Abstract
The transcription factor signal transducer and activator of transcription 3 (Stat3) is constitutively activated in a variety of cancers including squamous cell carcinoma of the head and neck (SCCHN). Previous investigations have demonstrated that activated Stat3 contributes to a loss of growth control and transformation. To investigate the therapeutic potential of blocking Stat3 in cancer cells, we developed a transcription factor decoy to selectively abrogate activated Stat3. The Stat3 decoy was composed of a 15-mer double-stranded oligonucleotide, which corresponded closely to the Stat3 response element within the c-fos promoter. The Stat3 decoy bound specifically to activated Stat3 and blocked binding of Stat3 to a radiolabeled Stat3 binding element. By contrast, a mutated version of the decoy that differed by only a single base pair did not bind the activated Stat3 protein. Treatment of head and neck cancer cells with the Stat3 decoy inhibited proliferation and Stat3-mediated gene expression, but did not decrease the proliferation of normal oral keratinocytes. Thus, disruption of activated Stat3 by using a transcription factor decoy approach may serve as a novel therapeutic strategy for cancers characterized by constitutive Stat3 activation.
STAT proteins perform the dual function of signal transduction and activation of transcription. STATs have been implicated in signaling by numerous cytokines, polypeptide growth factors, and oncoproteins. After activation, STAT proteins dimerize and translocate to the nucleus, where they bind to DNA-response elements and regulate gene expression (1). STAT proteins were initially described in the context of regulating physiologic cell signaling, contributing to such diverse processes as differentiation, proliferation, and apoptosis (2). An increasing number of studies have implicated STAT activation, particularly Stat3, in transformation and tumor progression (2).
Cumulative evidence supports a central role for aberrant STAT signaling in oncogenesis. Constitutive activation of Stat3 has been detected in many cancers, including, multiple myeloma, leukemias, lymphomas, mycoses fungoides, and brain, prostate, breast, lung, and head and neck cancers (3–7). Cells stably transformed by the v-Src oncoprotein have been found to harbor activated Stat3, thus linking Stat3 activation to Src-mediated oncogenesis (8). To directly address the role of Stat3 as an oncogene, a constitutively active mutant of Stat3 was generated and shown to induce transformation of fibroblasts and tumor formation in nude mice (9). Further investigation demonstrated that Stat3-transformed fibroblasts were resistant to apoptotic stimuli, indicating that cancers characterized by Stat3 activation may be less susceptible to chemotherapy and/or irradiation (10). The association of Stat3 activation with transformation and tumor progression suggests that Stat3 may be an attractive molecular target for cancer therapy.
Several strategies have been used to block the action of STAT proteins, including antisense methods, ectopic expression of dominant-negative mutants (11–13), inhibition of upstream kinases (14–16), and phosphotyrosyl peptides (17). An alternative approach to target the action of transcription factors, including STAT proteins, involves the use of double-stranded “decoy” oligonucleotides. The double-stranded DNA decoy closely corresponds to the response element within the promoter region of a responsive gene. By achieving a sufficient concentration of decoy in the target cells, the authentic interaction between a transcription factor and its endogenous response element in genomic DNA is impaired, with subsequent modulation of gene expression (18). This approach has been used successfully to target Stat6 activation resulting in preferential restriction of IL-4-driven T helper 2 cell activity (19).
The present study was undertaken to evaluate the ability of a double-stranded decoy oligonucleotide based on the Stat3 binding sequence, hSIE, to target activated Stat3 in a relevant tumor model (20). We previously demonstrated constitutive Stat3 activation downstream of an epidermal growth factor receptor (EGFR) autocrine growth pathway in squamous cell carcinoma of the head and neck (SCCHN) in vitro and in vivo (5, 21). Further investigation suggested that constitutive Stat3 activation contributed to tumor growth independent of the EGFR autocrine axis in SCCHN and may therefore serve as a therapeutic target (22). Based on these earlier results, we hypothesized that a Stat3 decoy would inhibit the binding of phosphorylated Stat3 dimers to the promoter region of Stat3 target genes, thereby inhibiting Stat3-mediated gene regulation (see Fig. 8, which is published as supporting information on the PNAS web site, www.pnas.org). We use cell lines established from patients with SCCHN to show that a Stat3 decoy selectively binds to activated Stat3 and blocks Stat3-mediated gene transcription in these cancer cells. Treatment with the Stat3 decoy formulation also inhibits the proliferation of SCCHN cells. By contrast, Stat3 decoy treatment of normal oral cells has no effect on cell growth. These results suggest a novel method for selectively blocking activated Stat3 in tumor cells that are characterized by constitutive activation of this oncogenic transcription factor.
Methods
Cells.
The SCCHN cell lines 1483 (23), UM-22b (24), and PCI-37a (25) were grown in DMEM (Cellgro, Washington, DC) with 15% FBS (GIBCO/BRL, Grand Island, NY), plus 100 units/ml penicillin and 100 units/ml streptomycin (GIBCO/BRL). Normal oral cells were grown as primary cultures derived from uvulopalatopharyngoplasty specimens as described (26).
Stat3 Decoy and Mutant Control Decoys.
Sense and antisense strands of Stat3 decoy and mutant control decoy oligonucleotides were designed and obtained from MWG Biotech (High Point, NC). The Stat3 decoy sequence was 5′-CATTTCCCGTAAATC-3′, 3′GTAAAGGGCATTTAC-5′ and the mutant control decoy sequence was 5′-CATTTCCTTAAATC-3′, 3′-GTAAAGGGAATTTAG-5′ (for the rest of the mutant decoy sequences, see Table 1, which is published as supporting information on the PNAS web site). Sense and antisense strands were dissolved in Tris⋅EDTA (pH 8.0) at a concentration of 900-1,200 μM. Each sense–antisense pair was annealed by heating to 90°C and decreasing the temperature by 5°C increments every 15 min. After 3 h the reaction mixture was held at a base temperature of 4°C.
Activation, Expression, Incorporation, and Reporter Assays.
All assays were performed by using standard methods (see Detailed Methods, which is published as supporting information on the PNAS web site, for details).
Results
Design of a Control Decoy Oligonucleotide.
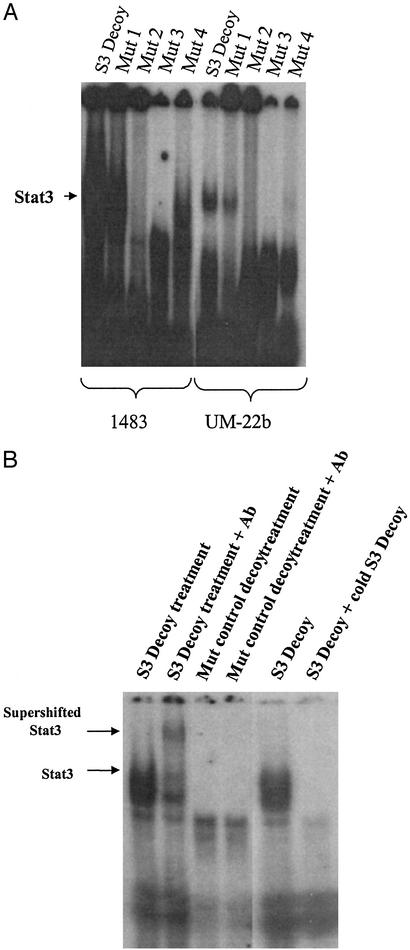
Earlier studies with transcription factor decoys have typically used a scrambled version of the decoy sequence as a control (27–32). We sought to design a control decoy with the greatest possible homology to the Stat3 decoy, but with no Stat3-specific DNA binding activity. Previous analysis of the c-fos promoter evaluated random mutations and reported their relative ability to induce SIF-binding activity (33). We systematically synthesized several possible permutation of these mutants and tested their ability to bind activated Stat3 by electrophoretic mobility-shift assay (EMSA) analysis (Fig. 1A).
Figure 1.
Binding of Stat3 decoy and mutant control decoys to activated Stat3. (A) Nuclear extracts (20 μg) were prepared from two representative SCCHN cell lines (1483 and UM-22b). EMSA was performed with radiolabeled hSIE Stat3 decoy duplex oligonucleotide and each of four single-nucleotide mutants of hSIE (see Table 1). (B) Supershift EMSA was performed with radiolabeled hSIE Stat3 decoy and mutant control decoy duplex oligonucleotide by using extracts from a representative SCCHN cell line (1483). Extracts were preincubated with rabbit antibody (C-20) or no antibody as indicated. Cold compete EMSA was performed with radiolabeled hSIE Stat3 decoy by using extracts from a representative SCCHN cell line (1483). A 400-fold excess of unlabeled hSIE Stat3 decoy was incubated with the radiolabeled hSIE Stat3 decoy as indicated.
The double or triple nucleotide mutants synthesized demonstrated variable degrees of binding to activated Stat3. Analysis of the single nucleotide mutants revealed that several mutants appeared to have little or no binding to activated Stat3. A single nucleotide mutant with no observable binding to activated Stat3 (Mutant 3) was further evaluated by supershift EMSA analysis using lysates from two representative SCCHN cell lines (Fig. 1B). In the supershift experiments, the bound proteins were incubated with Stat3-specific antisera to confirm that Stat3 was contained in the gel shift complex. The supershift EMSAs confirmed that a single-nucleotide mutant of the Stat3 decoy sequence (mutant control decoy) had negligible or no binding to activated Stat3 (Fig. 1B).
Incubation of SCCHN Cells with Stat3 Decoy Blocks Binding of Radiolabeled Stat3.
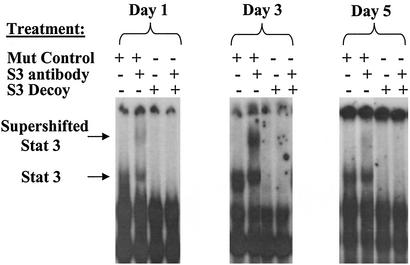
We hypothesized that treatment of cells with the Stat3 decoy would interfere with the binding of Stat3 to Stat3-specific DNA response elements. To test this hypothesis, a representative SCCHN cell line (1483) was treated with either Stat3 decoy or the mutant control decoy, and the cells were harvested at several time points and evaluated by EMSA. Protein lysates (20 μg) from the decoy-treated cells were subjected to supershift EMSA analysis with radiolabeled hSIE (Fig. 2). At all time points examined, the Stat3 decoy-treated cells demonstrated a marked decrease in formation of the hSIE-protein complex in the gel shift assay relative to mutant control decoy-treated cells, indicating that the Stat3 decoy interferes with activated Stat3. Although we cannot rule out the possibility that the Stat3 decoy present in the cell extracts was acting to “cold-compete” in the EMSA assay, subsequent experiments (see Fig. 7 below) demonstrated decreased Stat3 target gene expression following decoy treatment of whole cells, indicating that the decoy is functioning in vivo.
Figure 2.
Effect on cellular Stat3 protein after treatment with Stat3 decoy. Protein extracts were prepared from a representative SCCHN cell line (1483) after treatment with Stat3 decoy (25 μM) or mutant control decoy (25 μM). Cells were harvested on days 1, 3, and 5. Protein lysates (20 μg) were then subjected to EMSA analysis with radiolabeled hSIE with or without preincubation with Stat3 antibody.
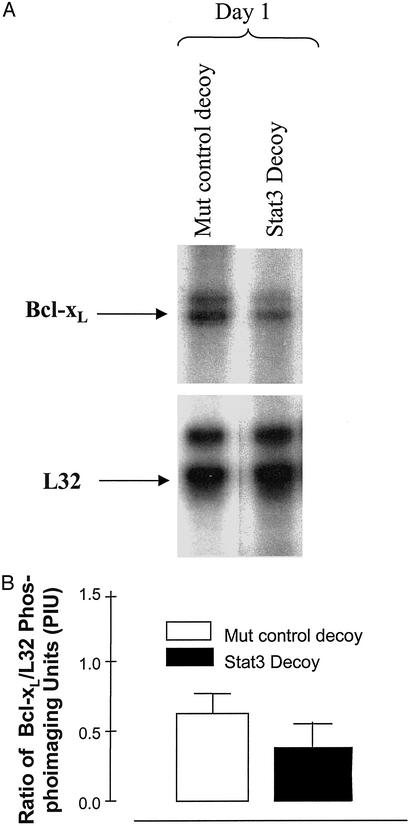
Figure 7.
Stat3 decoy decreases Bcl-xL RNA expression levels compared with mutant control decoy in a SCCHN cell line. Cells from a representative SCCHN cell line (1483) were treated with 25 μM Stat3 decoy in serum-free DMEM. Total RNA was harvested 1 day after treatment. RNase protection assays were then performed. (A) The Bcl-xL and L32 (loading control) bands were quantified by using phosphoimage analysis, and the ratio of Bcl-xL to L32 was calculated for each treatment group. (B) The percent decrease in the ratio of Bcl-xL to L32 in Stat3 decoy-treated cells compared with the mutant control decoy-treated cells was 40% after 24 h of Stat3 decoy treatment.
Changes in Stat3 activation levels can result from several mechanisms including alterations in steady state Stat3 protein levels or modulation of phosphorylated Stat3 protein. Stat3 decoy-induced abrogation of Stat3 activity was not associated with decreased steady-state expression levels of Stat3 protein or tyrosine phosphorylated Stat3 (see Fig. 9, which is published as supporting information on the PNAS web site). In addition to Stat3, Stat5 has been shown be constitutively activated in a variety of cancers including SCCHN (4, 34–37). To determine the specificity of the Stat3 decoy, Stat5 activation levels were examined in SCCHN cells treated with Stat3 decoy or mutant control decoy. In contrast to the effects on constitutive Stat3 activation, constitutive Stat5 activation was not abrogated by treatment of SCCHN cells with the Stat3 decoy. Similarly, Stat4 activation was not modulated by treatment with the Stat3 decoy (see Fig. 9).
Incorporation of Stat3 Decoy into Cells.
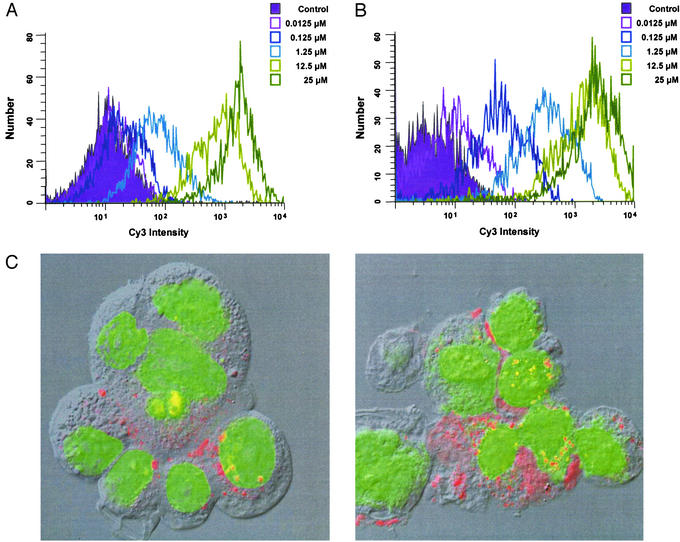
To determine the efficiency of Stat3 decoy incorporation into SCCHN cells, increasing concentrations of Cy3-labeled Stat3 decoy were added to SCCHN cells followed by flow cytometry. As shown in Fig. 3A, SCCHN cells incorporated the Stat3 decoy in a dose-dependent manner. Similar results were obtained on analysis of the mutant control decoy demonstrating that alteration of a single base pair of the Stat3 decoy sequence did not alter the incorporation kinetics of the oligonucleotide (data not shown). To verify that primary cultures established from normal oropharyngeal mucosa could serve as an appropriate comparison group for the decoy studies, the flow cytometry analysis was repeated after addition of labeled Stat3 decoy to these normal cells. As shown in Fig. 3B, the primary normal mucosal cells demonstrated nearly identical decoy uptake levels compared with the SCCHN cell lines. To further define the subcellular localization of the decoy formulations, optical imaging of decoy labeled with a fluorescent dye was performed in a representative SCCHN cell line (1483). By 6 h after incubation with the Stat3 decoy, the labeled decoy was detected in the nucleus, as well as in the cytosol of the SCCHN cells. Similar subcellular localization was observed with the mutant control decoy (data not shown). Confocal microscopy demonstrated similar intracellular localizalization of the labeled Stat3 decoy in both SCCHN and normal control cells (Fig. 3).
Figure 3.
Incorporation of Stat3 decoy into SCCHN and normal cells. (A) A representative SCCHN cell line (1483) was incubated with Cy3-labeled Stat3 decoy followed by flow cytometry. The cells were treated with Cy3-labeled Stat3 decoy for 2 h at 37°C, trypsinized, and washed extensively to eliminate surface-bound, noninternalized Cy3 signal. (B) Nearly identical uptake of the Stat3 decoy was observed in the normal mucosal epithelial cells. (C) Confocal microscopy demonstrating internalization of the Stat3 decoy in both SCCHN (Left) (1483) cells and normal mucosal epithelial (Right). Internalized Stat3 decoy (red) and nuclei (green) are overlaid with the corresponding differential interface contrast image.
Stat3 Decoy Inhibits SCCHN Cell Proliferation.
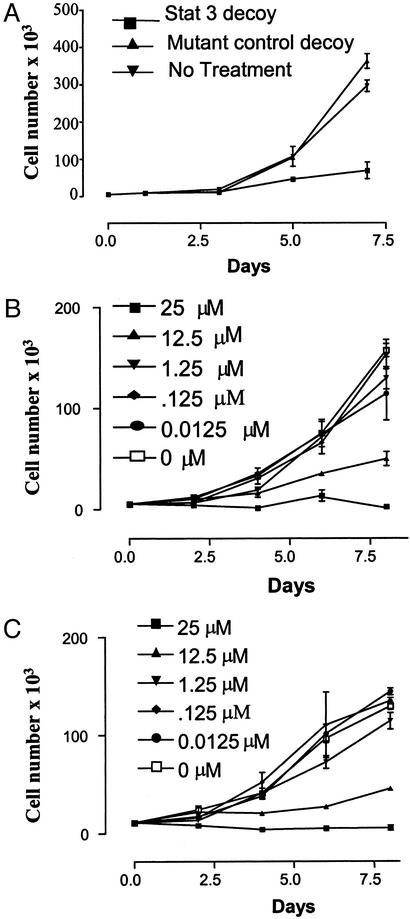
To determine the effects of the Stat3 decoy on SCCHN cell growth, cells were incubated with the Stat3 decoy, or the mutant control decoy, followed by cell count experiments (Fig. 4). Treatment with the Stat3 decoy (25 μM) resulted in potent growth inhibition of SCCHN cells in vitro. MTT assay also demonstrated up to 80% cytotoxicity resulting from Stat3 decoy treatment (data not shown). By contrast, the proliferation of SCCHN cells treated with the mutant control decoy was equivalent to no treatment (Fig. 4A). To determine whether the growth inhibitory effects of the Stat3 decoy were dose-dependent, SCCHN cells were incubated with a range of Stat3 decoy doses (0.0125–25 μM) and cell viability was determined at several time points. As shown in Fig. 4 B and C, the antiproliferative consequences of the Stat3 decoy were dose-dependent with maximum inhibition achieved with 25 μM Stat3 decoy. Other strategies have been shown to abrogate Stat3 including inhibition of upstream kinases or antisense oligonucleotides (11). To compare the relative potency of these Stat3 targeting strategies with the decoy, SCCHN cells were treated with an EGFR-specific tyrosine kinase inhibitor (PD153035), or phosphorothioated antisense oligonucleotides directed against the translation start site, or Stat3 decoy at doses previously optimized to abrogate Stat3 (11). Treatment with the Stat3 decoy was more growth inhibitory at several time points compared with the EGFR inhibitor or Stat3 antisense oligonucleotides (see Fig. 10, which is published as supporting information on the PNAS web site).
Figure 4.
The effect of Stat3 decoy on SCCHN proliferation. (A) SCCHN cells (1483) treated with 25 μM Stat3 decoy demonstrated growth inhibition compared with cells treated with mutant control decoy. (B and C) Representative SCCHN cell line (1483) (B) and (PCI-37a) (C) treated with 0.0125–25 μM Stat3 decoy demonstrated a dose-dependent inhibition of proliferation with maximum inhibition in both cell lines at a dose of 25 μM. Cells were harvested at the times indicated, and viable cells were counted by vital dye exclusion.
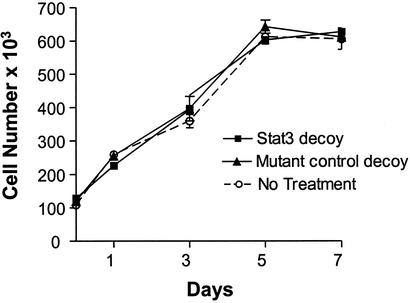
In addition to its role in transformation and tumor progression, Stat3 is important for a variety of critical functions in normal cells. We have previously demonstrated that Stat3 activation is restricted to SCCHN tumors and premalignant mucosa, and is not detected in normal epithelium from patients without cancer (5). To address the potential toxicity of Stat3 decoy-mediated Stat3 abrogation on noncancer cells, normal oral keratinocytes were treated with Stat3 decoy and mutant control decoy followed by growth determinations. As shown in Fig. 5, Stat3 decoy treatment had no effect on the growth of normal oral epithelial cells. This finding further highlights the potential therapeutic utility of the Stat3 decoy because, as shown in Fig. 3 B and C, these normal cells incorporated the decoy in a similar fashion to that observed in the SCCHN cells.
Figure 5.
Normal epithelial cells are resistant to the inhibitory effects of Stat3 decoy. Normal oral epithelial cells grown as primary cultures as described (26) were treated with Stat3 decoy or mutant control decoy and counted by vital dye exclusion at several time points after treatment.
Modulation of Stat3-Mediated Gene Expression.
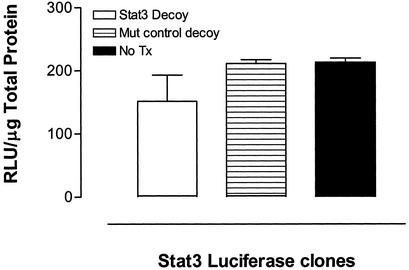
The biologic effects of activated Stat3 are likely mediated through regulation of Stat3 target genes. To examine the consequences of the Stat3 decoy on Stat3-mediated gene expression, we stably transfected SCCHN cells (UM-22b) with an hSIE-luciferase reporter, and then incubated the transfected cells with either Stat3 decoy or mutant control decoy. As shown in Fig. 6, Stat3 decoy treatment caused a 28% reduction in cellular luciferase activity, compared with treatment with a mutant control decoy.
Figure 6.
Stat3 decoy decreases luciferase activity in SCCHN stably expressing hSIE-luciferase activity. Cells from a representative cell line (UM-22b), which were stably transfected with a hSIE-luciferase construct (gift from R. Jove), were incubated with the Stat3 decoy or mutant control decoy and assayed 2 days later for luciferase activity. The luciferase activity units (RLU) were normalized to micrograms of total protein.
Stat3 response elements have been identified in the promoter regions of several genes that regulate cell growth and apoptosis. One such gene encodes the antiapoptotic protein Bcl-xL. To further investigate the consequences of the Stat3 decoy on Stat3-mediated gene expression, RNA was isolated from Stat3 decoy-treated SCCHN cells followed by RNase protection assay. As shown in Fig. 7, there was a 40% decrease in Bcl-xL levels after treatment with the Stat3 decoy compared with the mutant control decoy-treated cells.
Discussion
Cumulative evidence supports a role for aberrant Stat3 activation in transformation and tumor progression. We previously demonstrated increased Stat3 activation in head and neck carcinogenesis, where Stat3 contributes to the loss of growth control by an antiapoptotic mechanism (5). Targeting Stat3 with antisense oligonucleotides or dominant-negative mutants resulted in apoptosis and modulation of Stat3 regulated genes in several cancer-derived cell lines including multiple myeloma, melanoma, mycosis fungoides, and SCCHN (5, 38–40). The present study provides evidence that Stat3 activation can be targeted as an antitumor strategy. We used a novel decoy oligonucleotide approach to show selective abrogation of activated Stat3 accompanied by inhibition of tumor cell growth and abrogation of Stat3-mediated target gene expression. The lack of inhibitory effects on normal oral keratinocytes suggests that a Stat3 decoy strategy may selectively block the growth of cancer cells with relatively little toxicity to corresponding normal cells.
Decoy oligonucleotides have been proposed as a potential approach to block the action of transcription factors on gene expression (41). Treatment of cells with a decoy oligonucleotide, whose sequence closely corresponds to a transcription factor response element, may attenuate the authentic interaction of the transcription factor with the gene. The potential advantages of a decoy approach to target a transcription factor include (i) the potential drug targets (transcription factors) are readily identifiable; (ii) understanding the molecular structure of the target transcription factor is not essential; and (iii) synthesis of the decoy is relatively straightforward. Previous studies have demonstrated the potential efficacy of decoy oligonucleotides targeting other transcription factors in cancer treatment (27, 28, 42). In this study, we developed and optimized a decoy oligonucleotide targeting activated Stat3.
We used several approaches to examine the specificity of the Stat3 decoy formulation. In contrast to previous transcription factor decoy studies, we designed mutant control decoys that differed minimally from the Stat3 decoy. The control decoy we selected differed by only a single base pair compared with the Stat3 decoy yet demonstrated no binding to Stat3, and no abrogation of exogenous Stat3 activity was detected on EMSA after treatment of SCCHN cells with this control decoy. Furthermore, treatment of SCCHN cells with the Stat3 decoy inhibited the growth of cancer cells, whereas the mutant control decoy had no effect on SCCHN proliferation.
Stat3 activation has been reported to induce the expression of genes that control critical cellular functions including proliferation, immune response, survival, differentiation, and development. Homozygous deletion of Stat3 was embryonically lethal (43). The ubiquitous expression of Stat3 raises the possibility that blocking Stat3 may be harmful to normal cells. Analysis of tissue-specific Stat3-null cells revealed that keratinocytes lacking Stat3 demonstrated impaired would healing (44). However, disruption of Stat3 signaling with dominant-negative approaches in murine fibroblasts did not inhibit normal cell growth (39, 45). We previously reported that although Stat3 protein expression is similar in normal oral epithelium and oral cancer tissues, levels of activated Stat3 in cancers were dramatically elevated in SCCHN compared with normal mucosa (5). In the present study, the Stat3 decoy functioned by specifically abrogating activated Stat3 while having no effect on steady-state protein expression levels of Stat3 or phosphotyrosine Stat3. Furthermore, no anti-proliferative effect of the Stat3 decoy was observed in normal human oral keratinocytes. Therefore, abrogating activated Stat3 by using a transcription factor decoy approach may selectively impair cancer cell growth with minimal toxicity to normal cells. In this regard, brief inactivation of an oncogene was recently shown to have sustained antitumor effects without associated toxic in vivo (46). Prior studies of transcription factor decoys have reported that the decoy localized predominantly to the nucleus from 1 to 12 h after transfection (27, 30, 32). Use of the mutant control decoy verified that the specific effects of the Stat3 decoy were not caused by preferential kinetics of uptake or degree of incorporation.
Constitutive activation of Stat3 in cancer cells is thought to contribute to tumor progression by regulation of several genes that control cell proliferation and survival. Specifically, constitutively active Stat3 has been shown to enhance bcl-x and cyclin D1 transcription (9). Antisense oligonucleotdies and/or dominant-negative mutants targeting Stat3 blocked Stat3-dependent transcription and induced apoptosis in mycosis fungoides, melanoma, squamous cell carcinoma, and myeloma cell lines (5, 38–40). We used SCCHN cells stably expressing an hSIE-luciferase reporter and RNase protection assays to show that Stat3 decoy treatment of SCCHN cells decreased Stat3 target gene expression and Stat3-mediated gene transcription.
In this study, growth inhibition and alteration of target gene expression after treatment of SCCHN cells with the Stat3 decoy supports Stat3 as a potential therapeutic target. We assessed the incorporation of the decoy into SCCHN cells by using fluorescence-labeled flow cytometry and found that the decoy was detected in a high percentage (>90%) of the cells examined. Consistent with this observation, the Stat3 decoy demonstrated marked and reproducible effects on Stat3-mediated growth pathways. The profound effects of the decoy on proliferation suggests that in addition to direct incorporation, an extracellular or secreted mechanism may be involved. The overall efficiency of action of the decoy is likely affected by many complex interacting factors including intracellular levels of the Stat3 decoy, levels of activated Stat3, the availability of target genes, and the relative affinites of the DNA response elements for Stat3. In addition, there are likely multiple genes (in addition to Bcl-xL) that contribute to SCCHN growth. Abrogation of Stat3 (or specific target genes), below a threshold (<100%) may be sufficient to inhibit growth. Optimization of the Stat3 decoy to enhance delivery to SCCHN cells should result in more profound antitumor effects. We have previously demonstrated that constitutive Stat3 activation in SCCHN occurs downstream of the EGF receptor in vitro and in vivo (5, 11). Further investigation showed that SCCHN cells expressing constitutively activated Stat3 continued to proliferate in vitro and in vivo and were resistant to the antitumor effects of EGFR abrogation (22). A recent study reported that Stat3 activation levels in primary SCCHN tumors were associated with decreased survival rates (47). These cumulative results suggest that Stat3 blockade, alone or in combination with EGFR inhibition (e.g., with monoclonal antibodies or tyrosine kinase-specific inhibitors), may augment the antitumor effects of molecular targeting approaches for cancer therapy.
Supplementary Material
Acknowledgments
We are grateful to Dr. Richard Jove for providing the hSIE luciferase construct. This work was supported by National Institutes of Health Grant CA77308 (to J.R.G.), a Resident Research Award from the American Academy of Otolaryngology–Head and Neck Surgery (to P.L.L.), and a Medical Student Research grant from the Howard Hughes Medical Institute (to G.A.A.).
Abbreviations
- EGFR
epidermal growth factor receptor
- SCCHN
squamous cell carcinoma of the head and neck
- EMSA
electrophoretic mobility-shift assay
- STAT
signal transducer and activator of transcription
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Darnell J E., Jr Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 2.Bromberg J, Darnell J E., Jr Oncogene. 2000;19:2468–2473. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- 3.Garcia R, Bowman T L, Niu G, Yu H, Minton S, Muro-Cacho C A, Cox C E, Falcone R, Fairclough R, Parsons S, et al. Oncogene. 2001;20:2499–2513. doi: 10.1038/sj.onc.1204349. [DOI] [PubMed] [Google Scholar]
- 4.Gouilleux-Gruart V, Gouilleux F, Desaint C, Claisse J F, Capiod J C, Delobel J, Weber-Nordt R, Dusanter-Fourt I, Dreyfus F, Groner B, Prin L. Blood. 1996;87:1692–1697. [PubMed] [Google Scholar]
- 5.Grandis J R, Drenning S D, Zeng Q, Watkins S C, Melhem M F, Endo S, Johnson D E, Huang L, He Y, Kim J D. Proc Natl Acad Sci USA. 2000;97:4227–4232. doi: 10.1073/pnas.97.8.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang M, Page C, Reynolds R K, Lin J. Gynecol Oncol. 2000;79:67–73. doi: 10.1006/gyno.2000.5931. [DOI] [PubMed] [Google Scholar]
- 7.Bowman T, Garcia R, Turkson J, Jove R. Oncogene. 2000;19:2474–2488. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 8.Yu C L, Meyer D J, Campbell G S, Larner A C, Carter-Su C, Schwartz J, Jove R. Science. 1995;269:81–83. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- 9.Bromberg J F, Wrzeszczynska M H, Devgan G, Zhao Y, Pestell R G, Albanese C, Darnell J E., Jr Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 10.Shen Y, Devgan G, Darnell J E, Jr, Bromberg J F. Proc Natl Acad Sci USA. 2001;98:1543–1548. doi: 10.1073/pnas.041588198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grandis J R, Drenning S D, Chakraborty A, Zhou M Y, Zeng Q, Pitt A S, Tweardy D J. J Clin Invest. 1998;102:1385–1392. doi: 10.1172/JCI3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakajima K, Yamanaka Y, Nakae K, Kojima H, Ichiba M, Kiuchi N, Kitaoka T, Fukada T, Hibi M, Hirano T. EMBO J. 1996;15:3651–3658. [PMC free article] [PubMed] [Google Scholar]
- 13.Li L, Shaw P E. J Biol Chem. 2002;277:17397–17405. doi: 10.1074/jbc.M109962200. [DOI] [PubMed] [Google Scholar]
- 14.Fry D W, Kraker A J, McMichael A, Ambroso L A, Nelson J M, Leopold W R, Connors R W, Bridges A J. Science. 1994;265:1093–1095. doi: 10.1126/science.8066447. [DOI] [PubMed] [Google Scholar]
- 15.Kraker A J, Hartl B G, Amar A M, Barvian M R, Showalter H D, Moore C W. Biochem Pharmacol. 2000;60:885–898. doi: 10.1016/s0006-2952(00)00405-6. [DOI] [PubMed] [Google Scholar]
- 16.Turkson J, Bowman T, Adnane J, Zhang Y, Djeu J Y, Sekharam M, Frank D A, Holzman L B, Wu J, Sebti S, Jove R. Mol Cell Biol. 1999;19:7519–7528. doi: 10.1128/mcb.19.11.7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turkson J, Ryan D, Kim J S, Zhang Y, Chen Z, Haura E, Laudano A, Sebti S, Hamilton A D, Jove R. J Biol Chem. 2001;276:45443–45455. doi: 10.1074/jbc.M107527200. [DOI] [PubMed] [Google Scholar]
- 18.Nabel E G, Plautz G, Nabel G J. Science. 1990;249:1285–1288. doi: 10.1126/science.2119055. [DOI] [PubMed] [Google Scholar]
- 19.Wang L H, Yang X Y, Kirken R A, Resau J H, Farrar W L. Blood. 2000;95:1249–1257. [PubMed] [Google Scholar]
- 20.Sadowski H B, Shuai K, Darnell J E, Jr, Gilman M Z. Science. 1993;261:1739–1744. doi: 10.1126/science.8397445. [DOI] [PubMed] [Google Scholar]
- 21.Rubin Grandis J, Zeng Q, Drenning S D. Laryngoscope. 2000;110:868–874. doi: 10.1097/00005537-200005000-00016. [DOI] [PubMed] [Google Scholar]
- 22.Kijima T, Niwa H, Steinman R A, Drenning S D, Gooding W E, Wentzel A L, Rubin Grandis J. Cell Growth Differ. 2002;13:355–362. [PubMed] [Google Scholar]
- 23.Beckhardt R N, Kiyokawa N, Xi L, Liu T J, Hung M C, el-Naggar A K, Zhang H Z, Clayman G L. Arch Otolaryngol Head Neck Surg. 1995;121:1265–1270. doi: 10.1001/archotol.1995.01890110041008. [DOI] [PubMed] [Google Scholar]
- 24.Riser B L, Mitra R, Perry D, Dixit V, Varani J. Cancer Res. 1989;49:6123–6129. [PubMed] [Google Scholar]
- 25.Heo D S, Snyderman C, Gollin S M, Pan S, Walker E, Deka R, Barnes E L, Johnson J T, Herberman R B, Whiteside T L. Cancer Res. 1989;49:5167–5175. [PubMed] [Google Scholar]
- 26.Rubin Grandis J, Zeng Q, Tweardy D J. Nat Med. 1996;2:237–240. doi: 10.1038/nm0296-237. [DOI] [PubMed] [Google Scholar]
- 27.Park Y G, Nesterova M, Agrawal S, Cho-Chung Y S. J Biol Chem. 1999;274:1573–1580. doi: 10.1074/jbc.274.3.1573. [DOI] [PubMed] [Google Scholar]
- 28.Sharma H W, Narayanan R. Anticancer Res. 1996;16:589–596. [PubMed] [Google Scholar]
- 29.Sharma H W, Perez J R, Higgins-Sochaski K, Hsiao R, Narayanan R. Anticancer Res. 1996;16:61–69. [PubMed] [Google Scholar]
- 30.Sawa Y, Morishita R, Suzuki K, Kagisaki K, Kaneda Y, Maeda K, Kadoba K, Matsuda H. Circulation. 1997;96,(Suppl.):II-280–II-284. ; discussion, II-285. [PubMed] [Google Scholar]
- 31.Boccaccio C, Ando M, Tamagnone L, Bardelli A, Michieli P, Battistini C, Comoglio P M. Nature. 1998;391:285–288. doi: 10.1038/34657. [DOI] [PubMed] [Google Scholar]
- 32.Akimoto M, Hangai M, Okazaki K, Kogishi J, Honda Y, Kaneda Y. Exp Eye Res. 1998;67:395–401. doi: 10.1006/exer.1998.0531. [DOI] [PubMed] [Google Scholar]
- 33.Wagner B J, Hayes T E, Hoban C J, Cochran B H. EMBO J. 1990;9:4477–4484. doi: 10.1002/j.1460-2075.1990.tb07898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nieborowska-Skorska M, Wasik M A, Slupianek A, Salomoni P, Kitamura T, Calabretta B, Skorski T. J Exp Med. 1999;189:1229–1242. doi: 10.1084/jem.189.8.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olayioye M A, Beuvink I, Horsch K, Daly J M, Hynes N E. J Biol Chem. 1999;274:17209–17218. doi: 10.1074/jbc.274.24.17209. [DOI] [PubMed] [Google Scholar]
- 36.Weber-Nordt R M, Egen C, Wehinger J, Ludwig W, Gouilleux-Gruart V, Mertelsmann R, Finke J. Blood. 1996;88:809–816. [PubMed] [Google Scholar]
- 37.Leong P L, Xi S, Drenning S D, Dyer K F, Wentzel A L, Lerner E C, Smithgall T E, Grandis J R. Oncogene. 2002;21:2846–2853. doi: 10.1038/sj.onc.1205385. [DOI] [PubMed] [Google Scholar]
- 38.Catlett-Falcone R, Landowski T H, Oshiro M M, Turkson J, Levitzki A, Savino R, Ciliberto G, Moscinski L, Fernandez-Luna J L, Nunez G, et al. Immunity. 1999;10:105–115. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- 39.Niu G, Heller R, Catlett-Falcone R, Coppola D, Jaroszeski M, Dalton W, Jove R, Yu H. Cancer Res. 1999;59:5059–5063. [PubMed] [Google Scholar]
- 40.Nielsen M, Kaestel C G, Eriksen K W, Woetmann A, Stokkedal T, Kaltoft K, Geisler C, Ropke C, Odum N. Leukemia. 1999;13:735–738. doi: 10.1038/sj.leu.2401415. [DOI] [PubMed] [Google Scholar]
- 41.Bielinska A, Shivdasani R A, Zhang L Q, Nabel G J. Science. 1990;250:997–1000. doi: 10.1126/science.2237444. [DOI] [PubMed] [Google Scholar]
- 42.Romano M F, Lamberti A, Bisogni R, Tassone P, Pagnini D, Storti G, Del Vecchio L, Turco M C, Venuta S. Gene Ther. 2000;7:1234–1237. doi: 10.1038/sj.gt.3301216. [DOI] [PubMed] [Google Scholar]
- 43.Takeda T, Kurachi H, Yamamoto T, Homma H, Morishige K, Miyake A, Murata Y. J Endocrinol. 1997;153:R1–R3. doi: 10.1677/joe.0.153r001. [DOI] [PubMed] [Google Scholar]
- 44.Sano S, Itami S, Takeda K, Tarutani M, Yamaguchi Y, Miura H, Yoshikawa K, Akira S, Takeda J. EMBO J. 1999;18:4657–4668. doi: 10.1093/emboj/18.17.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turkson J, Bowman T, Garcia R, Caldenhoven E, De Groot R P, Jove R. Mol Cell Biol. 1998;18:2545–2552. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, Sundberg C D, Bishop J M, Felsher D W. Science. 2002;297:102–104. doi: 10.1126/science.1071489. [DOI] [PubMed] [Google Scholar]
- 47.Masuda M, Suzui M, Yasumatu R, Nakashima T, Kuratomi Y, Azuma K, Tomita K, Komiyama S, Weinstein I B. Cancer Res. 2002;62:3351–3355. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.