22). Primer extension preamplification was performed as described (22) using a random 15-mer oligo (Qiagen, Valencia, CA) and Expand High Fidelity DNA polymerase (Roche Diagnostics, Mannheim, Germany). The regions including the highly mutated clusters of B29 (nt 258-1324 and nt 2375–3800) and mb1 (nt 989-2223) were amplified by two rounds of seminested PCR with gene specific primers (see Table 8), PCR products were cloned by using a Zero Blunt TOPO PCR cloning kit (Invitrogen) following the manufacter's directions, and clones were sequenced as described above.
RNA Purification, RT-PCR Amplification, Cloning, and Sequencing.
RNA from selected cell lines was prepared using the RNeasy Mini kit in combination with QIAshredder (both from Qiagen). For amplification of B29 and mb1 200 ng of RNA was reverse transcribed with SuperScript II (GIBCO/BRL, Rockville, MD), and 2 μl of the cDNA was then amplified with PfuTurbo DNA polymerase (Stratagene) RT-PCR primers (see Table 8). RT-PCR products were cloned and sequenced, and sequences were aligned to GenBank accession numbers M89957 (B29; ref. 23) and M86921 (mb1, ref. 24) as described above.
Results
B29 Is Mutated in GC and Post-GC B Cells.
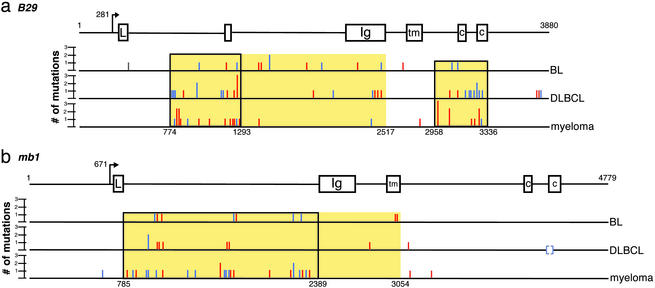
To determine whether B29 mutations could arise from mistargeted SHM, we analyzed genomic DNA sequences amplified from a variety of well-characterized malignant human B cell lines of GC (BL and DLBCL) through post-GC (PEL and myeloma) origins, which are known to have hypermutated IgV regions (see Table 5). B cell lines, particularly the BL cell lines used here (BL-2, BL-41, CL-01, and Ramos), have been used extensively in studies of SHM of Ig and non-Ig genes (5, 14, 25–27). Non-B and pre-B cell lines were included as negative controls. PCR amplification was performed employing two separate high-fidelity DNA polymerases to minimize the introduction of sequence alterations by PCR errors. To further reduce the impact of PCR artifacts, amplicons were directly sequenced. Multiple polymorphisms in B29 were found in all cell lines studied (see Table 6). B29 mutations were not detected in non-B (HeLa and Jurkat) or pre-B (BLIN-1 and Nalm6) cells that do not experience SHM (see Table 5). SHM occur almost entirely downstream of the transcriptional start site (28). To confirm that B29 sequence alterations were indeed SHM, a 459-bp fragment of the region upstream of the start of B29 transcription (nucleotides 23–482) was amplified from 31 cell lines, the amplicons were sequenced, and sequences were analyzed for alterations. No changes were found, verifying the fidelity of our protocol (see Table 6).
Numerous B29 mutations were detected downstream of the B29 transcriptional start site in essentially all (26/27 = 96%) of the GC- and post-GC-derived cell lines studied (Tables 1 and 5). Only one PEL line, BC-3, which carried no mutations in the VH region (11), had unmutated B29 (see Table 5). Mutation frequencies were calculated over two regions (clusters I and II, Fig. 1a) that contained the highest densities of B29 mutations (Table 1). Over this broad spectrum of GC-derived B cell lymphoma and myeloma lines, B29 mutations appeared as often as IgV SHM. Although the B29 mutations occurred at a lower frequency (e.g., 5–10%) than IgV regions, the frequency was ≈103 to 104 times that of spontaneous mutation (29).
Table 1.
Frequencies and occurrences of B29 and mb1 mutations in normal peripheral B cells and GC-derived B cell lines
| B cell |
B29
|
mb1
|
|||
|---|---|---|---|---|---|
| Mutation frequency/100-bp cluster I nt 774–1293, mean (range) | Mutation frequency/100-bp cluster II nt 2958–3336, mean (range) | Occurrence | Mutation frequency/100-bp intron 1 nt 785–2389, mean (range) | Occurrence | |
| Normal PBL B cells* | 0.03 (0–0.19) | 0.05 (0–0.26) | 20/60 | 0.03 (0–0.12) | 13/30 |
| BL | 0.11 (0–0.38) | 0.11 (0–0.53) | 5/5 | 0.09 (0–0.19) | 4/5 |
| DLBCL | 0.25 (0–0.77) | 0.26 (0–0.53) | 10/10 | 0.04 (0–0.06) | 7/10 |
| PEL | 0.19 (0–0.38) | 0.09 (0–0.26) | 2/3 | 0 (0) | 1/3 |
| Myeloma | 0.36 (0–1.16) | 0.33 (0–0.53) | 8/8 | 0.19 (0.06–0.25) | 8/8 |
The mutation frequencies were calculated as the number of mutations per 100 bp within the most densely mutated regions, B29: cluster I (nt 774–1293) and cluster II (nt 2958–3336); mb1: intron I (nt 785–2389). The mutation occurrence represents the number of samples with mutations relative to the total number of samples.
Mutation frequencies and occurrences were calculated based on clones derived from normal peripheral B cells.
Figure 1.
Distribution of B29 and mb1 mutations in B cell lymphomas and myelomas. (a) Mutations in B29 were located across the length of the gene but occurred more often in two regions (nt 774–2517 and 2958–3336). The transcriptional start site is located at nt 281. (b) mb1 mutations were found predominantly within ≈2.5 of the +1 (nt 785–3054). Transcription begins at nt 671. The regions of B29 and mb1 containing the majority of mutations are highlighted in yellow. The most densely mutated clusters of the B29 (774–1293 and 2958–3336) and mb1 (785–2389) genes are in boxes. Mutations located within SHM hotspots are shown in red, mutations found outside of hotspots are shown in blue; brackets indicate a larger deletion. Multiple mutations occurring at the same nucleotide are indicated by the height of the bar.
mb1 Is Also Widely Mutated.
We have also analyzed genomic sequences of the second BCR accessory protein, mb1, from the same spectrum of B cells used for B29 studies (see Table 7). We found that mb1 was also mutated in the majority (21 of 27 = 78%) of GC and post-GC-derived cell lines, but not in control non-B or pre-B lines (Tables 1 and 7). A single base pair deletion (ΔC610) was found in the mb1 promoter, which is unlikely to affect mb1 regulation (30). Although rare, SHM in the promoter or 5′ flanking sequences have previously been reported for VH regions (28, 31, 32). Alternatively, this deletion could arise from spontaneous mutation or from PCR artifacts. The latter is unlikely because the alteration was reproducible in independent PCR reactions and occurred at a rate greater than the reported PCR error rate. Overall, mb1 was less extensively mutated than B29, and contained fewer polymorphisms (see Table 7). Not all cell lines exhibiting B29 mutations had mutated mb1 (Tables 1 and 7) and the mutation frequency was approximately half that of B29, or 0.5–5% of that of Ig genes.
B29 and mb1 Mutations Are Characteristic of SHM.
The features of B29 and mb1 mutations identify them as SHM (Table 2). Like SHMs of IgV regions, mutations in B29 and mb1 were single nucleotide changes (except for one larger mb1 deletion in the DLBCL line, OCI Ly10, see Table 7) consisting primarily of base substitutions (B29, 75%; mb1, 65%) rather than insertions (B29, 10%; mb1, 18%) or deletions (B29, 15%; mb1, 16%; Table 2). Nearly all insertions (B29, 100%; mb1, 89%) were nucleotide duplications. Also characteristic of SHM, transitions were more prevalent than transversions in B29 (1.5:1) and greatly outnumbered transversions in mb1 (4:1, Table 2). B29 and mb1 mutations showed statistically significant associations with the SHM hotspot motifs RGYW/WRCY or WA (where R = A or G, Y = C or T and W = A or T; B29, 59%; mb1, 53%) and were biased toward G or C bases (B29, 64%; mb1, 73%; Table 2 and Fig. 2, which is published as supporting information on the PNAS web site).
Table 2.
The features of B29 and mb1 gene mutations are characteristic of SHM of Ig and non-Ig genes in GC-derived B cell lines
| Substitutions | Transitions/ transversions | Deletions | Insertions | Mutations within hotspots | Mutations of G/C nucleotides | |
|---|---|---|---|---|---|---|
| B29 | 62/83 (75%) | 1.5:1 | 13/83 (15%) | 8/83 (10%) | 49/83 (59%)* | 53/83 (64%) |
| mb1 | 32/49 (65%) | 4:1 | 8/49 (16%) | 9/49 (18%) | 26/49 (53%)* | 36/49 (73%) |
All mutations are point mutations and predominantly transitions, except for one deletion in mb1 in one DLBCL line. Mutations associated with SHM hotspots were defined as mutations occurring within or adjacent to the RGYW/WRCY or WA motifs.
Statistically significant (P < 0.001).
Distribution of SHM in B29 and mb1.
SHM of IgV regions typically begin ≈100 bp downstream of the transcriptional start site (+1), peak at ≈1 kb, and are rarely found at distances greater than 2.5 kb (28, 31, 32). Mutations in mb1 showed this characteristic distribution with >80% of the identified mb1 mutations falling in the large first intron, within 2.5 kb of the +1 (Fig. 1b). Because there are no known regulatory regions in this mb1 intron, mutations there are unlikely to affect mb1 expression or function. Of the nine mutations situated outside of intron one, one was found in the promoter region and five were mapped to introns two or three. Only three coding region mutations were found, of which two, in the transmembrane domain, are silent. The third coding region mutation is a 43-bp deletion crossing the intron four/exon five (cytoplasmic domain) boundary and removing the 3′ splice acceptor. Sequencing of mb1 cDNA showed that this cell line expressed one normal allele, and that splicing of the mutated allele introduced a frameshift (data not shown). This frameshift results in a substitution in the first immune receptor tyrosine-based activation motif (ITAM) of the mb1 cytoplasmic tail (L191T) and truncation of both the motif required for tethering Fyn tyrosine kinase to the resting BCR (DCSM) and the second ITAM. Deletion of the entire mb1 cytoplasmic domain has been shown to result in B cells expressing normal levels of surface BCR (33) but with receptors that were constitutively active (34). The presence of the second normal mb1 allele in the cell line expressing the truncated mb1 protein is expected to modulate any effect of this mb1 mutation.
In contrast to mb1 mutations, B29 mutations showed a bimodal distribution with 62% of the mutations occurring 450 bp to 2.2 kb from the start of transcription and a second cluster containing 29% of the mutations 2.7–3 kb downstream of the transcriptional +1 (Fig. 1a). Mutations in B29 occurred primarily outside of exons (66 of 82 = 80%), but a significant number (16 of 82 = 20%) were found in coding sequences in exons two, three, and six. Ten of the 16 coding region mutations were either silent or highly conserved amino acid substitutions. One more radical substitution (V26E), appearing in two different cell lines and located in the B29 Ig domain, is of unknown effect. A third mutation replaces threonine with alanine (T206A) in the region between the cytoplasmic ITAM motifs. Serine/threonine phosphorylation has been found to modulate the tyrosine phosphorylation of the mb1 cytoplasmic tail (35), and this mutation may disregulate B29 protein signaling. The fourth mutation substitutes histidine for proline in the transmembrane domain (P176H) and could interfere with BCR assembly or stable membrane insertion. The remaining two mutations are single nucleotide duplications predicted to lead to frameshifts and result in severely truncated B29 proteins (see Table 5). These frameshifts in exon three truncate portions of the Ig domain and all of the downstream transmembrane and cytoplasmic domains. Deletion of the B29 transmembrane domain has been shown to abrogate BCR surface assembly (10).
B29 and mb1 Are Mutated More Often than Other non-Ig Genes.
We found B29 and mb1 to be more widely mutated than any other known non-Ig gene (Tables 3 and 4). The frequencies of the B29 and mb1 mutations were as high or higher than the mutation frequencies of other non-Ig genes (Table 3). In contrast to these other non-Ig genes, B29 and mb1 were mutated in the large majority of human B cell lymphoma and myeloma lines analyzed here, which represent the major developmental stages of GC and post-GC B cell maturation (Tables 3 and 4). GC-derived BL and DLBCL cell lines all had mutations in B29 (100%), and most had mutations in mb1 (74%). Myeloma cell lines represent terminally differentiated plasma cells that have all experienced extensive hypermutation of their IgV regions (1) and all exhibited B29 and mb1 mutations. Thus, B29 and mb1 hypermutation occurred far more often than other non-Ig genes (Table 4). The only examples of non-Ig genes found to be mutated as widely as B29 and mb1 have been translocated into Ig loci (2).
Table 3.
Comparison of somatic hypermutation frequencies of Ig and non-Ig genes in normal peripheral B cells and GC-derived B cell malignancies
| Gene | Cell type
|
|||
|---|---|---|---|---|
| Normal PBL B | BL | DLBCL | Myeloma | |
| VH | 0–55 | 2.1–7.8* | 1–23.25 | 2.7–16.540 |
| Bcl6 | 0–0.23 | 0–0.045 | 0.06–2.95 | 0.06–0.25 |
| CD95/Fas | <0.004–0.036 | NG36 | NG5,36 | NG4,36 |
| RhoH/TTF5 | 0–0.008 | 0–0.02 | 0–0.4 | 0 |
| Pax55 | 0–0.03 | 0–0.005 | 0–0.3 | 0 |
| c-myc | 0.013 | 0.25,‡ | 0–0.65 | 0–0.0055 |
| Pim-15 | ND | 0 | 0–0.7 | 0 |
| B29* | 0–0.3† | 0–0.5 | 0–0.8 | 0–1.2 |
| mb1* | 0–0.1† | 0–0.2 | 0–0.06 | 0.06–0.3 |
All frequencies were calculated over the most densely mutated regions and recorded as mutations per 100 bp. ND, not done; NG, not given. Superscript numbers indicate the reference number of the source of these data.
This study.
PBL B cell-derived clones.
Translocated c-myc t (8,14).
Table 4.
Comparison of the occurrence of somatic hypermutation of Ig and non-Ig genes in normal peripheral B cells and GC-derived B cell malignancies
| Gene | Cell type
|
|||
|---|---|---|---|---|
| Normal PBL B | BL | DLBCL | Myeloma | |
| VH | 40%1 | 4/4 (100%)* | 100%1 | 100%1 |
| Bcl6 | 26/111 (23%)3 | 24/65 (37%)5,45 | 103/183 (56%)2,5,45 | 58/203 (29%)5,27 |
| CD95/Fas | 27/261 (10%)6 | 0/8 (0%)36 | 11/63 (17%)36 | 7/54 (13%)4,36 |
| RhoH/TTF5 | 4/64 (6%) | 3/20 (15%) | 13/28 (46%) | 0/13 (0%) |
| Pax55 | 4/38 (10%) | 1/10 (10%) | 16/28 (57%) | 0/12 (0%) |
| c-myc | 2/24 (8%)3 | 12/12 (100%)5,‡ | 12/37 (32%)5 | 2/10 (20%)5 |
| Pim15 | ND | 0/10 (0%) | 12/28 (43%) | 0/14 (0%) |
| B29* | 20/60 (33%)† | 5/5 (100%) | 10/10 (100%) | 8/8 (100%) |
| mb* | 13/30 (43%)† | 4/5 (80%) | 7/10 (70%) | 8/8 (100%) |
B29 and mb1 Are Mutated in Peripheral B Cells.
We used single-cell PCR to analyze selected regions of the B29 and mb1 genomic sequences from normal CD19+ peripheral B cells for mutations. Multiple clones were sequenced to ensure that both alleles of each gene were represented. In CD19− negative control PBLs both B29 and mb1 were unmutated (see Tables 5 and 7). Mutations in B29 and mb1, however, were detected in the CD19+ B cells from all donors analyzed, although at mutation frequencies lower than those of transformed B cell lines (Tables 1, 5, and 7). The occurrence of B29 (20 of 60 clones = 33%) and mb1 mutations (13 of 30 clones = 43%, Table 4) closely corresponded to the occurrence of IgV SHM (≈40%) of post-GC peripheral B cells (1). Characteristic of SHM, substitutions predominated (B29, 90%; mb1, 93%) with transitions exceeding transversions (B29, 5:1; mb1, 2:1). Hotspot motifs, however, were less strongly targeted (B29, 19%; mb1, 60%). Few (4 of 21) B29 mutations occurred in coding regions. Three of these were silent or highly conserved, and the fourth (E115G), in the Ig domain, is not likely to affect B29 function. Interestingly, pairs of clones were isolated for both B29 and mb1 that contained unique as well as shared mutations. This suggests that these genes have likely experienced multiple rounds of SHM (see Tables 5 and 7).
Discussion
We have determined that the genes encoding the critical BCR accessory proteins, B29 and mb1, are mutated in essentially all malignant GC and post-GC-derived B cell lines analyzed with somatically hypermutated IgV. Mutations were not found in cells outside the B lymphocyte lineage, or in B lineage cells that have not experienced SHM (e.g., pre-B cells or a PEL line, BC-3, with unmutated VH). The B29 and mb1 mutations exhibited the characteristic features of SHM in IgV and other somatically hypermutated non-Ig genes. Mutations in both genes showed an increased incidence of single base pair substitutions over deletions or insertions, prevalence of transitions over transversions, and preferential targeting to the SHM hotspots motifs, RGYW/WRCY, or WA. As with SHM in other genes, the majority of B29 and mb1 mutations were confined to a region ≈2.5 kb downstream of the transcriptional start site (+1). Mutations were infrequently detected in mb1 downstream of this region. B29, however, contained a second significant cluster of mutations located 2.7–3 kb from +1, over the segment containing the exons encoding the B29 cytoplasmic tail and ITAM signaling motif. Like B29, mutations in CD95/Fas also occurred in two clusters, a promoter proximal cluster, located 100–850 bp downstream of +1, and a distal cluster >15 kb from the start of transcription, encoding the CD95/Fas death domain (36). Mutations in the death domain occurred more often in myeloma cells and in normal peripheral B cells selected for resistance to CD95/Fas-mediated apoptosis (6). Mutations in the distal cluster that impaired death domain function conferred a selective advantage. Certain mutations in the B29 distal cluster that affect the cytoplasmic tail may similarly contribute to survival in B cells. Interestingly, half of the B29 alterations previously described in B-CLL cells were located in this region (7–9). Diminished BCR surface expression is postulated to provide a survival advantage by restricting the B cell responsiveness to BCR signals leading to apoptosis (37). Truncation of the B29 cytoplasmic domain resulted in reduced surface BCR and signaling (33, 38). In contrast, deletion of the mb1 cytoplasmic domain had no effect on membrane BCR levels (33). These latter findings indicate that the cytoplasmic domain of B29, but not mb1, plays a central role in regulating BCR surface display and function. Thus, the clustering of a subset of mutations over the distal region encoding the B29 cytoplasmic domain may be related to regulatory effects of B29 on BCR surface levels and function.
The B29 and mb1 mutations in these GC-derived B cell lines are unlikely to have occurred during malignant transformation or passage in culture. It is known that certain BL cell lines express the critical SHM protein activation induced cytidine deaminase (AID, refs. 25 and 39) and that BL and DLBCL cells exhibit ongoing SHM (1, 40). However, the mutation frequencies determined for the BL cell lines analyzed here are similar to those of the PEL and myeloma cell lines that do not express AID (data not shown). It has been shown reproducibly that the features of SHM in established B cell lines are highly similar to SHM in vivo (25, 41–43). We also determined that the B29 and mb1 genes from isolated normal peripheral B cells exhibited mutations with the features of SHM, though at lower mutation frequencies than those in transformed B cell lines. Mutations in B29 and mb1 were detected in 30% and 43%, respectively, of the clones that were generated from normal peripheral B cells. The occurrence of these mutations closely approximates the occurrence of SHM in the Ig genes (40%) of peripheral B cells (1).
Deletions and insertions were found to comprise a larger fraction of B29 (25%) and mb1 (34%) mutations compared with other somatically hypermutated non-Ig genes. Deletions and insertions accounted for only 2 - 9% of total mutations in the Bcl6, c-myc, Pax5, Pim1, and RhoHTTF oncogenes in DLBCL (5). However, deletions and insertions were frequently detected in somatically hypermutated IgV in GC-derived normal B cells and in B cell lymphomas. Such alterations were found in ≈4% of functionally rearranged IgV and ≈43% of nonfunctional IgV in normal GC B cells (44). Deletions and insertions were seen even more often in somatically hypermutated IgV of GC-derived B cell lymphomas, where ≈10% of functional and ≈54% of nonfunctional IgV had such alterations (40). The significantly higher incidence of deletions and insertions in nonfunctional versus functional somatically hypermutated IgV indicates that the manifestation of these alterations is strongly influenced by selection. Unlike deletions and insertions in IgV, which largely occur in coding sequences, the overwhelming majority of deletions and insertions in B29 and mb1 (90% and 94%, respectively) are located in introns or untranslated regions and are unlikely to have any selective advantage or disadvantage. Accordingly, the incidence of deletions and insertions in B29 and mb1 is likely to be related to the structural organization of these genes rather than an aberration of the SHM process.
It is notable that B29 and mb1 are mutated in a far broader spectrum of GC-derived B cell lines than other non-Ig genes, in which SHM is largely restricted to specific B cell malignancies. Other than B29 and mb1, Bcl6 is the most widely mutated non-Ig gene with alterations detected in ≈30–50% of different GC-derived lymphomas and myelomas (2, 5, 27, 45) and in ≈25% of normal GC B cells (3). SHM of CD95/Fas is more limited, seen in ≈15% of lymphomas and myelomas (4, 46) but only rarely in normal GC B cells (6). The oncogenes, c-myc, Pax5, Pim1, and RhoH/TTF are mutated in ≈40–50% of DLBCL, but not in normal GC B cells nor in other GC-derived B cell malignancies (5). The restricted distribution of SHM in these oncogenes was attributed to selective advantages in malignant transformation (2, 5, 45, 47). SHM is tightly coupled to transcription and directly correlated with transcriptional activity (48–51). These known somatically hypermutated oncogenes are transcribed at relatively low levels unless translocated into the Ig locus (52–54). In contrast, B29 and mb1 must be transcribed at levels comparable to Ig genes to fulfill the stoichiometric requirements for all these gene products in the BCR on the B cell surface. The B29 and mb1 promoters contain essentially the same array of transcription factor motifs present in Ig promoters and enhancers (30, 55, 56), and their transcriptional activities are estimated to be only moderately lower than Ig genes (≈20–30%, refs. 48 and 57). The similarities in transcriptional control sequences and comparable transcriptional activities of the B29, mb1, and Ig genes may explain the widespread accessibility of B29 and mb1 to the SHM machinery over the diverse spectrum of GC B cells.
Although the features of somatically hypermutated genes have been extensively characterized, the mechanisms of SHM and the complete spectrum of genes affected by this process remain to be resolved. It was previously presumed that certain cis-acting promoter/enhancer sequences or B cell-specific transcription factors were likely to be involved in targeting SHM to Ig genes. A substantial body of recent evidence, however, casts doubt on this hypothesis. Instead, it is now speculated that SHM could occur in any gene expressed in GC B cells. Whether these mutations could be detected would depend on the relative rate of transcription, positive and negative selection, and specific intrinsic features of the affected gene (e.g., hot spot motifs or chromatin structure; refs. 48, 49, and 51). It has been recently shown that expression of AID is sufficient for SHM in highly expressed genes in nonlymphoid cells (25, 42, 43). However, these in vitro systems may not reproduce SHM processes in vivo. AID transgene expression in transfected cells was estimated to be greater than in GC centroblasts undergoing SHM (25). Consequently, the highly expressed AID transgene was mutated in transfected cells, though the endogenous expressed AID gene was not mutated in DLBCL, in which SHM was detected in multiple oncogenes (5). It should also be noted that even with the overexpression of AID, the rates of SHM detected in transfected nonlymphoid cells were significantly lower that in IgV regions in vivo (25). A recent study examining the induction of SHM in a BL cell line also suggests that other B cell factors in addition to AID may be required for optimal SHM activity (39, 58). This issue will be resolved by further studies to establish the spectrum of genes affected by SHM and the features controlling their transcription.
Supplementary Material
Acknowledgments
We thank S. E. Henson, C. S. Malone, and M. A. Teitell for critical reading of this manuscript, and the members of the Wall laboratory for helpful discussions. This work was funded by National Institutes of Health Grants R01CA85841 and R01GM40185. Additional support was provided by National Institutes of Health Grants T32-CA009120-26 (to M.S.G.) and T32-AI07126-26 (to J.R.D.).
Abbreviations
- SHM
somatic hypermutation
- DLBCL
diffuse large B cell lymphoma
- GC
germinal center
- CLL
chronic lymphocytic leukemia
- BCR
B cell receptor
- BL
Burkitt lymphoma
- PBL
peripheral blood lymphocyte
- PEL
primary effusion lymphoma
- ITAM
immune receptor tyrosine-based activation motif
- AID
activation-induced cytidine deaminase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kuppers R, Klein U, Hansmann M L, Rajewsky K. N Engl J Med. 1999;341:1520–1529. doi: 10.1056/NEJM199911113412007. [DOI] [PubMed] [Google Scholar]
- 2.Migliazza A, Martinotti S, Chen W, Fusco C, Ye B H, Knowles D M, Offit K, Chaganti R S, Dalla-Favera R. Proc Natl Acad Sci USA. 1995;92:12520–12524. doi: 10.1073/pnas.92.26.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen H M, Peters A, Baron B, Zhu X, Storb U. Science. 1998;280:1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- 4.Landowski T H, Qu N, Buyuksal I, Painter J S, Dalton W S. Blood. 1997;90:4266–4270. [PubMed] [Google Scholar]
- 5.Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti R S, Kuppers R, Dalla-Favera R. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 6.Muschen M, Re D, Jungnickel B, Diehl V, Rajewsky K, Kuppers R. J Exp Med. 2000;192:1833–1840. doi: 10.1084/jem.192.12.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson A A, Talley J A, Do H N, Kagan H L, Kunkel L, Berenson J, Cooper M D, Saxon A, Wall R. Blood. 1997;90:1387–1394. [PubMed] [Google Scholar]
- 8.Payelle-Brogard B, Magnac C, Mauro F R, Mandelli F, Dighiero G. Blood. 1999;94:3516–3522. [PubMed] [Google Scholar]
- 9.Rassenti L Z, Kipps T J. Blood. 2000;95:2725–2727. [PubMed] [Google Scholar]
- 10.Gordon M S, Kato R M, Lansigan F, Thompson A A, Wall R, Rawlings D J. Proc Natl Acad Sci USA. 2000;97:5504–5509. doi: 10.1073/pnas.090087097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matolcsy A, Nador R G, Cesarman E, Knowles D M. Am J Pathol. 1998;153:1609–1614. doi: 10.1016/S0002-9440(10)65749-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bemark M, Neuberger M S. Oncogene. 2000;19:3404–3410. doi: 10.1038/sj.onc.1203686. [DOI] [PubMed] [Google Scholar]
- 13.Malone C S, Miner M D, Doerr J R, Jackson J P, Jacobsen S E, Wall R, Teitell M. Proc Natl Acad Sci USA. 2001;98:10404–10409. doi: 10.1073/pnas.181206898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerutti A, Zan H, Schaffer A, Bergsagel L, Harindranath N, Max E E, Casali P. J Immunol. 1998;160:2145–2157. [PMC free article] [PubMed] [Google Scholar]
- 15.Epstein A L, Levy R, Kim H, Henle W, Henle G, Kaplan H S. Cancer. 1978;42:2379–2391. doi: 10.1002/1097-0142(197811)42:5<2379::aid-cncr2820420539>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 16.Chang H, Blondal J A, Benchimol S, Minden M D, Messner H A. Leuk Lymphoma. 1995;19:165–171. doi: 10.3109/10428199509059672. [DOI] [PubMed] [Google Scholar]
- 17.Tweeddale M E, Lim B, Jamal N, Robinson J, Zalcberg J, Lockwood G, Minden M D, Messner H A. Blood. 1987;69:1307–1314. [PubMed] [Google Scholar]
- 18.Shou Y, Martelli M L, Gabrea A, Qi Y, Brents L A, Roschke A, Dewald G, Kirsch I R, Bergsagel P L, Kuehl W M. Proc Natl Acad Sci USA. 2000;97:228–233. doi: 10.1073/pnas.97.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashimoto S, Chiorazzi N, Gregersen P K. Immunogenetics. 1994;40:145–149. doi: 10.1007/BF00188178. [DOI] [PubMed] [Google Scholar]
- 20.Hashimoto S, Mohrenweiser H W, Gregersen P K, Chiorazzi N. Immunogenetics. 1994;40:287–295. doi: 10.1007/BF00189974. [DOI] [PubMed] [Google Scholar]
- 21.Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Pruss M, Reuter I, Schacherer F. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinmoller E, Liu Q, Sun Y, Schlake G, Hill K A, Weiss L M, Sommer S S. Lab Invest. 2002;82:443–453. doi: 10.1038/labinvest.3780437. [DOI] [PubMed] [Google Scholar]
- 23.Wood W J, Jr, Thompson A A, Korenberg J, Chen X N, May W, Wall R, Denny C T. Genomics. 1993;16:187–192. doi: 10.1006/geno.1993.1157. [DOI] [PubMed] [Google Scholar]
- 24.Ha H J, Kubagawa H, Burrows P D. J Immunol. 1992;148:1526–1531. [PubMed] [Google Scholar]
- 25.Martin A, Scharff M D. Proc Natl Acad Sci USA. 2002;99:12304–12308. doi: 10.1073/pnas.192442899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poltoratsky V, Woo C J, Tippin B, Martin A, Goodman M F, Scharff M D. Proc Natl Acad Sci USA. 2001;98:7976–7981. doi: 10.1073/pnas.141222198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasqualucci L, Migliazza A, Fracchiolla N, William C, Neri A, Baldini L, Chaganti R S, Klein U, Kuppers R, Rajewsky K, Dalla-Favera R. Proc Natl Acad Sci USA. 1998;95:11816–11821. doi: 10.1073/pnas.95.20.11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lebecque S G, Gearhart P J. J Exp Med. 1990;172:1717–1727. doi: 10.1084/jem.172.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bachl J, Olsson C. Eur J Immunol. 1999;29:1383–1389. doi: 10.1002/(SICI)1521-4141(199904)29:04<1383::AID-IMMU1383>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 30.Ha H, Barnoski B L, Sun L, Emanuel B S, Burrows P D. J Immunol. 1994;152:5749–5757. [PubMed] [Google Scholar]
- 31.Both G W, Taylor L, Pollard J W, Steele E J. Mol Cell Biol. 1990;10:5187–5196. doi: 10.1128/mcb.10.10.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogerson B J. Mol Immunol. 1994;31:83–98. doi: 10.1016/0161-5890(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 33.Reichlin A, Hu Y, Meffre E, Nagaoka H, Gong S, Kraus M, Rajewsky K, Nussenzweig M C. J Exp Med. 2001;193:13–23. doi: 10.1084/jem.193.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torres R M, Hafen K. Immunity. 1999;11:527–536. doi: 10.1016/s1074-7613(00)80128-4. [DOI] [PubMed] [Google Scholar]
- 35.Muller R, Wienands J, Reth M. Proc Natl Acad Sci USA. 2000;97:8451–8454. doi: 10.1073/pnas.97.15.8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muschen M, Rajewsky K, Kronke M, Kuppers R. Trends Immunol. 2002;23:75–80. doi: 10.1016/s1471-4906(01)02115-9. [DOI] [PubMed] [Google Scholar]
- 37.Matsuuchi L, Gold M R. Curr Opin Immunol. 2001;13:270–277. doi: 10.1016/s0952-7915(00)00215-6. [DOI] [PubMed] [Google Scholar]
- 38.Kraus M, Pao L I, Reichlin A, Hu Y, Canono B, Cambier J C, Nussenzweig M C, Rajewsky K. J Exp Med. 2001;194:455–469. doi: 10.1084/jem.194.4.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faili A, Aoufouchi S, Gueranger Q, Zober C, Leon A, Bertocci B, Weill J C, Reynaud C A. Nat Immunol. 2002;3:815–821. doi: 10.1038/ni826. [DOI] [PubMed] [Google Scholar]
- 40.Klein U, Goossens T, Fischer M, Kanzler H, Braeuninger A, Rajewsky K, Kuppers R. Immunol Rev. 1998;162:261–280. doi: 10.1111/j.1600-065x.1998.tb01447.x. [DOI] [PubMed] [Google Scholar]
- 41.Martin A, Bardwell P D, Woo C J, Fan M, Shulman M J, Scharff M D. Nature. 2002;415:802–806. doi: 10.1038/nature714. [DOI] [PubMed] [Google Scholar]
- 42.Petersen-Mahrt S K, Harris R S, Neuberger M S. Nature. 2002;418:99–103. [PubMed] [Google Scholar]
- 43.Yoshikawa K, Okazaki I M, Eto T, Kinoshita K, Muramatsu M, Nagaoka H, Honjo T. Science. 2002;296:2033–2036. doi: 10.1126/science.1071556. [DOI] [PubMed] [Google Scholar]
- 44.Goossens T, Klein U, Kuppers R. Proc Natl Acad Sci USA. 1998;95:2463–2468. doi: 10.1073/pnas.95.5.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Capello D, Vitolo U, Pasqualucci L, Quattrone S, Migliaretti G, Fassone L, Ariatti C, Vivenza D, Gloghini A, Pastore C, et al. Blood. 2000;95:651–659. [PubMed] [Google Scholar]
- 46.Gronbaek K, Straten P T, Ralfkiaer E, Ahrenkiel V, Andersen M K, Hansen N E, Zeuthen J, Hou-Jensen K, Guldberg P. Blood. 1998;92:3018–3024. [PubMed] [Google Scholar]
- 47.Kikuchi M, Miki T, Kumagai T, Fukuda T, Kamiyama R, Miyasaka N, Hirosawa S. Oncogene. 2000;19:4941–4945. doi: 10.1038/sj.onc.1203864. [DOI] [PubMed] [Google Scholar]
- 48.Tumas-Brundage K, Manser T. J Exp Med. 1997;185:239–250. doi: 10.1084/jem.185.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Storb U, Peters A, Klotz E, Kim N, Shen H M, Hackett J, Rogerson B, Martin T E. Immunol Rev. 1998;162:153–160. doi: 10.1111/j.1600-065x.1998.tb01438.x. [DOI] [PubMed] [Google Scholar]
- 50.Michael N, Martin T E, Nicolae D, Kim N, Padjen K, Zhan P, Nguyen H, Pinkert C, Storb U. Immunity. 2002;16:123–134. doi: 10.1016/s1074-7613(02)00261-3. [DOI] [PubMed] [Google Scholar]
- 51.Bachl J, Carlson C, Gray-Schopfer V, Dessing M, Olsson C. J Immunol. 2001;166:5051–5057. doi: 10.4049/jimmunol.166.8.5051. [DOI] [PubMed] [Google Scholar]
- 52.Chen W Y, Iida S, Louie D C, Dallafavera R, Chaganti R S K. Blood. 1998;91:603–607. [PubMed] [Google Scholar]
- 53.Gerbitz A, Mautner J, Geltinger C, Hortnagel K, Christoph B, Asenbauer H, Klobeck G, Polack A, Bornkamm G W. Oncogene. 1999;18:1745–1753. doi: 10.1038/sj.onc.1202468. [DOI] [PubMed] [Google Scholar]
- 54.Iida S, Rao P H, Ueda R, Chaganti R S K, Dalla-Favera R. Leuk Lymphoma. 1999;34:25–33. doi: 10.3109/10428199909083377. [DOI] [PubMed] [Google Scholar]
- 55.Ernst P, Smale S T. Immunity. 1995;2:427–438. doi: 10.1016/1074-7613(95)90024-1. [DOI] [PubMed] [Google Scholar]
- 56.Malone C S, Wall R. J Immunol. 2002;168:3369–3375. doi: 10.4049/jimmunol.168.7.3369. [DOI] [PubMed] [Google Scholar]
- 57.Hermanson G G, Briskin M, Sigman D, Wall R. Proc Natl Acad Sci USA. 1989;86:7341–7345. doi: 10.1073/pnas.86.19.7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Honjo T. Nat Immunol. 2002;3:800–801. doi: 10.1038/ni0902-800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.