Abstract
Chimeras of the Halobacterium salinarum transducers HtrI and HtrII were constructed to study the structural determinants for their specific interaction with the phototaxis receptors sensory rhodopsins I and II (SRI and SRII), respectively. Interaction of receptors and transducers was assessed by two criteria: phototaxis responses by the cells and transducer-modulation of receptor photochemical reaction kinetics in membranes. Coexpression of HtrI with SRII or HtrII with SRI did not result in interaction by either criterion. Each receptor was coexpressed with chimeric transducers in which various domains of the two transducers were interchanged. The results show that the presence of the two transmembrane helices of HtrI in a chimera is necessary and sufficient for functional transducer complexation with SRI, i.e., for wild-type SRI photoreactions and attractant and 2-photon repellent phototaxis responses. Additionally, a previously demonstrated chaperone-like facilitation of SRI folding or stability by HtrI was shown to depend only on the two transmembrane helices of HtrI in chimeric transducers. Similarly, the two transmembrane helices of HtrII specify interaction with the repellent receptor SRII according to motility analysis and laser-flash spectroscopy. The results support a model in which the membrane domains of the receptor/transducer complexes, consisting of the seven helices of the receptor interacting with the four-helix bundle of the transducer dimer, produce SRI- and SRII-specific signals to the flagellar motor by means of interchangeable cytoplasmic domains.
Keywords: halobacteria, phototaxis, seven-transmembrane helix receptor, helix–helix interaction
Halobacterium salinarum exhibits color-discriminating phototaxis responses through the use of two visual pigment-like photoreceptors, sensory rhodopsins I and II (SRI and SRII) (1). SRI mediates attractant responses to orange light (λ max 587 nm) and, by means of photoexcitation of its long-lived photoproduct M (λ max 373 nm), it also mediates repellent responses to near-UV light. SRII is a repellent receptor sensing blue light (λ max 487 nm). Each of the SRs interacts with a specific transducer protein [HtrI (2) and HtrII (3) for SRI and SRII, respectively], which in turn controls the activity of a histidine kinase/phosphoregulator two-component system that regulates the cells’ flagellar motors (4). Based on the function of the homologous components in eubacterial chemotaxis (5, 6), attractant and repellent responses from the SR/Htr complexes are believed to result from inhibition and activation, respectively, of the histidine kinase.
HtrI has been shown to be a dimer (7) and to interact physically with SRI both in the light (8–10) and in the dark (11), and probably these features can be generalized to the SRII–HtrII molecular complex. For both receptors, transducer complexation can be assessed by laser-flash kinetic spectroscopy because HtrI binding alters the kinetics and pH dependence of the SRI photocycle (8), and HtrII binding also alters SRII photocycle kinetics (12).
Viewed from the outside of the cell, four distinct domains of the Htr proteins are evident: (i) a periplasmic region that is small (<5 residues) in HtrI and large (≈250 residues) in HtrII; (ii) a membrane domain formed by two transmembrane hydrophobic helices, TM1 and TM2; (iii) a hydrophilic “linker” region of ≈200 residues extending from the inner membrane cytoplasmic surface to (iv) the methylation and signaling domain homologous to the domains of eubacterial chemotaxis receptor/transducers (13) that control the kinase activity. The methylation and signaling domain of HtrI are dispensable for the control of SRI photoreaction kinetics in a truncated transducer (14), and more extensive deletion analysis (15) established that the N-terminal 147 residues of HtrI, which contain the two transmembrane helices and ≈90 residues of the cytoplasmic linker, are sufficient for interaction with SRI. Deletion of the linker region in that study and in an independent investigation (16) resulted in loss of functional interaction with SRI. This negative result does not distinguish whether the cytoplasmic portion is required for receptor interaction or alternatively for proper folding or stability of the partial transducer proteins. The chimeras studied here were constructed to overcome this limitation, because they are full-length transducer molecules more likely to fold properly. Phototaxis responses of cells expressing the chimeras together with SRI or SRII were analyzed by motion analysis, and membranes isolated from these cells were studied by flash photolysis. The results demonstrate that TM1 and TM2 of the Htrs are the only determinants specifying interaction with the cognate SR proteins as well as signaling specificity.
MATERIALS AND METHODS
Bacterial Strains, Culture Conditions, and Transformation.
H. salinarum strain Pho81Wr− (SRI−HtrI−SRII−HtrII−;W indicates carotenoid-deficient and r−, lack of a restriction system that reduces transformation efficiency) (14) and its transformants were cultured in the dark at 37°C and 240 rpm on a gyratory shaker. Polyethylene glycol-mediated spheroplast transformation of halobacteria was performed as described (17) with the following modifications: (i) Polyethylene glycol-600 was purified by absorbing to ion exchange resin AG 501-X8 (Bio-Rad) according to the instructions provided by the manufacturer; (ii) spheroplasts were made from freshly grown cultures at A600 = 0.4; and (iii) DNA at the concentration of 200 ng/μl in TE buffer (10 mM Tris⋅HCl/1 mM EDTA, pH 8.0) was used directly for the transformation without first mixing with spheroplast solution.
Plasmid Construction.
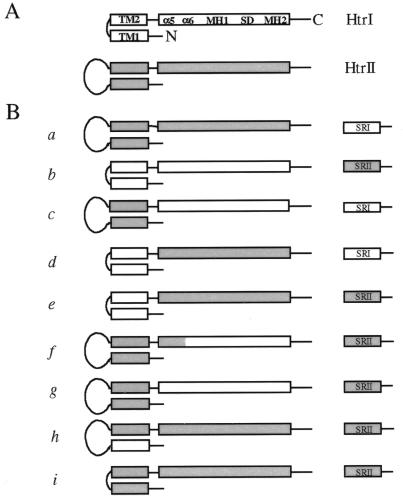
Htr chimeras were constructed by using recombinant PCR. Nine Htr–SR combinations were constructed (Fig. 1B). The promoter of the htrI–sopI operon was used in all cases. Construct a places wild-type htrII together with wild-type sopI, with the htrII stop codon and sopI initiation codon arranged as in wild-type htrII–sopII (3). In construct b, wild-type htrI is placed in front of sopII (the gene encoding SRII apoprotein) with the stop codon of htrI overlapping the initiation codon of sopII, as in the wild-type htrI–sopI pair (2). Construct c encodes HtrI (I61-A536) with the transmembrane and periplasmic region of HtrII (M1-A329). Construct d encodes HtrII (L330-Y765) following the transmembrane region of HtrI (M1-S60). Construct e is the same as d except sopII instead of sopI follows the transducer chimera gene. In the chimeras encoded by f and g, N-terminal regions of HtrII (M1-R368 and M1-A329, respectively) are fused to C-terminal portions of HtrI (Q100-A536 and I61-A536, respectively). In h, TM1 of HtrI (M1-T38) replaces that of HtrII (M1-R48). The region encoding the periplasmic part of HtrII (V47-R292) is deleted in construct i.
Figure 1.
(A) Schematic representation of the secondary structure of HtrI and HtrII. Transmembrane regions (TM1 and TM2), signaling domain (SD), methylation regions (MH1 and MH2), and α5 and α6 [as defined for E. coli Tar and Trg (5, 6, 13)] are labeled. The periplasmic domain of HtrII is drawn as a loop. N, C: N terminus and C terminus of the molecule. (B) Constructs used in this study. For each construct, the encoded Htr and SR proteins are shown. SRI and regions in Htr chimeras that derive from HtrI are not shaded; SRII and regions in Htr chimeras that derive from HtrII are shaded.
Synthetic oligonucleotides (BioServe Biotechnologies, Laurel, MD) used for the construction were designed for use with the megaprimer method (18). Plasmid pPR5 (19), which carries the htrII–sopII gene pair under control of the HtrI promoter, was used in all cases as the starting plasmid for the construction. PCR reactions were performed in a Programmable Thermal Controller-100 (MJ Research, Cambridge, MA) in most cases at 94°C, 1 min, 55°C, 1 min, and 72°C, 1 min for 31 cycles. Annealing temperatures were adjusted and 1%–6% formamide was included (20) in some of the PCR reactions to optimize the reaction condition. PCR fragments were purified from agarose gel by using a glass powder-based method (21) or QIAEX II (Qiagen, Chatsworth, CA). After digestion with appropriate enzymes, the fragment was replaced into pVJY1 (14) or pPR5 (19). Escherichia coli strain DH5α (Stratagene) was used for plasmid manipulation and amplification.
Membrane Preparation and Western Blotting Analysis.
Membranes were isolated from sonicated stationary-phase cells as described (22) and suspended in 4 M NaCl/25 mM Tris⋅HCl, pH 6.8. Polyclonal antibodies made against the conserved signaling domain of transducers [HC23 (23)] and against the C-terminal 24 residues of SRI (24) were used for detection of transducers and SRI, respectively, with the ECL Western blotting kit (Amersham Pharmacia). Linearity of the exposure was tested by comparing signals of serially diluted samples.
Motion Analysis.
Motility responses to SRI and SRII photoactivation were assayed by computer-assisted cell tracking and motion analysis as described (25). Pulse durations were controlled by a Uniblitz electronic shutter (Vincent, Rochester, NY). Phototaxis stimuli were delivered through an epiiluminator from a Nikon 100-W Hg/Xe lamp or from a 150-W tungsten/halogen lamp.
Spectroscopy.
Absorption spectra were measured by using an SLM– Aminco UV-Vis spectrophotometer (SLM–Aminco, Urbana, IL). Flash-induced absorbance changes of pigments in membrane suspensions (1-cm pathlength) were measured with a laboratory-constructed crossbeam kinetic spectrophotometer (26). The actinic flash was from a Nd-YAG pulse laser (532 nm, 6 ns duration, 40 mJ; Surelite I, Continuum, Santa Clara, CA). Flash photolysis data were fit by Sigma Plot (Jandel, San Rafael, CA).
SRII content and membranes were measured by difference spectroscopy. Reference membranes were prepared from Pho81Wr− transformed with a plasmid not carrying sop genes and grown under the same conditions as the experimental transformants. Experimental sample minus reference sample difference spectra were recorded from 350 nm to 700 nm, and the concentration of the reference, initially slightly higher than that of the sample, was adjusted by dilution with sample buffer until a horizontal line near zero occurred at the red side of the spectrum (600 nm to 700 nm). The height of the absorption peak at 487 nm above this line was taken as a measure of SRII absorption. The value obtained in this manner is reproducible and insensitive to slight differences in scattering between the reference and sample suspensions and is applicable to membrane preparations from strains deficient in carotenoids, such as Pho81W and its derivatives.
RESULTS
The Two Transmembrane Helices of HtrI Exhibit a Chaperone-Like Function for SRI Production.
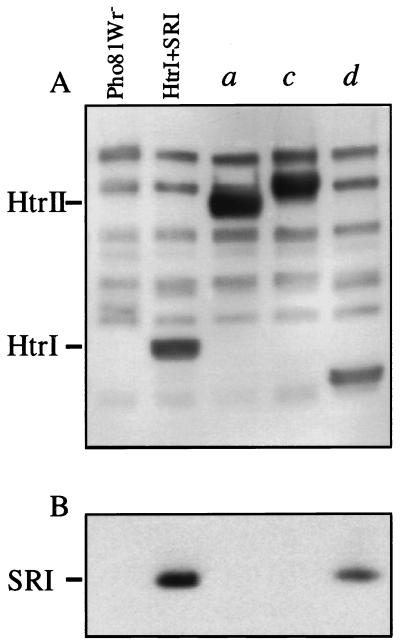
An immunoblot with polyclonal antibody directed against the conserved signaling domain sequence of Htrs shows that transducer expression was at similar levels from the wild-type htrI–sopI gene pair, and from constructs a, c, and d (Fig. 1), in which the gene encoding SRI apoprotein is preceded by genes encoding full-length HtrII (Fig. 2A, lane a), HtrII periplasmic and transmembrane domains with the HtrI cytoplasmic domain (lane c), and HtrII cytoplasmic domain with the HtrI transmembrane domain (lane d), respectively. Immunoblotting with antibody against the signaling domain of H. salinarum transducers demonstrated production of each of these transducers at readily detectable levels with relative migration positions correlating with their expected mass (Fig. 2A), as well as ≈8 other bands corresponding to other transducers (23).
Figure 2.
Immunoblots of membranes with antitransducer antibody (A) and anti-SRI antibody (B). Lanes contain membrane proteins from Pho81Wr− transformed with plasmids encoding the constructs noted above for each lane. First lane, Pho81Wr− transformed with vector alone and, second lane, with pVJY1 encoding wild-type HtrI and SRI (14). Construct a encodes wild-type HtrII and SRI. Constructs c and d encode chimeric transducers and SRI (see Fig. 1).
Immunoblot analysis with anti-SRI antibody, however, shows that the SRI apoprotein is produced at detectable levels only in the presence of wild-type HtrI and in the presence of the transducer chimera d (Fig. 2B). We previously observed that SRI apoprotein was produced at negligible levels in the absence of HtrI and that high-level production of SRI from a plasmid was rescued by HtrI expression in trans (15). The requirement for HtrI was eliminated when the SRI apoprotein was extended at its N terminus with the signal sequence of the bacteriorhodopsin protein. These results suggested a chaperone-like function for HtrI that facilitates membrane insertion or proper folding of the SRI apoprotein. The results in Fig. 2 demonstrate that HtrII does not provide this function for SRI, and that the N-terminal 60 residues of HtrI, which form primarily the two transmembrane helices of HtrI, are sufficient to confer the chaperone-like activity.
The Two Transmembrane Helices of HtrI Confer Functional Interaction with SRI.
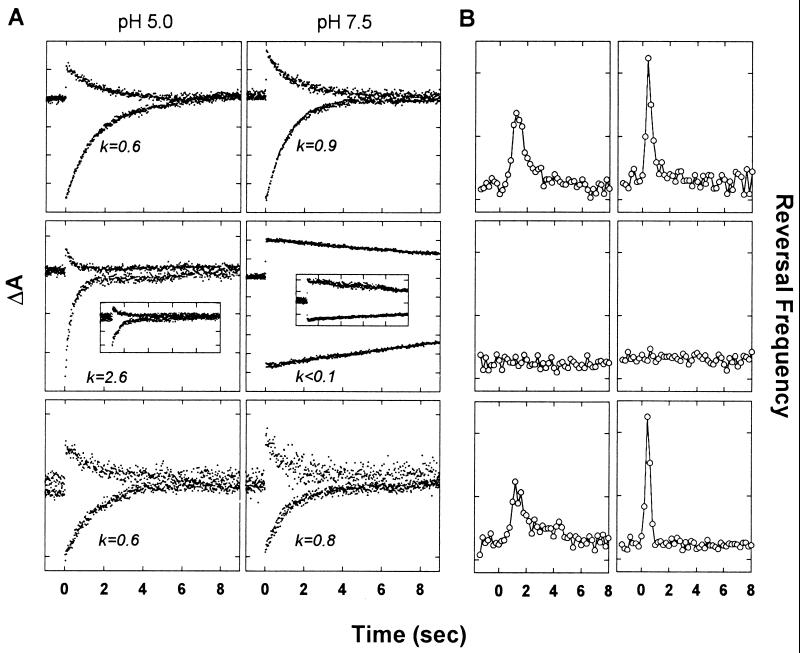
Flash-induced absorbance changes of SRI and phototaxis signaling were examined to test whether TM1 and TM2 confer functional interaction with the receptor. The complexation of SRI with HtrI results in modulation of the SRI photocycle kinetics and their pH sensitivity (8). Transducer-free SRI, which is produced at high levels when attached to the bacteriorhodopsin signal sequence, exhibits pH-dependent photoreactions. At pH 5.0, the return of the long-lived M intermediate occurred with a rate constant of 2.6 sec−1, calculated from the return of the depletion of the unphotolyzed state (λ max 587 nm) monitored at 590 nm (Fig. 3A Left Middle, lower trace). The laser-flash induced absorbance change at 400 nm (Left Middle, upper trace) exhibited comparable kinetics. These rate constants become <0.1 sec−1 at pH 7.5 (Fig. 3A). In contrast, SRI in complex with HtrI (Fig. 3A Top) exhibited similar rate constants of 0.6 and 0.9 0 sec−1, therefore largely independent of pH. Because construct a did not produce SRI, we replaced the htrI promoter with the high-efficiency bop promoter and truncated 15 residues of the C-terminal peptide of SRI (9) to increase its expression level. A pronounced effect of pH on the absorption transients, identical to that of transducer-free SRI, is shown by SRI expressed in the presence of HtrII (Fig. 3A Insets and Middle), indicating no interaction by this criterion. SRI and the chimera encoded by construct d exhibit normal interaction by this criterion; although the flash-induced absorption changes are smaller, reflecting the lesser amount of SRI produced (Fig. 2B), they exhibited rates indistinguishable from those of wild-type SRI–HtrI complex (Fig. 3A).
Figure 3.
(A) Transient absorption changes at 370 nm (top trace) and 590 nm (bottom trace) of Pho81Wr− membranes after a laser flash at time = 0. Each tick on the ordinate corresponds to 5 × 10−4 absorbance units. Rate constants in sec−1 of the 590-nm trace from single exponential fits are shown. Top, SRI and HtrI; Middle, transducer-free enSRI (24). The insets in the middle row show the flash photolysis data for the SRI–HtrII pair (see text); Bottom, SRI and the Htr chimera of construct d (Fig. 1). Leftmost column, pH 5.0; rightmost column, pH 7.5. (B) Motion analysis of Pho81Wr− transformants expressing various Htr–SRI pairs. Responses to each of two saturating stimuli (initiated at 0 sec) were recorded: Left, stepdown of 600 nm light for 4 sec; Right, stepup of 400 nm light for 100 msec in a constant 600-nm background. Each tick on the ordinate corresponds to 0.1 sec−1. Top, SRI and HtrI; Middle, SRI and HtrII [same cells as used for the flash photolysis data in the inset in Fig. 3A); note that the C-terminal 15 residues of SRI were removed to increase the SRI content since construct a (Fig. 1) did not produce detectable pigment]; Bottom, SRI and the Htr chimera of construct d (Fig. 1).
Phototaxis responses triggered by SRI photoactivation were assessed by cell tracking and motion analysis. The SRI–HtrI complex mediates attractant responses to orange light (assayed in Fig. 3B as a transient increase in swimming reversal frequency to a step-down in light intensity at 600 nm), and repellent responses resulting from excitation of its M photointermediate by near UV light (assayed in Fig. 3B as a transient increase in swimming reversal frequency to a pulse of 400 nm light in an orange background that generates M). Cells containing SRI and HtrII in their membranes did not exhibit responses to either stimulus, as evidenced by the lack of response by cells carrying the HtrII–SRI pair for which flash photolysis data is shown in the figure (Fig. 3 Middle). The lack of responses was confirmed in cells in which SRI was overexpressed 4-fold above wild-type levels by extending it at the N terminus with the bacteriorhodopsin signal sequence, and HtrII was expressed at the high levels used in this study (data not shown). Cells containing SRI and the chimera with the N-terminal 60 residues of HtrI exhibited wild-type responses to both types of SRI photostimuli (Fig. 3 Bottom).
Expression Levels of Htr Chimeras and SRII.
Immunoblots with polyclonal antibody directed against the conserved signaling domain sequence of Htrs showed that transducer expression was high (40–100% of wild type) from all of the constructs (Fig. 2 and Table 1). The content of SRII in membranes of the transformed Pho81Wr− strains was measured by absorption spectroscopy (Table 1). The presence of full-length HtrII in the membrane does not appear to be necessary for SRII stability. Therefore, SRII differs from SRI in the dependence of the latter on its transducer for a wild-type level of expression.
Table 1.
SRII and Htr expression in different constructs*
| Construct | b | e | f | g | h | i |
|---|---|---|---|---|---|---|
| SRII | 100 | 60 | 92 | 88 | 62 | 40 |
| Htr | 102 | 45 | 94 | 87 | 65 | 43 |
Expression is defined as % of the content of SRII and HtrII protein expressed from the htrII–sopII gene pair from the htrI promoter (19).
The Two Transmembrane Helices of HtrII Confer Functional Interaction with SRII.
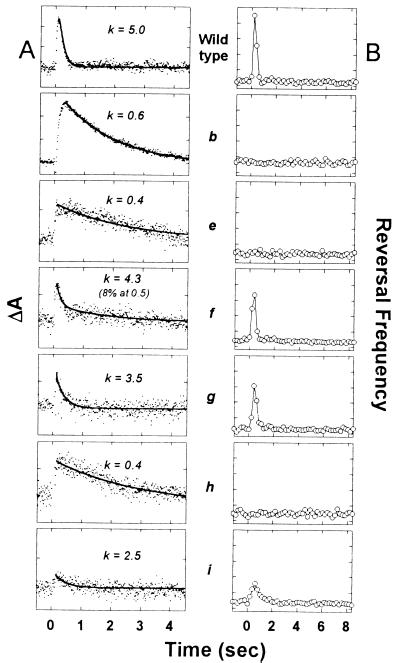
Flash-induced absorbance changes of SRII and phototaxis signaling were examined to test for interaction of the various chimeras with the receptor. The complexation of SRII with HtrII results in modulation of the SRII photocycle kinetics (12). The most pronounced effect of HtrII interaction is the acceleration of the decay of the final photocycle intermediate, the red-shifted species O (λ max 530 nm). The reported rate constants for O decay at 35°C are 0.7 sec−1 and 4.1 sec−1 in HtrII-free SRII and HtrII-complexed SRII, respectively (12). Similar values were obtained here for wild-type complex and SRII in the presence of HtrI (Fig. 4), indicating lack of interaction by this criterion. Slow O decay was also observed in the presence of the chimeras encoded by constructs e and h, which lack one or both transmembrane helices of HtrII. In contrast, constructs f, g, and i, which contain both TM1 and TM2 of HtrII, each produce the more rapid kinetics characteristic of transducer-complexed SRII. Therefore, by this criterion, those and only those chimeras containing the two transmembrane helices of HtrII interact with SRII.
Figure 4.
(A) Transient absorption changes at 540 nm of Pho81Wr− membranes containing the various SRII-encoding constructs after a laser flash at time = 0. Rate constants in sec−1 from single or double exponential fits are shown above each trace. Each tick on the ordinate corresponds to 5 × 10−4 absorbance units. (B) Motion analysis of Pho81Wr− transformants expressing various Htr–SRII pairs. A 1-sec pulse of 500 nm light was delivered at time = 0. Each tick on the ordinate corresponds to 0.1 sec−1. From top to bottom: wild-type SRII and HtrII; constructs b, e–i (Fig. 1).
Coexpression of HtrI together with SRII also did not result in functional interaction as assessed by motion analysis (Fig. 4). Because both HtrI and SRII were expressed at high levels, the lack of phototaxis indicates lack of functional interaction. Transformants expressing chimeras e or h exhibit no phototaxis response to SRII stimulation, whereas those expressing chimeras f, g, or i exhibit repellent responses. A clear correlation between chimera modulation of the SRII photocycle and ability to mediate SRII phototaxis is evident (Fig. 4).
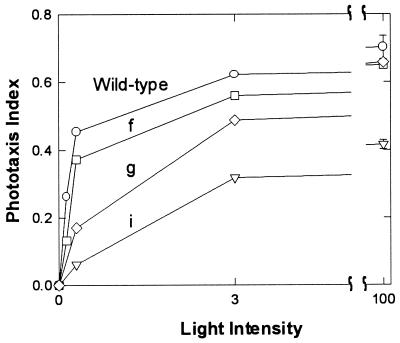
Fluence/response curves were measured to compare signaling efficiency by the chimeras that relay SRII signals. All three of the functional chimeras saturate in the same range as wild type, and their maximal responses (Fig. 5) are roughly proportional to the amount of SRII produced by the particular construct (Table 1). Therefore all three constructs exhibit near wild-type signaling efficiency.
Figure 5.
Fluence/response curves of H. salinarum strain Pho81Wr− expressing various constructs listed in Fig. 1. A 1-sec pulse of 500 nm light was used for stimulation of the cells. 100% intensity was 6 × 104 ergs⋅cm−2⋅sec−1 and light intensity was attenuated with neutral density filters. The phototaxis index is calculated in sec−1 as the integral of the swimming reversal frequency measured by motion analysis over the first 1 sec after the stimulus minus the integral over 1 sec starting from 6 sec after the stimulus was initiated.
We conclude from these data that the two transmembrane helices of HtrII are necessary and sufficient in a chimeric transducer to relay SRII phototaxis signals. The periplasmic domain of the HtrII protein can be removed (construct i) and the entire cytoplasmic domain can be substituted with that of HtrI (construct g) without loss of SRII-specific functional interaction. The removal of the periplasmic domain from HtrII without loss of coupling to SRII or phototaxis responses is consistent with the findings that the periplasmic domain contains a ligand-binding site for chemotaxis effectors and therefore might be expected to be dispensable for phototaxis (27), and that HtrII from Natronobacterium pharaonis lacks a periplasmic loop (28).
DISCUSSION
The results reported above establish that the two transmembrane helices of HtrI and HtrII define their specificity of interaction with SRI and SRII, respectively. All other transducer domains, i.e., those portions outside of the membrane, can either be removed or interchanged with little or no effect on the ability of the transducer to recognize and relay signals from its cognate receptor. In terms of the current model for the phototaxis signaling pathway, these findings imply that the membrane-embedded portion of the SR-Htr complexes produce signals altering CheA kinase activity by means of an interchangeable cytoplasmic domain.
It was previously shown that the N-terminal 147 residues of HtrI, which include TM1 and TM2 and about 90 residues of the cytoplasmic part, are sufficient for interaction with SRI as determined by flash photolysis of SRI in membranes containing shortened versions of HtrI (15). Mutations have been found in the cytoplasmic region of HtrI near TM2 that affect SRI photochemistry and function (29, 30). The results reported here strongly suggest that the receptor interaction sites are restricted to TM1 and TM2 of the transducer. Therefore the mutations in the cytoplasmic extension of TM2 probably influence SRI indirectly by altering HtrI structure at its SRI interaction sites in the membrane. Consistent with this, sequence comparison of the ≈90-residue region revealed no significant homology between HtrI and HtrII arguing against a common domain for nonselective interaction with SRI and SRII (7). Engineered cysteine crosslinking experiments and secondary structure prediction (7) indicate the 90-residue region to be a coiled coil involved in dimerization of HtrI. Note, however, that our results do not rule out that interaction of the receptor with transducer cytoplasmic loops occurs, as has been suggested by Krah et al. (16), although such an interaction, if it occurs, is not important to the specificity according to our data.
An interesting inference from this study is that, because only the transmembrane portions of the transducer proteins are responsible for specific interaction with the receptors, it seems likely that only the transmembrane parts of sensory rhodopsins interact with the transducers. Signal transmission would then occur by means of light-induced changes in helix–helix lateral packing interactions (31, 32). This mode of receptor/transducer coupling appears to be different from that of visual pigments and other seven-transmembrane-helix receptors, which interact with G-proteins through their large cytoplasmic loops connecting adjacent transmembrane segments (33).
However, recent studies have revealed that the detailed mechanism of SR photoactivation is remarkably similar to that of mammalian rhodopsins. In bovine rhodopsin and in SRI, light absorption triggers deprotonation of the retinylidene Schiff base on Helix G by means of a steric interaction of a methyl group on the retinal with the protein (34, 35). In rhodopsin and in SRII, the Schiff base proton has been shown to be transferred to a carboxylate on Helix C, and in both the visual pigment and SRII, disruption of the protonated Schiff base–Helix C carboxylate salt bridge by mutagenesis activates the receptor (19, 36). A resulting conformational change involving in both cases movement of Helix F (37, 38) is communicated to the transducer (the G-protein transducin and Htr, respectively) by protein–protein interaction.
A difference in the subsequent events that may underlie the apparently different mode of receptor-transducer interaction shown here is that the G-protein is a mobile component in the pathway, whereas the Htr is a tightly bound subunit of a molecular complex containing the SRI receptor. Activation of multiple G-protein molecules by a given receptor contributes to amplification of the signal, and cytoplasmic GTP binding controls the lifetime of the activated state of the G-protein (33). An analogous amplification step in the taxis signaling pathway occurs by means of the mobile component CheY, whose phosphorylation is controlled by a histidine kinase believed to be tightly bound to the Htr (1, 5, 39). Considering these differences in the signal transduction pathways, the function of the Htr protein is more analogous to that of the cytoplasmic domain of rhodopsin than to the G-protein and can be thought of as a “coreceptor” and the histidine kinase/CheY pair as a nucleotide-dependent “transducer” of the signal into the cytoplasm, analogous to the G-protein transducin.
Cryoelectron microscopy of bacteriorhodopsin shows that light causes a global conformational change in which the main feature is that the cytoplasmic end of Helix F is tilted and displaced toward the periphery of the protein, expanding the structure within the membrane on the cytoplasmic side and thereby facilitating proton uptake from the cytoplasm during the pumping process (40–43). A similar conformational change is indirectly indicated in the sensory rhodopsins by the demonstration of proton pumping by transducer-free SRI (44) and constitutive activation of SRII by a mutation that induces the conformational change in bacteriorhodopsin (19). An attractive hypothesis is that tilting of the cytoplasmic end of Helix F in the receptors is communicated to their transducers by way of contact with the TM2 helix, which has been shown to be the mobile signaling helix in the aspartate chemotaxis receptor (45, 46).
Acknowledgments
We thank Elena Spudich and Kwang-Hwan Jung for discussion and critical reading of the manuscript, Jun Sasaki for assistance with flash spectroscopy, and Bastianella Perazzona for help with immunoblot analysis. This work was supported by National Institutes of Health Grant RO1-GM27750 (to J. L. S.).
ABBREVIATIONS
- SRI and SRII
sensory rhodopsins I and II, respectively
- HtrI and HtrII
halobacterial transducers for SRI and SRII, respectively
- TM1
the N-terminal transmembrane segment of a transducer
- TM2
the second transmembrane segment of a transducer
References
- 1.Hoff W D, Jung K H, Spudich J L. Annu Rev Biophys Biomol Struct. 1997;26:223–258. doi: 10.1146/annurev.biophys.26.1.223. [DOI] [PubMed] [Google Scholar]
- 2.Yao V J, Spudich J L. Proc Natl Acad Sci USA. 1992;89:11915–11919. doi: 10.1073/pnas.89.24.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang W, Brooun A, Mueller M M, Alam M. Proc Natl Acad Sci USA. 1996;93:8230–8235. doi: 10.1073/pnas.93.16.8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rudolph J, Oesterhelt D. EMBO J. 1995;14:667–673. doi: 10.1002/j.1460-2075.1995.tb07045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stock J B, Surette M. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt F, editor. Washington, D. C.: Am. Soc. Microbiol.; 1996. pp. 123–145. [Google Scholar]
- 6.Falke J J, Bass R B, Butler S L, Chervitz S A, Danielson M A. Annu Rev Cell Dev Bol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X-N, Spudich J L. J Biol Chem. 1998;273:19722–19728. doi: 10.1074/jbc.273.31.19722. [DOI] [PubMed] [Google Scholar]
- 8.Spudich E N, Spudich J L. J Biol Chem. 1993;268:16095–16097. [PubMed] [Google Scholar]
- 9.Olson K D, Spudich J L. Biophys J. 1993;65:2578–2585. doi: 10.1016/S0006-3495(93)81295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krah M, Marwan W, Vermeglio A, Oesterhelt D. EMBO J. 1994;13:2150–2155. doi: 10.1002/j.1460-2075.1994.tb06491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan B, Spudich E N, Sheves M, Steinberg G, Spudich J L. J Phys Chem. 1997;101:109–113. [Google Scholar]
- 12.Sasaki J, Spudich J L. Biophys J. 1998;75:2435–2440. doi: 10.1016/S0006-3495(98)77687-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Moual H, Koshland D E., Jr J Mol Biol. 1996;261:568–585. doi: 10.1006/jmbi.1996.0483. [DOI] [PubMed] [Google Scholar]
- 14.Yao V J, Spudich E N, Spudich J L. J Bacteriol. 1994;176:6931–6935. doi: 10.1128/jb.176.22.6931-6935.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perazzona B, Spudich E N, Spudich J L. J Bacteriol. 1996;178:6475–6478. doi: 10.1128/jb.178.22.6475-6478.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krah M, Marwan W, Oesterhelt D. FEBS Lett. 1994;353:301–304. doi: 10.1016/0014-5793(94)01068-4. [DOI] [PubMed] [Google Scholar]
- 17.Cline S W, Doolittle W F. J Bacteriol. 1987;169:1341–1344. doi: 10.1128/jb.169.3.1341-1344.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen B, Przybyla A E. BioTechniques. 1994;17:657–659. [PubMed] [Google Scholar]
- 19.Spudich E N, Zhang W, Alam M, Spudich J L. Proc Natl Acad Sci USA. 1997;94:4960–4965. doi: 10.1073/pnas.94.10.4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarkar G, Kapelner S, Sommer S S. Nucleic Acids Res. 1990;18:7465. doi: 10.1093/nar/18.24.7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olson K D, Zhang X N, Spudich J L. Proc Natl Acad Sci USA. 1995;92:3185–3189. doi: 10.1073/pnas.92.8.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, Brooun A, McCandless J, Banda P, Alam M. Proc Natl Acad Sci USA. 1996;93:4649–4654. doi: 10.1073/pnas.93.10.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krebs M P, Spudich E N, Khorana H G, Spudich J L. Proc Natl Acad Sci USA. 1993;90:3486–3490. doi: 10.1073/pnas.90.8.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spudich J L, Spudich E N. In: Archaea: A Laboratory Manual. Robb F T, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 23–28. [Google Scholar]
- 26.Spudich E N, Sundberg S A, Manor D, Spudich J L. Proteins. 1986;1:239–246. doi: 10.1002/prot.340010306. [DOI] [PubMed] [Google Scholar]
- 27.Hou S, Brooun A, Yu H, Freitas T, Alam M. J Bacteriol. 1998;180:1600–1602. doi: 10.1128/jb.180.6.1600-1602.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seidel R, Scharf B, Gautel M, Kleine K, Oesterhelt D, Engelhard M. Proc Natl Acad Sci USA. 1995;92:3036–3040. doi: 10.1073/pnas.92.7.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung K -H, Spudich J L. Proc Natl Acad Sci USA. 1996;93:6557–6561. doi: 10.1073/pnas.93.13.6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jung K -H, Spudich J L. J Bacteriol. 1998;180:2033–2042. doi: 10.1128/jb.180.8.2033-2042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bormann B J, Engelman D M. Annu Rev Biophys Biomol Struct. 1992;21:223–242. doi: 10.1146/annurev.bb.21.060192.001255. [DOI] [PubMed] [Google Scholar]
- 32.Mackenzie K R, Prestegard J H, Engelman D M. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 33.Helmreich E J M, Hofmann K-P. Biochim Biophys Acta. 1996;1286:285–322. doi: 10.1016/s0304-4157(96)00013-5. [DOI] [PubMed] [Google Scholar]
- 34.Yan B, Nakanishi K, Spudich J L. Proc Natl Acad Sci USA. 1991;88:9412–9416. doi: 10.1073/pnas.88.21.9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shieh T, Han M, Sakmar T P, Smith S O. J Mol Biol. 1997;269:373–384. doi: 10.1006/jmbi.1997.1035. [DOI] [PubMed] [Google Scholar]
- 36.Rao R, Oprian D D. Annu Rev Biophys Biomol Struct. 1996;25:287–314. doi: 10.1146/annurev.bb.25.060196.001443. [DOI] [PubMed] [Google Scholar]
- 37.Farrens D L, Altenbach C, Yang K, Hubbell W L, Khorana H G. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 38.Spudich J L, Lanyi J K. Curr Opin Cell Biol. 1996;8:452–457. doi: 10.1016/s0955-0674(96)80020-2. [DOI] [PubMed] [Google Scholar]
- 39.Rudolph J, Tolliday N, Schmitt C, Schuster S C, Oesterhelt D. EMBO J. 1995;14:4249–4257. doi: 10.1002/j.1460-2075.1995.tb00099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subramaniam S, Gerstein M, Oesterhelt D, Henderson R. EMBO J. 1993;12:1–8. doi: 10.1002/j.1460-2075.1993.tb05625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lanyi J K. Nature (London) 1995;375:461–463. doi: 10.1038/375461a0. [DOI] [PubMed] [Google Scholar]
- 42.Vonck J. Biochemistry. 1996;35:5870–5878. doi: 10.1021/bi952663c. [DOI] [PubMed] [Google Scholar]
- 43.Subramaniam S, Faruqi A R, Oesterhelt D, Henderson R. Proc Natl Acad Sci USA. 1997;94:1767–1772. doi: 10.1073/pnas.94.5.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bogomolni R A, Stoeckenius W, Szundi I, Perozo E, Olson K D, Spudich J L. Proc Natl Acad Sci USA. 1994;91:10188–10192. doi: 10.1073/pnas.91.21.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen X, Koshland D E., Jr J Biol Chem. 1995;270:24038–24042. doi: 10.1074/jbc.270.41.24038. [DOI] [PubMed] [Google Scholar]
- 46.Chervitz S A, Falke J J. J Biol Chem. 1995;270:24043–24053. doi: 10.1074/jbc.270.41.24043. [DOI] [PMC free article] [PubMed] [Google Scholar]