Abstract
SecA, an essential component of the general protein secretion pathway of bacteria, is present in Escherichia coli as soluble and membrane-integral forms. Here we show by electron microscopy that SecA assumes two characteristic forms in the presence of phospholipid monolayers: dumbbell-shaped elongated structures and ring-like pore structures. The ring-like pore structures with diameters of 8 nm and holes of 2 nm are found only in the presence of anionic phospholipids. These ring-like pore structures with larger 3- to 6-nm holes (without staining) were also observed by atomic force microscopic examination. They do not form in solution or in the presence of uncharged phosphatidylcholine. These ring-like phospholipid-induced pore-structures may form the core of bacterial protein-conducting channels through bacterial membranes.
The Escherichia coli SecA, a homodimer of 102-kDa subunits in solution, along with SecYEG and other Sec proteins (1–6) are intrinsic components of the protein secretion machinery. SecA binds precursor proteins, hydrolyzes ATP, and uses the energy of hydrolysis to translocate proteins across the cytoplasmic membrane. SecA lacks hydrophobic stretches sufficiently long to span a membrane (7), but it binds to the membrane and interacts with SecYEG and acidic phospholipids. It may integrate into membrane either by itself or together with other Sec proteins (1, 4–6, 8–13). The prevailing model of protein translocation depicts the SecYEG complex as forming the essential translocation core channel through the membrane, and SecA being a peripheral protein which hydrolyzes ATP to insert and deinsert a 30-kDa domain into the membrane. By cycling on and off the membrane it pushes precursor proteins through the SecYEG channel (4, 5, 14). Several findings, however, contest this model (15). Blobel's group showed that reconstituted membranes containing <1% of the normal level of SecY are active in protein translocation, and suggested that SecY is not the obligatory receptor for SecA and that it may not be essential for protein translocation (16). We confirmed and extended these findings by showing that both SecE- and SecY-deficient membranes are capable of translocating some precursor proteins in vitro, indicating that neither SecE nor SecY is essential for translocating all proteins (17, 18). Moreover, we and others have shown that SecA, which is often referred to as the peripheral subunit of preprotein translocase (4, 5), integrates into membranes (6, 8, 13, 19, 20), a fraction of which does not cycle on and off the membrane during translocation (13), and possibly forms an integral part of the protein-conducting channel (6, 13, 17, 20, 21).
SecA's structure in solution has been studied in some detail (10, 11), and recently the three-dimensional crystals of SecA from Bacillus subtilis were obtained for x-ray structural analysis (22, 23). The structure of SecA within membranes, however, is not well understood. SecA has been shown to assume two membrane-integrated forms, one of which is membrane-specific (21), presumably because of its deep penetration into the lipids (9). Previously, the lipid layer technique and electron microscopy has been used to reveal the structures of several membrane-related soluble proteins and their interaction with membranes (24, 25). Here we used this technique to examine structures of E. coli SecA on phospholipid layers by electron microscopy. We also examined these structures by atomic force microscopy (AFM) (26, 27). We observed that SecA forms ring-like pore structures when it interacts with E. coli anionic phospholipids. These pores may form the central core of protein-conducting channels.
Experimental Methods
Purification of SecA.
SecA was purified as described (13) from lysates of E. coli BL21(λDE3)/pT7-SecA (1) by stepwise elution on a Pharmacia S-Sepharose column, followed by gel filtration chromatography on a Sephacyl-200 column. The final preparations contained virtually pure SecA as determined by Coomassie blue staining of samples separated by dodecyl sulfate-polyacrylamide gel electrophoresis. SecA preparations were kept at −86°C in small amounts in 50 mM NH4HCO3/1 mM DTT/1% lactose or lyophilized.
Electron Microscopy.
The previously described monolayer technique for studying the two-dimensional crystallization of proteins (28) was performed. Briefly, 15–20 μl samples of a solution of SecA (20–200 μg/ml) in 20 mM Tris⋅HCl (pH 7.3) buffer were added to small Teflon wells (4 mm in diameter and 0.5 mm deep) until the liquid surface bulged up. The surface was coated with 0.5–1.0 μl of E. coli phospholipids (from Avanti Polar Lipids) solutions (≈1 mg/ml) in chloroform/methanol (3:1 vol/vol) added from a syringe. The wells were incubated in a sealed humid atmosphere at <20°C for at least 6 h (typically, 4°C for 24 h). Then, the lipid monolayers at the air/liquid interfaces were picked off with hydrophobic carbon coated gold grids. After washing with several drops of incubation buffer (20 mM Tris⋅HCl, pH 7.3), grids were blotted and negatively stained with uranyl acetate solution (0.5–1% wt/vol). Samples of soluble SecA, 5 μg/ml, were also adsorbed for 30 s on grids with a glow-discharged carbon coat and negatively stained. The specimens were examined with a Philips CM120 transmission electron microscope with an accelerating voltage of 100 kV at a magnification of ×50,000. Interesting fields were recorded on Kodak SO-163 films.
Digitization of the Images and Single-Particle Analysis.
The best images (selected visually and by optical diffraction) were digitized at a step size of 25 μm per pixel by an AGFA Duoscan camera system. The numeric images were converted to SPIDER format and processed with spider image-processing software (29).
Six images containing a total of ≈1,000 dispersed particles were analyzed by interactive marking and extraction. Two-dimensionally ordered, packed particles were examined by automatic extraction: a reference by averaging small numbers of dispersed particles was cross-correlated with the image containing crystal-packing particles; peaks of the cross-correlation map were indicated, and the position of the peaks were used in the extraction of the particles. About 1,000 particles were extracted from one image after manually screening the automatic extracted particles.
The reference-free alignment procedure was performed on in-plane alignment of the dispersed particles by using the corresponding program of spider. They were then masked and classified by using the multivariance statistical analysis method (30). In practice, the hierarchy classification algorithm was used (31). For the particles extracted from the two-dimensionally ordered packing, shift alignment to the reference was first performed without angular rotation alignment. The particles were then masked with a circle to exclude the boundary elements and rotationally aligned by free alignment followed by the hierarchical classification.
Average and variance maps of each particle cluster were calculated and analyzed for the resolution and intraclass variance. For those projection images with intra p2 symmetry, the average between the 180° rotational image and the original image was calculated. For estimation of the resolutions of these average maps in each cluster, the different phase residual method was used (32) and resolutions in the range of 3.0–3.2 nm were obtained.
AFM.
All AFM slides were prepared as described by Butamante and Revetii (33) with minor modifications. Briefly, 10 μl samples of different mixtures in 10 mM Tris⋅HCl, pH 8.0, containing 50 mM KC1 and 2 mM MgC12 (TKM) were applied to freshly cleaved mica, which were held at room temperature for 10–15 min, rinsed four times with deionized water, and dried in a dessicator. To prepare lipid bilayers, extracted E. coli membrane lipids or 1,2-dioleoyl-sn-glycero-3 phosphochloine (phosphatidylcholine) in chloroform were vacuum dried, resuspended in TKM buffer and sonicated for 15 min in an ice bath (34). The mixture of SecA (1.7 μg) and the bilayer (20 μg) in 10 μl TKM buffer was vortexed for 10 s before applied to the mica. Goat anti-rabbit IgG-gold (purchased from EY Laboratories) at a concentration of 15 μg/ml in the same buffer as above, was used as a reference for measuring the sizes.
AFM images were obtained with a CP-Autoprobe (Park Scientific, Sunnyvale, CA) by using the noncontact mode in a scan rate of 0.5 Hz and drive force of 3–8%, and analyzed by using image processing software (spmlab nt version 5.01) according to the manufacturer's manual.
Chemicals and Reagents.
E. coli phospholipids mixtures, phosphatidylcholine, and phosphatidylserine were purchased from Avanti Polar Lipids. S-Sepharose and Sephacyl-200 were from Amersham Pharmacia. All others are reagents grade, and were purchased from commercial sources.
Results
Distinct SecA Structures On Interaction With Phospholipids.
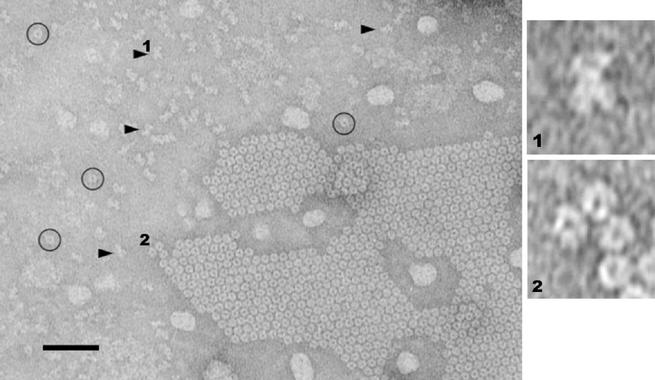
When soluble SecA was incubated underneath lipid monolayers of E. coli phospholipids at 4° for 24 h and examined by transmission electron microscopy, patches of ordered lattice packed particles, and two forms of dispersed particles (Fig. 1) were seen. The dispersed structures were a mixture of dumbbell-shaped (Fig. 1, arrows, 1) and ring-like shaped particles with pores (Fig. 1, circles, 2). The patches contained mostly ring-like particles with pores and a few horseshoe-shaped particles. The appearance of these ring-like particles depended on the incubation time, temperature and the protein concentration. They formed when the solution of SecA was more concentrated than 20 μg/ml and was held for >6 h at a temperature <20°C. On the other hand, the dumbbell-shaped structures could be found shortly after protein interaction with E. coli lipid monolayer in a wide range of subphase conditions. The time course of the appearance of dumbbell and ring-like structures suggested that the dumbbell-shaped structures may be intermediates in the formation of ring-like structures. Particles of SecA in solution in the absence of phospholipids exhibited no characteristic privileged orientation on hydrophilic carbon layered grids examined by electron microscopy (data not shown). No ring-like structures were seen in solution without phospholipids.
Figure 1.
SecA structures formed in the presence of E. coli lipid monolayers. SecA (100 μg/ml) in 20 mM Tris⋅HCl, pH 7.3, was incubated at 4°C for 24 h with E. coli lipid monolayers covered until picked up, and negatively stained. A patch of two-dimensional ordered-packing of ring-like particles is visible in the lower right quadrant. Some dispersed with ring-like pores are marked by circles. Dispersed dumbbell-shaped particles are marked by arrowheads. The scale bar represents 50 nm. (Right) Enlarged digitized images of dumbbell-shaped (1) and pored (2) particles.
This visual evidence for SecA forming two distinct structures on interaction with phospholipids is consistent with previous biochemical evidence based on proteolysis analysis that two forms of integral SecA [(SecAs and SecAm exist in E. coli membranes (21) and in liposomes (J. You and P.C.T., unpublished data)].
These particles were fairly homogeneous on negatively charged phospholipids. No such dumbbell-shaped or ring-like particles were observed when SecA interacts with nonnegatively charged phospholipids, such as phosphatidylcholine (data not shown). These results suggest that the dumbbell-shaped and ring-like particles might be the forms that anchor SecA onto the negatively charged phospolipid layers. Because the sum of its complement of Glu and Asp residues exceeds the sum of its Lys and Arg residues by 26 (7), SecA has a net negative charge in the buffer used (pH 7.3). Thus specific local positively charged areas on the surface of SecA probably interact with the negatively charged phospholipids in a privileged orientation.
Dumbbell-Shaped Structures.
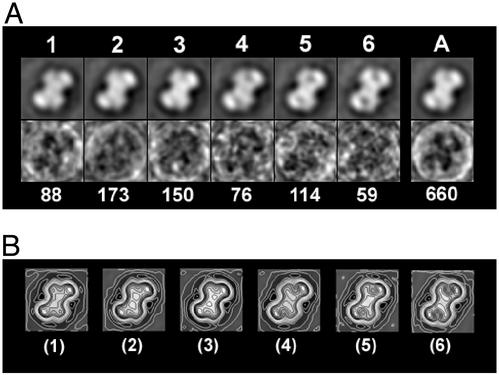
By single-particle analysis, the dispersed elongated dumbbell-shaped particles were selected, aligned, classified and averaged (29). Six clusters were classified. These varied with respect to the relative position of the two domains, the area and shape of the low-density gulf inside each domain, and the size of the particles. Particles in clusters 1–3 have only a shallow gulf in both ends of the “dumbbell”; those in clusters 4–6 have a deep gulf or a hole within the ends of the “dumbbell” (Fig. 2). The average maps reveal that dumbbell-shaped particles are composed of two main domains, arranged in a 2-fold symmetry (Fig. 2). A 2-nm-diameter gulf with low density can be observed between these two domains. The whole particle is “X” shaped with a lobe at each of its four corners, an architecture shown more clearly by the global variance map in which the dark areas represent less variety of structure among all particles (Fig. 2A, lane A). The particles measure 13–14 nm in their longest dimension and 8–9 nm wide, perpendicular to the maximum axis. The contour maps indicate that there is a high-density region in each domain located at the farthest lobe-like end of the particle and another high-density motif located in the middle of the whole particle extending to the other lobe of the domain (Fig. 2B). The different depths of the gulfs revealed by the classification of the particles would be consistent with the heterogeneous conformations of SecA proteins. This suggests the presence of flexible motifs within the molecules that may play a functional role in hydrolyzing ATP and/or translocating preproteins (23).
Figure 2.
Single-particle analysis of the dumbbell structures of SecA formed in the presence of E. coli lipid monolayers. A total of 660 dumbbell-shaped particles boxed from four films are aligned by reference-free alignments. The first alignment gives an average map, which was used as the reference for a second round alignment. The aligned particles were masked by a mask made from the reference map and were applied to multivariance statistical analysis (30). By hierarchical classification (31), the particles were classified into six clusters, from which the average maps and variance maps were calculated in A. The number above each average map is the cluster number; the number below the variance map is the number of particles included in that cluster. The global average and variance maps were also calculated as shown in lane A. The maximum density values in variance maps are < 5% of the average. The resolutions of average maps in each cluster were estimated to be in the range of 3.0–3.2 nm according to the different phase residual method (32). Two-fold symmetry applied average maps of each cluster were calculated. The average map of each cluster was rotated 180° in plane. Then the rotated images were averaged with the original ones. The small variances between the 180° rotational maps and their original ones indicate the presence of two-fold symmetry within each particle. The final maps were then contoured and shown in B. The image box length represents 17.5 nm.
The similarity of the outlines of particles as determined by electron microscopy and previously by small angle x-ray scattering (11) as well as three-dimensional SecA structures (23), and the presence of a 2-fold symmetry inside the particles suggest that the dumbbell-shaped particles on lipid layers may be dimers of similar shapes to those modeled from monomeric SecA found in solution by x-ray crystallography (23). The modeled dimeric SecA in solution measured 12 nm long along the longest dimension and 8 nm wide (23), results similar to those obtained by electron microscopy. The dumbbell-like projection averages and the modeled dimers derived by x-ray crystallography suggest that the two separate domains of each polypeptide nucleotide binding domain and preprotein cross-linking may correspond to the lobes on the images (22, 23).
Lipid-Specific Ring-Like Pore Structures.
The hexagonal ring-like pore structures of SecA on phospholipid layers is lipid-specific (Figs. 1 and 3). These particles exhibit distinct domains (Fig. 1, 2). The ring-like particles are more compressed than the dumbbell-shaped particles and most occur as a hexagonal lattice (Fig. 1). In addition to forming on E. coli lipids, these structures were observed to form in lipid mixtures containing phosphatidylcholine and negatively charged phosphatidylserine or phosphatidylglycerol, but not with phosphatidylcholine alone (data not shown). These results along with the absence of ring-like structures in the solution suggest that negatively charged phospholipids specifically induce formation of these characteristic ring-like pore structures. These structures may be the lipid-specific SecAM in membranes (21). The observation that ring-like structures were observed more often at 4°C than at room temperature is in agreement with the instability of the SecAM at high temperature in liposomes (J. You and P.C.T., unpublished data). Single-particle analysis of >1,000 of these particles revealed that the rings are quite homogeneous with an outer diameter of 8 nm with an inner 2-nm hole (Fig. 3).
Figure 3.
Single-particle analysis of the ring-like pore structures. About 30 dispersed hexagonally ring-like particles were boxed and averaged to give a reference. Then 1,077 particles in the lattice packing form were extracted automatically by a cross-correlation method and screened manually for final processing. Shift alignment to the reference is first performed without angular rotation alignment. The particles were then masked with a circle to exclude the boundary elements and rotationally aligned by free alignment followed with the hierarchical classification. The variances among the particles are so small that only the global average maps were calculated (1). The global average map of all of the particles with only shift alignment but without rotational alignment were also calculated (2). The difference between the rotationally aligned and unaligned average images is so small that the final projection map was calculated only from the global average map without rotational alignment (3). The resolution of the averages were determined to be 3.2 nm by different phase residual method (32). The image box length represents 11.7 nm.
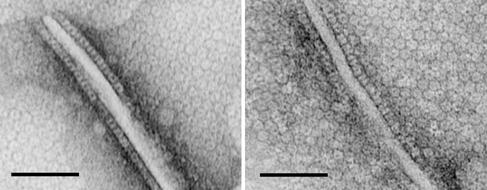
Sometimes while transferring lipid monolayers onto grids, self-attraction of the hydrophobic side forms ridges within the monolayers. In these regions the ring-like particles can be observed in a side-view (Fig. 4) as rectangular, cylindrically shaped particles of 8 nm by 5 nm with a central stain-filled tunnel. Such side-views allow the molecular sizes of the structures to be estimated from their volumes. Assuming the density of SecA protein to be 1.33–1.35 g/ml, the ring-like structures average 205,000 Da (ranging from 174 to 253 kDa), suggesting that they are SecA dimers (alternatively, one monomer may stack on the other).
Figure 4.
Side views of ring-like SecA on E. coli lipid layers with ridges. Two patches of two-dimensional ordered-packing of ring-like particles in ridges of E. coli lipid layers formed by the attraction of the monolayers' hydrophobic sides are shown. The experimental conditions were the same as in Fig. 1. Side views of the ring-like particles can be seen along the sides of the ridges. (Scale bar = 50 nm.) The height of the particles is estimated to be 5 nm.
AFM Images of Ring-like Pore Structures of SecA.
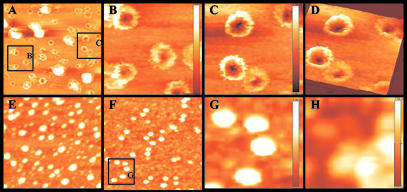
To eliminate the possibility that the observed pores in SecA were artifacts induced by fixing and staining, we examined surface topology of SecA by AFM, a technique that has been used to examine molecular interactions (33, 35), and to study the surface structures of porin channels (26, 27). Again we found that in the presence of E. coli phospholipid mixtures, SecA forms ring-like pore structures with diameters of about 8–9 nm, surrounding indented holes of 3–6 nm, the depths of which vary (Fig. 5 A–D). Such structures are not seen in the presence of detergents such as octyl glucoside (Fig. 5E), and they do not form in the absence of phospholipids (Fig. 5 F and G), or in the presence of noncharged phosphatidylcholine (Fig. 5H). Comparisons of the images of pore structures and soluble SecA allow estimation of the depth of the indented holes. SecA in the absence of lipids is ≈6.4-nm thick (Fig. 5G); the ring-like structures are ≈8.4–9 nm. Substraction suggests a minimal pore depth of ≈2–2.6 nm.
Figure 5.
Ring-like pore SecA structures observed by AFM. AFM images of SecA in various conditions. (A) Ring-like pore structure of SecA in E. coli lipid bilayers. (B and C) Zoom-in images of A. (D) Three-dimensional image of C. (E) SecA in lipids plus 2% octyl glucoside. (F) SecA in buffer without lipids. (G) Zoom-in images of F. (H) SecA in phosphatidylcholine. (Bars show the depth of the images: 9.02 nm for B, 8.36 nm for C, 6.38 nm for G, and 7.94 nm for H.)
The ring-like pore structures observed by AFM and electron microscopy are remarkably similar with the exception that the inside diameter of an indented hole appears smaller by electron microscopy. Presumably fixation and negative staining narrow the pore. We conclude that such ring-like pore structures of SecA are induced only in the presence of negatively charged phospholipids.
Discussion
Pore Structures of SecA On Interaction with Anionic Phospholipids.
Although the primary structure of SecA (7) predicts it to be a soluble protein, its association with, and integration into, membranes has long been observed (1–3, 6, 8, 12, 13, 19–21). Interaction with phospholipids increases the intrinsic ATPase activity of soluble SecA (2, 36), presumably because of conformational changes. Such induced changes allow SecA to penetrate deeply (9) and even to traverse lipid bilayers (37, 38). This interaction restricts the movement of both SecA and the lipid moiety in the bilayers (39), and depends on the anionic natures of the phospholipids (2, 9, 36, 40): both electrostatic and hydrophobic interactions are involved in the insertion (40).
The work presented here shows that interaction with phospholipids also induces E. coli SecA to undergo physical change. Two distinct structures are observed by electron microscopy when SecA interacts with an anionic phospholipid monolayer: a dumbbell-shaped and a ring-like structure. The dumbbell-shaped structures resemble soluble SecA in the absence of anionic phospholipids, and are similar to those observed by small angle x-ray scattering (11) as well as three-dimensional structures of SecA in solution (10, 22, 23). However, under current resolution (3 nm), it is not clear whether dumbbell-shaped structure is identical to the dimeric SecA in solution. It is possible that some subtle conformation changes happen when the protein interacts with phospholipids. The ring-like pore structures of SecA, however, are lipid-specific; they are observed only with anionic phospholipids layers. The relationship of the two structures is not yet clear, but based on the time of their appearance, the dumbbell-shaped structures may be intermediates in the formation of the ring-like structures. Based on side-view observations, the ring-like pore structures appear to be dimeric SecA (Fig. 4); their formation is enhanced by higher concentrations of SecA within membrane, a condition favoring the dimer formation (41). However, further work is needed to verify it.
Almost identical ring-like pore structures of SecA but with larger holes were observed with AFM, without staining. These ring-like pore structures are quite homogeneous with an estimated 8–9 nm diameter and an indented hole 3–6 nm wide and 2–2.6 nm deep. These structures may be related to the previously described lipid-specific SecAM with unique domains in membranes. Possibly the flexible domain (the preprotein cross-linking domain as mentioned in ref. 23) undergoes a conformational change to insert into the membrane, triggering the formation of a transmembrane pore in the other part of the SecA molecule.
The pore structure of SecA is remarkably similar to those of Bacillus SecYE (42) and eukaryotic Sec61p (43, 44). These were proposed to be the tunnels through which polypeptides tranverse membranes of Bacillus and eukaryotes. The SecA ring-like pore structures (2–6 nm) observed here both by electron microscopy and AFM, may be such transmembrane tunnels. The existence of these pore structures raises questions about the composition of the core of the bacterial translocation channel. Monomeric or dimeric E. coli SecYEG complexes themself alone do not form such large hole structures (45–47); SecA does, but only in the presence of negatively charged lipids.
Roles of SecYEG in Protein Translocation.
The prevailing model of Sec mediated protein translocation in bacteria (4, 5) depicts the SecYEG complex as forming the essential translocation channel, and SecA as being a peripheral protein that cycles on and off the membrane as it hydrolyses ATP thereby pushing precursor peptides across the translocation channel in a step-wise process (48, 49). The other Sec and related proteins confer specificity and efficiency. SecYEG allows SecA to bind to the membranes with higher affinity (50–52), and to shield it from phospholipids (53–55). Translocation through the SecYEG channel is depicted as being a dynamic process involving SecA cycling, coupled with the inversion of SecG topology in the membranes (56, 57). The SecYEG complex is also involved in proof-reading precursor signal peptides (58, 59), and enhancing the efficiency of the role of protonmotive force in the translocation process (60, 61). SecA interacts directly with SecY and SecG (54, 58), but the nature of its interaction with the SecYEG complex is not yet clear. Manting et al. (47) suggested that SecA recruits SecYEG complexes to form a tetrameric translocation channel; others provided evidence for a monomeric (62) or dimeric (46, 49, 63, 64) channel. Regardless of the detailed mechanism, the current prevailing model views SecYEG monomer as being too small to form a functional channel (45–47, 64); SecA must interact with it to enlarge the channel (4, 5, 47).
As stated in the Introduction, earlier biochemical findings indicated that E. coli SecYEG is not essential for the translocation of all proteins in vitro (16–18), that SecA plays a major structural role in the function of the channel (6, 8, 13, 17, 20, 21), and that the exposure of SecA to periplasmic side of the membrane is not dependent on protein translocation (20, 51). The observations that efficient SecA-dependent protein translocation occurs in SecY- and SecE deficient membranes in vitro strongly argue that SecYEG is not essential for protein translocation of certain precursors at least in vitro. It should be emphasized that this is not to say that SecYEG plays no role in protein translocation. On the contrary, the high-affinity binding of SecYEG to SecA makes translocation more efficient. It is reminiscent of the roles of protonmotive force in the process: it is not essential, but it enhances efficiency and lowers the requirement of SecA or ATP for translocation (65, 66).
Implication of the Role of SecA Pore Structures in Protein Translocation.
Previous observations that SecYEG alone forms a structure that is too small to be a functional channel (45, 47) unless SecA is recruited to form a supercomplex (45, 47), that protein translocation can occur in the absence of SecYEG (6, 17, 18), and the current observation that SecA forms a large pore structure on interaction with phospholipids indicate that SecA may play a more important structural role in forming a translocation channel than was previously realized. Although the observations of pore structures do not prove the physiological role of SecA, they suggest that SecA forms the central core of the translocation channel that has been shown to be opened by signal peptides (67, 68). The sizes of the SecA pore structures observed here (2–6 nm) appear to be large enough to serve as a conducting channel for 1.1-nm hydrated peptide chains (69).
It is also possible that there are two types of SecA-dependent protein conducting channels (67, 68): one in which SecA recruits SecYEG as high-affinity binding sites to form a supercomplex and the other in which SecA forms a channel without SecYEG as low-affinity binding sites (but perhaps with other membrane proteins such as YidC). It is to be noted that there is more SecA than SecYEG in the membranes (41, 70, 71). Recently, Or et al. (72) reported that dimeric SecA may dissociate during translocation, contradicting earlier findings that SecA is functional as dimers (73). However, this interpretation is questioned by Benach et al. (74) who reported that phospholipids induced monomerization but signal peptides induced oligomerization of SecA. Moreover, the drastic structural and conformational change of SecA observed here could also account for the lack of crosslinking of SecA dimers leading to the conclusion (72) that dimeric SecA dissociates into monomers. Regardless, these observations further complicate the current understanding of the mechanism of Sec-dependent protein translocation. More work is necessary to resolve the paradox.
Acknowledgments
We thank D. Oliver for providing the strains; J. Frank for providing the software spider; Z. Liu for helping to build the spider software; Z. Pan and W. Jiang for maintenance of the electron microscope; and J. Houghton and P. Kaur for comments. J. Ingraham provided stimulating insights and discussions in addition to editing; P.C.T. dedicates this article to him for his years of unyielding faith and friendship. This work was supported by a research grant from the National Nature Science Foundation of China (to S.-F.S.) and by National Institutes of Health Grant GM 34766 (to P.C.T.). Facility and equipment is supported by fund from Georgia Research Alliance, and Research Program Enhancement, Georgia State University.
Abbreviation
- AFM
atomic force microscopy
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Cabelli R J, Chen L, Tai P C, Oliver D B. Cell. 1988;55:683–692. doi: 10.1016/0092-8674(88)90227-9. [DOI] [PubMed] [Google Scholar]
- 2.Cunningham K, Lill R, Crooke E, Rice M, Moore K, Wickner W, Oliver D. EMBO J. 1989;8:955–959. doi: 10.1002/j.1460-2075.1989.tb03457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oliver D B, Beckwith J. Cell. 1981;25:765–772. doi: 10.1016/0092-8674(81)90184-7. [DOI] [PubMed] [Google Scholar]
- 4.Manting E H, Driessen A J. Mol Microbiol. 2000;37:226–238. doi: 10.1046/j.1365-2958.2000.01980.x. [DOI] [PubMed] [Google Scholar]
- 5.Wickner W, Leonard M R. J Biol Chem. 1996;271:29514–29516. doi: 10.1074/jbc.271.47.29514. [DOI] [PubMed] [Google Scholar]
- 6.Watanabe M, Blobel G. Proc Natl Acad Sci USA. 1993;90:9011–9015. doi: 10.1073/pnas.90.19.9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt M G, Rollo E E, Grodberg J, Oliver D B. J Bacteriol. 1988;170:3404–3414. doi: 10.1128/jb.170.8.3404-3414.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabelli R J, Dolan K M, Qian L P, Oliver D B. J Biol Chem. 1991;266:24420–24427. [PubMed] [Google Scholar]
- 9.Ulbrandt N D, London E, Oliver D B. J Biol Chem. 1992;267:15184–15192. [PubMed] [Google Scholar]
- 10.Weaver A J, McDowall A W, Oliver D B, Deisenhofer J. J Struct Biol. 1992;109:87–96. doi: 10.1016/1047-8477(92)90040-h. [DOI] [PubMed] [Google Scholar]
- 11.Shilton B, Svergun D I, Volkov V V, Koch M H, Cusack S, Economou A. FEBS Lett. 1998;436:277–282. doi: 10.1016/s0014-5793(98)01141-7. [DOI] [PubMed] [Google Scholar]
- 12.Hendrick J P, Wickner W. J Biol Chem. 1991;266:24596–24600. [PubMed] [Google Scholar]
- 13.Chen X, Xu H, Tai P C. J Biol Chem. 1996;271:29698–29706. doi: 10.1074/jbc.271.47.29698. [DOI] [PubMed] [Google Scholar]
- 14.Economou A, Wickner W. Cell. 1994;78:835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt M G, Kiser K B. Microbes Infect. 1999;1:993–1004. doi: 10.1016/s1286-4579(99)80517-6. [DOI] [PubMed] [Google Scholar]
- 16.Watanabe M, Nicchitta C V, Blobel G. Proc Natl Acad Sci USA. 1990;87:1960–1964. doi: 10.1073/pnas.87.5.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y B, Lian J, Tai P C. J Bacteriol. 1997;179:7386–7393. doi: 10.1128/jb.179.23.7386-7393.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y B, Yu N, Tai P C. J Biol Chem. 1997;272:13660–13665. doi: 10.1074/jbc.272.21.13660. [DOI] [PubMed] [Google Scholar]
- 19.Rajapandi T, Oliver D. Mol Microbiol. 1996;20:43–51. doi: 10.1111/j.1365-2958.1996.tb02487.x. [DOI] [PubMed] [Google Scholar]
- 20.Ramamurthy V, Oliver D. J Biol Chem. 1997;272:23239–23246. doi: 10.1074/jbc.272.37.23239. [DOI] [PubMed] [Google Scholar]
- 21.Chen X, Brown T, Tai P C. J Bacteriol. 1998;180:527–537. doi: 10.1128/jb.180.3.527-537.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weinkauf S, Hunt J F, Scheuring J, Henry L, Fak J, Oliver D B, Deisenhofer J. Acta Crystallogr D. 2001;57:559–565. doi: 10.1107/s0907444901001202. [DOI] [PubMed] [Google Scholar]
- 23.Hunt J F, Weinkauf S, Henry L, Fak J J, McNicholas P, Oliver D B, Deisenhofer J. Science. 2002;297:2018–2026. doi: 10.1126/science.1074424. [DOI] [PubMed] [Google Scholar]
- 24.Wang H W, Sui S. J Struct Biol. 2001;134:46–55. doi: 10.1006/jsbi.2001.4364. [DOI] [PubMed] [Google Scholar]
- 25.Mosser G, Mallouh V, Brisson A. J Mol Biol. 1992;226:23–28. doi: 10.1016/0022-2836(92)90120-9. [DOI] [PubMed] [Google Scholar]
- 26.Scheuring S, Ringler P, Borgnia M, Stahlberg H, Muller D J, Agre P, Engel A. EMBO J. 1999;18:4981–4987. doi: 10.1093/emboj/18.18.4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muller D J, Engel A. J Mol Biol. 1999;285:1347–1351. doi: 10.1006/jmbi.1998.2359. [DOI] [PubMed] [Google Scholar]
- 28.Uzgiris E E, Kornberg R D. Nature. 1983;301:125–129. doi: 10.1038/301125a0. [DOI] [PubMed] [Google Scholar]
- 29.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 30.Frank J, Verschoor A, Boublik M. J Mol Biol. 1982;161:107–133. doi: 10.1016/0022-2836(82)90281-9. [DOI] [PubMed] [Google Scholar]
- 31.Frank J, Bretaudiere J P, Carazo J M, Verschoor A, Wagenknecht T. J Microsc. 1988;150:99–115. doi: 10.1111/j.1365-2818.1988.tb04602.x. [DOI] [PubMed] [Google Scholar]
- 32.Frank J, Verschoor A, Boublik M. Science. 1981;214:1353–1355. doi: 10.1126/science.7313694. [DOI] [PubMed] [Google Scholar]
- 33.Bustamante C, Rivetti C. Annu Rev Biophys Biomol Struct. 1996;25:395–429. doi: 10.1146/annurev.bb.25.060196.002143. [DOI] [PubMed] [Google Scholar]
- 34.Tian G, Wu H C, Ray P H, Tai P C. J Bacteriol. 1989;171:1987–1997. doi: 10.1128/jb.171.4.1987-1997.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wyman C, Rombel I, North A K, Bustamante C, Kustu S. Science. 1997;275:1658–1661. doi: 10.1126/science.275.5306.1658. [DOI] [PubMed] [Google Scholar]
- 36.Lill R, Dowhan W, Wickner W. Cell. 1990;60:271–280. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- 37.Ahn T, Kim H. Biochem Biophys Res Commun. 1994;203:326–330. doi: 10.1006/bbrc.1994.2185. [DOI] [PubMed] [Google Scholar]
- 38.Ahn T, Kim H. J Biol Chem. 1996;271:12372–12379. doi: 10.1074/jbc.271.21.12372. [DOI] [PubMed] [Google Scholar]
- 39.Keller R C, Snel M M, de Kruijff B, Marsh D. FEBS Lett. 1995;358:251–254. doi: 10.1016/0014-5793(94)01439-8. [DOI] [PubMed] [Google Scholar]
- 40.Breukink E, Demel R A, de Korte-Kool G, de Kruijff B. Biochemistry. 1992;31:1119–1124. doi: 10.1021/bi00119a021. [DOI] [PubMed] [Google Scholar]
- 41.Woodbury R L, Hardy S J, Randall L L. Protein Sci. 2002;11:875–882. doi: 10.1110/ps.4090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer T H, Menetret J F, Breitling R, Miller K R, Akey C W, Rapoport T A. J Mol Biol. 1999;285:1789–1800. doi: 10.1006/jmbi.1998.2413. [DOI] [PubMed] [Google Scholar]
- 43.Beckmann R, Bubeck D, Grassucci R, Penczek P, Verschoor A, Blobel G, Frank J. Science. 1997;278:2123–2126. doi: 10.1126/science.278.5346.2123. [DOI] [PubMed] [Google Scholar]
- 44.Hanein D, Matlack K E, Jungnickel B, Plath K, Kalies K U, Miller K R, Rapoport T A, Akey C W. Cell. 1996;87:721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- 45.Collinson I, Breyton C, Duong F, Tziatzios C, Schubert D, Or E, Rapoport T, Kuhlbrandt W. EMBO J. 2001;20:2462–2471. doi: 10.1093/emboj/20.10.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Breyton C, Haase W, Rapoport T A, Kuhlbrandt W, Collinson I. Nature. 2002;418:662–665. doi: 10.1038/nature00827. [DOI] [PubMed] [Google Scholar]
- 47.Manting E H, van Der Does C, Remigy H, Engel A, Driessen A J. EMBO J. 2000;19:852–861. doi: 10.1093/emboj/19.5.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Wolk J P, de Wit J G, Driessen A J. EMBO J. 1997;16:7297–7304. doi: 10.1093/emboj/16.24.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uchida K, Mori H, Mizushima S. J Biol Chem. 1995;270:30862–30868. doi: 10.1074/jbc.270.52.30862. [DOI] [PubMed] [Google Scholar]
- 50.Hartl F U, Lecker S, Schiebel E, Hendrick J P, Wickner W. Cell. 1990;63:269–279. doi: 10.1016/0092-8674(90)90160-g. [DOI] [PubMed] [Google Scholar]
- 51.Fekkes P, van der Does C, Driessen A J. EMBO J. 1997;16:6105–6113. doi: 10.1093/emboj/16.20.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dapic V, Oliver D. J Biol Chem. 2000;275:25000–25007. doi: 10.1074/jbc.M001100200. [DOI] [PubMed] [Google Scholar]
- 53.Eichler J, Brunner J, Wickner W. EMBO J. 1997;16:2188–2196. doi: 10.1093/emboj/16.9.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Joly J C, Wickner W. EMBO J. 1993;12:255–263. doi: 10.1002/j.1460-2075.1993.tb05651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Voorst F, van der Does C, Brunner J, Driessen A J, de Kruijff B. Biochemistry. 1998;37:12261–12268. doi: 10.1021/bi9809021. [DOI] [PubMed] [Google Scholar]
- 56.Nishiyama K, Suzuki T, Tokuda H. Cell. 1996;85:71–81. doi: 10.1016/s0092-8674(00)81083-1. [DOI] [PubMed] [Google Scholar]
- 57.Nagamori S, Nishiyama K, Tokuda H. J Biochem (Tokyo) 2002;132:629–634. doi: 10.1093/oxfordjournals.jbchem.a003266. [DOI] [PubMed] [Google Scholar]
- 58.van der Wolk J P, Fekkes P, Boorsma A, Huie J L, Silhavy T J, Driessen A J. EMBO J. 1998;17:3631–3639. doi: 10.1093/emboj/17.13.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flower A M, Doebele R C, Silhavy T J. J Bacteriol. 1994;176:5607–5614. doi: 10.1128/jb.176.18.5607-5614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nishiyama K, Fukuda A, Morita K, Tokuda H. EMBO J. 1999;18:1049–1058. doi: 10.1093/emboj/18.4.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nouwen N, de Kruijff B, Tommassen J. Proc Natl Acad Sci USA. 1996;93:5953–5957. doi: 10.1073/pnas.93.12.5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yahr T L, Wickner W T. EMBO J. 2000;19:4393–4401. doi: 10.1093/emboj/19.16.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Driessen A J, Manting E H, van der Does C. Nat Struct Biol. 2001;8:492–498. doi: 10.1038/88549. [DOI] [PubMed] [Google Scholar]
- 64.Bessonneau P, Besson V, Collinson I, Duong F. EMBO J. 2002;21:995–1003. doi: 10.1093/emboj/21.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen L, Tai P C. Proc Natl Acad Sci USA. 1985;82:4384–4388. doi: 10.1073/pnas.82.13.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Geller B L, Movva N R, Wickner W. Proc Natl Acad Sci USA. 1986;83:4219–4222. doi: 10.1073/pnas.83.12.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simon S M, Blobel G, Zimmerberg J. Proc Natl Acad Sci USA. 1989;86:6176–6180. doi: 10.1073/pnas.86.16.6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simon S M, Blobel G. Cell. 1992;69:677–684. doi: 10.1016/0092-8674(92)90231-z. [DOI] [PubMed] [Google Scholar]
- 69.Hamman B D, Chen J C, Johnson E E, Johnson A E. Cell. 1997;89:535–544. doi: 10.1016/s0092-8674(00)80235-4. [DOI] [PubMed] [Google Scholar]
- 70.Mizushima S, Tokuda H, Matusuyama S. In: Membrane Biogenesis and Protein Targeting. Neupert I W, Lill R, editors. Amsterdam: Elsevier Science; 1992. pp. 21–32. [Google Scholar]
- 71.Seoh H K, Tai P C. J Bacteriol. 1997;179:1077–1081. doi: 10.1128/jb.179.4.1077-1081.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Or E, Navon A, Rapoport T. EMBO J. 2002;21:4470–4479. doi: 10.1093/emboj/cdf471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Driessen A J. Biochemistry. 1993;32:13190–13197. doi: 10.1021/bi00211a030. [DOI] [PubMed] [Google Scholar]
- 74.Benach J, Chou Y T, Fak J J, Itkin A, Nicolae D D, Smith P C, Wittrock G, Floyd D L, Golsaz C M, Gierasch L M, Hunt J F. J Biol Chem. 2003;278:3628–3638. doi: 10.1074/jbc.M205992200. [DOI] [PubMed] [Google Scholar]