Abstract
Procyclins are abundant, glycosylphosphatidylinositol (GPI)-anchored proteins on the surface of procyclic (insect) form trypanosomes. To investigate whether trypanosomes are able to survive without a procyclin coat, all four procyclin genes were deleted sequentially. Bloodstream forms of the null mutant exhibited no detectable phenotype and were able to differentiate to procyclic forms. Initially, differentiated null mutant cells were barely able to grow, but after an adaptation period of 2 mo in culture they proliferated at the same rate as wild-type trypanosomes. Analysis of these culture-adapted null mutants revealed that they were covered by free GPIs. These were closely related to the mature procyclin anchor in structure and were expressed on the surface in numbers comparable with that of procyclin in wild-type cells. However, free GPIs were smaller than the procyclin anchor, indicative of a lower number of poly-N-acetyllactosamine repeats, and a proportion contained diacylphosphatidic acid. Free GPIs are also expressed by wild-type cells, although to a lesser extent. These have been overlooked in the past because they partition in a solvent fraction (chloroform/water/methanol) that is normally discarded when GPI-anchored proteins are purified.
INTRODUCTION
Glycosylphosphatidylinositol (GPI)-anchored proteins, lipophosphoglycans, and glycosylinositol phospholipids (GIPLs) are abundant components of the surface coat of many protozoan parasites (McConville and Ferguson, 1993; Ferguson, 1999; Ilgoutz and McConville, 2001). A variety of important functions have been attributed to these molecules, including protection against the host's adaptive immune responses (Cross, 1996), stimulation of a proinflammatory response (Almeida et al., 2000; Ropert and Gazzinelli, 2000), binding to host tissues (Lekutis et al., 2001; Sacks, 2001), and protection against proteolysis (Acosta-Serrano et al., 2001; Sacks, 2001). In addition, the lipid moieties of GPI anchors can modulate host-signaling pathways (Tachado et al., 1997).
Throughout its life cycle, the protozoan parasite Trypanosoma brucei is coated by several million copies of GPI-anchored proteins. The metacyclic form (the infective stage that is transmitted by the tsetse fly) and the bloodstream form in the mammalian host are covered by a coat of variant surface glycoproteins (VSGs). In the mammal, the parasite evades the immune system by periodically replacing the existing VSG coat by a different variant, a phenomenon known as antigenic variation (Cross, 1996). The VSG coat forms a physical barrier that prevents the binding of antibodies to invariant molecules that are embedded in the plasma membrane (Ziegelbauer and Overath, 1993; Salmon et al., 1994; Nolan et al., 2000). When bloodstream forms are ingested by the tsetse fly, they differentiate to procyclic forms in the insect midgut. During this process the VSG coat is progressively replaced by a new set of GPI-anchored glycoproteins known as procyclins. These are characterized by internal dipeptide (EP) or pentapeptide (GPEET) repeats, which confer an extended structure to the polypeptide backbone (Roditi et al., 1989; Treumann et al., 1997). T. brucei encodes three EP isoforms (EP1, EP2, and EP3) and a single copy of GPEET (reviewed in Ruepp et al., 1997). We have recently shown that the composition of the procyclin coat changes during development, both in culture and in the tsetse fly. Several hours after initiating synchronous differentiation of bloodstream forms to procyclic forms, all species of procyclin can be detected at similar levels (Vassella et al., 2001). Between 1 and 3 days after triggering differentiation, GPEET is the major form on the surface (Acosta-Serrano et al., 2001; Vassella et al., 2001) but is replaced by the glycosylated isoforms EP1 and EP3 within a few days (Acosta-Serrano et al., 2001; Vassella et al., 2001).
The change from the VSG to the procyclin coat is accompanied by an alteration in the structure of the GPI anchor. Both anchors consist of the common core structure ethanolamine-PO4-Manα1–2Manα1–6Manα1–4GlcN-phosphatidylinositol, but the lipid moiety is diacylglycerol in the VSG anchor (Ferguson et al., 1985, 1988) and acyl-2-lyso phosphatidic acid in the procyclin anchor (Field et al., 1991). In addition, the procyclin anchor contains a fatty acid linked to the myo-inositol ring, rendering the anchor resistant to phosphatidylinositol-specific phospholipase C (PI-PLC) (Roberts et al., 1988), and, unlike the VSG anchor, is extensively decorated with branched poly-N-acetyllactosamine repeats that are capped by sialic acid residues (Treumann et al., 1997). It has been postulated that the branched side chains of the anchor form a dense glycocalyx that contributes to the protective function of the coat against digestive enzymes in the fly midgut (McConville and Ferguson, 1993).
Two different approaches have been taken to removing the procyclin coat to investigate its function. We previously deleted all EP genes from procyclic culture forms, but we were unable to eliminate the final GPEET gene (Ruepp et al., 1997), suggesting that trypanosomes without a procyclin coat might not be viable under these conditions. The EP null mutant exhibited no phenotype in culture, but it was considerably less effective than the wild-type at establishing heavy infections in the tsetse fly (Ruepp et al., 1997). By disrupting GPI10, a key gene in the GPI anchor biosynthetic pathway, Nagamune et al. (2000) recently generated mutant procyclic forms that were incapable of attaching GPI anchors to proteins. These cells were unable to grow unless cultured in nonadherent flasks, but it is not clear whether this phenotype was due to a lack of procyclins or to some other GPI-anchored protein(s).
In this article, we describe the construction of a procyclin null mutant in bloodstream form trypanosomes. When triggered to differentiate, this mutant was able to shed the VSG coat and express procyclic-specific markers. Freshly differentiated cells were very fragile and barely able to grow, but they became more robust as they were passaged, finally proliferating at the same rate as wild-type trypanosomes. An analysis of these culture-adapted cells revealed that the null mutant had compensated for the lack of a procyclin coat by expressing free GPIs on its surface.
MATERIALS AND METHODS
Trypanosomes
Monomorphic bloodstream forms of T. brucei 427 (Cross and Manning, 1973) were cultured in HMI 9 medium (Hirumi and Hirumi, 1989) supplemented with 10% heat-inactivated fetal bovine serum at 37°C/5% CO2. Procyclic forms were cultured in modified DTM (Vassella and Boshart, 1996) supplemented with 15% heat-inactivated fetal bovine serum at 27°C. Bloodstream form trypanosomes were triggered to differentiate to the procyclic form by the addition of 6 mM cis-aconitate to the culture medium and lowering the incubation temperature to 27°C as described previously (Brun and Schönenberger, 1981).
Constructs and Stable Transformation of Trypanosomes
Two constructs, pCorleone-hyg and pKOP, that were designed to delete tandemly linked procyclin genes from the GPEET/PAG3 or EP/PAG1 and EP/PAG2 loci, respectively, are described previously (Ruepp et al., 1997; Vassella et al., 2000). For construction of pCorleone-neo, the neomycin-resistance gene was excised from pKON (Ruepp et al., 1997) and cloned between the HindIII and BamHI sites of pCorleone-hyg, thereby replacing the hygromycin-resistance gene. For construction of pKOPAC, the neomycin-resistance gene in pKON was replaced by the puromycin-resistance gene (PAC) as follows: pKON was linearized by cleavage with HindIII, the ends were repaired by treatment with Klenow, and the neomycin-resistance gene was released by cleavage with BamHI. The PAC gene was excised from pBluescript(KS+) (Ruepp et al., 1997) by cleavage with AgeI and BamHI and ligated to the linearized vector.
Stable transformation of bloodstream forms was performed as described previously (Li and Gottesdiener, 1996) by using 10 μg of plasmid DNA digested with KpnI and NotI (pKOP and pKOPAC) or with SalI and XbaI (pCorleone-hyg and pCorleone-neo) to release the insert. Individual clones were selected in microtiter plates with 0.1 μg/ml puromycin, 1.5 μg/ml phleomycin, 1.0 μg/ml hygromycin, or 1.0 μg/ml neomycin.
Southern Blot Analysis
Genomic DNA was extracted from individual clones as described previously (Young et al., 1982). One microgram of DNA was digested with PstI and separated on 0.8% agarose gels. Southern blot analysis was performed using standard procedures (Sambrook et al., 1989). A multiprime-labeled probe (Amersham, Dübendorf, Switzerland) used for hybridization was generated from the coding region of GPEET.
Western Blot Analysis and Immunofluorescence
Total cell lysates were separated on 12% polyacrylamide gels and transferred to Immobilon-P membranes (Millipore, Bedford, MA). Rabbit anti-VSG 221 antiserum (obtained from M.L. Cardoso de Almeida, Department of Microbiology, Universidade Federal de São Paulo, São Paulo, Brazil) was used at a dilution of 1:2000 and rat anti-CAP 5.5 antiserum (Hertz-Fowler et al., 2001), provided by K. Gull (School of Biological Sciences, University of Manchester, Manchester, United Kingdom) at 1:30.
For immunofluorescence, cells were fixed with 2% formaldehyde at 4°C (Vassella et al., 1997). The anti-EP monoclonal antibody TRBP1/247 (Richardson et al., 1986, 1988) was used at a dilution of 1:500 and rabbit polyclonal anti-GPEET K1 antiserum (Ruepp et al., 1997) at 1:500. Tetramethylrhodamine B isothiocyanate-conjugated anti-mouse antibody (Sigma) was used at 1:400 and fluorescein isothiocyanate-conjugated anti-rabbit antibody (Sigma) at 1:2000.
Labeling of Trypanosomes, Extraction, and SDS-PAGE
Metabolic labeling of procyclic form trypanosomes with [3H]ethanolamine or [3H]myristic acid (Bütikofer et al., 1997) was performed for 16–18 h. Surface labeling with [3H]borohydride was performed as described previously (Pollevick et al., 2000).
For extraction of GPI-anchored molecules, 2–4 × 108 radioactively labeled trypanosomes were harvested by centrifugation and washed with phosphate-buffered saline. The wet pellet was extracted twice with 10 ml of chloroform/methanol (used at 2:1 volume ratios) and three times with 5 ml of chloroform/methanol/water (CMW) used at 10:10:3 volume ratios). The pooled CMW-soluble fractions were dried under nitrogen, partitioned between 1 ml of butan-1-ol and 1 ml of water, and the resulting butanol and water phases were dried under nitrogen. The CMW-insoluble fraction was extracted sequentially with 1 ml of 9% (vol/vol) butan-1-ol in water at room temperature, with 100 μl of 0.1% (wt/vol) Triton X-100 in 20 mM Tris-HCl, pH 7.5, by boiling for 10 min and with 1% (wt/vol) SDS by boiling for 10 min (Bütikofer et al., 1997). Digestion of extracts with Pronase (Sigma), SDS-PAGE, and autoradiography were performed as described previously (Bütikofer et al., 1997).
Enzymatic and Chemical Treatment of Trypanosomal Extracts and Thin Layer Chromotography (TLC)
[3H]Myristic acid-labeled extracts were treated with PI-PLC, GPI-specific phospholipase D (GPI-PLD), strong base (Bütikofer et al., 1997), or nitrous acid (Field and Menon, 1992) as described previously. Products released from [3H]myristate-labeled GPIs by cleavage with GPI-PLD were incubated for 2 h at 37°C in 20 mM Tris-HCl, pH 8, 2 mM CaCl2, and 0.02% Triton X-100 in the presence of phospholipase A2 (PLA2) from Crotalus adamanteus (Sigma) or phospholipase B (PLB) from Vibrio sp. (Sigma). Released hydrophobic products were extracted with water-saturated butanol, dried under nitrogen, and separated by one-dimensional TLC (Field and Menon, 1992) in solvent system I (chloroform/methanol/water, 10:10:3, by volume) or system II (chloroform/methanol/acetic acid/water, 25:15:4:2, by volume). For quantification of radiolabeled lipids on TLC plates, areas containing radioactivity were scraped, extracted with methanol/6 N HCl (50:3, by volume), and counted (Bütikofer and Brodbeck, 1993).
Sialic Acid Analysis and trans-Sialidase Activity
For lectin binding, 106 trypanosomes were harvested, washed twice with trypanosome dilution buffer containing 5 mM KCl, 80 mM NaCl, 1 mM MgSO4, 20 mM Na2HPO4, 2 mM NaH2PO4 and 20 mM glucose (pH 7.7), and fixed with 4% formaldehyde for 18 h at 4°C. The washed cells were incubated with 1 μg/ml Texas Red-conjugated Maackia amurensis lectin (E. Y. Labs, San Mateo, CA) or with 2 μg/ml Texas Red-conjugated peanut agglutinin from Arachis hypogaea (E. Y. Labs) for 20 min on ice followed by two further washing steps. Sialic acids were cleaved from the surface of trypanosomes before lectin binding by treating 106 formaldehyde-fixed cells with 200 U of α2–3 neuraminidase (recombinant enzyme from Salmonella typhimurium; New England Biolabs, Beverly, MA) in 100 μl of 50 mM sodium citrate (pH 6.0), 100 mM NaCl, and 100 μg/ml bovine serum albumin for 2 h at 37°C. Lectin binding was measured by quantitative three-dimensional fluorescence microscopy as described previously (Grünenfelder et al., 2002).
Sialic acid content was measured by fluorometric reversed phase high-performance liquid chromatography (HPLC) (Hara et al., 1989; Engstler et al., 1997). Briefly, 5 × 108 cells were washed five times with 50 ml of ice-cold trypanosome dilution buffer and incubated for 60 min at 80°C in 250 μl of 0.1 M HCl. After centrifugation at 10,000 × g for 10 min, aliquots of the supernatant (20 μl) were derivatized with 1,2-diamino-4,5-methylenedioxybenzole for 60 min at 56°C in the dark. Sialic acids were analyzed by fluorometric HPLC by using an RP-18 cartridge (25 × 4 cm; Merck, Darmstadt, Germany) as described previously (Engstler et al., 1993).
trans-Sialidase activity was measured as sialidase activity (Engstler et al., 1992, 1997). Briefly, 108 trypanosomes were lysed with 0.4% Triton CF-54 in 50 mM bis-Tris-HCl, pH 7.0, 1 mM phenylmethylsulfonyl fluoride, 0.1 mM N-tosyl-l-lysine chloromethyl ketone, 0.1 mM EDTA, and the extract was incubated in the presence of 0.1 mM 4-methylumbelliferyl-α-d-N-acetylneuraminic acid (MU-Neu5Ac) for 30 min at 37°C. After centrifugation at 10,000 × g for 5 min, the supernatant was adjusted to pH 10 by the addition of 5 volumes of 0.15 M glycine, 0.04 M Na2CO3, and 0.06 M NaCl (pH 10). Fluorescence intensity was measured in a F-4010 fluorescence spectrophotometer (Hitachi, Tokyo, Japan).
RESULTS
Construction of a Procyclin Null Mutant in Bloodstream Form Trypanosomes
Procyclins are the major surface glycoproteins of procyclic form trypanosomes. They are not detectably expressed in the bloodstream form, however, and therefore unlikely to be essential for this stage. To obtain viable procyclin null mutant clones, we adopted the strategy of deleting all procyclin genes from bloodstream form trypanosomes. Bloodstream forms can be triggered to differentiate to procyclic forms by the addition of cis-aconitate to the culture medium and lowering the incubation temperature from 37 to 27°C (Brun and Schönenberger, 1981), allowing us to investigate whether these cells are able to develop to the procyclic form and to survive without a procyclin coat.
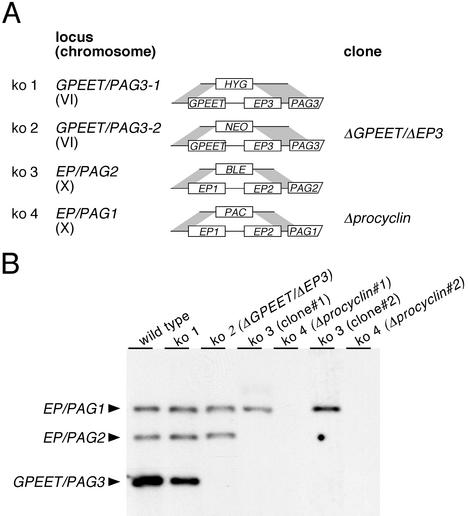
Procyclin genes occur in tandem repeats on chromosomes VI and X (Roditi and Clayton, 1999). To delete pairs of procyclin genes, locus-specific targeting constructs were made that contained antibiotic resistance genes cloned between sequences flanking the procyclin genes (Figure 1A). The constructs pCorleone-hyg (Vassella et al., 2000) and pCorleone-neo were designed to delete the GPEET and EP3 genes from the GPEET/PAG3 locus. Two further constructs, pKOP (Ruepp et al., 1997) and pKOPAC, contained flanking sequences from the EP/PAG1 or EP/PAG2 loci, respectively, and were used to delete the EP1 and EP2 genes. Bloodstream form trypanosomes of the stock 427 were first subjected to two consecutive rounds of electroporation and selection by using the constructs pCorleone-hyg and pCorleone-neo (Figure 1A). Southern blot analysis of a hygromycin and neomycin double-resistant clone, ΔGPEET/ΔEP3, revealed that both copies of GPEET and EP3 genes were deleted (Figure 1B). This clone was stably transformed with the construct pKOP, and two independent phleomycin-resistant clones were selected and subjected to a fourth round of transformation with the construct pKOPAC and selection with puromycin. In two clones, Δprocyclin 1 and 2, which were derived from independent third and fourth rounds of transformation, all procyclin genes were deleted (Figure 1B). These bloodstream form clones had a population doubling time indistinguishable from that of the wild type and no detectable alterations in cell morphology (our unpublished data).
Figure 1.
Deletion of procyclin genes. (A) Schematic depiction of the procyclin loci together with the constructs used to delete tandemly linked procyclin genes by homologous recombination. Each construct contains the promoter and 5′-untranslated region (UTR) of the first procyclin gene from the GPEET/PAG3 or EP/PAG1 locus, respectively, followed by an antibiotic-resistance gene. The 3′-flanking region in each construct consists of the last 19 base pairs of the 3′-UTR and downstream intergenic region of the second procyclin gene of the same locus. Each construct is flanked on both sides by locus-specific sequences for site-directed integration into the GPEET/PAG3 loci or the EP/PAG1 and EP/PAG2 loci, respectively (Ruepp et al., 1997; Vassella et al., 2000; see MATERIALS AND METHODS). The 3′-UTR downstream of the resistance gene is truncated, giving rise to high levels of expression in bloodstream form trypanosomes (Schürch et al., 1997; Vassella et al., 2000). (B) Southern blot analysis of procyclin knockout (ko) mutants. Genomic DNA was digested with PstI, which separates the EP/PAG1 and the EP/PAG2 loci from the two copies of GPEET/PAG3. A GPEET-specific probe was used for hybridization under conditions that allow cross-hybridization to the EP genes.
Procyclin Null Mutant Clones Require an Adaptation Period to Establish Proliferating Procyclic Form Cultures
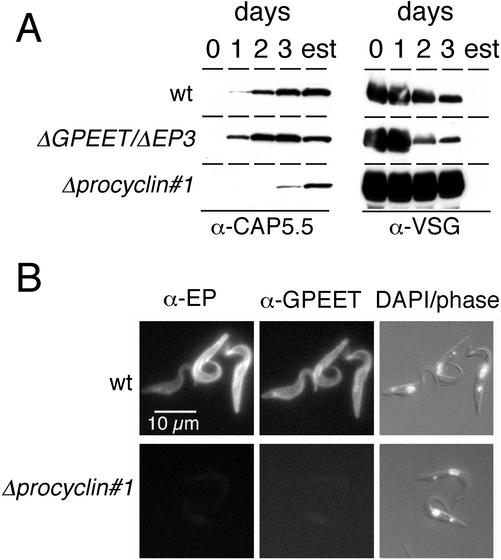
To investigate whether null mutants were able to differentiate to procyclic forms, they were exposed to cis-aconitate and a temperature shift. To monitor differentiation, expression of cytoskeletal-associated protein CAP5.5, which is considered to be a late marker of differentiation (Matthews and Gull, 1994), was analyzed by immunoblotting with a specific antiserum. As shown in Figure 2A, procyclin null mutant cells were able to express high levels of CAP5.5, but the kinetics of appearance of this protein was delayed relative to the wild type and the ΔGPEET/ΔEP3 mutant. CAP5.5 occurred with similar kinetics in the two independent Δprocyclin clones 1 and 2 (our unpublished data), suggesting that the delay in differentiation of these cells was not due to clonal variability. In keeping with the expression profile of CAP5.5, release of the VSG also occurred with delayed kinetics in the two null mutant clones (Figure 2A). Immunofluorescence analysis revealed that 97% of the wild-type cells had shed the VSG coat within 48 h. In contrast, >90% of the null mutant cells were still positive for VSG after 48 h, decreasing to ∼60% by days 4–6 and 14% by day 8. Δprocyclin cells which expressed CAP5.5 and had lost VSG had the morphology of procyclic forms and had repositioned the kinetoplast correctly (Figure 2B). EP and GPEET procyclins were not detected in these cells by immunofluorescence microscopy (Figure 2B) or immunoblotting (our unpublished data), confirming that all procyclin genes were deleted.
Figure 2.
Expression of stage-specific markers by differentiating trypanosomes. (A) Expression of CAP5.5 and release of VSG in trypanosomes exposed to the differentiation signal during a time-course experiment or in established procyclic forms (est). At the times indicated, aliquots were removed and analyzed by SDS-PAGE and immunoblotting. (B) Immunofluorescence analysis of established procyclic forms of the wild-type and a culture-adapted procyclin null mutant (Δprocyclin#1). Cells were fixed with formaldehyde and labeled with a polyclonal anti-GPEET antiserum and a monoclonal anti-EP antibody. Cells were counterstained with 4,6-diamidino-2-phenylindole (DAPI) to visualize nuclei and kinetoplasts. Corresponding images are from the same microscopic field.
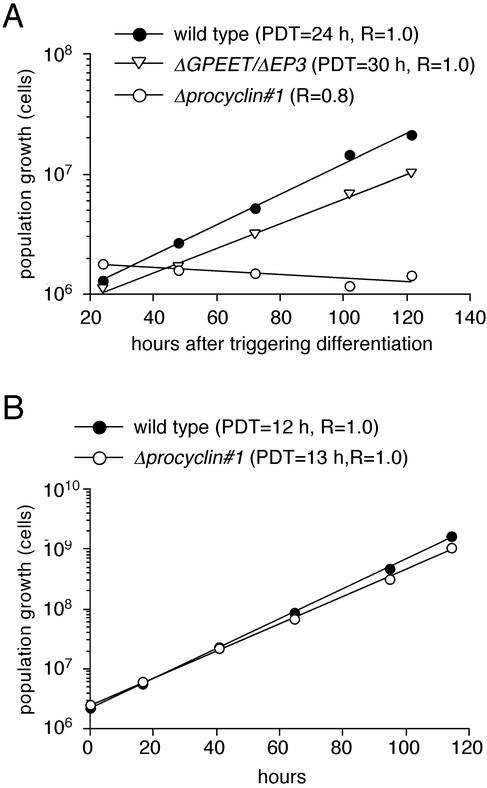
Analysis of the different cultures revealed that, in contrast to the wild-type and the ΔGPEET/ΔEP3 clone, freshly differentiated Δprocyclin cells were unable to proliferate, but remained at a constant cell density for a period of up to 2 mo (Figure 3A). To our surprise, however, the null mutant cells resumed growth after this period and exhibited the same population doubling time as the wild type when cultured under the same conditions (Figure 3B). The two independent null mutant clones showed similar growth properties during the adaptation period. In summary, these results indicate that the procyclin coat is not required for the survival of procyclic form trypanosomes in vitro, but null mutant cells require an adaptation/selection period to be able to grow exponentially.
Figure 3.
Population growth of procyclin mutants upon triggering differentiation with cis-aconitate (A) or after long-term culture as procyclic forms (B). The population growth was calculated as cell density multiplied by the cumulative dilution factors. The population doubling time (PDT) was calculated from the slope of the linear regression of the growth curves.
Are Procyclin Null Mutants Naked?
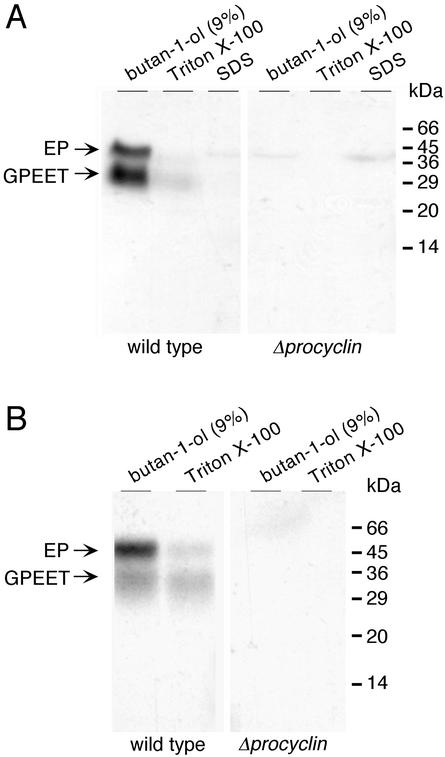
To investigate whether culture-adapted procyclic forms of the null mutant express an alternative glycoprotein coat, cells were metabolically labeled with [3H]ethanolamine or [3H]myristate, which are incorporated into GPI-anchored proteins (Field and Menon, 1992). After extensive delipidation, the insoluble cell pellet was extracted sequentially with 9% butan-1-ol, 0.1% Triton X-100, and 1% SDS, and aliquots of the different fractions were analyzed by SDS-PAGE followed by autoradiography. In [3H]ethanolamine-labeled wild-type cells, most radioactivity was incorporated into GPEET and EP procyclins. As shown previously (Bütikofer et al., 1997), these were soluble in 9% butan-1-ol (Figure 4A, left). The only additional product in wild-type cells was a faint band of ∼40 kDa in butan-1-ol and SDS fractions, which constitutes a convenient internal marker for the labeling reaction. It has been suggested that this protein is most likely to be elongation factor EF-1α (Rosenberry et al., 1989), which is posttranslationally modified with ethanolamine residues. Apart from the 40-kDa protein, no further radiolabeled products were detected in the different extracts of the null mutant (Figure 4A, right). Consistent with these results, incubating null mutant cells with [3H]myristic acid did not yield any labeled proteins in the butanol or Triton X-100 extracts (Figure 4B).
Figure 4.
Analysis of GPI-anchored proteins. Procyclic forms of the wild-type and the procyclin null mutant were labeled with [3H]ethanolamine (A) or [3H]myristic acid (B) and the delipidated (CMW-insoluble) fractions were extracted sequentially with 9% (vol/vol) butan-1-ol, 0.1% (wt/vol) Triton X-100, and 1% (wt/vol) SDS. Then 3 × 108 equivalents of [3H]ethanolamine-labeled cells and 108 equivalents of [3H]myristic acid-labeled cells were analyzed by SDS-PAGE and autoradiography.
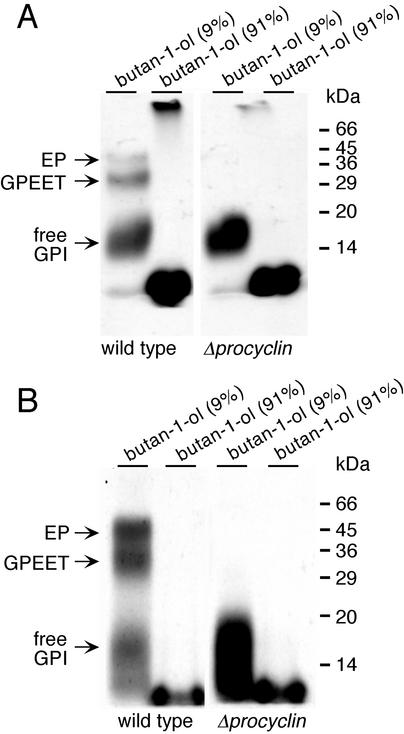
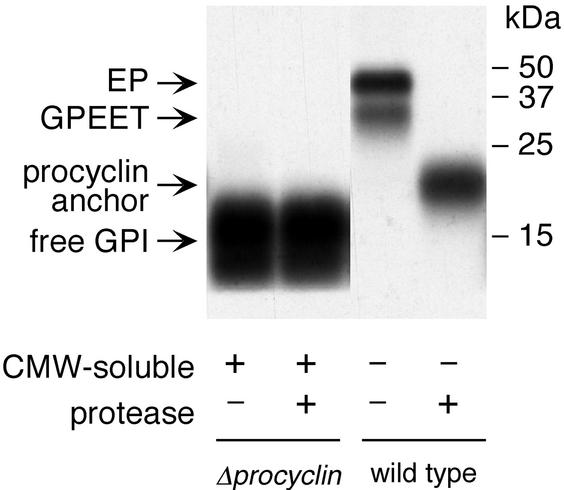
To investigate whether null mutant cells, which lack the major GPI acceptors, express nonprotein-linked (free) GPIs, the pooled CMW-soluble extracts of [3H]ethanolamine or [3H]myristic acid-labeled cells were dried and subsequently partitioned between butan-1-ol and water (see MATERIALS AND METHODS). When aliquots of the butan-1-ol phase (91% butan-1-ol), containing the GPI precursor lipids (Field and Menon, 1992), and the aqueous phase (9% butan-1-ol) were also analyzed by SDS-PAGE and autoradiography, we detected radiolabeled products in the aqueous phase with an apparent molecular mass of 14–20 kDa. These were labeled with both [3H]ethanolamine and [3H]myristic acid (Figure 5, A and B, right), suggesting the presence of GPI-anchored molecules. The same species were also detected in wild-type trypanosomes, although the amount of labeled products per cell equivalent was approximately fourfold lower in these cells than in the null mutant (Figure 5, A and B, left). To investigate whether these molecules contain protein, they were treated extensively with Pronase. This had no effect on the mobility of the labeled products on SDS-polyacrylamide gels (Figure 6, compare lanes 1 and 2), strongly suggesting that they are free GPIs. The Pronase-treated procyclin anchor and the free GPI differed not only in their electrophoretic mobility (Figure 6, lanes 2 and 4) but also in physicochemical properties. Whereas the free GPI was completely soluble in CMW, the Pronase-digested procyclin anchor was only partially soluble (our unpublished data). It has been shown previously that the C-terminal glycine residue of mature procyclin, to which the GPI anchor is attached, is not removed by Pronase (Bütikofer et al., 1997), but this alone is unlikely to account for the differences in the mobility and solubility of the two molecules. A more probable explanation is that the two GPIs differ in their carbohydrate or lipid moieties.
Figure 5.
Analysis of the CMW fraction of labeled trypanosomes. Wild-type trypanosomes (lanes 1 and 2) and null mutant cells (lanes 3 and 4) were labeled with [3H]ethanolamine (A) or [3H]myristic acid (B). The CMW-soluble extract was dried under nitrogen and partitioned between butan-1-ol and water. The lower 9% (vol/vol) butan-1-ol phase and the upper 91% (vol/vol) butan-1-ol phase were dried under nitrogen and analyzed by SDS-PAGE and autoradiography. The labeled material at the bottom of the gel in the 91% butan-1-ol fraction most probably consists of GPI precursors or phospholipids.
Figure 6.
Protease digestion of the free GPI and the procyclin anchor. Wild-type and procyclin null mutant cells were labeled with [3H]ethanolamine and extracted as described in the legends to Figures 4 and 5. The 9% (vol/vol) butan-1-ol extracts of the CMW-soluble (+) fraction containing the free GPI and the CMW-insoluble (−) fraction containing the procyclins were incubated for 24 h in the presence (+) or absence (−) of Pronase and analyzed by SDS-PAGE and autoradiography.
Free GPI Is Related to Procyclin Anchor
To provide further evidence for the GPI structure of the radiolabeled molecule(s), extracts of [3H]myristic acid-labeled trypanosomes were treated with GPI-PLD, PI-PLC, or with nitrous acid. Cleaved lipids were extracted with butan-1-ol, and the percentage of radioactivity released into the organic phase was quantified by scintillation counting. In agreement with published results (Bütikofer et al., 1997), 58 and 69% of radioactivity of the procyclin anchor was released into the butan-1-ol phase after treatment with GPI-PLD and nitrous acid, respectively (Table 1). The free GPI was also sensitive to cleavage with GPI-PLD (62% release) and nitrous acid (65% release) confirming the identity of a free GPI molecule (Table 1). Procyclins contain a fatty acid residue linked to the inositol ring that renders the GPI anchor insensitive to cleavage with PI-PLC (Roberts et al., 1988). Both the procyclin anchor and the free GPI were resistant to cleavage with this enzyme, whereas the VSG anchor, in which the inositol ring is not acylated (Ferguson et al., 1988), was sensitive (Table 1). Thus, the two GPIs seem to be structurally related.
Table 1.
Enzymatic and chemical treatment of cell extracts of trypanosomes labeled with [3H]myristic acid
| GPI-anchored moleculea | Release of radiolabel (%)b
|
|||||||
|---|---|---|---|---|---|---|---|---|
| GPI-PLD | PI-PLC | Nitrous acid | Untreated | |||||
| Free GPI | 62 ± 7 | n = 15 | 9 ± 3 | n = 4 | 65 ± 20 | n = 3 | 9 ± 1 | n = 4 |
| Procyclin | 58 ± 6 | n = 5 | 5 | n = 1 | 69 | n = 1 | 1 | n = 1 |
| VSG 221 | 93 | n = 2 | 86 | n = 1 | 78 | n = 2 | n.d. | |
Extraction of free GPI or procyclin molecules was performed as described in the legends to Figures 4 and 5, and labeling of bloodstream form trypanosomes (T. brucel 427) and extraction of VSG 221 was performed as described previously (Bütikofer et al., 1996).
Percentage of recovery of radiolabel after extraction with water-saturated butanol.
Acyl-lyso- and -diacyl-phosphatidic Acid Are Major Components of Free GPI
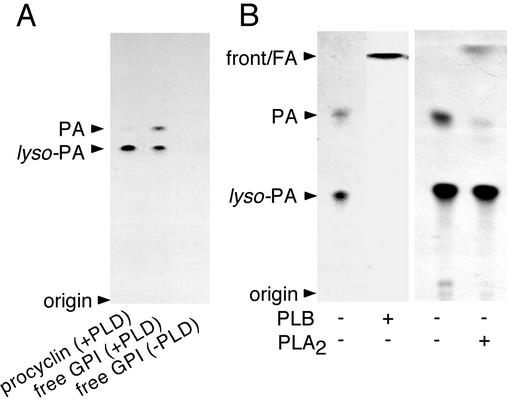
The major product of the procyclin anchor released into the butanol phase after treatment with GPI-PLD is acyl-2 lyso-phosphatidic acid (Field et al., 1991). However, analysis of GPI-PLD–cleaved material of the free GPI by TLC revealed two products, one of which comigrated with acyl-lyso-phosphatidic acid from the procyclin anchor (Figure 7A, compare lanes 1 and 2). No radiolabeled material was recovered in the butanol phase in the absence of GPI-PLD (Figure 7A, lane 3). When the TLC plate was exposed for a longer period, it became apparent that the faster migrating, unknown cleavage product was also present in extracts from the procyclin anchor (Figure 7A, lane 2). Quantification of the GPI-PLD–treated products by scintillation counting of material scraped from TLC plates revealed that 29 ± 10% (n = 4) of the radiolabeled free GPI and 6 ± 1% (n = 3) of the radiolabeled procyclin anchor consisted of the unknown cleavage product.
Figure 7.
Analysis of the products released from the free GPI by cleavage with GPI-PLD by TLC. The free GPIs and the procyclins were extracted from [3H]myristic acid-labeled null mutant or wild-type cells as described in the legends to Figures 4 and 5. (A) Products released from the labeled molecules by cleavage with GPI-PLD. (B) GPI-PLD-released products subsequently treated with phospholipase B (PLB +), phospholipase A2 (PLA2 +), or untreated (−). Samples were spotted onto Silica Gel-60 TLC plates and developed in solvent system I (A) or solvent system II (B) (see MATERIALS AND METHODS). Treatment of the free GPI with GPI-PLD and PLB gave rise to 90% of radioactivity in free fatty acids (FA), 5% in diacylphosphatidic acid (PA), and 5% in acyl-2 lyso-phosphatidic acid (lyso-PA). Treatment of the free GPI with GPI-PLD and PLA2 gave rise to 12% of radioactivity in FA, 2% in PA, and 86% in lyso-PA.
PLB cleaves fatty acids bound in ester linkage to sn-1 and sn-2 positions of the glycerol moiety of a phospholipid. When the products, released from the free GPI by cleavage with GPI-PLD, were subsequently treated with PLB, or subjected to strong base hydrolysis, the radioactivity was released quantitatively from both GPI-PLD cleavage products and comigrated with fatty acids at the front of TLC plates (Figure 7B, lanes 1 and 2). In addition, the faster migrating compound, but not lyso-phosphatidic acid, was sensitive to cleavage with PLA2 (Figure 7B, lanes 3 and 4), which specifically releases ester-linked fatty acid residues from the sn-2 positions. Together, these results indicate that the faster migrating lipid is diacyl-phosphatidic acid, whereas the slower migrating form is acyl-2-lyso-phosphatidic acid.
Free GPIs Are Exposed on the Surface of the Null Mutant
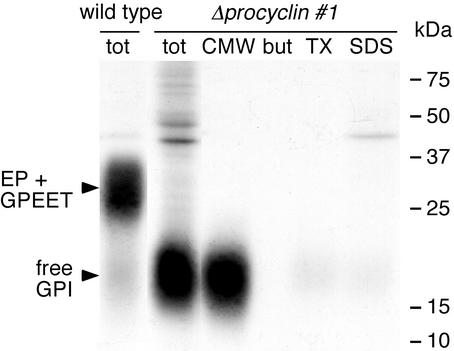
To investigate whether the free GPI is exposed on the surface of the null mutant, living procyclic forms were incubated in serum-free medium, resulting in the removal of sialic acid residues by the endogenous trans-sialidase (Pollevick et al., 2000). The cells were subsequently treated with galactose oxidase and the oxidized terminal β-galactose residues were reduced with [3H]borohydride. Under these conditions, wild-type and null mutant cells incorporated similar amounts of radioactivity (∼6 × 105-labeled molecules/cell). Although procyclins were the major tritium-labeled product in the wild type, most of the radioactivity in null mutant cells was incorporated into free GPIs. These were soluble in CMW and migrated with the same apparent molecular mass as the metabolically labeled products in the null mutant (Figure 8). These results indicate that wild-type and mutant cells express similar numbers of GPI-anchored molecules on their surfaces. In contrast to Δprocyclin cells, however, only a minority of free GPIs are exposed on the surface of wild-type cells.
Figure 8.
Galactose oxidase treatment and sodium [3H]borohydride labeling of living wild-type or null mutant cells. No cell death was observed during the labeling period. Total cell extracts (tot) and the 9% (vol/vol) butan-1-ol extract of the CMW-soluble fraction (CMW) or the CMW-insoluble fraction sequentially extracted with 9% (vol/vol) butan-1-ol (but), 0.1% (wt/vol) Triton X-100 (TX), and 1% (wt/vol) SDS (SDS) were analyzed by SDS-PAGE and autoradiography. Each lane contains equal numbers of cell equivalents from the different fractions.
Procyclins are the major acceptors of sialic acids, which are transferred from external proteins to the GPI anchor moiety by a cell surface-associated trypanosomal trans-sialidase (Engstler et al., 1993; Pontes de Carvalho et al., 1993). Because free GPI is on the surface of the null mutant, it is possible that this molecule contains sialic acids. Cells were fixed with formaldehyde and incubated with Texas Red-conjugated M. amurensis lectin, which binds sialic acids. Wild-type and null mutant cells bound similar numbers of lectin molecules (Table 2). To verify the specificity of this reaction, fixed cells were treated with neuraminidase (which cleaves sialic acids) before incubation with the lectin. Under these conditions, cells showed minimal lectin binding, similar to untreated bloodstream forms, which contain no sialic acids (Engstler et al., 1993). Direct measurements of the sialic acid content by fluorometric reversed phase HPLC confirmed that wild-type and null mutant cells contained similar amounts of sialic acid (Table 2). Removal of sialic acids by neuraminidase exposes terminal β-galactose residues, which are specific ligands of peanut agglutinin. Neuraminidase-treated null mutant or wild-type cells, but not untreated cells, bound Texas Red-conjugated peanut agglutinin, demonstrating that sialic acids are linked to β-galactose residues in both the wild-type and the null mutant. Furthermore, because terminal β-galactose residues of the free GPI in desialylated null mutant cells are the major acceptors for the borohydride-labeling reaction (Figure 8), we conclude that the free GPI is also the major acceptor for sialic acids in these cells.
Table 2.
Comparison of the sialic acid content of the wild type and Δprocyclin (clone 1)
| Clone (stage) | Fluorescence intensitya
|
Sialic acid contentd
|
|||
|---|---|---|---|---|---|
| MAAb
|
PNAc
|
||||
| Neurame | Neurame | ng/107 cells | |||
| Δprocyclin (procyclic) | 2017 ± 241 | 91 ± 17 | 391 ± 31 | 3075 ± 278 | 108 ± 9 |
| Wild type (procyclic) | 2546 ± 189 | 72 ± 30 | 397 ± 42 | 2911 ± 239 | 116 ± 4 |
| Wild type (bloodstream) | 67 ± 24 | n.d. | n.d. | n.d. | 2 |
MAA, Maackia amurensis lectin; n.d., not determined; PNA, peanut agglutinin.
Determined by quantitative fluorescence microscopy. The mean (±SD) from 200–300 segments is presented.
Texas Red-conjugated M. amurensis lectin used at 1 μg/ml.
Texas Red-conjugated peanut agglutinin from A. hypogaea used at 2 μg/ml.
Determined by fluorimetric reversed phase HPLC. The mean (±SD) from four independent measurements is presented.
Cells were treated with 200 U of α2-3 neuraminidase (neuram) before lectin binding.
The free GPI is up-regulated on the surface of the null mutant, but does this also hold true for other GPI-anchored molecules? To compare the expression levels of trans-sialidase (which has been shown to contain a GPI anchor; Engstler et al., 1993) in wild-type and null mutant cells, the activity of this enzyme per cell equivalent was determined as sialidase activity. As shown in Table 3, both clones exhibited similar enzyme activities, suggesting that trans-sialidase is present at similar levels in wild-type and null mutant cells. Thus, in contrast to the free GPI, trans-sialidase seems not to be up-regulated on the surface of the null mutant.
Table 3.
Comparison of trans-sialidase activity in Δprocyclin and wt cells
| Clone (stage) |
trans-Sialidase activitya
|
|
|---|---|---|
| RFU/107 cellsb | n | |
| Null mutant (procyclic) | 1153 ± 89 | 6 |
| Wild type (procyclic) | 1263 ± 109 | 6 |
| Wild type (bloodstream) | 12 ± 8 | 6 |
Measured as sialidase activity by using the substrate MU-Neu5Ac.
The mean (±SD) of the cleaved product determined as relative fluorescence units (RFU).
DISCUSSION
To generate procyclin null mutants, four sequential rounds of stable transformation were undertaken in bloodstream form trypanosomes. Null mutants showed no detectable phenotype at this stage of the life cycle and were also capable of differentiating to the procyclic form as indicated by appropriate changes in morphology, repositioning of the kinetoplast, loss of the VSG coat, and expression of CAP 5.5. However, the null mutants released the VSG coat more slowly than the parental line implying that, under normal circumstances, procyclins contribute to this process. These might disrupt the dense packing of the coat, rendering the VSG molecules more susceptible to the proteases involved in shedding (Bülow et al., 1989; Ziegelbauer et al., 1993).
Initially, in contrast to wild-type cells, the Δprocyclin mutants were barely able to proliferate as procyclic forms, but after a period of 2 mo in culture they grew with the same population doubling time as wild-type cells. Because the same phenomenon was observed with two independent clones, this was unlikely to be due to unrelated secondary mutations. In light of these results, we can now explain why we were previously unsuccessful in generating total procyclin knockouts in procyclic form trypanosomes (Ruepp et al., 1997). Because null mutants are considerably more fragile and require a period of adaptation before they can proliferate, they would tend to be overgrown by cells that had integrated the construct incorrectly and retained a procyclin gene.
Our data demonstrate unequivocally that trypanosomes can survive without any form of procyclin. Although GPI10 null mutants, which are unable to attach GPI anchors to proteins, were negative for procyclins by flow cytometry (Nagamune et al., 2000), several proteins could be immunoprecipitated from the cell pellet and culture supernatant with anti-EP antibodies. The simplest explanation is that these represent unanchored forms of the protein, some of which are secreted, although it is surprising that they were much larger than the polypeptide precursors (11–14 kDa) or the mature N-glycosylated form after chemical removal of the anchor (Mr ∼15 kDa by SDS-PAGE) (Bütikofer et al., 1997). It also cannot be excluded that, in GPI10 null mutants, some procyclin precursors are attached to the cell surface by the hydrophobic C terminus, as was recently shown to be the case for prions in platelets derived from patients with a defect in GPI synthesis (Holada et al., 2002). These might have been overlooked because several antibodies directed against GPI-anchored molecules, including the commercially available monoclonal antibody against EP procyclin used by Nagamune and colleagues, react very poorly with the protein after removal of the GPI lipid moiety (Bütikofer et al., 2001).
One possible way of compensating for the lack of procyclins might be to increase the expression of other GPI-anchored proteins. Δprocyclin mutants incorporated radiolabeled GPI precursors to the same extent as wild-type cells, but the radiolabel could not be detected in the butanol and Triton X-100 fractions where GPI-anchored proteins normally partition (Ferguson et al., 1993; Bütikofer et al., 1997). Furthermore, the surface trans-sialidase, which is GPI anchored (Engstler et al., 1993; Pontes de Carvalho et al., 1993), exhibited the same level of activity in wild-type and Δprocyclin cells. The null mutants were not naked, however, because they expressed free GPIs on their surface. These were closely related to the procyclin anchor in structure (same core structure, acylated inositol, phosphatidic acid), and, as judged by [3H]borohydride labeling of living cells and sialic acid content, were present on the surface in numbers comparable with that of procyclins in wild-type cells. However, free GPIs were smaller and less hydrophilic than procyclin anchors were after protease treatment, indicative of a lower number of poly-N-acetyllactosamine repeats, and a considerably higher proportion of them contained diacylphosphatidic acid.
Surface-associated GPIs are abundant, if enigmatic, molecules in other organisms ranging from mammals (van't Hof et al., 1995; Singh et al., 1996) to protozoan parasites (Lederkremer et al., 1991; Ralton and McConville, 1998). Several recent studies indicate that free GPIs are not essential for the viability of Leishmania mexicana in culture (Garami and Ilg, 2001; Garami et al., 2001). However, the fact that the density of GPI molecules on the surface of Δprocyclin mutants was similar to that of wild-type cells suggests that export of the anchor is required for growth of T. brucei procyclic forms. Consistent with these results, the GPI10 null mutant, which is unable to transfer the third mannose residue to the GPI precursor, and is thus unable to attach GPI anchors to proteins (Nagamune et al., 2000), is now known to express free GPIs (Ferguson, personal communication). This would not have been apparent from the biosynthetic labeling experiments performed previously with this mutant (Nagamune et al., 2000) because the N-acetyllactosamine residues prevent the free GPIs from migrating on TLC plates.
GPI lipid moieties might be important for the physicochemical properties of the plasma membrane, whereas the sialic acid residues would provide negative charges on the cell surface that might compensate, in part, for the absence of the highly acidic procyclin polypeptide. It has also been proposed that the elaborate side chains form a glycocalyx that protects underlying proteins from digestion by tsetse fly proteases (McConville and Ferguson, 1993). Although free GPIs are mainly intracellular in wild-type cells (compare Figures 5 and 8), a subpopulation might be exported to the cell surface to tile the spaces between procyclin molecules. In Leishmania, GPI-anchored proteins and free GPIs are transported with different kinetics, suggesting that these molecules are packaged into different transport vesicles (Ralton et al., 2002). Newly differentiated Δprocyclin cells might fail to grow because they require GPIs on their surface, but are unable to export them in sufficient quantities. Alternatively, transport might be necessary to prevent the accumulation of toxic amounts of intracellular GPIs. At present, we can only speculate about the nature of the alterations that occur during the adaptation period. One possibility is that vesicles loaded with free GPIs become capable of bypassing a retention signal, so that they can be transported to the plasma membrane.
ACKNOWLEDGMENTS
We thank Keith Gull (University of Manchester) and Lucia Cardoso de Almeida (Universidade Federal de São Paulo) for antibodies, Monika Boschung for technical assistance and Mike. Ferguson (University of Dundee, Dundee, United Kingdom) for critical reading of the manuscript. M.E. thanks Michael Boshart for support. This research was supported by Swiss National Science Foundation grants to I.R. to E.V. and to P.B. M.E. was supported by the Deutsche Forschungsgemeinschaft.
Abbreviations used:
- CMW
chloroform/methanol/water
- GPI
glycosylphosphatidylinositol
- GPI-PLD
GPI-specific phospholipase D
- PI-PLC
phosphatidylinositol-specific phospholipase C
- PLA2
phospholipase A2
- PLB
phospholipase B
- TLC
thin layer chromatography
- VSG
variant surface glycoprotein
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–10–0694. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–10–0694.
REFERENCES
- Acosta-Serrano A, Vassella E, Liniger M, Kunz Renggli C, Brun R, Roditi I, Englund PT. The surface coat of procyclic Trypanosoma brucei: programmed expression and proteolytic cleavage of procyclin in the tsetse fly. Proc Natl Acad Sci USA. 2001;98:1513–1518. doi: 10.1073/pnas.041611698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida IC, Camargo MM, Procopio DO, Silva LS, Mehlert A, Travassos LR, Gazzinelli RT, Ferguson MA. Highly purified glycosylphosphatidylinositols from Trypanosoma cruzi are potent proinflammatory agents. EMBO J. 2000;19:1476–1485. doi: 10.1093/emboj/19.7.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun R, Schönenberger M. Stimulating effect of citrate and cis-aconitate on the transformation of Trypanosoma brucei bloodstream forms to procyclic forms in vitro. Z Parasitenkd. 1981;66:17–24. doi: 10.1007/BF00941941. [DOI] [PubMed] [Google Scholar]
- Bülow R, Nonnengässer C, Overath P. Release of the variant surface glycoprotein during differentiation of bloodstream to procyclic forms of Trypanosoma brucei. Mol Biochem Parasitol. 1989;32:85–92. doi: 10.1016/0166-6851(89)90132-1. [DOI] [PubMed] [Google Scholar]
- Bütikofer P, Boschung M, Brodbeck U, Menon AK. Phosphatidylinositol hydrolysis by Trypanosoma brucei glycosylphosphatidylinositol phospholipase C. J Biol Chem. 1996;271:15533–15541. doi: 10.1074/jbc.271.26.15533. [DOI] [PubMed] [Google Scholar]
- Bütikofer P, Brodbeck U. Partial purification and characterization of a (glycosyl) inositol phospholipid-specific phospholipase C from peanut. J Biol Chem. 1993;268:17794–17802. [PubMed] [Google Scholar]
- Bütikofer P, Malherbe T, Boschung M, Roditi I. GPI-anchored proteins: now you see 'em, now you don't. FASEB J. 2001;15:545–548. doi: 10.1096/fj.00-0415hyp. [DOI] [PubMed] [Google Scholar]
- Bütikofer P, Ruepp S, Boschung M, Roditi I. ‘GPEET’ procyclin is the major surface protein of procyclic culture forms of Trypanosoma brucei brucei strain 427. Biochem J. 1997;326:415–423. doi: 10.1042/bj3260415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross GA. Antigenic variation in trypanosomes: secrets surface slowly. Bioessays. 1996;18:283–291. doi: 10.1002/bies.950180406. [DOI] [PubMed] [Google Scholar]
- Cross GAM, Manning JC. Cultivation of Trypanosoma brucei ssp. in semi-defined and defined media. Parasitology. 1973;67:315–331. doi: 10.1017/s0031182000046540. [DOI] [PubMed] [Google Scholar]
- Engstler M, Reuter G, Schauer R. Purification and characterization of a novel sialidase found in procyclic culture forms of Trypanosoma brucei. Mol Biochem Parasitol. 1992;54:21–30. doi: 10.1016/0166-6851(92)90091-w. [DOI] [PubMed] [Google Scholar]
- Engstler M, Reuter G, Schauer R. The developmentally regulated trans-sialidase from Trypanosoma brucei sialylates the procyclic acidic repetitive protein. Mol Biochem Parasitol. 1993;61:1–13. doi: 10.1016/0166-6851(93)90153-o. [DOI] [PubMed] [Google Scholar]
- Engstler M, Wirtz E, Cross GA. Generation of constitutive and inducible trans-sialylation dominant-negative phenotypes in Trypanosoma brucei and Trypanosoma cruzi. Glycobiology. 1997;7:955–964. doi: 10.1093/glycob/7.7.955. [DOI] [PubMed] [Google Scholar]
- Ferguson MA. The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosome research. J Cell Sci. 1999;112:2799–2809. doi: 10.1242/jcs.112.17.2799. [DOI] [PubMed] [Google Scholar]
- Ferguson MA, Haldar K, Cross GA. Trypanosoma brucei variant surface glycoprotein has a sn-1,2-dimyristyl glycerol membrane anchor at its COOH terminus. J Biol Chem. 1985;260:4963–4968. [PubMed] [Google Scholar]
- Ferguson MA, Homans SW, Dwek RA, Rademacher TW. Glycosyl-phosphatidylinositol moiety that anchors Trypanosoma brucei variant surface glycoprotein to the membrane. Science. 1988;239:753–759. doi: 10.1126/science.3340856. [DOI] [PubMed] [Google Scholar]
- Ferguson MA, Murray P, Rutherford H, McConville MJ. A simple purification of procyclic acidic repetitive protein and demonstration of a sialylated glycosyl-phosphatidylinositol membrane anchor. Biochem J. 1993;291:51–55. doi: 10.1042/bj2910051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field M, Menon AK. Biosynthesis of glycosylphosphatidylinositol membrane protein anchors. In: Hooper NM, Turner AJ, editors. Lipid Modification of Proteins: A Practical Approach. Oxford, United Kingdom: IRL Press; 1992. [Google Scholar]
- Field MC, Menon AK, Cross GA. A glycosylphosphatidylinositol protein anchor from procyclic stage Trypanosoma brucei: lipid structure and biosynthesis. EMBO J. 1991;10:2731–2739. doi: 10.1002/j.1460-2075.1991.tb07821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garami A, Ilg T. Disruption of mannose activation in Leishmania mexicana: GDP-mannose pyrophosphatase is required for virulence, but not for viability. EMBO J. 2001;20:3657–3666. doi: 10.1093/emboj/20.14.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garami A, Mehlert A, Ilg T. Glycosylation defects and virulence phenotypes of Leishmania mexicana phosphomannomutase and dolicholphosphate-mannose synthase gene deletion mutants. Mol Cell Biol. 2001;21:8168–8183. doi: 10.1128/MCB.21.23.8168-8183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünenfelder CG, Engstler M, Weise F, Schwarz H, Stierhof Y-D, Boshart M, Overath P. Accumulation of a GPI-anchored protein at the cell surface requires sorting at multiple intracellular levels. Traffic. 2002;3:547–559. doi: 10.1034/j.1600-0854.2002.30805.x. [DOI] [PubMed] [Google Scholar]
- Hara S, Yamaguchi M, Takemori Y, Furuhata K, Ogura H, Nakamura M. Determination of mono-O-acetylated N-acetylneuraminic acids in human and rat sera by fluorometric high-performance liquid chromatography. Anal Biochem. 1989;179:162–166. doi: 10.1016/0003-2697(89)90218-2. [DOI] [PubMed] [Google Scholar]
- Hertz-Fowler C, Ersfeld K, Gull K. CAP5.5, a life-cycle-regulated, cytoskeleton-associated protein is a member of a novel family of calpain-related proteins in Trypanosoma brucei. Mol Biochem Parasitol. 2001;116:25–34. doi: 10.1016/s0166-6851(01)00296-1. [DOI] [PubMed] [Google Scholar]
- Hirumi H, Hirumi K. Continuous cultivation of Trypanosoma brucei bloodstream forms in a medium containing a low concentration of serum protein without feeder cell layers. J Parasitol. 1989;75:985–989. [PubMed] [Google Scholar]
- Holada K, Simak J, Risitano AM, Maciejewski J, Young NS, Vostal JG. Activated platelets of patients with paroxysmal nocturnal hemoglobinuria express cellular prion protein. Blood. 2002;100:341–343. doi: 10.1182/blood.v100.1.341. [DOI] [PubMed] [Google Scholar]
- Ilgoutz SC, McConville M. Function and assembly of the Leishmania surface coat. Int J Parasitol. 2001;31:899–908. doi: 10.1016/s0020-7519(01)00197-7. [DOI] [PubMed] [Google Scholar]
- Lederkremer RM, Lima C, Ramirez MI, Ferguson MA, Homans SW, Thomas-Oates J. Complete structure of the glycan of lipopeptidophophoglycan from Trypanosoma cruzi epimastigotes. J Biol Chem. 1991;266:23670–23675. [PubMed] [Google Scholar]
- Lekutis C, Ferguson DJP, Grigg ME, Camps M, Boothroyd JC. Surface antigens of Toxoplasma gondii: variations on a theme. Int J Parasitol. 2001;31:1285–1292. doi: 10.1016/s0020-7519(01)00261-2. [DOI] [PubMed] [Google Scholar]
- Li FS, Gottesdiener KM. An efficient method for stable transfection of bloodstream form Trypanosoma brucei. Nucleic Acids Res. 1996;24:534–535. doi: 10.1093/nar/24.3.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews KR, Gull K. Evidence for an interplay between cell cycle progression and the initiation of differentiation between life cycle forms of African trypanosomes. J Cell Biol. 1994;125:1147–1156. doi: 10.1083/jcb.125.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConville MJ, Ferguson MA. The structure, biosynthesis and function of glycosylated phophatidylinositols in the parasitic protozoa and higher eukaryotes. Biochem J. 1993;294:305–324. doi: 10.1042/bj2940305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamune K, et al. Critical roles of glycosylphosphatidylinositol for Trypanosoma brucei. Proc Natl Acad Sci USA. 2000;97:10336–10341. doi: 10.1073/pnas.180230697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan DP, Jackson DG, Biggs MJ, Brabazon ED, Pays A, Van Laethem F, Paturiaux-Hanocq F, Elliot JF, Voorheis HP, Pays E. Characterization of a novel alanine-rich protein located in surface microdomains in Trypanosoma brucei. J Biol Chem. 2000;275:4072–4080. doi: 10.1074/jbc.275.6.4072. [DOI] [PubMed] [Google Scholar]
- Pollevick GD, Di Noia JM, Salto ML, Lima C, Leguizamon MS, de Lederkremer RM, Frasch AC. Trypanosoma cruzi surface mucins with exposed variant epitopes. J Biol Chem. 2000;275:27671–27680. doi: 10.1074/jbc.M000253200. [DOI] [PubMed] [Google Scholar]
- Pontes de Carvalho LC, Tomlinson S, Vandekerckhove F, Bienen EJ, Clarkson AB, Jiang MS, Hart GW, Nussenzweig V. Characterization of a novel trans-sialidase of Trypanosoma brucei procyclic trypomastigotes and identification of procyclin as the main sialic acid acceptor. J Exp Med. 1993;177:465–474. doi: 10.1084/jem.177.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralton JE, McConville MJ. Delineation of three pathways of glycosylphosphatidylinositol biosynthesis in Leishmania mexicana. Precursors from different pathways are assembled on distinct pools of phosphatidylinositol and undergo fatty acid remodeling. J Biol Chem. 1998;273:4245–4257. doi: 10.1074/jbc.273.7.4245. [DOI] [PubMed] [Google Scholar]
- Ralton JE, Mullin KA, McConville M. Intracellular trafficking of glycosylphosphatidylinositol (GPI)-anchored proteins and free GPIs in Leishmania mexicana. Biochem J. 2002;363:365–375. doi: 10.1042/0264-6021:3630365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JP, Beecroft RP, Tolson DL, Liu MK, Pearson TW. Procyclin: an unusual immunodominant glycoprotein surface antigen from the procyclic stage of African trypanosomes. Mol Biochem Parasitol. 1988;31:203–216. doi: 10.1016/0166-6851(88)90150-8. [DOI] [PubMed] [Google Scholar]
- Richardson JP, Jenni L, Beecroft RP, Pearson TW. Procyclic tsetse fly midgut forms and culture forms of African trypanosomes share stage- and species-specific surface antigens identified by monoclonal antibodies. J Immunol. 1986;136:2259–2264. [PubMed] [Google Scholar]
- Roberts WL, Santikarn S, Reinhold VN, Rosenberry TL. Structural characterization of the glycoinositol phospholipid membrane anchor of human erythrocyte acetylcholinesterase by fast atom bombardment mass spectrometry. J Biol Chem. 1988;263:18776–18784. [PubMed] [Google Scholar]
- Roditi I, Clayton C. An unambiguous nomenclature for the major surface glycoproteins of the procyclic form of Trypanosoma brucei. Mol Biochem Parasitol. 1999;103:99–100. doi: 10.1016/s0166-6851(99)00124-3. [DOI] [PubMed] [Google Scholar]
- Roditi I, et al. Procyclin gene expression and loss of the variant surface glycoprotein during differentiation of Trypanosoma brucei. J Cell Biol. 1989;108:737–746. doi: 10.1083/jcb.108.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropert C, Gazzinelli RT. Signaling of immune system cells by glycosylphosphatidylinositol (GPI) anchor and related structures derived from parasitic protozoa. Curr Opin Microbiol. 2000;3:395–403. doi: 10.1016/s1369-5274(00)00111-9. [DOI] [PubMed] [Google Scholar]
- Rosenberry TL, Krall JA, Dever TE, Haas R, Louvard D, Merrick WC. Biosynthetic incorporation of [3H]ethanolamine into protein synthesis elongation factor 1 alpha reveals a new post-translational protein modification. J Biol Chem. 1989;264:7096–7099. [PubMed] [Google Scholar]
- Ruepp S, Furger A, Kurath U, Renggli CK, Hemphill A, Brun R, Roditi I. Survival of Trypanosoma brucei in the tsetse fly is enhanced by the expression of specific forms of procyclin. J Cell Biol. 1997;137:1369–1379. doi: 10.1083/jcb.137.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks DL. Leishmania-sand fly interactions controlling species-specific vector competence. Cell Microbiol. 2001;3:189–196. doi: 10.1046/j.1462-5822.2001.00115.x. [DOI] [PubMed] [Google Scholar]
- Salmon D, Geuskens M, Hanocq F, Hanocq-Quertier J, Nolan D, Ruben L, Pays E. A novel heterodimeric transferrin receptor encoded by a pair of VSG expression site-associated genes in T. brucei. Cell. 1994;78:75–86. doi: 10.1016/0092-8674(94)90574-6. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T, editors. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schürch N, Furger A, Kurath U, Roditi I. Contributions of the procyclin 3′ untranslated region and coding region to the regulation of expression in bloodstream forms of Trypanosoma brucei. Mol Biochem Parasitol. 1997;89:109–121. doi: 10.1016/s0166-6851(97)00107-2. [DOI] [PubMed] [Google Scholar]
- Singh N, Liang L-N, Tykocinski ML, Tartakoff AM. A novel class of cell surface glycolipids of mammalian cells. J Biol Chem. 1996;271:12879–12884. doi: 10.1074/jbc.271.22.12879. [DOI] [PubMed] [Google Scholar]
- Tachado SD, Gerold P, Schwarz R, Novakovic S, McConville M, Schofield L. Signal transduction in macrophages by glycosylphosphatidylinositols of Plasmodium, Trypanosoma, and Leishmania: activation of protein tyrosine kinases and protein kinase C by inositolglycan and diacylglycerol moieties. Proc Natl Acad Sci USA. 1997;94:4022–4027. doi: 10.1073/pnas.94.8.4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treumann A, Zitzmann N, Hulsmeier A, Prescott AR, Almond A, Sheehan J, Ferguson MA. Structural characterization of two forms of procyclic acidic repetitive protein expressed by procyclic forms of Trypanosoma brucei. J Mol Biol. 1997;269:529–547. doi: 10.1006/jmbi.1997.1066. [DOI] [PubMed] [Google Scholar]
- van't Hof W, Rodriguez-Boulan E, Menon AK. Nonpolarized distribution of glycosylphosphatidylinositols in the plasma membrane of polarized Madin-Darby canine kidney cells. J Biol Chem. 1995;270:24150–24155. doi: 10.1074/jbc.270.41.24150. [DOI] [PubMed] [Google Scholar]
- Vassella E, Acosta-Serrano A, Studer E, Lee SH, Englund PT, Roditi I. Multiple procyclin isoforms are expressed differentially during the development of insect forms of Trypanosoma brucei. J Mol Biol. 2001;312:597–607. doi: 10.1006/jmbi.2001.5004. [DOI] [PubMed] [Google Scholar]
- Vassella E, Boshart M. High molecular mass agarose matrix supports growth of bloodstream forms of pleomorphic Trypanosoma brucei strains in axenic culture. Mol Biochem Parasitol. 1996;82:91–105. doi: 10.1016/0166-6851(96)02727-2. [DOI] [PubMed] [Google Scholar]
- Vassella E, Strässer K, Boshart M. A mitochondrion-specific dye for multicolour fluorescent imaging of Trypanosoma brucei. Mol Biochem Parasitol. 1997;90:381–385. doi: 10.1016/s0166-6851(97)00171-0. [DOI] [PubMed] [Google Scholar]
- Vassella E, van Den Abbeele J, Bütikofer P, Renggli Kunz C, Furger A, Brun R, Roditi I. A major surface glycoprotein of Trypanosoma brucei is expressed transiently during development and can be regulated post-transcriptionally by glycerol or hypoxia. Genes Dev. 2000;14:615–626. [PMC free article] [PubMed] [Google Scholar]
- Young JR, Donelson JE, Majiwa PA, Shapiro SZ, Williams O. Analysis of genomic rearrangements associated with two variable antigen genes of Trypanosoma brucei. Nucleic Acids Res. 1982;10:803–819. doi: 10.1093/nar/10.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelbauer K, Overath P. Organization of two invariant surface glycoproteins in the surface coat of Trypanosoma brucei. Infect Immun. 1993;61:4540–4545. doi: 10.1128/iai.61.11.4540-4545.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelbauer K, Stahl B, Karas M, Stierhof YD, Overath P. Proteolytic release of cell surface proteins during differentiation of Trypanosoma brucei. Biochemistry. 1993;32:3737–3742. doi: 10.1021/bi00065a028. [DOI] [PubMed] [Google Scholar]