Abstract
Motor proteins have been implicated in various aspects of mitosis, including spindle assembly and chromosome segregation. Here, we show that acentrosomal Arabidopsis cells that are mutant for the kinesin, ATK1, lack microtubule accumulation at the predicted spindle poles during prophase and have reduced spindle bipolarity during prometaphase. Nonetheless, all abnormalities are rectified by anaphase and chromosome segregation appears normal. We conclude that ATK1 is required for normal microtubule accumulation at the spindle poles during prophase and possibly functions in spindle assembly during prometaphase. Because aberrant spindle morphology in these mutants is resolved by anaphase, we postulate that mitotic plant cells contain an error-correcting mechanism. Moreover, ATK1 function seems to be dosage-dependent, because cells containing one wild-type allele take significantly longer to proceed to anaphase as compared with cells containing two wild-type alleles.
INTRODUCTION
In eukaryotes, the transfer of genetic material to daughter cells relies on the proper formation of the microtubule-based spindle. This bipolar apparatus aligns and segregates chromosomes by arranging microtubules in an antiparallel manner, such that the slower-growing minus-ends are anchored at the spindle poles and the faster-growing plus-ends are facing the spindle equator (Euteneur and McIntosh, 1981; Euteneur et al., 1982). In some cell types, bipolar spindle assembly is mediated by centrosomes, which are composed of two centrioles surrounded by pericentriolar material that nucleates microtubules, leading to the formation of an astral microtubule array (for a review see Doxsey, 2001). Microtubule asters are considered to be the building blocks of the incipient spindle poles in centrosome-containing cells.
Most mammalian meiotic cells, as with all higher plants cells, are acentrosomal and consequently, microtubule nucleation and spindle assembly occur in a different manner than in cells that have centrosomes (Smirnova and Bajer, 1992). In the current model of acentrosomal spindle assembly, microtubules are proposed to be nucleated around chromosomes and then self-organized into a bipolar array (Compton, 2000; Wittmann et al., 2001). Motor proteins, which hydrolyze ATP and move along microtubules, are key players in this reorganization process (Merdes and Cleveland, 1997; Waters and Salmon, 1997; Wittmann et al., 2001). In a centrosome-free environment, motor proteins can induce the formation of microtubule asters (McNiven and Porter, 1988; Maniotis and Schliwa, 1991; Verde et al., 1991; Nedelec et al., 1997). In particular, in acentrosomal Xenopus egg extracts, the minus-end directed microtubule motor, dynein, focuses microtubule minus-ends into spindle poles (Heald et al., 1996, 1997) and the plus-end directed kinesin, Eg5, is necessary for the establishing a bipolar array (Walczak et al., 1998). Moreover, the minus-end directed Drosophila kinesin, NCD, is thought to function in spindle pole formation, since ncd mutants have multipolar spindles with broad poles (Hatsumi and Endow, 1992).
Despite these detailed observations in acentrosomal mammalian cells, the molecular mechanisms guiding bipolar spindle assembly in acentrosomal plant cells are not well understood. Plant mitosis differs from mitosis in other acentrosomal cell types due to the presence of a microtubule-based preprophase band (PPB), which appears before nuclear envelope breakdown and functions in spindle alignment and in predicting the future cell division site. Also, plant cells have a phragmoplast that appears in telophase and guides vesicle deposition during cytokinesis. Furthermore, a majority of microtubules appear to be nucleated along the nuclear envelope (Lambert, 1993; Stoppin et al., 1994; Azimzadeh et al., 2001), not around chromosomes. This is supported by cytological studies done in acentrosomal Hemanthus endosperm cells, which revealed that in prophase, microtubules radiate from finite regions along the nuclear envelope, termed microtubule converging centers (Smirnova and Bajer, 1994, 1998). Microtubule converging centers are hypothesized to be microtubule nucleation sites, which subsequently self-organize to form the spindle poles before nuclear envelope breakdown (Smirnova and Bajer, 1994). Similar microtubule patterning is also observed in acentrosomal Allium cells (Wick and Duniec, 1984), and this pathway of bipolar spindle assembly might be universal among somatic plant cells (Baskin and Cande, 1990).
Although differences exist between acentrosomal animal and plant mitoses, motor proteins are hypothesized to function in bipolar spindle assembly in plants as well (Smirnova and Bajer, 1998). There are at least 61 kinesins in the Arabidopsis genome based upon homology in the kinesin motor domain, most of whose function has not been identified (Reddy and Day, 2001). Recently, an Arabidopsis minus-end directed kinesin (Marcus et al., 2002), ATK1 (formerly known as KATA), was shown to be required for bipolar spindle assembly during male meiosis; mutant cells of ATK1 have a broad spindle that lacks focused poles in metaphase I and II (Chen et al., 2002). As a result, chromosome segregation is abnormal and the plant has reduced male fertility. Here, we show that in acentrosomal Arabidopsis cells, ATK1 also functions in mitotic bipolar spindle assembly. Mutants of ATK1 lack microtubule focusing at the spindle poles during prophase and have abnormal bipolar spindles in prometaphase. However, unlike the mutant phenotype observed during male meiosis, the anaphase spindle and chromosome segregation appear normal. These data reveal that an error-correcting mechanism exists in mitotic cells, but not in male meiotic cells. Furthermore, ATK1 function seems to be dosage dependent, whereby cells containing a single wild-type allele take longer to proceed to anaphase than cells with two wild-type alleles.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana wild-type and mutant atk1-1 plants were of the Landsberg erecta ecotype. Plants were germinated in sterile Petri dishes on Arabidopsis media (Granger and Cyr, 2001a) and grown on an 8 h/16 h day/night cycle at a temperature ranging from 20 to 24°C. The atk1-1 mutant contains a Ds element in the first intron of the ATK1 gene. The Ds insertion contains a transcriptional termination sequence leading to the production of truncated mRNAs that are usually unstable (Chen et al., 2002).
Studies done in planta used plants expressing a recombinant green fluorescent protein::microtubule binding domain (GFP::MBD; Granger and Cyr, 2001a) in the atk1-1 background (Chen et al., 2002). The plants were generated by crossing homozygous GFP::MBD plants (Columbia ecotype) to homozygous atk1-1 plants. The resulting F1 generation (all heterozygous for both genes) was grown in soil in a greenhouse and allowed to self-pollinate, and the resulting F2 seed was collected. The F2 generation (predicted to have a 1:2:1 genotype ratio of the mutant allele) was plated on Arabidopsis media (supplemented with 50 μg/ml kanamycin for selection) and placed in a growth chamber set at the same conditions as above.
RNA Extraction and Analysis
Seven-day-old wild-type and atk1-1 roots were excised from plants, immediately flash frozen in liquid nitrogen, and ground using a pestle attached to a drill press. The final weight of each tissue after grinding was ∼100 mg. RNA extraction was done using the RNeasy Plant Mini Kit (Qiagen,Valencia, CA) and the final RNA concentration was estimated spectroscopically at 260 nm.
Reverse-transcription PCR (RT-PCR) was done with the C. therm Polymerase One-Step RT-PCR System (Roche Diagnostics; Manheim, Germany) with the following primers made to the published sequence of ATK1: forward, 5′-GTGGACATGGCTTCTCGCAACCAG-3′ and reverse, 5′-AAGCTTGTGTTCTC-3′. Thirty minutes of reverse transcription at 60°C, followed by 30 cycles at a 55°C annealing temperature was used for amplification, and the final product was electrophoresed on a 1% agarose gel. To quantitatively control for RNA loading, all samples were adjusted after fluoremetric analysis to give equal band intensity on a 1% agarose gel when RT-PCR was done for rRNA.
Tissue Fixation and Immunofluorescence Microscopy
Seven-day-old roots of known genotypes were fixed with 4% formaldehyde (freshly prepared from paraformaldehyde) and 0.1% glutaraldehyde in PMEG buffer(50 mM Pipes, 5 mM EGTA, 1 mM MgSO4, and 1% glycerol) for 30 min. The fixed plants were then rinsed three times in PM buffer (50 mM Pipes, 1 mM MgSO4, and 1 mM EGTA, pH 6.9) and stored overnight at 4°C. The next day, ∼2 mm of the root tip was subjected to cell wall degrading enzymes for 20 min (1% cellulase and 0.1% pectolyase in H2O) and then rinsed twice with PM. Roots were then squashed onto slides that were coated with poly-l-lysine (Sigma P1524) and then blocked with 3% BSA for 1 h in a humidity chamber. After a brief rinse with PBS (phosphate-buffered saline), cells were incubated with 0.1% Triton X-100 for 20 min in a humidity chamber and briefly rinsed again in PBS. Next, cells were incubated with the DMA1-FITC–conjugated antitubulin antibody (Sigma Corp., St. Louis, MO) for 2 h in a humidity chamber. After rinsing three times in PBS, a fluorescent mounting media solution (0.1 M Tris, pH 9.0, 50% glycerol, and 1 mg/ml phenylenediamine), supplemented with 1 μl of Hoescht 33258 (for chromosome staining) was added. Cells were viewed with a 100× Neofluor oil lens (NA = 1.25) mounted on a Zeiss Axioskop (Zeiss Corp, Thornwood, NY). The microscope was equipped with a variable intensity 100-W mercury lamp (at 50% intensity to view microtubules and 5% intensity for chromosomes), 450–490 nm excitation filter, 500 nm dichroic, and 520–560 nm emission filter to view microtubules and a G365 excitation filter, 400 nm dichroic, and a LP420 emission filter for chromosomes. Images were collected using a digital camera (Orca, Hamamatsu Corp., Bridgewater, NJ) and used for mitotic analysis and mitotic index studies. The Student's t test was used to evaluate significant differences.
In Planta Microscopy
Temporal analysis studies done in planta used a 40× Achroplan lens (NA = 0.8) mounted on a Zeiss Axiovert microscope, which was equipped with a 450–490 nm excitation filter and 520–590 nm emission filter. The roots were illuminated using a mercury arc lamp at 20% intensity and images were captured with a Hamamatsu Orca CCD camera (Hamamatsu Corp.) running on the Esee program (ISee Image Systems, Raleigh, NC). Images were taken in 30-s intervals and time measurements were then recorded.
Spatial analysis studies done in planta used a 40× C-Apochromat lens (NA = 1.2) mounted on Zeiss LSM 410 confocal microscope (488 nm excitation, 515–565 nm emission). Four-second scan times with four-line averaging was done to obtain better spatial resolution at the expense of temporal resolution. Higher scan times were not possible because of photobleaching of the cell. Images were processed using Adobe Photoshop 7.0 (Adobe Corp., Mountain View, CA).
Genotype Identification
The F2 generation of the GFP::MBD plants, in the ATK1/atk1-1 background, was predicted to have a 1:2:1 genotype ratio of the mutant and wild-type alleles. Therefore, all in planta data were collected without knowing the genotype of individual plants and subsequent genotypic analysis using PCR amplification of genomic DNA was done. To extract genomic DNA, a 20-d-old leaf was ground in 200 μl of extraction buffer (200 mM Tris-HCl, pH 7.5, 250 mM NaCl, 25 mM EDTA, pH 8.0, and 0.5% SDS), and then 200 μl of phenol was added. This solution was vortexed and spun in a microcentrifuge at 14,000 × g for 5 min, and the aqueous layer was collected. Next, 200 μl of isopropanol was added and the solution was respun in a microcentrifuge at 14,000 × g for 5 min. The pellet was collected and washed with 70% ethanol, vacuum dried for 5 min, and resuspended in 20 μl H2O. To determine the genotype of the plant, two PCR reactions, with 30 cycles each, were done. The first reaction used primers made specifically to the ATK1 genomic sequence and would only amplify ATK1 genomic DNA if there were no Ds insertion present. They are as follows: forward 5′-TCCACTTTCCTTCG-CTTGTCAAA-3′ and reverse 5′-ATTGTCGGTAACCCGAATTTAGG. The second reaction used one primer made specifically to the Ds insertion (Chen et al., 2002) in conjunction with the forward primer used in the first reaction. This primer set would amplify the Ds insertion if it were present in the ATK1 gene. Thus, on the basis of the band patterning on an agarose gel, we could identify the genotype of each plant. This technique also confirmed the predicted 1:2:1 ATK1 genotype ratio of the plants (unpublished data).
RESULTS
ATK1 Is Expressed in Mitotically Dividing Wild-type Roots
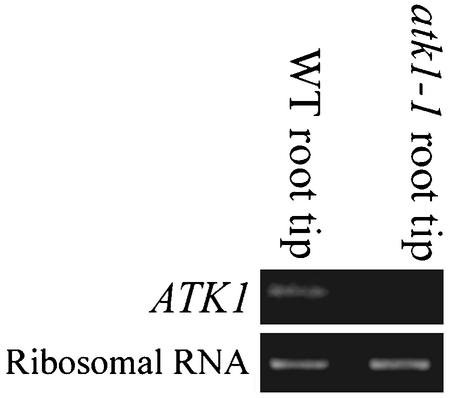
Previously, ATK1 was shown to function in male meiosis based on the observations that atk1-1 mutant plants had abnormal meiotic spindles and reduced male fertility (Chen et al., 2002). Despite this meiotic phenotype, ATK1 is expressed in meiotic and nonmeiotic tissue of wild-type flowers (Chen et al., 2002). This raises the possibility that ATK1 function is not limited to meiosis but may also play a role in mitosis. Therefore, ATK1 expression in Arabidopsis roots, which contain a high percentage of mitotically dividing cells, was investigated. Using RT-PCR with primers specific to ATK1, wild-type root tips had the predicted 0.5 kb on an agarose gel (Figure 1). The ATK1 mutant, atk1-1, which has a Ds insertion in the ATK1 gene that disrupts the ATK1 transcript, lacked this band when the same primer set was used (Figure 1). As a control for RNA loading, amplification of ubiquitously expressed rRNA was adjusted to give equal band intensity in the two tissues (Figure 1).
Figure 1.
ATK1 expression. Using RT-PCR with primers specific to ATK1, a 0.5-kb band was amplified in wild-type (WT) root tips, but not in homozygous atk1-1 roots (top lanes). As a control for RNA loading, RT-PCR amplification of rRNA in the two samples was adjusted to give equal intensity of a 0.4-kb band, when run on a 1% agarose gel (bottom lanes).
atk1-1 Cells Lack Spindle Bipolarity in Metaphase
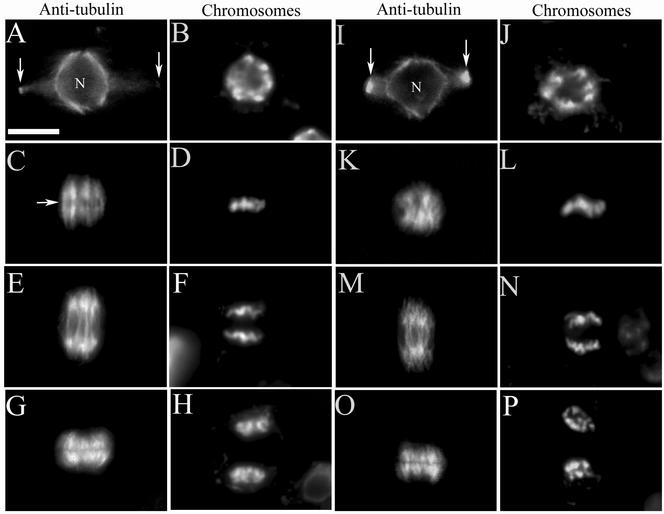
Using antitubulin antibodies, immunofluorescence was performed in wild-type and atk1-1 root mitotic cells (Figure 2). In prophase, both wild-type and mutant cells exhibited a perinuclear microtubule array and preprophase band (Figures 2A and 2I, respectively) with condensed chromosomes (Figure 2, B and J, respectively). In metaphase, a typical wild-type cell had a prominent bipolar spindle with a distinct zone of microtubule clearing that represented the metaphase plate (Figure 2C) and chromosomes aligned linearly along the metaphase plate (Figure 2D). In contrast, the metaphase spindle in a typical mutant cell lacked obvious spindle bipolarity (Figure 2K). Furthermore, chromosomes were not typically aligned along the metaphase plate (Figure 2L). After metaphase, both wild-type and mutant cells had a normal anaphase spindle (Figure 2, E and M, respectively) and in telophase, both developed a phragmoplast. (Figure 2, G and O, respectively). Chromosome segregation also appeared normal during anaphase and telophase in wild-type (Figure 2, F and H, respectively) and mutant cells (Figure 2, N and P, respectively). Overall, these data show that mutant cells lack spindle bipolarity during early mitosis; however, anaphase occurs normally.
Figure 2.
Root mitosis of wild-type and mutant plants. Immunofluorescence was performed using antitubulin antibodies in dividing root cells, in both wild-type (A, C, E, and G) and homozygous mutant (I, K, M, and O) cells; the adjacent image shows the same cell stained for chromosomes (B, D, F, and H for wild-type; J, L, N, and P for mutant). In wild-type (A) and mutant cells (I), the preprophase microtubule band (arrows) and perinuclear microtubule array are present in prophase (nucleus = N). A wild-type metaphase cell (C) has a well-defined equatorial zone of microtubule clearing (arrow) and a distinct bipolar spindle with chromosomes aligned linearly along the metaphase plate (D). A mutant metaphase cell lacks an equatorial zone of microtubule clearing (K) and chromosomes do not align linearly along the metaphase plate (L); as a result there is an indistinct bipolar spindle axis. Wild-type and mutant cells have a normal anaphase spindle (E and M, respectively) and chromosomes segregate normally (F and N, respectively). Wild-type and mutant cells in telophase (G and O, respectively) show a normal microtubule-based phragmoplast and the start of chromosome decondensation (H and P, respectively). Scale bar, 10 μm.
atk1-1 Cells Contain a Significantly Higher Percentage of Cells in Prometaphase/Metaphase
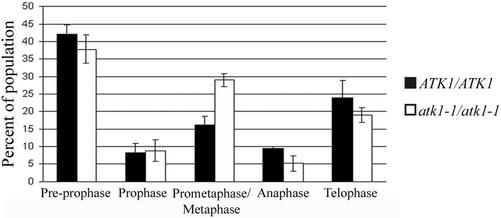
We hypothesized that mutant cells with abnormal mitotic spindles take longer to progress through mitosis. To test this, the mitotic index of mutant cells and wild-type cells was compared using chromosomes as the marker for mitotic stage. The results show that mutant cells contain a significantly higher percentage of cells in prometaphase/metaphase than wild-type cells (p < 0.05; Figure 3). The percentages of cells in all other stages of mitosis were not significantly different between mutant and wild-type cells (p > 0.05; Figure 3). These results suggest that mutant cells have an extended prometaphase/metaphase and supports the hypothesis that spindle formation takes longer in mutant cells than in wild-type cells.
Figure 3.
Mitotic indices of wild-type and mutant cells. The percentage of cells in preprophase, prophase, anaphase, and telophase in wild-type and mutant cells are not statistically different (p > 0.05 for all). However, mutant cells have significantly more cells in prometaphase/metaphase as compared with wild-type cells (p < 0.05). Error bars represent SE. The Student's t test was used to calculate p values.
atk1-1 Cells Have a Greater Percentage of Prometaphase Spindles than Wild-type Cells
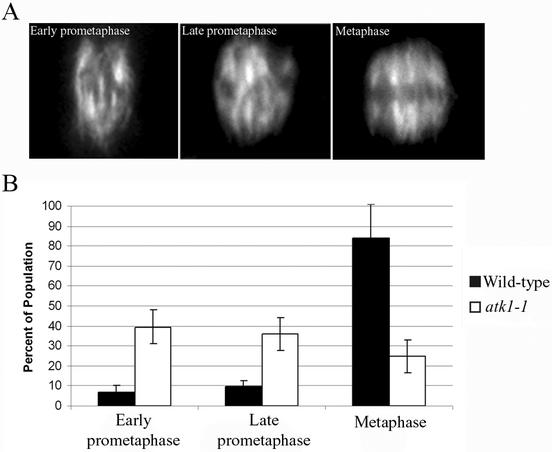
To extend our characterization of the mutant phenotype, spindles were classified into three categories: early prometaphase, late prometaphase, and metaphase. This allowed us to pinpoint which stage of spindle formation in the mutant is abnormally longer. The criteria used to classify the spindles are as follows: 1) early prometaphase spindles have little or no bipolarity (because mitotic plant spindles lack fusiform poles or asters, we define bipolarity as the presence of two half-spindles, symmetrically opposed about the spindle equatorial plane), are relatively narrow, and lack an equatorial zone of microtubule clearing (representing the metaphase plate); 2) late prometaphase spindles are relatively wider than early prometaphase spindles but still lack a distinct equatorial zone of microtubule clearing, and in many cases bipolarity is more evident; 3) metaphase spindles are clearly bipolar, having a distinct and linear zone of microtubule clearing that defines the equatorial plane (Figure 4A). The results show that mutant cells had a significantly higher percentage of early prometaphase and late prometaphase spindles as compared with wild-type cells, but a significantly lower percentage of metaphase spindles than wild-type cells (p < 0.05 for all; Figure 4B). These results suggests that mutant cells have an extended prometaphase and a shortened metaphase.
Figure 4.
Quantitative analysis of prometaphase/metaphase spindles. Spindles were classified into three categories: early prometaphase, late prometaphase, and metaphase (top). The results were quantitated in a histogram (bottom). Early prometaphase spindles typically lack bipolar character and an equatorial zone of microtubule clearing. Late prometaphase spindles exhibit more bipolarity than early prometaphase spindles, although an equatorial zone of microtubule clearing is still not robust. Metaphase spindles are bipolar and have a distinct equatorial zone of microtubule clearing (top panel). Mutant cells have significantly less early prometaphase and late prometaphase spindles and more metaphase spindles (bottom panel; p < 0.05). Error bars, SE. The Student's t test was used to calculate p values.
Temporal Analysis Studies Done in Plants Confirms that atk1-1 Cells Take Longer to Proceed to Anaphase
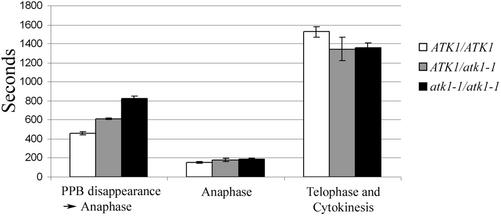
Plants expressing a microtubule reporter gene (Granger and Cyr, 2001) were crossed with atk1-1 plants for in planta analysis of mitosis. Temporal studies using these hybrid lines revealed that it takes longer to proceed from PPB disappearance to the onset of anaphase in the mutant genotypes (i.e., ATK1/atk1-1 and atk1-1/atk1-1 cells) than in ATK1/ATK1 cells (p < 0.05; Figure 5). Furthermore, when comparing ATK1/atk1-1 and atk1-1/atk1-1 genotypes to each other, atk1-1/atk1-1 cells took longer to proceed to anaphase than ATK1/atk1-1 cells (p < 0.05). All other time point measurements did not reveal a significant difference between the genotypes (p > 0.05). Thus, these data show that that cells having more than one mutant allele take longer to proceed to anaphase, but events after anaphase are not temporally abnormal.
Figure 5.
In planta temporal analysis of mitosis. For each genotype, time intervals for various stages of mitosis were quantified. The amount of time it took to proceed from PPB disappearance to the onset of anaphase was greater in ATK1/atk1-1 plants (mean = 608.7 ± 28.7 s) compared with ATK1/ATK1 plants (mean = 455.3 ± 64.7 s). However, atk1-1/atk1-1 plants took significantly longer (mean = 825.6 ± 106 s; p < 0.05), compared with the other genotypes. For all genotypes, the time it took for anaphase and from the completion of anaphase to the disappearance of the phragmoplast was not statistically different (p > 0.05). Error bars, SE. The Student's t test was used to calculate p values.
atk1-1 Mutants Have Abnormal Spindle Formation In Planta
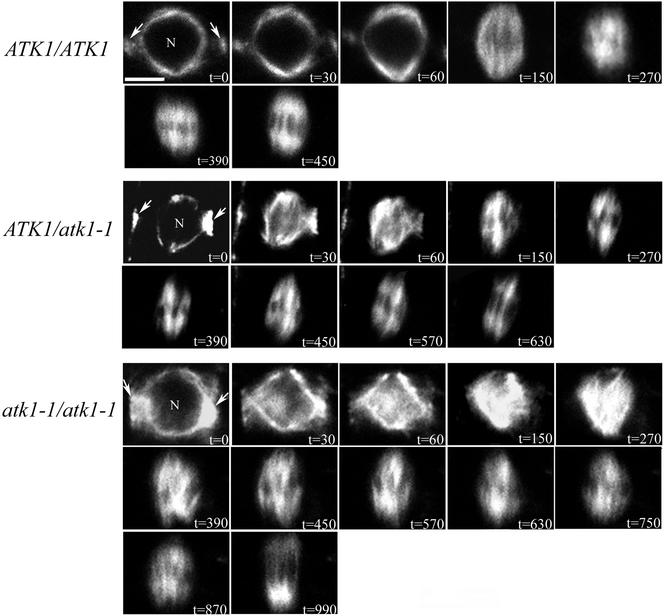
Confocal microscopy was used to provide better spatial resolution of in planta mitotic events. For all cells, the contrast was purposely left unchanged throughout the time course to accurately depict an increase in microtubule density. In preprophase, all genotypes had a perinuclear microtubule array and PPB at t = 0 s (Figure 6). At t = 30 s and t = 60 s in ATK1/ATK1 plants, perinuclear microtubules accumulate in a polar manner with a prominent clearing near the PPB (i.e., formation of the incipient spindle poles is evident); however, at these time points in the mutant and heterozygote genotypes, perinuclear microtubules were globally distributed without accumulating at the future spindle poles. Also, in the mutant cells there was a dense buildup of microtubules in the former nuclear region and the PPB had not narrowed to the same degree as in wild-type cells. At t = 150 s in ATK1/ATK1 cells, a bipolar prometaphase spindle was observed (i.e., there are two half-spindles, symmetrically opposed about the spindle equatorial plane) and microtubules radiated from the spindle poles, and at t = 270 s, the spindle decreased in size and had reduced bipolarity. By t = 390 s, a mature bipolar spindle with a distinct equatorial zone of microtubule clearing was observed and at t = 450 s, an anaphase spindle was observed. In contrast, at t = 150 s in ATK1/atk1-1 cells, the prometaphase spindle was microtubule organization and bipolarity. From t = 180 s to t = 570, the spindle became increasingly bipolar and a normal anaphase spindle was observed at t = 630 s. In atk1-1/atk1-1 cells, at t = 150 s and t = 270 s, microtubule density continues to increase throughout the prometaphase spindle, but at t = 390 s, microtubule density began to attenuate, and a disorganized prometaphase spindle was observed. From t = 390 s till t = 870 s, the spindle became more bipolar and a normal anaphase spindle was observed at t = 990 s. Overall, these data show that mutant and heterozygote genotypes had an abnormal prophase and prometaphase spindle and took longer to proceed to anaphase; however, spindle abnormalities are completely rectified by anaphase. Furthermore, the observation that during prophase in ATK1/atk1-1 and atk1-1/atk1-1 cells, the PPB did not narrow to the same degree as wild-type cells, is addressed in a future correspondence (unpublished results). It should also be noted that a similar mutant phenotype was observed in immunopreparations that did not contain the GFP::MBD transgene (unpublished data).
Figure 6.
In planta spatial analysis of microtubules in mitosis. Shown here is a time course of mitosis. At t = 0 s, all genotypes have a normal perinuclear array (N = nucleus) and a PPB (arrows). In ATK1/ATK1 cells at t = 30 s and t = 60 s, microtubules increase at the incipient spindle poles, and by t = 150 s, microtubules radiate from the spindle poles. The beginning of a bipolar spindle is seen at t = 150 s, and by t = 450 s, anaphase typically occurs. In contrast, at t = 30 s and t = 60 s in both ATK1/atk1-1 and atk1-1/atk1-1 genotypes, microtubule accumulation is not specific to the spindle pole regions and a high density of microtubules are observed in the former nuclear region. At t = 150 s in ATK1/atk1-1 cells, a disorganized spindle is seen, which resolves into a mature spindle at t = 570 s, and anaphase typically occurs by t = 630 s. In atk1-1/atk1-1 cells at t = 150 s and at t = 270 s, microtubules continue to accumulate in the former nuclear region, and a mature spindle is not seen until t = 870 s. Anaphase typically occurs by t = 990 s. Scale bar, 10 μm.
atk1-1 Cells Have Abnormal Spindle Pole Formation
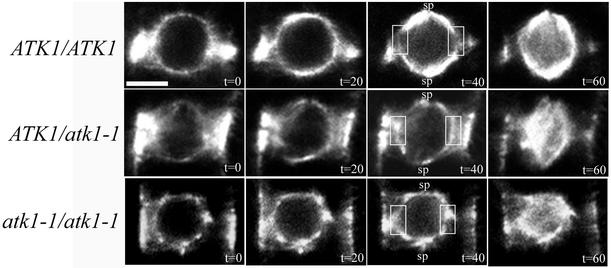
An in planta time course study of prophase and prometaphase was done to examine the early events of spindle formation in all three ATK1 genotypes (Figure 7). At t = 20 s, wild-type cells in prophase exhibited increased microtubule accumulation in the polar regions distal to the PPB axis (i.e., where the incipient spindle poles form). This continued at t = 40 s, but there was also a concomitant clearing of microtubules in the areas of the perinuclear array that was proximal to the PPB (Figure 7, boxed area). In prometaphase, at t = 60 s, microtubules extended into the previously cleared zone along the perinuclear array and microtubules also appeared within the nucleus. In prophase, at t = 20 s or t = 40 s, both mutant genotypes did not have significant microtubule accumulation at the spindle poles. Furthermore, there was no microtubule clearing at t = 40 s in the area of the perinuclear array proximal to the PPB (Figure 7, boxed area). Both heterozygous and mutant genotypes were in prometaphase at t = 60 s and exhibited an increase of microtubule density along the perinuclear array and within the nucleus. Thus, these data show microtubule clearing in the region of the perinuclear array proximal to the PPB is abnormal in the mutant and heterozygous genotypes.
Figure 7.
Spindle pole formation in wild-type and mutant plants. For each genotype a time course was done to look at the early events of spindle pole formation, starting at preprophase and ending at early prometaphase. In ATK1/ATK1 plants at t = 20 s, the perinuclear microtubule array dims in the region proximal to the PPB, and a clearing is observed at t = 40 s (boxed region). Furthermore, there is a concomitant increase in microtubules at the spindle poles (sp), and by t = 60 s, the microtubules emanating from the poles have extended into the clearing zone along the perinuclear array. In the mutant genotypes at t = 40 s, microtubule clearing in the region of the perinuclear array that is proximal to the PPB is not seen (boxed region) and microtubule localization is not increasing at the spindle poles (sp), but is in other areas along the perinuclear array. This continues at t = 60 s, though microtubules also appear in the nucleus. Scale bar, 10 μm.
DISCUSSION
ATK1 Functions during Spindle Pole Formation
The role of centrosomes in organizing microtubules during spindle assembly has been questioned, because numerous studies show that when centrosomes are genetically or physically removed, spindles still form (Megraw et al., 1999; Vaizel-Ohayon and Schejter, 1999; Vidwans and O'Farrell, 1999; Bonaccorsi et al., 2000; Khodjakov et al., 2000). Rather, it has been proposed that motor proteins are fundamental to the organization of the bipolar array, and in particular, minus-end directed motor proteins are essential for spindle pole formation (Walczak et al., 1998; Mountain and Compton, 2000; Wittmann et al., 2001). The data presented here support this notion by showing that mutants of the minus-end directed kinesin, ATK1, have abnormal spindle pole formation (Figure 7). Specifically, they lack microtubule accumulation at the spindle pole sites during prophase and instead, accumulate microtubules nonspecifically along the nuclear envelope. Therefore, we conclude that in wild-type cells, ATK1 is required for microtubule accumulation at the spindle pole sites during prophase. Notably, this phenotype is not due to a cryptic gene in the Col/Ler hybrid, because fixed tissue studies of heterozygotes in the Ler background, had the same phenotype as homozygous mutants (unpublished data).
One way that ATK1 could assist in microtubule accumulation is by transporting microtubule polymers to the spindle pole sites by tracking along anchored microtubules. This would be consistent with the current model of spindle pole formation, whereby minus-end directed motors focus microtubules at the spindle poles because of the polarity of their motor activity and ability to cross-link microtubules (Mountain and Compton, 2000; Sharp et al., 2000b; Wittmann et al., 2001). Related to this model as well as to ATK1 function is the observation that when spindle pole formation is occurring in plant cells, there is a concomitant clearing of microtubules observed in the region of the nuclear envelope proximal to the PPB (Figure 7; Wick and Duniec, 1984; Granger and Cyr, 2001b; Dixit and Cyr, 2002). This clearing was not observed in ATK1 mutant cells (Figure 7). Therefore, we speculate that this clearing, at least in part, occurs due to the activity of ATK1. Moreover, because ATK1 appears to function in spindle pole formation, it is possible that ATK1 transports microtubule polymers from this region to the spindle poles; however, we cannot rule out the possibility that microtubule dynamicity per se also contributes to this phenomona.
Does ATK1 also Function in Prometaphase?
It has been proposed that minus-end directed motors not only focus microtubules during spindle pole formation, but also generate inward-directed forces during spindle assembly that counteract opposing forces created by plus-end directed motors (Saunders and Hoyt, 1992; O'Connell et al., 1993; Endow and Komma, 1997). This leads to a balance of forces within the spindle, which are thought to maintain steady state spindle structures (Endow, 1999; Sharp et al., 2000b). Here we observed that during prometaphase, mutant spindles lacked bipolarity and had a dense buildup of seemingly disorganized microtubules in the former nuclear region (Figure 6). If ATK1 does counteract forces produced by plus-end directed motors, then this phenotype could be caused by a misbalance of forces within the spindle. This was the case with mutants of the minus-end directed kinesin, NCD, which have spindles that appear to push outward (Endow and Komma, 1997; Sharp et al., 2000a) and conversely, the opposite effect (i.e., collapse inward) was observed in mutants of plus-end directed kinesins (Saunders and Hoyt, 1992; O'Connell et al., 1993). Furthermore, immunofluorescence data revealed that ATK1 localizes to the spindle midzone during metaphase, which would be consistent with a role in spindle assembly (Liu et al., 1996). Alternatively, this mutant phenotype might occur as a consequence of the aberrant spindle pole formation observed during prophase. Predictably, a lack of microtubule focusing at the spindle poles would result in a disorganized spindle. Both of these hypotheses are equally probable and it is also possible that this phenotype is a combination of both.
An Error-correcting Mechanism during Mitotic Bipolar Spindle Formation
Experiments done with Haementhus endosperm cells show that spindle abnormalities that occur at prophase, such as multipolarity or apolarity, are rectified during prometaphase (Smirnova and Bajer, 1998). This has been interpreted as evidence for an error-correcting mechanism in bipolar spindle formation. The results presented here support this hypothesis by showing that spindle defects in mutant cells are less apparent as prometaphase proceeds and are completely resolved by anaphase (Figure 6). Furthermore, chromosome segregation also appears normal (Figure 2), and roots do not appear to have any gross morphological defects (unpublished data). This differs from male meiosis, where spindle abnormalities in the ATK1 mutant are seen before, during, and after anaphase, and consequently chromosomes segregate abnormally. Thus, we propose that an error-correcting mechanism only exists in mitosis and not in male meiosis.
If an error-correcting mechanism does exist in mitotic plant cells, then what is its molecular basis? One possibility is that abnormalities in the spindle are targeted and removed by proteins that can either depolymerize or sever microtubules. In other organisms, proteins that bind to microtubules and induce their disassembly have been identified. One example of this is the Xenopus kinesin, XKCM1, which targets microtubule protofilaments and induces their depolymerization (Walczak et al., 1996; Niederstrasser et al., 2002). To our knowledge, plant homologues of XKCM1 have not been identified; however, a plant microtubule-associated protein has been shown to sever microtubules (Burk et al., 2001; McClinton et al., 2001). This protein, named AtKTN1, is a homologue of the katanin p60 subunit, which severs microtubules at their minus-ends in mitotic cells (McNally and Thomas, 1998; McNally et al., 2000), and therefore, could have a similar role in plant mitosis. Another potential error-correcting mechanism could involve the activity of another minus-end directed kinesin, which acts as a functional substitute of ATK1. One likely candidate is the ATK1 homologue, ATK5, which has 83% amino acid identity (91% similar) to ATK1 across the full-length of the protein. This gene is expressed in the root (unpublished data), and experiments regarding a functional redundancy between ATK1 and ATK5 are underway. Although, these mechanisms are speculative, the data presented here imply that mitotic cells have an error-correcting mechanism used to rectify spindle abnormalities.
ATK1 Functions in a Dosage-dependent Manner
The results obtained here reveal that mutant cells have a prolonged prometaphase as compared with wild-type cells (Figures 3 and 4). In particular, cells having one wild-type allele (ATK1/atk1-1 genotype) took longer to proceed to anaphase than cells having two wild-type alleles, and cells that do not have any wild-type alleles (atk1-1/atk1-1 genotype) took the longest (Figure 5). Moreover, in planta spatial analysis confirms this pattern (Figure 6). From this we conclude that the dosage of ATK1 is important to gene function. To our knowledge, the only other kinesin whose function is sensitive to dosage is NOD1 (Knowles and Hawley, 1991; Murphy and Karpen, 1995), which is thought to be essential to the transmission of achiasmate chromosomes in meiosis (Carpenter, 1973; Zhang and Hawley, 1990). The reason why ATK1 could function in a dosage-dependent manner remains unclear; however, it could be related to the finding that ATK1 functions in a cooperative manner (i.e., force generation requires >4 ATK1 molecules bound to a microtubule; Marcus et al., 2002). Predictably, the cooperativity of ATK1 would be sensitive to the cellular concentration of ATK1 and thus, by removing one wild-type allele (i.e., introducing a mutant allele), the probability of an optimal stoichiometry between this motor and microtubules is reduced.
In summary, the data presented here show that ATK1 functions in mitosis because mutants of ATK1 have abnormal spindle morphologies. Specifically, ATK1 appears to be required for the accumulation of microtubules at the spindle poles during prophase. Because all abnormalities are rectified by anaphase and chromosomes appear to segregate normally, it is likely that these mitotic cells contain an error-correcting mechanism. One limitation of these experiments was the inability to observe in vivo chromosome localization. It is possible that chromosome movement to the metaphase plate is affected by the ATK1 mutation, because chromosomes in mutant cells were aligned nonlinearly along the metaphase plate (Figure 2). Future in planta experiments investigating chromosome movement in the mutant background will likely address this topic.
ACKNOWLEDGMENTS
We thank Deborah Fisher for critically reviewing this manuscript. This work was supported by U.S. Department of Agriculture grant 2001-35301-10570.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–09–0586. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–09–0586.
REFERENCES
- Azimzadeh J, Traas J, Pastuglia M. Molecular aspects of microtubule dynamics in plants. Curr Opin Plant Biol. 2001;4:513–519. doi: 10.1016/s1369-5266(00)00209-0. [DOI] [PubMed] [Google Scholar]
- Baskin TI, Cande WZ. The structure and function of the mitotic spindle in flowering plants. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:277–315. [Google Scholar]
- Bonaccorsi S, Giansanti MG, Gatti M. Spindle assembly in Drosophila neuroblasts and ganglion mother cells. Nat Cell Biol. 2000;2:54–56. doi: 10.1038/71378. [DOI] [PubMed] [Google Scholar]
- Burk DH, Liu B, Zhong RQ, Morrison WH, Ye ZH. A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell. 2001;13:807–827. [PMC free article] [PubMed] [Google Scholar]
- Carpenter ATC. A meiotic mutant defective in distributive disjunction in Drosophila melanogaster. Genetics. 1973;73:393–428. doi: 10.1093/genetics/73.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CB, Marcus A, Li WX, Hu Y, Calzada JPV, Grossniklaus U, Cyr RJ, Ma H. The Arabidopsis ATK1 gene is required for spindle morphogenesis in male meiosis. Development. 2002;129:2401–2409. doi: 10.1242/dev.129.10.2401. [DOI] [PubMed] [Google Scholar]
- Compton DA. Spindle assembly in animal cells. Annu Rev Biochem. 2000;69:95–114. doi: 10.1146/annurev.biochem.69.1.95. [DOI] [PubMed] [Google Scholar]
- Dixit R, Cyr RJ. Spatio-temporal relationship between nuclear-envelope breakdown and pre-prophase band disappearance in cultured tobacco cells. Protoplasma. 2002;219:116–121. doi: 10.1007/s007090200012. [DOI] [PubMed] [Google Scholar]
- Doxsey S. Re-evaluating centrosome function. Nat Rev Mol Cell Biol. 2001;2:688–698. doi: 10.1038/35089575. [DOI] [PubMed] [Google Scholar]
- Endow S. Microtubule motors in spindle and chromosome motility. Eur J Biochem. 1999;262:12–17. doi: 10.1046/j.1432-1327.1999.00339.x. [DOI] [PubMed] [Google Scholar]
- Endow SA, Komma DJ. Spindle dynamics during meiosis in Drosophila oocytes. J Cell Biol. 1997;137:1321–1336. doi: 10.1083/jcb.137.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euteneur U, McIntosh JR. Structural polarity of kinetochore microtubules in PtK1 cells. J Cell Biol. 1981;89:338–345. doi: 10.1083/jcb.89.2.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euteneur U, Jackson WT, McIntosh JR. Polarity of spindle microtubules in Hemanthus endosperm. J Cell Biol. 1982;94:644–653. doi: 10.1083/jcb.94.3.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger CL, Cyr RJ. Spatiotemporal relationships between growth and microtubule orientation as revealed in living root cells of Arabidopsis thaliana transformed with green-fluorescent-protein gene construct GFP-MBD. Protoplasma. 2001a;216:201–214. doi: 10.1007/BF02673872. [DOI] [PubMed] [Google Scholar]
- Granger CL, Cyr RJ. Use of abnormal preprophase bands to decipher division plane determination. J Cell Sci. 2001b;114:599–607. doi: 10.1242/jcs.114.3.599. [DOI] [PubMed] [Google Scholar]
- Hatsumi M, Endow S. Mutants of the microtubule motor protein, nonclaret disjunctional, affect spindle structure and chromosome movement in meiosis and mitosis. J Cell Sci. 1992;101:547–559. doi: 10.1242/jcs.101.3.547. [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Habermann A, Karsenti E, Hyman A. Spindle assembly in Xenopus egg extracts: Respective roles of centrosomes and microtubule self-organization. J Cell Biol. 1997;138:615–628. doi: 10.1083/jcb.138.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Cole RW, Oakley BR, Rieder CL. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol. 2000;10:59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- Knowles BA, Hawley RS. Genetic-analysis of microtubule motor proteins in Drosophila—a mutation at the Ncd locus is a dominant enhancer of Nod. Proc Natl Acad Sci USA. 1991;88:7165–7169. doi: 10.1073/pnas.88.16.7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert A-M. Microtubule-organizing centers in higher plants. Curr Opin Cell Biol. 1993;5:116–122. doi: 10.1016/s0955-0674(05)80016-x. [DOI] [PubMed] [Google Scholar]
- Liu B, Cyr RJ, Palevitz BA. A kinesin-like protein, KatAp, in the cells of Arabidopsis and other plants. Plant Cell. 1996;8:119–132. doi: 10.1105/tpc.8.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniotis A, Schliwa M. Microsurgical removal of centrosomes blocks cell reproduction and centriole generation in BSC-1 cells. Cell. 1991;67:495–504. doi: 10.1016/0092-8674(91)90524-3. [DOI] [PubMed] [Google Scholar]
- Marcus AI, Ambrose JC, Blickley L, Hancock WO, Cyr RJ. Arabidopsis thaliana protein, ATK1, is a minus-end directed kinesin that exhibits non-processive movement. Cell Motil Cytoskel. 2002;52:144–150. doi: 10.1002/cm.10045. [DOI] [PubMed] [Google Scholar]
- McClinton RS, Chandler JS, Callis J. cDNA isolation, characterization, and protein intracellular localization of a katanin-like p60 subunit from Arabidopsis thaliana. Protoplasma. 2001;216:181–190. doi: 10.1007/BF02673870. [DOI] [PubMed] [Google Scholar]
- McNally FJ, Thomas S. Katanin is responsible for the M-phase microtubule-severing activity in Xenopus eggs. Mol Biol Cell. 1998;9:1847–1861. doi: 10.1091/mbc.9.7.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally KP, Bazirgan OA, McNally FJ. Two domains of p80 katanin regulate microtubule severing and spindle pole targeting by p60 katanin. J Cell Sci. 2000;113:1623–1633. doi: 10.1242/jcs.113.9.1623. [DOI] [PubMed] [Google Scholar]
- McNiven MA, Porter KR. Organization of microtubules in centrosome-free cytoplasm. J Cell Biol. 1988;106:1593–1605. doi: 10.1083/jcb.106.5.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw TL, Li KJ, Kao LR, Kaufman TC. The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development. 1999;126:2829–2839. doi: 10.1242/dev.126.13.2829. [DOI] [PubMed] [Google Scholar]
- Merdes A, Cleveland DW. Pathways of spindle pole formation: different mechanisms; conserved components. J Cell Biol. 1997;138:953–956. doi: 10.1083/jcb.138.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountain V, Compton DA. Dissecting the role of molecular motors in the mitotic spindle. Anat Rec. 2000;261:14–24. doi: 10.1002/(SICI)1097-0185(20000215)261:1<14::AID-AR5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Murphy TD, Karpen GH. Interactions between the Nod(+) kinesin-like gene and extracentromeric sequences are required for transmission of a Drosophila minichromosome. Cell. 1995;81:139–148. doi: 10.1016/0092-8674(95)90378-x. [DOI] [PubMed] [Google Scholar]
- Nedelec FJ, Surrey T, Maggs AC, Leibler S. Self-organization of microtubules and motors. Nature. 1997;389:305–308. doi: 10.1038/38532. [DOI] [PubMed] [Google Scholar]
- Niederstrasser H, Salehi-Had H, Gan EC, Walczak C, Nogales E. XKCM1 acts on a single protofilament and requires the C terminus of tubulin. J Mol Biol. 2002;316:817–828. doi: 10.1006/jmbi.2001.5360. [DOI] [PubMed] [Google Scholar]
- O'Connell MJ, Meluh PB, Rose MD, Morris NR. Suppression of the bimC4 mitotic spindle defect by deletion of klpA, a gene encoding a KAR3-related kinesin- like protein in Aspergillus nidulans. J Cell Biol. 1993;120:153–162. doi: 10.1083/jcb.120.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy ASN, Day I. Kinesins in the Arabidopsis genome: A comparative analysis among eukaryotes. BMC Genomics. 2001;2:2. doi: 10.1186/1471-2164-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders WS, Hoyt MA. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- Sharp DJ, Brown HM, Kwon M, Rogers GC, Holland G, Scholey JM. Functional coordination of three mitotic motors in Drosophila embryos. Mol Biol Cell. 2000a;11:241–253. doi: 10.1091/mbc.11.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Rogers GC, Scholey JM. Microtubule motors in mitosis. Nature. 2000b;407:41–47. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- Smirnova E, Bajer A. Spindle poles in higher plant mitosis. Cell Motil Cytoskel. 1992;23:1–7. doi: 10.1002/cm.970230102. [DOI] [PubMed] [Google Scholar]
- Smirnova E, Bajer A. Early stages of spindle formation and independence of chromosome and microtubule cycles in Hemanthus endosperm. Cell Motil Cytoskel. 1998;40:22–37. doi: 10.1002/(SICI)1097-0169(1998)40:1<22::AID-CM3>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Smirnova EA, Bajer AS. Microtubule converging centers and reorganization of the interphase cytoskeleton and the mitotic spindle in higher plant Hemanthus. Cell Motil Cytoskel. 1994;27:219–233. doi: 10.1002/cm.970270304. [DOI] [PubMed] [Google Scholar]
- Stoppin V, Vantard M, Schmit A-C, Lambert A-M. Isolated plant nuclei nucleate microtubule assembly: The nuclear surface in higher plants has centrosome-like activity. Plant Cell. 1994;6:1099–1106. doi: 10.1105/tpc.6.8.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaizel-Ohayon D, Schejter ED. Mutations in centrosomin reveal requirements for centrosomal function during early Drosophila embryogenesis. Curr Biol. 1999;9:889–898. doi: 10.1016/s0960-9822(99)80393-5. [DOI] [PubMed] [Google Scholar]
- Verde F, Berrez J-M, Antony C, Karsenti E. Taxol-induced microtubule asters in mitotic extracts of Xenopus eggs: requirement for phosphorylated factors and cytoplasmic dynein. J Cell Biol. 1991;112:1177–1187. doi: 10.1083/jcb.112.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidwans SJ, O'Farrell PH. Cytoskeleton: centrosome-in absentia. Curr Biol. 1999;9:R764–R766. doi: 10.1016/S0960-9822(00)80007-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE, Mitchison TJ, Desai A. XKCM1: A Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Vernos I, Mitchison TJ, Karsenti E, Heald R. A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr Biol. 1998;8:903–913. doi: 10.1016/s0960-9822(07)00370-3. [DOI] [PubMed] [Google Scholar]
- Waters JC, Salmon ED. Pathways of spindle assembly. Curr Opin Cell Biol. 1997;9:37–43. doi: 10.1016/s0955-0674(97)80149-4. [DOI] [PubMed] [Google Scholar]
- Wick SM, Duniec J. Immunofluorescence microscopy of tubulin and microtubule arrays in plant cells. II. Transition between the pre-prophase band and the mitotic spindle. Protoplasma. 1984;122:45–55. [Google Scholar]
- Wittmann T, Hyman A, Desai A. The spindle: a dynamic assembly of microtubules and motors. Nat Cell Biol. 2001;3:E28–E34. doi: 10.1038/35050669. [DOI] [PubMed] [Google Scholar]
- Zhang P, Hawley RS. The genetic analysis of distributive segregation in Drosophila melanogaster. II. Further genetic-analysis of the Nod locus. Genetics. 1990;125:115–127. doi: 10.1093/genetics/125.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]