Abstract
The noncovalent association of transmembrane α-helices is a fundamental event in the folding of helical membrane proteins. In this work, a system (TOXCAT) is developed for the study of transmembrane helix–helix oligomerization in a natural membrane environment. This assay uses a chimeric construct composed of the N-terminal DNA binding domain of ToxR (a dimerization-dependent transcriptional activator) fused to a transmembrane domain (tm) of interest and a monomeric periplasmic anchor (the maltose binding protein). Association of the tms results in the ToxR-mediated activation of a reporter gene encoding chloramphenicol acetyltransferase (CAT). The level of CAT expression indicates the strength of tm association. The assay distinguishes between a known dimerizing tm and a mutant in which dimerization is disrupted. In addition, modulation of the chimera concentration shows that the dimerization exhibits concentration dependence in membranes. TOXCAT also is used to select oligomeric tms from a library of randomized sequences, demonstrating the potential of this system to reveal novel oligomerization motifs. The TOXCAT system has been used to investigate glycophorin A tm-mediated dimerization. Although the overall sensitivity of glycophorin A tm dimerization to mutagenesis is found to be similar in membranes and in detergent micelles, several significant differences exist. Mutations to polar residues, which are generally disruptive in SDS, exhibit sequence specificity in membranes, demonstrating both the limitations of detergent micelles and the wider range of application of the TOXCAT system.
The environment presented by the lipid bilayer imposes substantial constraints on the structures of the transmembrane segments of integral membrane proteins, providing a thermodynamic rationale for the formation of stable transmembrane α-helices. The establishment of tertiary and quaternary structure then comprises interactions between preformed helical transmembrane domains (tms) (1). However, the study of helix–helix association in the folding of integral membrane proteins is technically difficult because of the necessity for solubilizing membranes or detergent micelles. Here, we present a method to investigate transmembrane helix association in a biological membrane.
Although the environment’s influence on secondary structure formation is well conceptualized, less is known about the forces that stabilize interactions between transbilayer α-helices. Transmembrane helix interactions are governed by the formation of helix–helix contacts and by interactions between the protein and its lipid environment. These environmental influences on folding are poorly understood because of the inability of most experimental systems to directly report helix–helix interactions in their native environment, a natural membrane. Typically, folding studies of membrane proteins have used detergents to provide a convenient membrane-like environment, although the extent to which observations made in detergent micelles accurately reflect helix–helix interactions in lipid membranes is not known.
The dimerization of the glycophorin A (GpA) tm in detergent micelles (2) provides a convenient example of membrane protein folding. Site-directed mutagenesis (3), computational modeling (4), and solution NMR (5) have demonstrated that the association between GpAtm monomers is mediated by helix–helix contacts involving a seven-residue motif, presented on one face of each transmembrane α-helix. The dimer interface is characterized by tightly packed surfaces formed by complementary ridges and grooves that allow close approach of the helices at a right-handed crossing angle (5). The specificity of the interaction is such that seemingly conservative mutations of the side chains contributing to the interface can disrupt the dimer, whereas hydrophobic mutations at noninterfacial positions generally have no effect (3, 6). The GpAtm dimerization motif is sufficient to drive the association of helices in a detergent environment, even when all noninterfacial residues are mutated to leucine (7). In addition, a peptide corresponding to the GpAtm dimerizes in synthetic lipid bilayers (8). Although the energy terms contributed by the GpAtm helix dimer contacts have been studied in detergent micelles (3, 5, 6, 9), little is known about the influence of environment on transmembrane helix–helix association.
To study tm association in a natural membrane environment, we have developed the TOXCAT assay system, which is based on the dimerization-dependent ToxR transcriptional activation domain (10, 11). TOXCAT provides substantial advantages over previous implementations of ToxR (11, 12), exhibiting heightened sensitivity to changes in dimerization affinity, tunable expression level, and the ability to apply selective pressure to isolate strongly oligomerizing tms. Application of this system to investigate the effects of mutagenesis of the GpAtm dimerization domain in a natural membrane has demonstrated significant environmental influences on the association of transmembrane α-helices.
MATERIALS AND METHODS
General.
Media were prepared as described in Sambrook et al. (13). All genes cloned using PCR were confirmed by sequence analysis. Ampicillin (Amp) was used at 200 μg/ml.
Vectors and Constructs.
pMAL-c2 and –p2 were obtained from New England Biolabs. The ctx∷chloramphenicol acetyltransferase (CAT) reporter construct was cloned by PCR amplifying the ctx promoter from genomic DNA isolated from Escherichia coli FHK12 (kindly provided by H. Kolmar, George-August University, Göttingen, Germany; ref. 11), using primers that add HindIII sites at both termini. The resulting product was ligated into the HindIII site of pkk232–8 (Stratagene), and the BamHI site was deleted to generate plasmid pkkctxΔB. ToxR′(tm) maltose binding protein (MBP) chimerae were cloned as follows: pkktgm, the SmaI/XbaI fragment of pHKToxR′(TMneu)MalE (kindly provided by H. Kolmar; ref. 11) was cloned as a blunt fragment into the Klenow-filled AatII site of pkkctxΔB; pccKAN, to facilitate cloning, the kanamycin resistance cassette from pACYC177 was ligated as an NheI/BamHI fragment into pkktgm; pccTNM, for expression of a ToxR-MBP chimera with no tm, NotI linkers were ligated into 3′ filled NheI/BamHI sites on pkktgm; pLTMKAN, for expression of ToxR′(tm)MBP chimerae under inducible control, the lac promoter (PCR amplified from pBS/KS+; Stratagene) and ToxR′ (PCR amplified from pHKToxR′(TMneu)MalE) were subcloned into the SmaI site of pBS/KS+. The EcoRV/NheI fragment containing the cytoplasmic domain of ToxR downstream of the lac promoter was ligated into the SmaI/NheI sites of a modified pccKAN (with deleted NdeI, SmaI, and NruI sites). The GpAtms for wild-type (wt) and G83I mutant sequences were cloned into the NheI/BamHI sites of pLTMKAN as described below.
tms.
tms corresponding to residues L75–T87 of the wt GpAtm were PCR-amplified and cloned into the NheI/BamHI sites of pccKAN as NheI/DpnII fragments by using an NheI site incorporated in the primer. wt GpAtm and mutants V80F, V80Y, A82Y, and G83Y were amplified from pT7SN/GpA99 clones (3). All other tms were amplified from pT7SN/GpA99 by using primers that contain a mismatch to generate the desired mutation.
Inducible Expression of Chimerae.
Expression of ToxR′(tm)MBP chimerae was induced by addition of isopropyl β-d-thiogalactoside to MM39 cells grown in M9 medium containing 0.4% glucose and 100 μg/ml of carbenicillin. Chimera concentration was measured by using immunoblotting followed by densitometry. Purified MBP was used as a concentration standard (kindly provided by L. Hanakahi, Yale University, New Haven, CT). The surface area of the E. coli cytoplasmic membrane was estimated by using values reviewed by Cronan et al. (14) and Neidhardt (15). To decrease leaky expression from the lac promoter in the absence of inducer, pLTMwt was cotransformed with pRG1 (kindly provided by T. Griffin, Yale University), a pACYC177-based plasmid that confers kanamycin resistance and overproduces the lac repressor LacI.
Library Preparation.
Library inserts were generated by Klenow-catalyzed extension of primer AS1 (5′-TCGTGCGGTGATCAG-3′) annealed to R2L [5′-ACACACCGCAGGCTAGCVBTVBTCTCTTAVBTVBTTTGCTTVBTVBTCTATTAVBTCTGATCGCCCTAACGGATATC-3′ where V and B indicate equimolar mixtures of (ACG) or (CGT), respectively]. The resulting product was ligated as an NheI/DpnII fragment into the NheI/BamHI sites of pccKAN. The pool of ligation products was transformed into E. coli DH5α and plated as a lawn on Luria–Bertani (LB) agar containing Amp. The lawn was resuspended, diluted between 10- and 107-fold, and plated on LB agar containing Amp and varying concentrations of chloramphenicol (CAM).
malE Complementation on Agar Plates.
ToxR′(tm)MBP chimera-expressing MM39 cells (kindly provided by J. Beckwith, Harvard Medical School, Boston) were cultured on M9 agar plates containing 0.4% maltose, 1% ion agar, and Amp.
Proteolysis of Spheroplasts.
Protease sensitivity assays were carried out as described in Chen and Kendall (16) with the following alterations. MM39 cells expressing ToxR′(GpAwt)MBP were grown in LB medium containing Amp to a final OD600 of 0.6. Lysis was carried out by using 3× freeze/thaw and mechanical disruption by 10× passage through a 23-gauge hypodermic needle. Proteinase K was added to 125 μg/ml.
Disk Diffusion Assay.
MM39 cells were grown to an OD600 of 0.6, diluted 6× with LB medium, and plated on 20 ml of LB/Amp agar plates. A disk, prepared by drying 60 μl of 90 mg/ml CAM in ethanol onto a Whatman 3MM filter paper disk (2.4cm diameter), was placed in the center of the plate. The diameter of the clear zone of inhibition of growth around the disk is reported.
CAT Assays.
To generate cell-free extracts, 200 μl of an 0.6 OD600 culture of MM39 cells was pelleted, resuspended in 0.5 ml of 100 mM Tris⋅HCl, pH 8.0, and lysed by addition of 20 μl of 100 mM EDTA, 100 mM DTT, 50 mM Tris⋅HCl, pH 8.0, and a small drop of toluene (from a drawn-out Pasteur pipette) at 30°C for 30 min. CAT assays were performed either as described in Sambrook et al. (13) or by using Quan-T-CAT (Amersham).
Immunoblots.
Immunoblotting was carried out by using standard methods and detected by using the ECL kit (Amersham). α-glucose-6-phosphate dehydrogenase antibodies were kindly provided by J. Beckwith.
RESULTS
TOXCAT Application to Gptm-Mediated Dimerization.
GpAtm was used to test the utility of the TOXCAT system for studying helix–helix interactions in the inner membrane of E. coli. According to the model for ToxR-mediated transcriptional activation (11), introduction of an intramembranous dimerization domain, such as the GpAtm, is expected to drive dimerization of the ToxR cytoplasmic domain, resulting in activation of the CAT gene and subsequent resistance to CAM. Mutations in the GpAtm that disrupt dimerization should yield relatively low levels of CAT, resulting in CAM-sensitive cells. Because CAM arrests bacterial growth without killing, a graded response in apparent survivorship is allowed.
CAM Resistance to Report Helix Interactions.
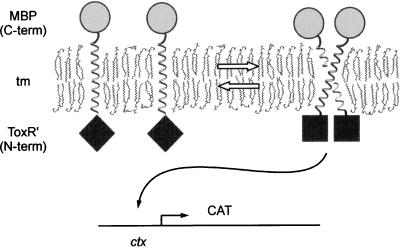
The TOXCAT system uses the chimeric construct described by Kolmar et al. (11), composed of the ToxR N-terminal transcriptional activation domain (ToxR′) fused to a transmembrane segment of interest and a C-terminal MBP domain (Fig. 1). The resulting chimera—ToxR′(tm)MBP—is constitutively expressed at low levels by the toxR promoter. tm-mediated dimerization of the chimera in the E. coli inner membrane results in transcriptional activation of a reporter gene driven by the ctx promoter, in this case CAT (Fig. 1). The extent of tm-induced dimerization can be measured in two ways: (i) acquired resistance to the antibiotic CAM in vivo, or (ii) direct quantitation of CAM acetylation by CAT in vitro. Incorporation of the reporter construct into the expression plasmid provides several copies of the reporter gene per cell, as opposed to previous systems, which used a single-copy chromosomally integrated reporter gene (10–12). This increase in reporter gene copy number is expected to provide greater sensitivity to low-level dimerization, as well as an enhanced upper range of measurement.
Figure 1.
The TOXCAT assay for tm oligomerization. tm-mediated oligomerization of the ToxR cytoplasmic domains (⧫) results in activation of CAT at the ctx promoter by dimerized N-terminal ToxR cytoplasmic domains (■). The periplasmic C-terminal MBP domain (•) anchors the chimera in the inner membrane. ToxR′: the cytoplasmic DNA binding domain of ToxR.
Dimerization Assays.
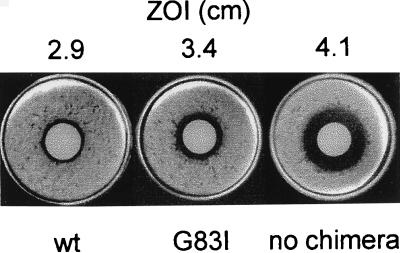
Disk diffusion assays (17) demonstrate that tm-mediated dimerization confers CAM resistance in this system (Fig. 2). Dimerization of the wt GpAtm (GpAwt) results in cells that are highly resistant to CAM relative to cells expressing no chimera, whereas cells expressing a chimera with a transmembrane point mutation (GpAG83I) are much less resistant to CAM. In SDS micelles, the G83I mutant does not dimerize (3). It is clear from the NMR structure of the GpAtm (5) that introduction of an isoleucine into position 83 would result in steric clashes with the backbone at position 84 on the opposing monomer, shifting the dimerization equilibrium toward monomer. The relative disk diffusion assay results for GpAwt and GpAG83I are consistent with these observations, suggesting that association of transmembrane α-helices is solely responsible for activation of CAT. Furthermore, these experiments demonstrate that antibiotic selection can distinguish tms with differing dimerization affinities.
Figure 2.
Selection assay for tm dimerization. Disk diffusion assays of cells expressing ToxR′(GpAwt)MBP (wt), ToxR′(GpAG83I)MBP (G83I), or no chimera. The zone of inhibition (ZOI) is shown above each assay.
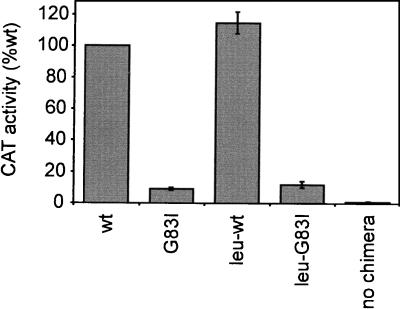
Assays of CAT activity in cell-free extracts (Fig. 3) provide a quantitative measurement of tm association. Dimerization of the wt GpAtm causes strong activation of the ctx promoter, resulting in high levels of CAT expression, whereas activation by the mutant G83I is reduced 10-fold. The observation that position G83 is sensitive to mutation in membranes and in SDS suggests that similar structural contacts are being made in both environments. To verify that the same interface is being used, all noninterfacial residues were simultaneously mutated to leucine, while motif residues (L75, I76, G79, V80, G83, V84, and T87) were preserved. As observed in SDS (7), mutation of all nonmotif residues to leucine has no effect on dimerization, and sensitivity to the G83I mutation is maintained.
Figure 3.
Quantitative assay of tm dimerization. CAT activity relative to wt for cells expressing the wt and G83I constructs in both the native sequence context (wt and G83I) and with noninterfacial leucines (leu-wt and leu-G83I), or with no chimera. Error bars represent the SD of 2–3 independent measurements.
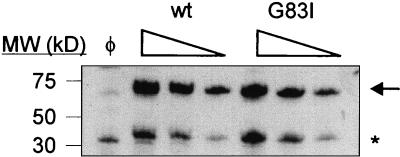
Because concentration differences should affect the relative amount of dimerization, the expression levels of ToxR′(GpAtm)MBP chimerae were determined by immunoblotting (Fig. 4). Although some proteolysis results in a high mobility band corresponding to cleaved MBP, the majority of the chimera is full length. Examination of serial 2-fold dilutions reveals no significant difference in the levels of chimera expression from either construct, demonstrating that both chimerae are being assayed in the same concentration range. Dimerization of ToxR chimerae was not observed in SDS/PAGE, probably because of the steric hindrance imposed by SDS-denatured soluble domains at each end of the transmembrane helix. Lemmon et al. (18) observed destabilization of the GpAtm dimer upon shortening of the linker between the soluble domains and tms. In the ToxR′(GpAtm)MBP chimerae, short linker lengths and the substantially large MBP domain inhibit association.
Figure 4.
Comparison of chimera expression levels in MM39 cells. Anti-MBP immunoblots of serial 2-fold dilutions of cells expressing ToxR′(tm)MBP chimera with wt or mutant (G83I) transmembrane domains or untransformed cells (φ). Arrow indicates full-length chimera. * indicates proteolyzed chimera. For undiluted samples, 4 μl of 6.0 OD600 units of cells were loaded.
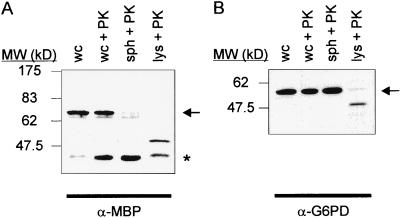
Dimerization of soluble ToxR′ chimerae also can activate transcription at the ctx promoter (11). To verify that intramembranous interactions are responsible for the observed CAT activation, the membrane insertion and orientation of ToxR′(GpAtm)MBP chimerae was measured. The localization of the MBP domain in E. coli MM39, a malE deletion strain, was probed directly by using proteolysis. In this assay, the protease sensitivity of a chimera is assayed in cells that have been stripped of their outer membrane and cell wall (spheroplasts), exposing the periplasmic contents to an exogenous protease, and permitting degradation of periplasmic proteins. Cytoplasmic proteins such as glucose-6-phosphate dehydrogenase are protected from the protease by the inner membrane (Fig. 5B). Immunoblots show almost complete degradation of the ToxR′(GpAwt)MBP chimera’s MBP domain in protease-treated spheroplasts (Fig. 5A), indicating that >90% of the MBP domain of the chimera is located in the periplasm and demonstrating the transmembrane topology of the chimera.
Figure 5.
Transmembrane topology of ToxR′(GpAwt)MBP. Immunoblots of a spheroplast proteolysis assay using MM39 cells expressing ToxR′(GpAwt)MBP, using antibodies directed against (A) MBP (α-MBP) or (B) glucose-6-phosphate dehydrogenase (α-G6PD). wc, whole cells. wc+PK, whole cells treated with Proteinase K. sph+PK, spheroplasts treated with Proteinase K. lys+PK, lysed spheroplasts treated with Proteinase K. Arrows denote full-length protein. ∗ indicates MBP liberated by proteolysis.
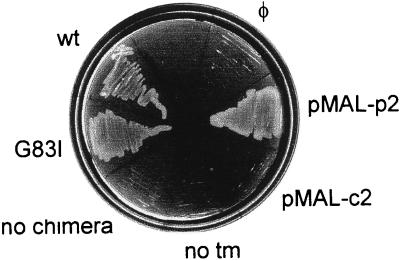
Another assay for the topology of ToxR′(tm)MBP chimerae in the E. coli inner membrane exploits the function of MBP in the maltose transport pathway. MM39 cells, which lack endogenous MBP, cannot transport maltose into the cytoplasm for metabolism, and consequently cannot grow on media in which the only available carbon source is maltose. If the chimeric ToxR′(tm)MBP are correctly inserted in the inner membrane, the periplasmic MBP domain will complement the MM39 malE-deficient phenotype and support growth on maltose. To demonstrate the requirement for periplasmic (vs. cytoplasmic) localization of MBP in the complementation assay, pMAL-p2 or pMAL-c2 (expressing MBP in the periplasm or cytoplasm, respectively) were transformed into MM39 cells and cultured on M9-maltose. As shown in Fig. 6, cells expressing periplasmic MBP (pMAL-p2, and either wt or G83I mutant ToxR′(GpAtm)MBP chimerae) grow on M9-maltose. Cells that lack MBP (expressing no chimera) or that express cytoplasmic MBP (pMAL-c2, or a ToxR′MBP chimera with no tm) fail to grow. These experiments demonstrate that both the wt and mutant tm-containing chimerae express their MBP domains in the periplasm, whereas the genetic response to dimerization (CAT activation) demonstrates cytoplasmic localization of the ToxR′ domain, consistent with correct membrane insertion and topology.
Figure 6.
malE complementation to test of ToxR′(tm)MBP chimera topology. malE-deficient E. coli MM39 transformed with various expression constructs were cultured on M9 agar containing 0.4% maltose. wt, ToxR′(GpAwt)MBP. G83I, ToxR′(GpAG83I)MBP. no chimera, cells expressing no chimera. no tm, pkkTNM, a ToxR′-MBP chimera with no tm. pMAL-c2, cytoplasmic accumulation of MBP. pMAL-p2, periplasmic accumulation of MBP. φ, untransformed cells.
The ToxR-Mediated Response Is Concentration Dependent.
As with any reversible dimerization event, intramembranous helix–helix interactions should exhibit concentration dependence. Although ToxR′(tm)MBP chimerae differing in only a single amino acid are expressed at comparable levels, the expression of chimerae containing significantly different tms can vary (data not shown), which complicates direct comparison of dimerization propensities. To overcome this obstacle, the ToxR′(tm)MBP chimera was placed under the control of the inducible lac promoter.
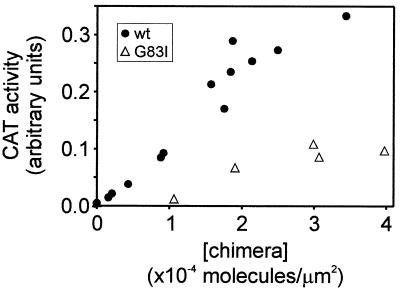
Expression of ToxR′(GpAtm)MBP chimerae containing wt or G83I mutant tms was induced with varying amounts of isopropyl β-d-thiogalactoside and quantitated by using immunoblotting and densitometry. CAT activity measurements demonstrate that dimerization depends on the expressed ToxR′(GpAtm)MBP concentration for both chimerae (Fig. 7). Also, as previously observed, the G83I mutation in the GpAtm decreases dimerization substantially relative to wt at all concentrations examined.
Figure 7.
Concentration-dependent dimerization of ToxR′(tm)MBP chimerae. E. coli MM39 expressing ToxR′(GpAwt)MBP (wt) or ToxR′(GpAG83I)MBP (G83I) under inducible control were induced at various levels and measured for CAT activity (expressed in arbitrary units) and chimera expression level (expressed as molecules of chimera per membrane surface area). •, ToxR′(GpAwt)MBP. ▵, ToxR′(GpAG83I)MBP.
Tunable control of the ToxR chimera provides significant advantages to the investigation of transmembrane dimerization. The regulation of chimera expression levels permits the comparison of associations at specified chimera concentrations. In addition, the use of an inducible promoter will allow dimerization to be measured as a function of chimera concentration, permitting a quantitative assessment of association in vivo.
Sequence Specificity of the GpAtm in a Natural Membrane.
The sequence specificity of GpAtm oligomerization was assayed by using the TOXCAT system and compared with previous assessments of dimerization in SDS micelles (3). Fig. 8 shows the CAT activities of a series of ToxR′(GpAtm)MBP mutants relative to ToxR′(GpAwt)MBP. Where available, the effect of each mutation on dimerization in SDS micelles is presented (3). Expression levels (immunoblots, Fig. 8) and correct membrane insertion (by malE complementation, data not shown) were measured for each mutant. As with G83I, expression levels were consistent, although a small and variable amount of cleaved MBP was observed (data not shown). Although this degradation may be related to the oligomeric state of the chimera, with monomeric species exhibiting heightened sensitivity to endogenous proteases (19), there was no correlation between dimerization (as reported by CAT activity) and the quantity of the high mobility species.
Figure 8.
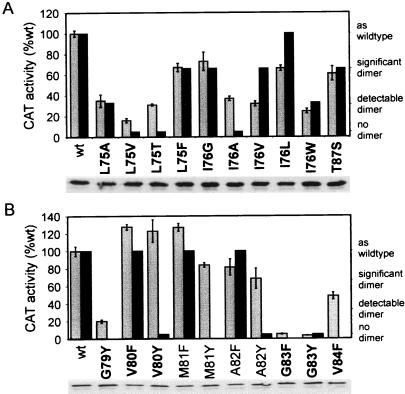
Mutations of GpAtm modulate dimerization in E. coli membranes. Numbering for transmembrane residues is taken from the full-length GpA sequence. Light bars represent 2–3 independent CAT assays (left axis), and SD is shown by error bars; dark bars are values for SDS/PAGE (right axis) from Lemmon et al. (3) except for M81F and A82F (K. R. MacKenzie, personal communication). Interfacial substitutions are shown in bold. Anti-MBP immunoblots of each mutant are shown below. (A) Mutations of interfacial residues L75, I76, and T87. (B) Mutagenesis of G79-V84 to F or Y, representing both interfacial and noninterfacial positions over a full turn of α-helix.
The relative dimerization of mutants at interfacial residues (L75, I76, G79, V80, G83, V84, and T87) varies from highly disruptive (G83Y) to greater than wt stability (V80F). The results of mutation in membranes follows the general trend of those observed in SDS micelles. For example, mutants of I76 and L75 exhibited a broad range of dimerization affinities in both systems (Fig. 8A), whereas substitutions at residues G79 and G83 always disrupted dimerization (Figs. 2, 3, and 9B).
Figure 9.
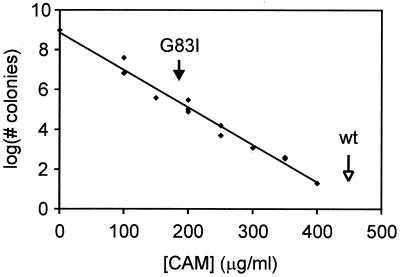
Selection of oligomeric tms from a randomized library. The logarithm of the number of colonies forming at varying CAM concentrations is reported. Arrows indicate the resistance of chimerae containing wt (open arrow) and G83I (filled arrow). R2 for the correlation is 0.98.
In contrast to the general agreement between data obtained in detergent micelles and membranes for hydrophobic substitutions, mutations to polar residues yielded strikingly different results. To directly test the effects of polar side chains, both phenylalanine and tyrosine were substituted at several contiguous positions. This pair of amino acids differs only in the presence of a hydroxyl group and should make similar van der Waals contacts at the dimer interface. If the polarity of the hydroxyl group does not play a role, mutation to either phenylalanine or tyrosine would be expected to have the same effect on dimerization. In all cases, however, mutation to tyrosine is disruptive when assayed in SDS micelles (even away from the dimer interface), whereas mutation to phenylalanine disrupts dimerization only at interfacial positions of the dimerization motif. In membranes, mutations to F and Y have almost identical effects at V80, A82, and G83, indicating that the addition of this polar group is not inherently disruptive in membranes. There is a significant difference between M81F and M81Y, however. Given that M81 is located away from the dimer interface, the effects of these mutations are not yet understood. Additional confirmation of the behavior of polar mutations in natural membranes is evident from mutant L75T. This mutation results in a moderately dimeric phenotype in vivo, as opposed to the complete disruption of the dimer seen in SDS micelles.
These data suggest that many properties of tm oligomerization are indeed conserved between the environments provided by detergent micelles and natural membranes. However, they also demonstrate that environment can significantly affect tm oligomerization. This observation underscores the importance of assaying folding in a native-like environment, especially when polar groups are present in the transmembrane helix.
Oligomeric Sequences Can Be Selected from a Library of tms.
As demonstrated by the disk diffusion assay (Fig. 2), the increased CAM resistance conferred by tm-mediated oligomerization can be used to select transmembrane sequences with a strong propensity to associate. Selection was applied to a library of ToxR′(tm)MBP chimerae containing randomized tm dimerization motifs. Sequences simultaneously were randomized by using a set of nine residues (glycine, alanine, valine, leucine, isoleucine, serine, threonine, proline, and arginine) at the seven interfacial positions identified in the GpA right-handed crossing of α-helices. Residues occupying noninterfacial positions were fixed as leucine.
Bacteria transformed with this library were plated on medium containing varying concentrations of CAM. Fig. 9 shows the number of colonies forming as a function of CAM concentration. The logarithmic decay in the number of colonies with increasing CAM concentration indicates that only a small subset of sequences encode strongly dimerizing tms. The observation that colony number decreases as a continuous function of CAM concentration suggests that a broad and continuous range of helix–helix affinities exists in this population of transmembrane sequences. These results demonstrate that the selection of novel transmembrane oligomerization domains from a library of randomized sequences is feasible by using TOXCAT.
DISCUSSION
The TOXCAT transmembrane dimer assay system was designed to discriminate between strongly and weakly associating transmembrane α-helices in a natural membrane—the E. coli cytoplasmic membrane. The accuracy of the ToxR report is reflected in the general agreement between this system and previous results in detergent micelles (3, 7). The results demonstrate that transcriptional activation of the CAT reporter gene depends on the extent of tm-driven dimerization, and that the GpAtm dimerization interface identified in detergent micelles also mediates association in natural membranes.
Other applications of the ToxR protein to the study of GpAtm-mediated dimerization (12, 20) have indicated the applicability of this general approach to the study of transmembrane helix–helix association. Our enhancement of this system has resulted in a more controlled assay with greater sensitivity than previously observed. TOXCAT also enables genetic selection. By using a single-copy, chromosomally integrated reporter, Langosch et al. (12) have measured only a slight difference between the wt GpAtm and mutants previously shown to be mostly disruptive (L75A) or completely disruptive (I76A) in SDS micelles. In our system, L75A and I76A are significantly destabilized relative to wt (35% and 37% of the wt signal, respectively). The observed increase in sensitivity is probably a consequence of the increased cellular concentration and mobility of the plasmid-borne reporter construct, allowing a more accurate comparison of association affinities. Furthermore, an ability to control the chimera concentration within the membrane may enable the measurement of association constants for the dimerization in a natural environment, providing insight into the relative energies associated with tm oligomerization in vivo.
Because protein-environment interactions may differ between detergents and membranes, it may be possible to use TOXCAT to study interactions that do not persist in detergent micelles. The failure of SDS/PAGE to accurately report the phenotypes of GpAtm mutations to residues with polar side chains illustrates the potential differences between these systems. The favorable free energy of inserting a hydrophobic α-helix across the membrane is sufficient to compensate for the cost of burying a polar side chain away from water (21). When compared with a membrane, the detergent micelle provides a significantly different environment, in that polar head groups form a shell around the hydrophobic portion of the micelle. The free energy cost of burying a tm containing a polar side chain in the hydrophobic interior of a detergent micelle could be eliminated by breaking the helix and exposing the polar groups on the side chain and the newly exposed main-chain polar groups to the external aqueous environment (9, 22). Such a deformation of the helix monomer in SDS micelles is expected to disrupt dimerization by distorting the helix backbone. The observations (3) that (i) most hydrophobic to polar mutations were disruptive in SDS, and (ii) mutations to strongly polar residues not only disrupted dimerization, but also caused anomalous monomer migration in SDS/PAGE both are consistent with the notion of polar mutation-induced helix deformation.
As a direct test of the ability of tms to accommodate polar groups, mutations of hydrophobic to polar residues were made in the GpAtm, assayed for dimerization in the E. coli inner membrane, and compared with previous results in SDS micelles (3). The use of ToxR to measure the response of GpAtm dimerization to mutation has revealed that polar side chains can be incorporated into tms without a serious disruption of structure, even at the dimer interface (L75T and V80Y). It has been postulated (23) that the folding of integral membrane proteins may involve the burial of polar groups into the protein interior. Although some transmembrane regions (24–26) may use polar groups at their association interface, the precise role of these residues has not been confirmed. The ability of this system to tolerate mutations to residues with polar side chains may facilitate further investigation of this hypothesis.
Although mutations to hydrophobic residues affect dimerization similarly in membrane and micelle environments, significant differences do exist. As with polar mutations, these inconsistencies may be caused by different protein-environment interactions. Energy terms describing the change in entropy of both protein and “solvent” upon dimerization are expected to differ between detergent micelles and lipid bilayers, because of the additional degrees of freedom available to individual micelles relative to one another. Differences also may arise from the potential for detergent acyl chains to insert between protein helices in orientations that are not accessible to lipid chains. Biological membranes provide additional rationales for the observed changes in association, because factors such as controlled tm orientation, volume exclusion by other proteins (27), and lipid acyl chain heterogeneity all are expected to affect helix–helix association.
Application of the TOXCAT assay to the study of transmembrane helix association in a natural membrane broadens both the scope of the kinds of interactions that can be studied and the range of observable energies. The high sensitivity of the system, coupled with its ability to measure stabilizing mutations (V80F, Fig. 8) make it ideally suited for analyzing the effects of mutation. The fact that substitutions of more polar amino acids are allowed significantly enhances the scope of the assay as well. Continued application and development should test in membranes ideas of helix–helix interactions based on studies in micelles.
Although TOXCAT is applicable to the study of interactions between individual transmembrane helices, perhaps its greatest advantage lies in the ability to select oligomerizing tms from a population of different sequences. An initial demonstration of this application comes from our observation that increasing concentrations of CAM select for strongly decreasing numbers of oligomerizing sequences from a randomized library of tms. The variation in CAM resistance reveals a range of helix affinities in the randomized library. Further characterization of isolates from different CAM concentrations should reveal novel oligomerization motifs and may identify existing oligomerization domains in naturally occurring membrane proteins. Selection also can be used in the isolation of pseudo-revertants that compensate for a given disruptive mutation to help distinguish between computationally derived models (28). By applying selective pressure to randomized tm libraries we can simultaneously assay a much greater number of sequences, thus extending our ability to probe the association interface.
Acknowledgments
We thank K. R. MacKenzie for the use of unpublished mutation results (Fig. 8); Drs. J. Beckwith, T. Griffin, H. Kolmar, L. Hanakahi, and N. Maizels for generous donation of reagents; the Engelman lab and L. Hanakahi for critical review of this manuscript, and the National Institutes of Health, National Science Foundation, and National Foundation for Cancer Research for funding.
ABBREVIATIONS
- CAT
chloramphenicol acetyltransferase
- MBP
maltose binding protein
- GpA
glycophorin A
- tm
transmembrane domain
- wt
wild type
- CAM
chloramphenicol
- Amp
ampicillin
- LB
Luria–Bertani
References
- 1.Popot J L, Engelman D M. Biochemistry. 1990;29:4031–4037. doi: 10.1021/bi00469a001. [DOI] [PubMed] [Google Scholar]
- 2.Furthmayr H, Marchesi V T. Biochemistry. 1976;15:1137–1144. doi: 10.1021/bi00650a028. [DOI] [PubMed] [Google Scholar]
- 3.Lemmon M A, Flanagan J M, Treutlein H R, Zhang J, Engelman D M. Biochemistry. 1992;31:12719–12725. doi: 10.1021/bi00166a002. [DOI] [PubMed] [Google Scholar]
- 4.Adams P D, Engelman D M, Brunger A T. Proteins. 1996;26:257–261. doi: 10.1002/(SICI)1097-0134(199611)26:3<257::AID-PROT2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 5.MacKenzie K R, Prestegard J H, Engelman D M. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 6.Fleming K G, Ackerman A L, Engelman D M. J Mol Biol. 1997;272:266–275. doi: 10.1006/jmbi.1997.1236. [DOI] [PubMed] [Google Scholar]
- 7.Lemmon M A, Treutlein H R, Adams P D, Brunger A T, Engelman D M. Nat Struct Biol. 1994;1:157–163. doi: 10.1038/nsb0394-157. [DOI] [PubMed] [Google Scholar]
- 8.Adair B D, Engelman D M. Biochemistry. 1994;33:5539–5544. doi: 10.1021/bi00184a024. [DOI] [PubMed] [Google Scholar]
- 9.MacKenzie K R, Engelman D M. Proc Natl Acad Sci USA. 1998;95:3583–3590. doi: 10.1073/pnas.95.7.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ottemann K M, Mekalanos J J. Mol Microbiol. 1995;15:719–731. doi: 10.1111/j.1365-2958.1995.tb02380.x. [DOI] [PubMed] [Google Scholar]
- 11.Kolmar H, Hennecke F, Gotze K, Janzer B, Vogt B, Mayer F, Fritz H J. EMBO J. 1995;14:3895–3904. doi: 10.1002/j.1460-2075.1995.tb00061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langosch D, Brosig B, Kolmar H, Fritz H J. J Mol Biol. 1996;263:525–530. doi: 10.1006/jmbi.1996.0595. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 14.Cronan J E, Gennis R B, Maloy S R. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt F C, editor. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1987. pp. 31–55. [Google Scholar]
- 15.Neidhardt F C. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhard F C, editor. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1987. pp. 3–6. [Google Scholar]
- 16.Chen H, Kendall D A. J Biol Chem. 1995;270:14115–14122. doi: 10.1074/jbc.270.23.14115. [DOI] [PubMed] [Google Scholar]
- 17.Ericsson, H., Hogman, C. & Wickman, K. (1954) Scand. J. Clin. Lab. Invest.11, Suppl., 23–36. [PubMed]
- 18.Lemmon M A, Flanagan J M, Hunt J F, Adair B D, Bormann B J, Dempsey C E, Engelman D M. J Biol Chem. 1992;267:7683–7689. [PubMed] [Google Scholar]
- 19.Leeds J A, Beckwith J. J Mol Biol. 1998;280:799–810. doi: 10.1006/jmbi.1998.1893. [DOI] [PubMed] [Google Scholar]
- 20.Brosig B, Langosch D. Protein Sci. 1998;7:1052–1056. doi: 10.1002/pro.5560070423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engelman D M, Steitz T A. Cell. 1981;23:411–422. doi: 10.1016/0092-8674(81)90136-7. [DOI] [PubMed] [Google Scholar]
- 22.Ibel K, May R P, Kirschner K, Szadkowski H, Mascher E, Lundahl P. Eur J Biochem. 1990;190:311–318. doi: 10.1111/j.1432-1033.1990.tb15578.x. [DOI] [PubMed] [Google Scholar]
- 23.Engelman D M, Zaccai G. Proc Natl Acad Sci USA. 1980;77:5894–5898. doi: 10.1073/pnas.77.10.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manolios N, Bonifacino J S, Klausner R D. Science. 1990;249:274–277. doi: 10.1126/science.2142801. [DOI] [PubMed] [Google Scholar]
- 25.Bargmann C I, Hung M C, Weinberg R A. Cell. 1986;45:649–657. doi: 10.1016/0092-8674(86)90779-8. [DOI] [PubMed] [Google Scholar]
- 26.Machamer C E, Grim M G, Esquela A, Chung S W, Rolls M, Ryan K, Swift A M. Mol Biol Cell. 1993;4:695–704. doi: 10.1091/mbc.4.7.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grasberger B, Minton A P, DeLisi C, Metzger H. Proc Natl Acad Sci USA. 1986;83:6258–6262. doi: 10.1073/pnas.83.17.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams P D, Lee A S, Brunger A T, Engelman D M. Ann NY Acad Sci. 1998;853:178–185. doi: 10.1111/j.1749-6632.1998.tb08265.x. [DOI] [PubMed] [Google Scholar]