Abstract
Deinococcus radiodurans R1 (DEIRA) is a bacterium best known for its extreme resistance to the lethal effects of ionizing radiation, but the molecular mechanisms underlying this phenotype remain poorly understood. To define the repertoire of DEIRA genes responding to acute irradiation (15 kGy), transcriptome dynamics were examined in cells representing early, middle, and late phases of recovery by using DNA microarrays covering ≈94% of its predicted genes. At least at one time point during DEIRA recovery, 832 genes (28% of the genome) were induced and 451 genes (15%) were repressed 2-fold or more. The expression patterns of the majority of the induced genes resemble the previously characterized expression profile of recA after irradiation. DEIRA recA, which is central to genomic restoration after irradiation, is substantially up-regulated on DNA damage (early phase) and down-regulated before the onset of exponential growth (late phase). Many other genes were expressed later in recovery, displaying a growth-related pattern of induction. Genes induced in the early phase of recovery included those involved in DNA replication, repair, and recombination, cell wall metabolism, cellular transport, and many encoding uncharacterized proteins. Collectively, the microarray data suggest that DEIRA cells efficiently coordinate their recovery by a complex network, within which both DNA repair and metabolic functions play critical roles. Components of this network include a predicted distinct ATP-dependent DNA ligase and metabolic pathway switching that could prevent additional genomic damage elicited by metabolism-induced free radicals.
The Gram-positive aerobic bacterium Deinococcus radiodurans R1 (DEIRA) has an extraordinary resistance to γ-radiation and a wide range of other DNA-damaging conditions, including desiccation and oxidizing agents (1, 2). Ionizing radiation induces DNA double-stranded breaks (DSBs) that are the most lethal form of DNA damage (3). After acute exposures to 10 kGy, early stationary phase (ESP) DEIRA can reassemble its 3.285-Mbp genome, which consists of four haploid genomic copies per cell (4), from hundreds of DNA DSB fragments without lethality or induced mutagenesis (5, 6). Also remarkable is DEIRA's ability to grow at 60 Gy/h without any discernable effect on its growth rate (7). Because most organisms, generally, can tolerate so few DSBs (8), radiation-induced DSBs and their repair have been difficult to study. In DEIRA, however, there are so many DSBs in fully viable irradiated cells after high-dose irradiation that the steps in DSB repair can be monitored directly in mass culture (5, 9–11). This characteristic has been exploited and used to examine the timing of DNA recombination (5, 10, 12) after high-dose irradiation and has revealed the sequential action of RecA-independent and -dependent pathways during repair (11).
Comparative genomic and experimental analyses support the view that DEIRA's extreme radiation resistance phenotype is complex, likely determined collectively by an assortment of protection and DNA repair systems. Remarkably, the number of genes identified in DEIRA that are known to be involved in DNA repair is less than the number reported for Escherichia coli (13), and most of the DNA repair genes identified in DEIRA have functional homologs in other prokaryotic species (13, 14). These findings suggest that the organism's extreme resistance phenotype may be attributable to still unknown genes and pathways. Despite these efforts, the molecular mechanisms underlying its resistance remain poorly understood. Thus, a systematic genome-wide examination of the genes and pathways involved in cell recovery would be useful for a further understanding of how DEIRA responds to and recovers from irradiation. Here we report the analysis of genomic expression within cells recovering from 15 kGy by using whole-genome DNA microarrays. We find that the hallmark components of DEIRA's recovery encompass differential regulation of systems involved in information storage and processing, metabolism, and many uncharacterized genes that respond to high-dose irradiation.
Materials and Methods
Cell Growth, Irradiation, and Mutant Construction.
DEIRA strain R1 was grown at 32°C in liquid nutrient-rich medium TGY (1% tryptone/0.1% glucose/0.5% yeast extract) or on TGY solid medium (7). In liquid culture, cell density was determined at 600 nm by a Beckman Coulter spectrophotometer. For high-dose irradiation exposure, 150 ml of an early stationary phase (ESP) DEIRA culture [OD600 = 1.0, ≈1 × 108 colony-forming units (cfu)/ml] was divided in half. Half of the culture (75 ml) was irradiated on ice to a total dose of 15 kGy (Model 109 60Cobalt gamma cell irradiation unit, J. L. Shepherd and Associates, San Fernando, CA). The nonirradiated control culture was incubated on ice for the same length of time (≈98 min) as the culture being irradiated, followed by harvesting through brief (≈1 min) centrifugation (≈3,500 × g, ≈2°C). Control cells were washed and then frozen in RNAlater solution (Ambion, Austin, TX) that had been maintained on ice and then stored at −80°C. After exposure to 15 kGy, the irradiated cell culture (75 ml) was diluted 20-fold by using fresh TGY medium (at a final volume of 1.5 liters) and incubated at 32°C in an orbital shaker. At the indicated recovery time points (0, 0.5, 1.5, 3, 5, 9, 12, 16, and 24 h), ≈1 × 109 cells were harvested, washed, and frozen in RNAlater solution. Cell viability and cell numbers were determined by plate assay and hemocytometer count, respectively, as described (5). Three independent cell cultures and irradiation treatments of the same kind were performed and served as biological replicates for gene expression experiments, as well as for determining irradiation resistance profiles. To confirm the predicted involvement of one uncharacterized DEIRA gene implicated in postirradiation recovery (DR0070), a mutant was generated using previously developed DEIRA disruption protocols (15).
Nucleic Acid Extraction, Microarray Fabrication, and Data Analysis.
Protocols for DEIRA genomic DNA and total cellular RNA extraction and manipulation were as published (7, 11, 16, 18). Microarray construction was based on the genomic sequence data and annotation provided by the Institute for Genomic Research (Rockville, MD), where a total of 3,187 putative ORFs were assigned to the DEIRA genome (14). For PCR, gene-specific primers were selected using the primegens software (ref. 17; http://compbio.ornl.gov/structure/primegens). Generally, a full or nearly full sequence of a gene was selected as a probe on microarrays if it was <75% similar to all other genes. For genes having higher than 75% similarity to another gene, a maximum internal region showing <75% was selected as a probe. For genes where no primers could be obtained under the cutoff value of 75% similarity, a cutoff value of 85% was used. In total, 3,046 pairs of gene-specific primers were designed and then synthesized by MWG Biotec (High Point, NC).
Each gene was amplified four times in a 96-well plate. The amplified PCR products (400 μl) were pooled together and purified as described (18). The purified PCR products were visualized by gel electrophoresis in the presence of ethidium bromide, and their respective sizes were verified by comparison to the expected product length, yielding a collection of 2,976 distinct ORFs. Microarray fabrication, hybridization, probe labeling, image acquisition, and processing were carried out as described (18, 19). Because nonirradiated ESP DEIRA cells inoculated into fresh medium yield a fully grown culture in the time taken by the 15 kGy-induced growth lag, our expression analyses compared total RNA derived from recovering DEIRA with total RNA from nonirradiated control cells. The ratios of the irradiated samples to the nonirradiated control were normalized using the Pooled-Common-Error model provided by the statistical analysis software ARRAYSTAT V.2.0 (Imaging Research, St. Catherine's, ON, Canada). The outliers, represented by the data points that were not consistently reproducible and had a disproportionately large effect on the statistical result, were removed. A standard t test was performed so that a two-tailed probability of a mean deviating from 1.0 could be calculated and used to determine the significance for each data point. Identification of groups of genes exhibiting similar expression patterns was performed using the pairwise average-linkage hierarchical clustering algorithm (20) provided in cluster software (http://rana.stanford.edu/). The results of hierarchical clustering were visualized using treeview software (http://rana.stanford.edu/). The complete microarray data set for the recovery time course can be found in table A at www.esd.ornl.gov/facilities/genomics/TableA.pdf; Supporting Text, Figs. 5–9, and Tables 1–4 are published as supporting information on the PNAS web site, www.pnas.org (all of the supporting information can also be found at the Oak Ridge National Laboratory's Environmental Sciences Division web site, www.esd.ornl.gov/facilities/genomics/functional_genomics.html).
Results
Cell Growth, Inhibition, and Recovery.
After a dose of 15 kGy, DEIRA recovery typically progresses through three phases (5, 9, 10, 21): (i) early phase (0–3 h), where cell growth is inhibited and recA is induced, but there is little evidence of DNA repair; (ii) mid phase (3–9 h), where growth inhibition and recA expression continue, but with progressive DNA repair; and (iii) late phase (9–24 h), where recA is repressed and cell growth is restored. Consistent with these reports (5, 6, 9–11), after the exposure of DEIRA to 15 kGy, ≈150 DSBs per haploid genome were inflicted (data not shown). As expected, after the 9-h lag in growth, cells grew exponentially and reached stationary phase 15 h later (Fig. 5). To separate potentially damage-induced genes from cell-growth-related genes, we focused on the gene expression changes that occurred during the early and mid phases of recovery.
Quality of Microarray Hybridization Data.
Because microarray hybridization exhibits inherent high variability, experimental replications are essential for obtaining reliable results (22). In this study, samples were taken at nine time intervals over a period of 24 h. At each time point, three replicated samples were obtained. Each microarray slide contained two duplicate sets of gene fragments, and the RNA obtained from each sample was hybridized with two microarrays by using fluorescent-dye reversal. Thus, 12 data points were available for each time point and enabled the use of statistical tests to determine significant changes in gene expression. Only those genes with statistically significant differences were further analyzed.
Three additional methods were used to test the robustness of the microarray hybridization data: (i) correlation of gene expression with predicted operon organization, (ii) real-time quantitative PCR, and (iii) confirmation of the involvement of an uncharacterized induced gene in postirradiation recovery.
Operons are the principal form of gene coregulation in prokaryotes, so the expression patterns of genes within an operon are expected to be strongly correlated. Consistently, our analysis of 435 genes within 141 predicted operons showed highly significant correlation of their expression patterns compared with genes in “random operons” (see Supporting Text and Fig. 6). An example of expression patterns for genes located in one predicted operon is shown in Fig. 6A. Further supporting coregulation of functionally linked genes in DEIRA, we found that dispersed genes encoding different subunits of several distinct enzymatic complexes are also similarly regulated (Fig. 6B).
Seven genes that were up-regulated immediately on exposure to irradiation were evaluated by real-time quantitative PCR. Except for a single gene, DR0007, the expression patterns of all other genes were similar (r = 0.729) to those detected by microarray hybridization (Tables 1 and 2).
The uncharacterized gene DR0070 encodes a hypothetical protein (199 aa in length) that is unique to DEIRA and that was highly expressed after radiation (Table 3). To confirm that this gene is involved in radiation resistance, DR0070 was disrupted. The radiation resistance of a mutant (MD891) with a confirmed homozygous disruption for DR0070 was compared with wild-type DEIRA (Fig. 7). Although MD891 shows no metabolic or growth deficiencies in the presence or absence of chronic γ-radiation (60 Gy/h), it is substantially more sensitive to acute irradiation than wild-type DEIRA (Fig. 7A).
General Patterns of Expression in Response to Irradiation.
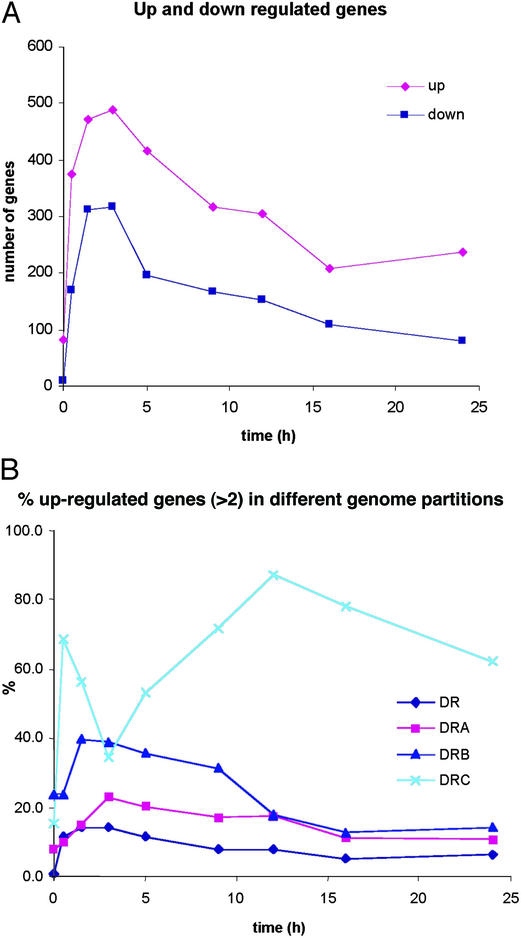
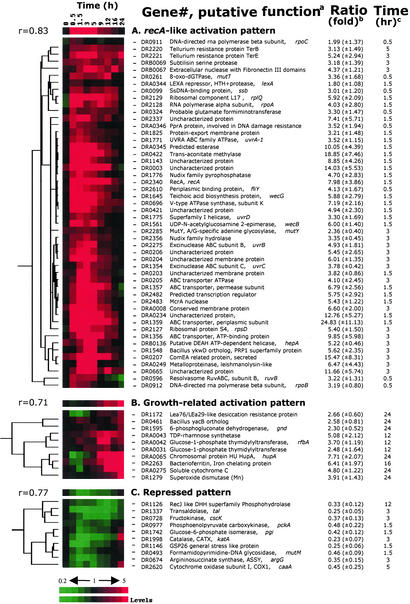
For genes with statistically significant expression ratios showing at least a 2-fold change, we found that 832 genes (28% of the genome) were induced and 451 genes (15%) were repressed at least at one time point during DEIRA recovery. Fig. 1A specifically shows differential regulation in the early and mid phases of recovery and illustrates that this time interval is an active period of coordinated gene expression, despite a highly fragmented genome. Within this large pool of significantly expressed genes, we operationally identified a subgroup that consists of genes with substantially greater expression levels and likely includes key players in the irradiation response. Table 3 lists genes and operons with recA-like expression patterns; DEIRA recA is critical to genomic restoration after irradiation (9–12), and its induction is considered a dominant marker for the onset of homologous recombination (ref. 21; Fig. 2). Table 4 lists the irradiation-response patterns of genes involved in replication, repair, and recombination functions in DEIRA.
Figure 1.
(A) Activation and repression of DEIRA genes as a function of time postirradiation. The data are for 2-fold activation or repression; up, activation; down, repression. (B) Activation of gene expression in response to irradiation in the four genomic partitions of DEIRA.
Figure 2.
Hierarchical clustering analyses of expression profile patterns. Gene expression patterns are displayed graphically. Three distinct patterns are sorted according to the hierarchical clustering analyses, i.e., recA-like activation pattern (A), growth-related activation pattern (B), and repressed patterns (C). The top row represents the general pattern of the selected group where a Pearson correlation coefficient (r) is shown on the left side. All displayed graphs are organized in a row/column format. Each row of colored strips represents a single gene whose expression levels are color-recorded sequentially in every column of the same row that represents recovery time intervals. Red denotes up-regulation and green indicates down-regulation. Black indicates the control level. The variation in transcript abundance is positively correlated with the color darkness. a, Gene numbers are offered for tracking the primary information of the gene of interest; b, the maximum (for recA-like and growth-related activation pattern) or minimum (for the repressed pattern) expression level for each of the exhibited genes over the 24-h recovery period is presented as the dye intensity ratio of the irradiated sample to the nonirradiated control at c, the indicated time interval. Values in parentheses show the standard deviation for each mean expression ratio.
A comparison of the percentage of responding genes for each of the four genomic partitions of DEIRA [DR_Main (2.65 Mbp), DRA (412 kbp), DRB (177 kbp), and DRC (46 kbp); ref. 14] unexpectedly showed that the majority of DRC genes were dramatically up-regulated during the mid and late phases of recovery, in marked contrast to the three other genomic partitions (Fig. 1B). Specifically, of the 41 DRC genes, 38 had expression ratios >2 in at least one time point, and almost all of the DRC genes were activated in the late phase.
To investigate whether genes of a particular functional group are significant contributors to DEIRA's γ-radiation-induced transcription dynamics, we constructed expression profiles for broad functional categories of genes by using the functional assignments in the COG database (www.ncbi.nlm.nih.gov/cgi-bin/COG/palox?fun=all). For most functional groups, we found that the maximum response occurred concurrently around the 3-h postirradiation time point (Fig. 8).
Induction of DNA Repair and Associated Systems.
About 23 of 71 (≈32%) DEIRA genes implicated in DNA repair, recombination, and replication showed patterns of expression similar to that of recA; the products of these genes likely comprise the core of its irradiation-response regulon (Table 3, Fig. 2). The remaining repair genes might respond differently, because their basal expression levels are near their maximum under normal cellular conditions, as proposed for yeast (23), or they might not be involved in the irradiation response. Only a few of the affected damage-response genes remained induced throughout recovery [e.g., genes for single-stranded DNA-binding protein, uracil DNA glycosylase, and 8-oxo-dGTPase, mutT; Fig. 2, Table 4]. Among genes with a recA-like expression pattern in DEIRA, orthologs of several are known to be activated during the error-prone DNA repair (SOS) response of E. coli and Bacillus subtilis, including recA, ssb, uvrABCD, and ruvB (Fig. 2). Interestingly, unlike SOS response in other bacteria, both DNA gyrase subunits (DR0906 and DR1943) were induced with the recA-like expression pattern, suggesting that regulation of DNA supercoiling is important for DNA repair.
Only a few of the pathways that are involved in direct damage reversal and base excision repair responded to irradiation. Specifically, we observed induction of the MutT ortholog (8-oxo-dGTPase), MutY (A-G mismatch DNA glycosylase), and uracil DNA glycosylase, but not many others (Table 4). The McrA-like family of predicted nucleases is expanded in DEIRA (13), and we found that at least two genes containing a domain of this family (DR2483 and DRA0057) showed high levels of induction in the early phase. Also, three genes encoding domains of a recently described VSR-like nuclease family, which might be involved in very short patch repair (24), were activated (DR0221, DR2566, and DRB0135; Table 4). The ComEA system, which has been identified through its role in DNA transformation competence (25), might be involved in the export of damaged DNA in irradiated DEIRA cells. Two genes of this system were induced: DR0207 (secreted protein ComEA), which is among the top 10 most strongly induced genes (Table 3), and DR0361 (metallobetalactamase family enzyme ComE). However, the role of these genes in the radiation resistance of DEIRA is unclear and is currently the subject of investigation.
Specific Response of Metabolic Gene Systems.
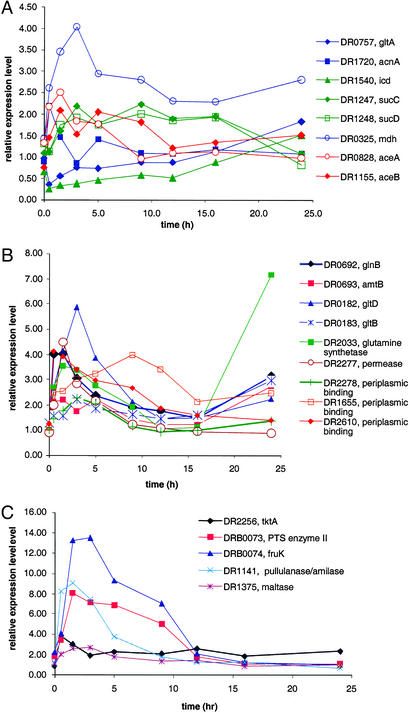
Among energy metabolism pathways, the V-type ATP synthase (DR0694–DR0702) genes were induced, as expected given the increased demand for energy during recovery. We also observed a strong induction of genes involved in cell wall biogenesis, which was expected given the need to restore membrane and associated ATP generation (Figs. 2 and 8A). Unexpectedly, we found that the genes encoding the tricarboxylic acid (TCA) cycle were repressed in the early and mid phases, whereas genes encoding the glyoxylate shunt were activated in this interval (Fig. 3A). In the late phase of recovery, glycolysis and the TCA cycle were progressively induced. It seems likely that the coinduction of genes for antioxidant proteins, in particular, superoxide dismutases (DR1546 and DR1279) and catalase (DR1998), contributes to the removal of the toxic free radicals. Specific induction of nitrogen metabolism genes, including the nitrogen regulatory protein P-II (GlnB homolog, DR0692; Fig. 3B) and genes involved in the utilization of exogenous and intracellular carbohydrates (Fig. 3C), was detected before 9 h had elapsed. The operon coding for enzymes of sulfur metabolism, i.e., sulfite reductase, APS kinase, PAPS reductase, and sulfate adenylyltransferase (DRA0013–DRA0016), remained induced throughout recovery, with the peak of expression occurring between 3 and 5 h (Table 3).
Figure 3.
(A) Expression patterns of selected genes for TCA cycle and glyoxylate bypass enzymes. Eight representative genes are shown in this figure, and table A lists the results of the remaining genes. Expression patterns for the following genes are shown: gltA, citrate synthase II; acnA, aconitase; icd, isocitrate dehydrogenase; sucC, succinyl-CoA synthetase, β-chain; sucD, succinyl CoA synthetase, α-chain; mdh, malate dehydrogenase; aceA, isocitrate lyase; and aceB, malate synthase. Genes specific to the TCA cycle are green, glyoxylate bypass genes are red, and genes shared by both TCA cycle and glyoxylate bypass are blue. Genes not represented here include: three subunits of 2-oxoglutarate dehydrogenase complex [lpd, lipoamide dehydrogenase (DR2526); sucA, 2-oxoglutarate dehydrogenase (DR0287); and sucB, dihydrolipoamide succinyltransferase (DR0083)] are substantially down-regulated like icd (DR1540). Malate synthase (DRA0277) of the glyoxylate bypass is slightly down-regulated. Four subunits of fumarate dehydrogenase complex [sdhB, fumarate/succinate dehydrogenase type iron sulfur protein (DR0951); sdhA, succinate dehydrogenase Rossman fold oxidoreductase (DR0952); shhC and sdhD, small succinate dehydrogenase associated proteins (DR0953 and DR0954); and fumC, fumarase (DR2627)] do not show significant changes in their expression levels during postirradiation recovery. (B) Expression patterns of the genes for nitrogen metabolism. Gene abbreviations: glnB, P-II nitrogen metabolism regulatory protein; amtB, ammonium transporter protein; gltD and gltB, glutamate synthase subunits. (C) Expression patterns of the genes for carbohydrate utilization. Gene abbreviations: tktA, transketolase; fruK, 1-phosphofructokinase.
Genes encoding two ribonucleoside/ribonucleotide reductases (DRB0107–DRB0109 and DR2374), the enzymes responsible for the final step of deoxyribonucleotide biosynthesis from ribonucleotide precursors, were induced in the early phase. The one encoded by DRB0107–DRB0109 remained extremely active throughout recovery, whereas DR2374 was repressed at the end of the mid phase.
Induction of Cell Cleaning and Stress-Response Genes.
Cellular mechanisms known to deal with by-products of γ-radiation include hydrolysis, modification, and direct damage reversal. Our data support at least partial activation of all these processes (Fig. 2, Table 4). Pyrophosphatases of the MutT/Nudix superfamily are thought to support one of the major house-cleaning systems of the cell (26). Specific expansion of this family in DEIRA was previously reported (13, 14), and 21 of these genes were functionally characterized (27). Five genes of this family with higher specificity to deoxynucleotide triphosphates (27), including the MutT ortholog, are induced in the early phase (see Fig. 2, Table 4, and table A). We also found early-phase activation of several genes involved in DNA degradation; export systems including protein-export membrane protein SecG (DR1825) and MDR-type exporter (DR2098); and transporters of a major facilitator family with undefined activities (Table 3).
A strong correlation has been reported between desiccation resistance and γ-radiation resistance in DEIRA (1, 28). Several putative proteins homologous to plant desiccation resistance proteins were identified in the DEIRA genome (13). Disruption of two of these genes, DR1172 and DRB0118, showed that they are important for desiccation resistance, but not for radiation resistance (28). Consistently, we found that these two genes are not induced in the early-mid phase interval (table A). Additionally, five uncharacterized genes were reported to be induced by both irradiation (3 kGy) and desiccation (29). We found the same genes to be activated in the early phase after 15 kGy (DR0003, DR0070, DR0423, and DRA0346; no data were obtained for DR0326; table A, Table 3). DRA0346 was identified in DEIRA previously as a DNA repair-related protein [I. Narumi, unpublished work; see GenBank, accession no. O32504] and is a predicted DNA-binding protein (E.V.K., unpublished work). We have also shown that disruption of DR0070 renders DEIRA much less resistant to γ-rays (Fig. 7A).
Consistent with our previous predictions (13), several genes encoding proteins from expanded families related to the stress response (terE, PR1, and dinB/yfiT families) are strongly induced throughout the early and mid phases (see Supporting Text and Table 4).
Poorly Characterized Genes Implicated in Cell Recovery.
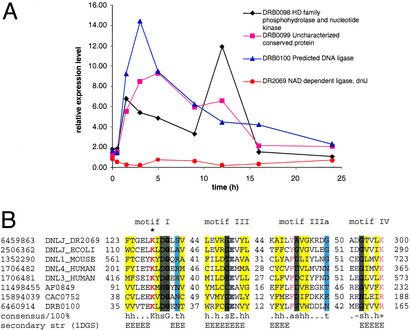
Approximately 48% of poorly characterized genes were highly expressed in at least one phase of recovery. Of this group, ≈40% showed the recA-like expression profile (Figs. 2 and 8A, Table 3). Many of these uncharacterized genes form probable operons with functions related to DNA repair, stress response, and various regulatory pathways. Examination of operonic context thus allows functional predictions to be made for some of the uncharacterized genes (30). Three genes encoded in one operon (DRB0098–DRB0100) are particularly interesting because of their potential direct involvement in DNA repair (Fig. 4). One of these genes (DRB0098) contains a HD-hydrolase family phosphatase domain and a polynucleotide kinase domain and resembles a similarly configured human protein that plays an important role in DSB repair (13, 31). The second (DRB0100) is a diverged homolog of ATP-dependent DNA ligases that shows the strongest similarity to eukaryotic DNA ligase III and retains all catalytic residues (32, 33), suggesting that this is a distinct, active DNA ligase (Fig. 4). The third protein remains functionally undefined. We found that expression of each of these genes was induced 5- to 10-fold at 1.5–5 h (Fig. 4). Strikingly, the typical bacterial NAD-dependent DNA ligase of DEIRA is repressed during this phase (Fig. 4).
Figure 4.
Expression of a predicted operon coding for components of a potential distinct DNA repair system. (A) Expression pattern of the genes in the predicted repair operon and the NAD-dependent DNA ligase. (B) Multiple alignment of the conserved motifs common to all DNA ligases, including the predicted distinct ATP-dependent DNA ligase DRB0100 from DEIRA. The alignment was constructed manually on the basis of high-scoring sequence pairs generated by PSI-BLAST searches (47) and previously described conservation of DNA ligase motifs (32, 33). Sequences are denoted by gene identifiers and gene names from the GenBank database: NAD-dependent DNA ligase from DEIRA (DR2069), E. coli (DNLJ_ECOLI), DNA ligase IV from human (DNL4_HUMAN), DNA ligase III from human (DNL3_HUMAN), DNA ligase I from mouse (DNL3_MOUSE), predicted ATP-dependent DNA ligases from DEIRA (DRB0100), Archaeoglobus fulgidus (AF0849), and Clostridium acetobutylicum (CAC0752). The positions of the first and the last residue of the aligned region in the corresponding protein are indicated for each sequence. The numbers within the alignment represent less conserved regions that are not shown. Invariant amino acid residues are in bold type. Alignment coloring is based on the consensus shown underneath the alignment; h, hydrophobic residues (A, C, F, L, I, M, V, W, Y, T, S, and G; yellow background); t, turn-forming residues (A, C, S, T, D, E, N, V, G, and P; cyan background); s, small residues (A, C, S, T, D, V, G, and P; green background); p, positively charged residues (R and K; red); a, aromatic residues (W, Y, F, and H; magenta); −, negatively charged residues (E and D; blue). The secondary structure elements are shown according to the structure of the NAD-dependent DNA ligase from Thermus filiformis, 1DGS (48). H, extended conformation, α-helix; E, extended conformation, β-strand. The catalytic lysine is marked by an asterisk.
Consistent with observations made about activation of some known export systems, we found high induction of the poorly characterized ABC transporter (DR1356–DR1359), which also might be involved in the export of damaged products (Table 3). Up-regulated kinases with unknown specificity (e.g., DR2467, DR0394, DR0609, and DR1564) might participate in regulating a variety of cellular processes (table A, Table 3). One of these uncharacterized kinases is the ortholog of a predicted kinase from Listeria that affects DNA topology when expressed in E. coli (13, 34).
Discussion
We examined changes in genomic expression in DEIRA recovering from an acute exposure to 15 kGy, which is about its D37 (irradiation dose yielding 37% survival) under our standard conditions (5, 9–11). During the early and mid phases of recovery, no cell growth was observed, but within this interval many genes of diverse functional groups were induced (Table 3). In contrast, many genes for enzymes of general metabolism were not affected in this interval (table A, Fig. 2), but were induced in the late phase. Collectively, this strongly supports the idea that most of the expression dynamics observed in the 9-h lag phase are a consequence of the effects of irradiation and not due to changes in nutrient or growth conditions resulting from transfer of irradiated DEIRA to fresh medium. Given the apparent complexity of DEIRA's irradiation response and the good correlation observed between its predicted and experimentally determined patterns of gene expression, the use of microarrays to identify DEIRA genes responsible for its radiation resistance holds promise as a useful approach (e.g., Fig 7).
After an exposure dose of 15 kGy, ≈150 DSBs are inflicted randomly over DEIRA's four circular genomic partitions (14), followed by extensive exonucleolytic DNA degradation (35). In acutely irradiated cells, this causes a substantial lowering of the copy number of the more heavily damaged, larger genomic partitions compared with smaller ones in the mid and late phases of recovery (12). We find that expression levels determined for DEIRA's four genomic partitions fit this model, where expression of DRC is the highest followed by DRB, DRA, and DR_Main, respectively (Fig. 1B). Within the broader context of partition-specific expression, we observed differences in the expression of functionally grouped genes that parallel the physiology of the early, mid, and late phases of recovery (Fig. 2). Based on expression data, the majority of DEIRA's irradiation-induced genes are functionally uncharacterized or have only general functional predictions assigned to them. Several striking examples include DR0003, DR0052, DR0140, DR0665, DR1143, and DR2337 (Table 4). However, some of these genes might not encode a protein at all. For example, DRA0234 is only 171 bp in length, shows no similarity to any protein sequences, and has a transcript that is predicted to form a stable stem–loop structure (predicted using the mfold program, www.bioinfo.rpi.edu/applications/mfold/old/rna/form1.cgi; data not shown). This gene, and perhaps other similar ones, might encode uncharacterized regulatory RNAs, as recently described in other bacterial systems (36).
Although the specific functions of many DEIRA genes induced by irradiation remain unclear, some readily allow an interpretation. Perhaps the most notable example is the prediction of a distinct ATP-dependent DNA ligase (Fig. 4). This ligase, in cooperation with two other proteins encoded in the same predicted operon, is probably the dominant ligase during postirradiation repair, whereas the typical bacterial NAD-dependent DNA ligase is down-regulated. Another example is a probable operon that consists of the predicted 2′–5′ RNA ligase (LigT) and the recA gene; both genes have similar expression profiles (Fig. 6). Furthermore, this gene organization is shared by Thermotoga maritima. Because RNA is also heavily damaged on irradiation, it is possible that the 2′–5′ RNA ligase is involved in the degradation of specific structures formed in damaged RNA (37). Alternatively, it could have an unrecognized function in DNA repair, perhaps in functional association with RecA, as suggested by colocalization of these genes within the same operon.
Generally, our expression data support the hypothesis that irradiated DEIRA strongly suppresses oxidative stress, perhaps as a mechanism to prevent additional loss of genome integrity. Notably, during the early and mid phases, when biosynthetic and energy demands are expected to be high, DEIRA represses its TCA cycle, particularly the O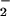 radical-generating step (i.e., sdhB; Fig. 3A; ref. 38). Furthermore, DEIRA appears to minimize its biosynthetic energy demands by inducing transporters of exogenous peptides and other secondary metabolites on which radiation resistance is highly dependent (7), and correspondingly shows increased expression of extra- and intracellular proteases and nucleases (Table 3). This helps explain the observation that de novo biosynthesis of amino acids, nucleotides, and coenzymes remains unchanged or is even down-regulated, at least within the early and mid phases (Figs. 2 and 8). In contrast, the glyoxylate bypass is strongly induced (Fig. 3A) and could provide some biosynthetic intermediates needed for recovery without generating free radicals. The gene for predicted transaconitate methylase (DR0422) is one of the most strongly induced in the early phase of recovery. Transaconitate is not a normal metabolite in most bacteria and is an attenuator of aconitase, a key enzyme of the TCA cycle (39). It seems possible that transaconitate is produced as a result of irradiation, and induction of DR0422 (Table 4) might be required for transaconitate detoxification. During the late phase, the TCA cycle was slowly induced, and genes involved in scavenging free radicals (e.g., sodA and katA) were correspondingly induced.
radical-generating step (i.e., sdhB; Fig. 3A; ref. 38). Furthermore, DEIRA appears to minimize its biosynthetic energy demands by inducing transporters of exogenous peptides and other secondary metabolites on which radiation resistance is highly dependent (7), and correspondingly shows increased expression of extra- and intracellular proteases and nucleases (Table 3). This helps explain the observation that de novo biosynthesis of amino acids, nucleotides, and coenzymes remains unchanged or is even down-regulated, at least within the early and mid phases (Figs. 2 and 8). In contrast, the glyoxylate bypass is strongly induced (Fig. 3A) and could provide some biosynthetic intermediates needed for recovery without generating free radicals. The gene for predicted transaconitate methylase (DR0422) is one of the most strongly induced in the early phase of recovery. Transaconitate is not a normal metabolite in most bacteria and is an attenuator of aconitase, a key enzyme of the TCA cycle (39). It seems possible that transaconitate is produced as a result of irradiation, and induction of DR0422 (Table 4) might be required for transaconitate detoxification. During the late phase, the TCA cycle was slowly induced, and genes involved in scavenging free radicals (e.g., sodA and katA) were correspondingly induced.
DEIRA has orthologs of 11 of 26 genes that comprise the SOS regulon in E. coli (40), and 7 of these genes respond to irradiation in DEIRA. However, it is notable that DEIRA does not have a homolog of the error-prone DNA polymerase umuC, a central gene of the SOS response in many bacteria (13). In agreement with recent reports (41), our expression data show that lexA (DRA0344) and the second lexA paralog (DRA0074) are not significantly induced after irradiation (Table 4). Taken together, these data suggest that a specific damage-response regulon is likely to exist in DEIRA, but that it is distinct from the classical E. coli-type SOS response in regard to the genes involved and the mode of regulation. The observed weak activation of the lexA paralogs (1.8-fold at 1.5–3 h) suggests that lexA might regulate other transcriptional regulators involved in the damage response or could selectively regulate a small subset of damage-response genes. Notwithstanding these possibilities, we searched for candidate transcriptional regulators that could play a major role in the response to DNA damage. Among >80 transcriptional regulators predicted to exist in DEIRA, we see significant induction of only a few corresponding genes (Fig. 9), with the largest induction ratio numbers observed for DRC0012, DR0171, DR2574, and DR2482. The induction of the DRC0012 regulator might be fortuitous, because it is encoded on the small plasmid DRC and belongs to the CsdG/RcsA family, whose members have not been identified as stress-response regulators. DR2482 contains a DNA-binding domain similar to those of sigA-like sigma factors (42) and, therefore, might function as a DNA-damage-specific sigma subunit of RNA polymerase. DR2574 belongs to the Xre family of transcriptional regulators, some of which were found to regulate stress response in bacteria (43, 44). Finally, DR0171 belongs to a specific DEIRA family of transcriptional regulators and has been shown to be involved directly in γ-radiation resistance (13, 45, 46). Therefore, we consider DR0171 and DR2574, which displayed recA-like expression patterns, to be two primary candidates for regulating the DNA irradiation response in DEIRA.
The present analysis of the transcriptome dynamics of DEIRA in response to acute irradiation revealed the complexity that could be expected of such a unique recovery process and, importantly, led to the identification of numerous previously unsuspected candidates for experimental analysis of genes and mechanisms that underlie the exceptional radiation resistance of this bacterium.
Supplementary Material
Acknowledgments
This research was funded by U.S. Department of Energy (Office of Biological and Environmental Research, Office of Science) Grants DE-FG02-01ER63220 from the Genomes to Life Program, DE-FG02-97ER62492 from the Natural and Accelerated Bioremediation Research Program, and ERKP385 from Microbial Genome Program, and by the Laboratory Directed and Research Development Program at Oak Ridge National Laboratory. Oak Ridge National Laboratory is managed by the University of Tennessee–Battelle LLC for the Department of Energy under Contract DOE-AC05-00OR22725.
Abbreviations
- DEIRA
Deinococcus radiodurans strain R1
- DSB
double-strand break
- SOS
error-prone DNA repair
- TCA
tricarboxylic acid
Footnotes
To whom correspondence should be addressed regarding Deinococcus biology and reprints. E-mail: mdaly@usuhs.mil.
References
- 1.Mattimore V, Battista J R. J Bacteriol. 1996;178:633–637. doi: 10.1128/jb.178.3.633-637.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Minton K W. Mutat Res. 1996;363:1–7. doi: 10.1016/0921-8777(95)00014-3. [DOI] [PubMed] [Google Scholar]
- 3.Hutchinson F. Prog Nucleic Acid Res Mol Biol. 1985;32:115–154. doi: 10.1016/s0079-6603(08)60347-5. [DOI] [PubMed] [Google Scholar]
- 4.Hansen M T. J Bacteriol. 1978;134:71–75. doi: 10.1128/jb.134.1.71-75.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daly M J, Ling O, Minton K W. J Bacteriol. 1994;176:7506–7515. doi: 10.1128/jb.176.24.7506-7515.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin J, Qi R, Aston C, Jing J, Anantharaman T S, Mishra B, White O, Daly M J, Minton K W, Venter J C, Schwartz D C. Science. 1999;285:1558–1562. doi: 10.1126/science.285.5433.1558. [DOI] [PubMed] [Google Scholar]
- 7.Venkateswaran A, McFarlan S C, Ghostal D, Minton K W, Vasilenko A, Makarova K S, Wackett L P, Daly M J. Appl Environ Microbiol. 2000;66:2620–2626. doi: 10.1128/aem.66.6.2620-2626.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krasin F, Hutchinson F. J Mol Biol. 1977;116:81–98. doi: 10.1016/0022-2836(77)90120-6. [DOI] [PubMed] [Google Scholar]
- 9.Daly M J, Ouyang L, Fuchs P, Minton K W. J Bacteriol. 1994;176:3508–3517. doi: 10.1128/jb.176.12.3508-3517.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daly M J, Minton K W. J Bacteriol. 1995;177:5495–5505. doi: 10.1128/jb.177.19.5495-5505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daly M J, Minton K W. J Bacteriol. 1996;178:4461–4471. doi: 10.1128/jb.178.15.4461-4471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daly M J, Minton K W. Gene. 1997;187:225–229. doi: 10.1016/s0378-1119(96)00755-x. [DOI] [PubMed] [Google Scholar]
- 13.Makarova K S, Aravind L, Wolf Y I, Tatusov R L, Minton K W, Koonin E V, Daly M J. Microbiol Mol Biol Rev. 2001;65:44–79. doi: 10.1128/MMBR.65.1.44-79.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White O, Eisen J A, Heidelberg J F, Hickey E K, Peterson J D, Dodson R J, Haft D H, Gwinn M L, Nelson W C, Richardson D L, et al. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Markillie L M, Varnum S M, Hradecky P, Wong K K. J Bacteriol. 1999;181:666–669. doi: 10.1128/jb.181.2.666-669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou J, Fries M R, Chee-Sanford J C, Tiedje J M. Int J Syst Bacteriol. 1995;45:500–506. doi: 10.1099/00207713-45-3-500. [DOI] [PubMed] [Google Scholar]
- 17.Xu D, Li G, Wu L, Zhou J, Xu Y. Bioinformatics. 2002;18:1432–1436. doi: 10.1093/bioinformatics/18.11.1432. [DOI] [PubMed] [Google Scholar]
- 18.Thompson D K, Beliaev A S, Giometti C S, Tollaksen S L, Khare T, Lies D P, Nealson K H, Lim H, Yates J, III, Brandt C C, et al. Appl Environ Microbiol. 2002;68:881–892. doi: 10.1128/AEM.68.2.881-892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beliaev A S, Thompson D K, Khare T, Lim H, Brandt C C, Li G, Murray A E, Heidelberg J F, Giometti C S, Yates J, III, et al. OMICS. 2002;6:39–60. doi: 10.1089/15362310252780834. [DOI] [PubMed] [Google Scholar]
- 20.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carroll J D, Daly M J, Minton K W. J Bacteriol. 1996;178:130–135. doi: 10.1128/jb.178.1.130-135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee M L, Kuo F C, Whitmore G A, Sklar J. Proc Natl Acad Sci USA. 2000;97:9834–9839. doi: 10.1073/pnas.97.18.9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Birrell G W, Brown J A, Wu H I, Giaever G, Chu A M, Davis R W, Brown J M. Proc Natl Acad Sci USA. 2002;99:8778–8783. doi: 10.1073/pnas.132275199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aravind L, Makarova K S, Koonin E V. Nucleic Acids Res. 2000;28:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inamine G S, Dubnau D. J Bacteriol. 1995;177:3045–3051. doi: 10.1128/jb.177.11.3045-3051.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bessman M J, Frick D N, O'Handley S F. J Biol Chem. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- 27.Xu W, Shen J, Dunn C A, Desai S, Bessman M J. Mol Microbiol. 2001;39:286–290. doi: 10.1046/j.1365-2958.2001.02267.x. [DOI] [PubMed] [Google Scholar]
- 28.Battista J R, Park M J, McLemore A E. Cryobiology. 2001;43:133–139. doi: 10.1006/cryo.2001.2357. [DOI] [PubMed] [Google Scholar]
- 29.Battista J R, Howell H A, Park M J, Earl A, SN P. Genome. 2002. DOE Contractor-Grantee Workshop IX, Oakland, CA), pp. 88. [Google Scholar]
- 30.Aravind L. Genome Res. 2000;10:1074–1077. doi: 10.1101/gr.10.8.1074. [DOI] [PubMed] [Google Scholar]
- 31.Jilani A, Ramotar D, Slack C, Ong C, Yang X M, Scherer S W, Lasko D D. J Biol Chem. 1999;274:24176–24186. doi: 10.1074/jbc.274.34.24176. [DOI] [PubMed] [Google Scholar]
- 32.Aravind L, Koonin E V. J Mol Biol. 1999;287:1023–1040. doi: 10.1006/jmbi.1999.2653. [DOI] [PubMed] [Google Scholar]
- 33.Doherty A J, Suh S W. Nucleic Acids Res. 2000;28:4051–4058. doi: 10.1093/nar/28.21.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez-Campillo M, Dramsi S, Gomez-Gomez J M, Michel E, Dehoux P, Cossart P, Baquero F, Perez-Diaz J C. Mol Microbiol. 1995;18:801–811. doi: 10.1111/j.1365-2958.1995.18050801.x. [DOI] [PubMed] [Google Scholar]
- 35.Moseley B E, Evans D M. J Gen Microbiol. 1983;129:2437–2445. doi: 10.1099/00221287-129-8-2437. [DOI] [PubMed] [Google Scholar]
- 36.Eddy S R. Nat Rev Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- 37.Arn E A, Abelson J N. J Biol Chem. 1996;271:31145–31153. doi: 10.1074/jbc.271.49.31145. [DOI] [PubMed] [Google Scholar]
- 38.Henle E S, Linn S. J Biol Chem. 1997;272:19095–19098. doi: 10.1074/jbc.272.31.19095. [DOI] [PubMed] [Google Scholar]
- 39.Lauble H, Kennedy M C, Beinert H, Stout C D. J Mol Biol. 1994;237:437–451. doi: 10.1006/jmbi.1994.1246. [DOI] [PubMed] [Google Scholar]
- 40.Courcelle J, Khodursky A, Peter B, Brown P O, Hanawalt P C. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narumi I, Satoh K, Kikuchi M, Funayama T, Yanagisawa T, Kobayashi Y, Watanabe H, Yamamoto K. J Bacteriol. 2001;183:6951–6956. doi: 10.1128/JB.183.23.6951-6956.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campbell E A, Muzzin O, Chlenov M, Sun J L, Olson C A, Weinman O, Trester-Zedlitz M L, Darst S A. Mol Cell. 2002;9:527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- 43.Gaur N K, Oppenheim J, Smith I. J Bacteriol. 1991;173:678–686. doi: 10.1128/jb.173.2.678-686.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Labie C, Bouche F, Bouche J P. J Bacteriol. 1989;171:4315–4319. doi: 10.1128/jb.171.8.4315-4319.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Battista J R. Annu Rev Microbiol. 1997;51:203–224. doi: 10.1146/annurev.micro.51.1.203. [DOI] [PubMed] [Google Scholar]
- 46.Makarova K S, Aravind L, Daly M J, Koonin E V. Genetica. 2000;108:25–34. doi: 10.1023/a:1004035424657. [DOI] [PubMed] [Google Scholar]
- 47.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee J Y, Chang C, Song H K, Moon J, Yang J K, Kim H K, Kwon S T, Suh S W. EMBO J. 2000;19:1119–1129. doi: 10.1093/emboj/19.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.