Abstract
Pancreatic ductal adenocarcinoma (PDAC) remains an important cause of malignancy-related death and is the eighth most common cancer with the lowest overall 5-year relative survival rate. To identify new molecular markers and candidates for new therapeutic regimens, we investigated the gene expression profile of microdissected cells from 11 normal pancreatic ducts, 14 samples of PDAC, and 4 well-characterized pancreatic cancer cell lines using the Affymetrix U133 GeneChip set. RNA was extracted from microdissected samples and cell lines, amplified, and labeled using a repetitive in vitro transcription protocol. Differentially expressed genes were identified using the significance analysis of microarrays program. We found 616 differentially expressed genes. Within these, 140 were also identified in PDAC by others, such as Galectin-1, Galectin-3, and MT-SP2. We validated the differential expression of several genes (e.g., CENPF, MCM2, MCM7, RAMP, IRAK1, and PTTG1) in PDAC by immunohistochemistry and reverse transcription polymerase chain reaction. We present a whole genome expression study of microdissected tissues from PDAC, from microdissected normal ductal pancreatic cells and pancreatic cancer cell lines using highdensity microarrays. Within the panel of genes, we identified novel differentially expressed genes, which have not been associated with the pathogenesis of PDAC before.
Keywords: Pancreatic cancer, microarray, microdissection, IRAK1, MCM7
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is an important cause of malignancy-related deaths. In the United States, it ranks fifth among the leading causes of cancer death, accounting for approximately 30,000 deaths annually [1]. Apart from surgery, there is no effective therapy; but even most of the resected patients usually die within 1 year postoperatively. In the past years, several cancer-related genes have been identified in PDAC. The most frequently affected are K-ras, DPC4, p53, and p16 [2–5], all of which appear to play a role in the development of PDAC. However, considering the complexity of the genome, it is most likely that most of the molecular changes causing pancreatic cancer still need to be elucidated [6].
Recently, DNA microarray technology has been applied to a number of tumors of, for example, the breast [7], colon [8], prostate [9], esophagus [10], stomach [11], and pancreas [12–17]. These studies generated large sets of new class II cancer genes revealing dysregulation at the level of gene expression [18]. However, most of these studies were performed on whole tissue samples or cell lines. In cell lines, in vitro conditions may induce changes in gene expression that are not present in vivo. PDAC specimens contain different cell types, including ductal, acinar, islet, inflammatory, and nerve cells, and fibrocystic elements. When whole tissues are used in such studies, expression profiles may represent both the tumor and the adjacent non-neoplastic tissue. Therefore, microdissection is the method of choice to generate a true picture of gene expression changes [19]. Improvement in the array technology made it possible to examine virtually every gene at single gene level by expression profiling. In this study, we used microdissected tumor tissue and microdissected normal ductal epithelium for RNA extraction and subsequent analysis using the Affymetrix U133 set (GeneChips A and B; Affymetrix, Santa Clara, CA), which contains 45,000 fragments corresponding to 33,000 known genes and 6000 expressed sequence tags (ESTs), and therefore approximately the whole genome.
We identified genes whose expression levels differed significantly between malignant and benign pancreatic cells of the ductal phenotype to generate a set of genes that may be used as diagnostic markers or as targets for new therapeutic approaches. Furthermore, we validated genes of this set to prove the appropriateness of our approach.
Materials and Methods
Patients and Tissues
Freshly frozen tissue samples of PDAC (n = 14) were obtained from surgical specimens from patients who were operated at the Department of Visceral, Thoracic, and Vascular Surgery, University Hospital Carl Gustav Carus, Technical University of Dresden (Dresden, Germany) and the Department of General Surgery, University of Kiel (Kiel, Germany) between 1996 and 2003. The clinical data of these patients are shown in Table 1. Normal pancreatic tissue was obtained from 11 patients who underwent pancreatic resection for other pancreatic diseases. These tissues were histologically normal tissues with no visible dysplastic changes in the ducts and were taken from the distal parts of the resected pancreas. Prior to surgery, all patients had given informed consent, which had been approved by the local ethics committee. Immediately after surgical removal, the specimens were sectioned and microscopically evaluated. Suitable samples of tumor tissue or normal tissue were snap frozen in liquid nitrogen and stored at -80°C until further processing.
Table 1.
Clinicopathologic Data of 14 Patients with PDAC.
| Code | Origin | Gender | Age (years) | Histology | T | N | M | Grading |
| PDT14 | Dresden | Female | 57 | PDAC, liver metastasis | X | X | 1 | X |
| PT10 | Dresden | Female | 62 | PDAC | 3 | 1b | 0 | 2 |
| PT55 | Dresden | Male | 66 | PDAC with anaplastic component | 3 | 1b | 0 | 3 |
| 39B | Dresden | Male | 59 | PDAC | 2 | 0 | 0 | 2 |
| 33B | Dresden | Female | 73 | PDAC | 3 | 1b | 0 | 3 |
| 35B | Dresden | Male | 74 | PDAC | 2 | 0 | 0 | 3 |
| 56A | Dresden | Female | 71 | PDAC | 3 | 1a | 0 | 3 |
| PDT12 | Dresden | Female | 74 | PDAC | 3 | 1b | 0 | 1 |
| PDT9 | Dresden | Male | 67 | PDAC, liver metastasis | X | X | 1 | 2 |
| PDT5 | Dresden | Male | 61 | PDAC, liver metastasis | X | X | 1 | 3 |
| PT44 | Dresden | Male | 64 | PDAC involving ampulla of Vater | 3 | 0 | 0 | 3 |
| PKT5 | Kiel | Female | 69 | PDAC | 3 | 1 | 0 | 2 |
| PKT9 | Kiel | Male | 71 | PDAC | 3 | 1 | 0 | 3 |
| PKT4 | Kiel | Female | 66 | PDAC | 3 | 1 | 0 | 2 |
X: at time of operation.
Microdissection
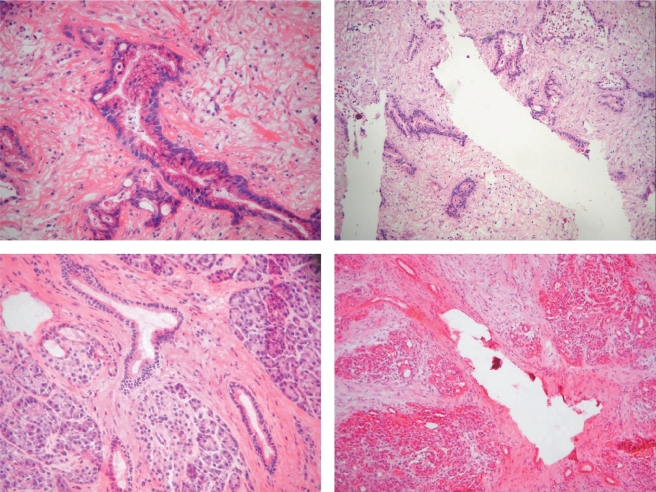
Frozen tissue specimens were cut into 10-µm-thick sections and immediately fixed on slides in 70% ethanol. The sections were briefly stained with hematoxylin and eosin (H&E), and coverslipped. Suitable areas for microdissection were marked on these slides serving as a template. The tissue blocks were serially cut to 5-µm-thin sections, briefly fixed in 70% RNase-free ethanol, and stained with H&E. PDAC cells and normal ductal cells were dissected manually using a sterile injection needle (Figure 1). The estimated cellularity was 10,000 to 11,000 cells per microdissected sample. The cellularity of the dissections was approximately 95%. These cells were pooled in ice-cooled guanadine thiocyanate (GTC) buffer (Promega, Heidelberg, Germany) for further RNA preparation.
Figure 1.
Manual microdissection of pancreatic tissue. Left: Before microdissection; right: after microdissection; upper panel: pancreatic ductal adenocarcinoma; lower panel: normal ductal epithelia.
Cell Culture
The pancreatic cell lines Colo357, PancTUI, PT45, Panc89, CAPAN2, HPAF-II, BxPC3, CAPAN1, PaCa44, CFPAC-1, PT64, PT89, PT96, PT115, PT101, PT103, R89, ASPC1, MiaPaCa2, and Panc1 were cultured in RPMI 1640 supplemented with 10% fetal bovine serum, 2 mM glutamine, nonessential amino acids (5 ml/l), penicillin (10,000 U/ml), and streptomycin (10 mg/ml), and passaged before they reached confluency. All cell culture materials were obtained from Invitrogen (Karlsruhe, Germany).
RNA Preparation and Array Hybridization
Poly A+ RNA from the microdissected surgical specimens and cell cultures was prepared using the PolyATtract 1000 kit (Promega) according to the manufacturer's recommendations. For each sample, cDNA synthesis and repetitive in vitro transcription were performed three times, as described previously [15]. In brief, first-strand cDNA synthesis was initiated using the Affymetrix T7-oligo-dT promoter-primer combination at 0.1 mM. The second-strand cDNA synthesis was generated with internal priming. In vitro transcription was performed using Ambion's Megascript kit (Ambion, Huntington, UK), as recommended by the manufacturer. From the generated aRNA, a new first-strand synthesis was initiated using 0.025mMof a random hexamer as primer. After completion, the second-strand synthesis was performed using the Affymetrix T7-oligo-dT promoter-primer combination as primer at a concentration of 0.1 mM. A second in vitro transcription was performed and then the procedure was repeated one additional time. During the last in vitro transcription, biotin-labeled nucleotides were incorporated into the aRNA, as recommended by the Affymetrix protocol. RNA amplification after each round of amplification was 50 to 100, and the correlation coefficient of gene expression profiles between the starting RNA and the amplified RNA is 0.77 to 0.79 [20]. Hybridization and detection of the labeled aRNA on the U133 A/B Affymetrix GeneChip set were performed according to Affymetrix's instructions.
Chip Design and Bioinformatics Analysis
The U133 A/B Affymetrix GeneChip set used in this study consists of more than 44,000 probe sets resembling roughly 33,000 genes and 6000 ESTs. The Cel Files obtained from the Affymetrix MAS 5.0 software were used for further analysis. The files were loaded into dChip 1.3 (http://www.dchip.org) then normalized, and expression values as well as absolute calls were calculated using the PM/MM model [21]. The expression values and absolute calls were exported and further explored using SAM ("http://www-stat.stanford.edu/∼tibs/SAM/) [22] and Excel (Microsoft, Redmond, WA). We scored genes as differentially expressed if they met the following criteria: a fold change > 2 and a q value < 15%, or presence call in at least of 60% of one tissue type but not within the other type (Figure 2).
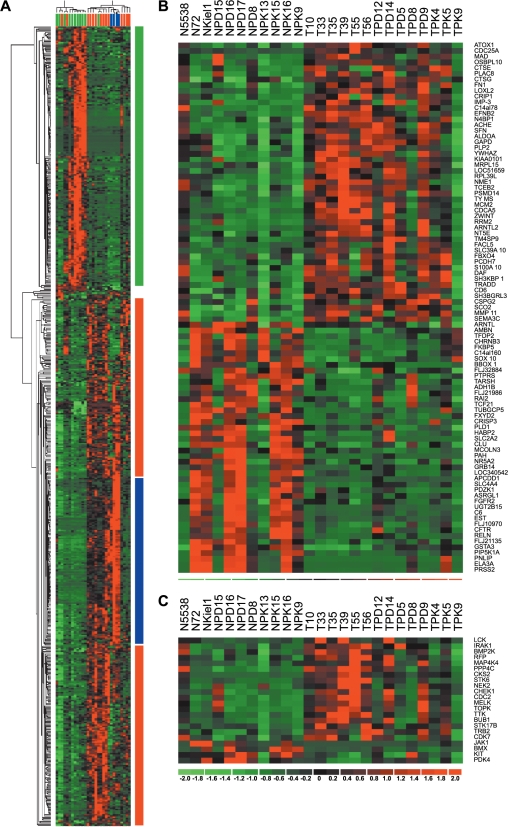
Figure 2.
Analysis of gene expression in PDAC. (A) Hierarchical clustering of 14 microdissected PDACs, 11 microdissected normal ductal cells, and 4 established pancreatic tumor cell lines using the 616 differential gene set and a Euclidian distance matrix. The different colors reflect the predominant expression of the genes (green: normal tissue; blue: cell lines and PDAC; red: PDAC). The individual samples are colored (green: normal; blue: cell lines; red: PDAC). (B) Heat map of signature genes in PDAC. Genes were identified using the KNN method with a LOO validation step. (C) Heat map of protein kinase expression found in the set of differentially expressed genes. Genes that are upregulated appear in red, and those that are downregulated appear in green, with the expression value reflected by the intensity of the color.
To identify signature genes, normalized gene expression values were loaded into Genecluster2 (http://www-genome.wi.mit.edu/cancer/software/genecluster2/gc2.html) [23]. Low expression values were floored to 10 and only those probesets fulfilling the criteria of Max/Min > 3 and Max-Min > 200 were used. Signature genes were identified using the K-nearest neighbor (KNN) method with a leave-one-out (LOO) validation step. Hierarchical clustering was performed using dChip (Figure 2).
Reverse Transcription Polymerase Chain Reaction (RT-PCR)
RNA from normal pancreatic tissue and pancreatic tumor tissue was isolated using the “Micro to Mini Total RNA Purification Kit” (Invitrogen) according to the manufacturer's procedure. Reverse transcription using random hexamers and “Superscript” reverse transcriptase (Invitrogen) followed by PCR amplification (58°C annealing temperature, 27 cycles) was performed under standard PCR conditions. Primers for PCR amplification were synthesized (MWGBiotech, Ebersberg, Germany) (Table 2). PCR products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining.
Table 2.
Primer and Probe Sequences Used for the Verification of Differential Expression.
| Gene | Sense 5′-3′ | Antisense 5′-3′ |
| GAPDH | CCAGCCGAGCCACATCGCTC | ATGAGCCCCAGCCTTCTCCAT |
| G6PD | ACGTGATGCAGAACCACCTACTG | ACGACGGCTGCAAAAGTGGCG |
| CCNB1 | GATATCTATCAGTATCTCAGGCAGCTG | ATACTTGGAAGCCAAGAGCAGAGC |
| CCNB2 | GATATCTATCAGTATCTCAGGCAGCTG | ATACTTGGAAGCCAAGAGCAGAGC |
| CENPF | GCGGCAGAAAAGAAACAGAC | TCTTCTGTGTCGATGCCAAG |
| CFLAR | ATGGACAGAAAAGCTGTGGAG | CTTCAGGTCTATTCTGTGGATG |
| EFNB2 | CTGCTGGATCAACCAGGAAT | CTGTTGCCGTCTGTGCTAGA |
| HOXC6 | ACAGACCTCAATCGCTCAGG | GGTACCGCGAGTAGATCTGG |
| LOXL2 | CAGACCACCTACCTGGAGGA | GTTGTGGATCTGGGAGGAGA |
| MADD | TGTGCAGGACCTGAAGACTG | ACAAAGACGCCTCGAACTGT |
| osf-2 | TGCATTATTCACAGGTGCCAG | ACTCTCCAGTGTTCTGAGTC |
| PTTG1 | AAGGAAAATGGAGAACCAGGC | GCTTGGCTGTTTTTGTTTGAGG |
| RAMP | ACAGCAGCAGGTGATCAAACAGC | GAAGGAGCAGAGTCCTTTTGAATTCTG |
| PLAG1 | TTTCCTTGCCAACTGTGTGAC | CTTTGTTAGGGTCGTGTGTATG |
| ECT2 | ACTAGCTTGGCAGACTCTTC | ATCCTGAAAGTCCGTGACTAC |
| UP | CGGAAAACGGACCTTAACAA | GATACGCCTGCTTGTCCTTC |
Total RNA from the pancreatic cell lines was isolated using the RNeasy RNA isolation kit (Qiagen, Hilden, Germany) and transcribed into cDNA as described above. Quantitative PCR detection was applied using an ABI Sequence Detection System 7700 and the Sybr Green Master Mix (ABI, Weiterstadt, Germany) according to the manufacturer's recommendation.
Immunohistochemistry
For immunohistochemistry, 10 cases of PDAC were randomly selected and 5-µm-thin sections were prepared. Mouse monoclonal antibodies against human CENPF (clone D8; Abcam Ltd., Cambridge, UK), human MCM2 (clone CRCT2; Novocastra, Newcastle, UK), and human MCM7 (clone CRCT7; Novocastra) were stained with the ABC method as described previously [24]. The staining intensity was semiquantitatively assessed using a cutoff point of more than 10% positive cells with either cytoplasmic or nuclear staining. The staining intensity was recorded as weak or strong. Normal pancreatic parenchyma was also screened for the presence of protein expression in acinar, ductal, and endocrine cells.
Results
Using microdissection, tissues from 14 patients with PDAC and normal ductal epithelia from 11 patients were obtained. Gene expression profiles of these tissues were acquired and compared.
The combination of fold change (cutoff: two-fold), SAM q value (cutoff < 15%), and absolute call (cutoff: present in at least 60% of one tissue type, absent in the other) analysis yielded a total of 616 differentially expressed genes. Overall, 204 genes were underexpressed and 412 were overexpressed in PDAC compared to microdissected normal ductal cells. In Table 3, all genes with a fold change of 3 or more and a q value < 5% (118 in total, 66 overexpressed and 52 underexpressed) are shown. Hierarchical clustering using this set of genes of the samples revealed three distinct clusters of primary tumors, benign tissues, and cell lines showing the diversity between primary tissue samples and established cell lines (Figure 2A).
Table 3.
Differentially Expressed Genes in Microdissected PDAC and Microdissected Normal Ductal Cells Using High-Density Microarrays.
| Affymetrix ID | Unigene ID | Gene Symbol | Fold Change | Reference PDAC | Reference Cancer | N Array | T Array | Reference Array in PDAC |
| (A) Upregulated genes | ||||||||
| 218741_at | Hs.208912 | MGC861 | 18.15 | 0 | 0 | |||
| 218542_at | Hs.14559 | C10orf3 | 9.03 | 0 | 2 | [17,31] | ||
| 208511_at | Hs.350968 | PTTG3 | 8.41 | X | 0 | 0 | ||
| 203454_s_at | Hs.279910 | ATOX1 | 8.13 | 0 | 0 | |||
| 202478_at | Hs.155418 | GS3955 | 7.86 | 0 | 1 | [17] | ||
| 203819_s_at | Hs.79440 | KOC1 | 7.42 | X | X | 0 | 2 | [16,31] |
| 221521_s_at | Hs.433180 | PSF2 | 6.97 | 0 | 0 | |||
| 205909_at | Hs.99185 | POLE2 | 6.59 | 0 | 0 | |||
| 200755_s_at | Hs.7753 | CALU | 5.53 | 0 | 0 | |||
| 204170_s_at | Hs.83758 | CKS2 | 5.45 | X | 0 | 4 | [15,17,32,37] | |
| 212398_at | Hs.263671 | RDX | 5.24 | X | 0 | 0 | ||
| 204822_at | Hs.169840 | TTK | 5.18 | X | 0 | 0 | ||
| 209642_at | Hs.98658 | BUB1 | 5.17 | X | X | 0 | 0 | |
| 210052_s_at | Hs.9329 | C20orf1 | 4.91 | X | 0 | 0 | ||
| 202107_s_at | Hs.57101 | MCM2 | 4.83 | X | 0 | 0 | ||
| 209172_s_at | Hs.77204 | CENPF | 4.77 | X | 0 | 0 | ||
| 203108_at | Hs.194691 | RAI3 | 4.74 | X | 0 | 5 | [14–17,31] | |
| 201467_s_at | Hs.406515 | NQO1 | 4.69 | X | 0 | 3 | [17,37,38] | |
| 218960_at | Hs.63325 | TMPRSS4 | 4.64 | 0 | 0 | |||
| 208932_at | Hs.2903 | PPP4C | 4.54 | 0 | 0 | |||
| 214710_s_at | Hs.23960 | CCNB1 | 4.46 | X | 0 | 1 | [16] | |
| 212992_at | Hs.57548 | LOC113146 | 4.42 | 0 | 0 | |||
| 208079_s_at | Hs.250822 | STK6 | 4.38 | X | 0 | 2 | [15,32] | |
| 208002_s_at | Hs.8679 | BACH | 4.27 | 0 | 0 | |||
| 201890_at | Hs.75319 | RRM2 | 4.24 | X | 0 | 0 | ||
| 218886_at | Hs.52256 | FLJ20624 | 4.24 | 0 | 0 | |||
| 206074_s_at | Hs.57301 | HMGA1 | 4.11 | X | X | 0 | 1 | [32] |
| 203764_at | Hs.77695 | DLG7 | 4.01 | 0 | 0 | |||
| 207165_at | Hs.72550 | HMMR | 3.98 | X | X | 0 | 0 | |
| 204507_s_at | Hs.278540 | PPP3R1 | 3.86 | 0 | 0 | |||
| 218585_s_at | Hs.126774 | RAMP | 3.79 | X | 0 | 0 | ||
| 219148_at | Hs.104741 | TOPK | 3.68 | X | 0 | 0 | ||
| 201292_at | Hs.156346 | TOP2A | 3.68 | X | 0 | 2 | [16,31] | |
| 202589_at | Hs.29475 | TYMS | 3.65 | X | 0 | 0 | ||
| 209405_s_at | Hs.289108 | FAM3A | 3.61 | 0 | 0 | |||
| 206858_s_at | Hs.820 | HOXC6 | 3.55 | X | 0 | 0 | ||
| 203213_at | Hs.334562 | CDC2 | 3.52 | X | X | 0 | 2 | [14,16] |
| 220060_s_at | Hs.121553 | FLJ20641 | 3.48 | 0 | 0 | |||
| 202784_s_at | Hs.18136 | NNT | 3.43 | 0 | 0 | |||
| 203939_at | Hs.153952 | NT5E | 3.38 | 0 | 0 | |||
| 205167_s_at | Hs.656 | CDC25C | 3.36 | X | 0 | 0 | ||
| 204026_s_at | Hs.42650 | ZWINT | 3.34 | 0 | 0 | |||
| 220658_s_at | Hs.222024 | ARNTL2 | 3.31 | 0 | 2 | [16,31] | ||
| 203234_at | Hs.77573 | UP | 3.31 | 0 | 0 | |||
| 210809_s_at | Hs.136348 | osf-2 | 3.31 | X | 0 | 3 | [13,31,37] | |
| 202998_s_at | Hs.83354 | LOXL2 | 3.28 | X | 0 | 1 | [17] | |
| 203878_s_at | Hs.155324 | MMP11 | 3.28 | X | 0 | 2 | [13,16] | |
| 204962_s_at | Hs.1594 | CENPA | 3.25 | 0 | 0 | |||
| 202668_at | Hs.30942 | EFNB2 | 3.25 | X | 0 | 0 | ||
| 201195_s_at | Hs.184601 | SLC7A5 | 3.13 | X | 0 | 1 | [31] | |
| 204351_at | Hs.2962 | S100P | 3.12 | X | X | 0 | 7 | [13,14,16,17,31,37,38] |
| 202705_at | Hs.194698 | CCNB2 | 3.07 | X | 0 | 0 | ||
| 201850_at | Hs.82422 | CAPG | 3.05 | 0 | 3 | [14,16,31] | ||
| 205081_at | Hs.423190 | CRIP1 | 3.02 | X | 0 | 1 | [13] | |
| 200085_s_at | Hs.172772 | TCEB2 | 3.02 | 0 | 0 | |||
| 204825_at | Hs.184339 | MELK | 3 | 0 | 0 | |||
| 202870_s_at | Hs.82906 | CDC20 | Only in T | X | X | 0 | 2 | [15,17] |
| 203596_s_at | Hs.27610 | RI58 | Only in T | X | 0 | 0 | ||
| 203870_at | Hs.109268 | FLJ12552 | Only in T | 0 | 0 | |||
| 204948_s_at | Hs.9914 | FST | Only in T | X | 0 | 0 | ||
| 205653_at | Hs.100764 | CTSG | Only in T | 0 | 0 | |||
| 210252_s_at | Hs.82548 | MADD | Only in T | X | 0 | 0 | ||
| 218355_at | Hs.279766 | KIF4A | Only in T | 0 | 0 | |||
| 218663_at | Hs.193602 | HCAP-G | Only in T | X | 0 | 0 | ||
| 219182_at | Hs.6853 | FLJ22167 | Only in T | 0 | 0 | |||
| 49452_at | Hs.234898 | EST | Only in T | 0 | 0 | |||
| (B) Downregulated genes | ||||||||
| 225016_at | Hs.374481 | DRAPC1 | 0.1 | X | 1 | 0 | [17] | |
| 206464_at | Hs.27372 | BMX | 0.1 | X | 1 | 0 | [17] | |
| 220275_at | Hs.114648 | ERG-1 | 0.1 | 0 | 0 | |||
| 217452_s_at | Hs.181353 | B3GALT2 | 0.1 | 0 | 0 | |||
| 206131_at | Hs.1340 | CLPS | 0.04 | 3 | 0 | [15,17,37] | ||
| 205910_s_at | Hs.406160 | CEL | 0.04 | 1 | 0 | [37] | ||
| 206827_s_at | Hs.302740 | TRPV6 | 0.04 | X | 0 | 0 | ||
| 210168_at | Hs.1282 | C6 | 0.04 | X | 1 | 0 | [13] | |
| 205971_s_at | Hs.74502 | CTRB1 | 0.07 | X | 1 | 0 | [15] | |
| 207077_at | Hs.169234 | ELA2B | 0.07 | 1 | 0 | [13] | ||
| 205719_s_at | Hs.1870 | PAH | 0.08 | 0 | 0 | |||
| 206694_at | Hs.73923 | PNLIPRP1 | 0.08 | X | 1 | 0 | [37] | |
| 216687_x_at | Hs.150207 | UGT2B15 | 0.09 | X | 0 | 0 | ||
| 203924_at | Hs.89552 | GSTA1 | 0.09 | X | 1 | 0 | [13] | |
| 206297_at | Hs.8709 | CTRC | 0.09 | 0 | 0 | |||
| 206212_at | Hs.89717 | CPA2 | 0.11 | 2 | 0 | [17,37] | ||
| 206446_s_at | Hs.21 | ELA2A | 0.11 | 1 | 0 | [17] | ||
| 210080_x_at | Hs.181289 | ELA3A | 0.12 | 0 | 0 | |||
| 213071_at | Hs.80552 | DPT | 0.12 | 0 | 0 | |||
| 206311_s_at | Hs.992 | PLA2G1B | 0.13 | X | X | 2 | 0 | [13], [15] |
| 219564_at | Hs.50151 | KCNJ16 | 0.14 | 0 | 0 | |||
| 210262_at | Hs.2042 | TPX1 | 0.14 | 0 | 0 | |||
| 207636_at | Hs.158308 | SERPINI2 | 0.14 | X | X | 0 | 0 | |
| 216699_s_at | Hs.123107 | KLK1 | 0.14 | X | 3 | 0 | [37], [19] | |
| 215563_s_at | Hs.278657 | MSTP9 | 0.15 | 0 | 0 | |||
| 214324_at | Hs.53985 | GP2 | 0.16 | 1 | 0 | [37] | ||
| 211738_x_at | Hs.425790 | ELA3B | 0.16 | 0 | 0 | |||
| 208498_s_at | Hs.274376 | AMY1A | 0.17 | X | 0 | 0 | ||
| 205771_s_at | Hs.12835 | AKAP7 | 0.18 | 0 | 0 | |||
| 205869_at | Hs.241395 | PRSS1 | 0.18 | X | 0 | 0 | ||
| 205912_at | Hs.102876 | PNLIP | 0.19 | 2 | 0 | [17,37] | ||
| 208450_at | Hs.113987 | LGALS2 | 0.19 | X | 2 | 0 | [13,17] | |
| 205509_at | Hs.180884 | CPB1 | 0.19 | 0 | 0 | |||
| 211766_s_at | Hs.143113 | PNLIPRP2 | 0.19 | 0 | 0 | |||
| 229963_at | Hs.47209 | EST | 0.19 | 0 | 0 | |||
| 207434_s_at | Hs.19520 | FXYD2 | 0.19 | 1 | 0 | [13] | ||
| 205799_s_at | Hs.239106 | SLC3A1 | 0.21 | 0 | 0 | |||
| 205923_at | Hs.12246 | RELN | 0.22 | X | 0 | 0 | ||
| 206262_at | Hs.2523 | ADH1C | 0.23 | X | 1 | 0 | [17] | |
| 206610_s_at | Hs.1430 | F11 | 0.23 | 0 | 0 | |||
| 205363_at | Hs.9667 | BBOX1 | 0.23 | 0 | 0 | |||
| 203908_at | Hs.5462 | SLC4A4 | 0.25 | 2 | 0 | [13,17] | ||
| 204359_at | Hs.48998 | FLRT2 | 0.25 | X | 0 | 0 | ||
| 213436_at | Hs.75110 | CNR1 | 0.27 | X | 0 | 0 | ||
| 205380_at | Hs.15456 | PDZK1 | 0.29 | X | 1 | 0 | [17] | |
| 206204_at | Hs.83070 | GRB14 | 0.29 | X | 0 | 0 | ||
| 244402_at | Hs.110 | KIAA0436 | 0.31 | 0 | 0 | |||
| 206010_at | Hs.241363 | HABP2 | 0.32 | X | 2 | 0 | [37] | |
| 208741_at | Hs.23964 | SAP18 | Only in N | X | 0 | 0 | ||
| 209773_s_at | Hs.75319 | RRM2 | Only in N | X | 0 | 0 | ||
| 214373_at | Hs.356686 | PPP4R2 | Only in N | 0 | 0 | |||
| 235132_at | Hs.296995 | EST | Only in N | 0 | 0 | |||
Differentially expressed genes with their Unigene Cluster ID, gene symbol, fold change tumor/normal, and references concerning the stated differential expression in previous expression profiling experiments in PDAC [13–17,31,32,37–39].
CF T/N: the change folds of the mean expression values of 14 microdissected PDAC against the mean of 11 microdissected normal pancreatic ductal tissues.
N array: overexpressed in the normal pancreatic tissue in the specified paper.
T array: overexpressed in PDAC in the specified paper.
X: a reference found January 2004 in PubMed database. Only genes with a fold change of at least three are included in the table.
We applied KNN analysis with LOO validation to identify pancreatic cancer signature genes. We detected a set of 104 probesets, which enabled us to discriminate between tumor tissue and normal tissue with a specificity of 73% (three normal tissues within the tumor group) and a sensitivity of 93% (one tumor tissue within the normal group; Figure 2B).
Analysis of the differentially expressed genes using the Geneontology system of molecular function (http://www.geneontology.org) revealed several distinct groups (data not shown). Interestingly, 23 of 616 genes were grouped into the family of protein kinases (Figure 2C). Of these, 19 were overexpressed in PDAC, whereas four were underexpressed. Within the group of overexpressed protein kinases, we identified STK6 (STK15/Aurora), which already has been reported by others [25], and IRAK1, an inducer of NF-κB [26]. Moreover, the protein kinase KIT that is implicated in the development of gastrointestinal stromal tumors was found to be downregulated in PDAC. This implies that KIT, in contrast to STK6/STK15, might not be a target in PDAC therapy.
Comparison of Expression Data
To interpret our results in a general context, we analyzed the data already published on the 616 genes. We found that 163 of 616 genes already have been reported by other groups, which analyzed PDAC with microarrays (Table 3). One hundred four of the upregulated genes in our set have been reported before. Among them was S100P, which has been found in seven independent experiments. Fifty-nine from the downregulated genes of the set have been found in other expression profiling experiments. Among them were GATM and KLK1, which have been found in four and three other investigations, respectively. No discrepant finding concerning the direction of differential expression between our results and those of others has been found.
Interestingly, the majority of genes (453 of 616) was not reported by other groups, possibly showing the advantages of the microdissection approach. This is especially true for genes that are downregulated in PDAC because of the composition of normal pancreatic tissue where the ductal epithelia comprise only around 5% of the cells.
Verification of Differential Expression
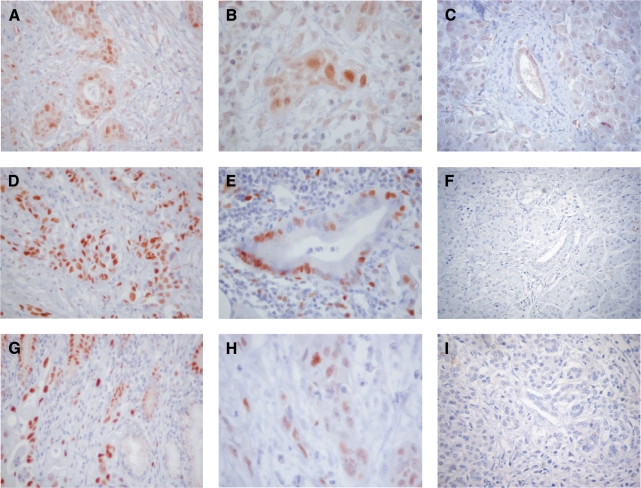
From the 616 differentially expressed genes, we selected several for further validation by immunohistochemistry and/or RT-PCR. We found that for the majority of genes, the differential expression could be confirmed by another method. Immunohistochemical analysis revealed that CENPF as well as MCM2 and MCM7 were expressed in 9 of 10 PDACs with mainly strong nuclear expression (Table 4, Figure 3), whereas the normal pancreas lacked specific reactions. In particular, normal duct epithelia, as well as acinar cells, were unreactive.
Table 4.
Results of Immunohistochemical Stainings in PDAC.
| Gene Product | PDAC Nuclear | PDAC Cytoplasmic | Normal Duct Cells | Acinar Cells | Endocrine Cells |
| CENPF | 9/10 | 2/10 | 0/10 | 1/10 | 0/10 |
| MCM2 | 7/10 | 1/10 | 1/10 | 0/10 | 0/10 |
| MCM7 | 9/10 | 0/10 | 1/10 | 5/10 | 1/10 |
Figure 3.
Immunohistochemical analysis of differentially expressed genes found in PDAC by microarray analysis. CENPF (A–C), MCM7 (D–F), and MCM2 (G–I) are highly expressed in the nucleus of PDACs of different grades (D: G1; A and E: G2; B, C, and F: G3; C, F, and I: normal pancreas). Original magnification, x100 (A, D, and E). Original magnification, x200 (C). Original magnification, x400 (B and F).
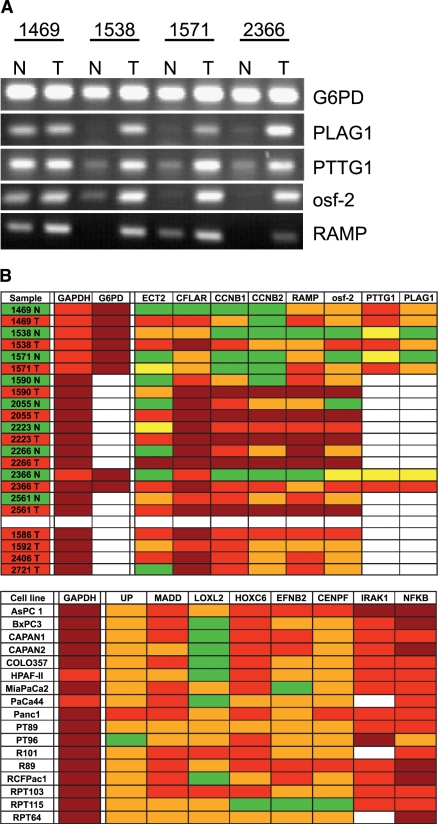
For RT-PCR analysis, 9 normal tissues and 13 tissues from PDAC (nine of them as corresponding pairs) from individual tumors have been investigated for expression of the selected genes (Figure 4, A and B). Out of 62 PCR reactions from the normal/tumor tissue pairs, 51 reactions showed an upregulation of gene expression in tumor tissues when compared to normal tissues, whereas 11 PCR reactions did not show this outcome. This is because not all individual tumor samples showed an upregulation of the tested gene transcripts, whereas others did. Although some genes are clearly upregulated in virtually every tumor sample (e.g., RAMP), other ones are not (e.g., CCNB2). These results confirm our approach. However, it indicates the fact that gene expression is heterogeneous within tumors. This is also underlined by our observation that genes identified by gene expression profiling are heterogeneously expressed in analyzed cell lines, such as LOXL2, which is expressed mainly in the primary cell lines. We also found a high expression of IRAK1 in all cell lines tested (Figure 4B).
Figure 4.
Validation of differentially expressed genes using RT-PCR. (A) RT-PCR analysis of five genes. RNA from tumor (T) and corresponding normal tissue (N) from four patients with a PDAC was isolated, and RT-PCR analysis for G6PD (control), PLAG1, PTTG, osf-2, and RAMP1 was performed. PCR products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. (B) Overview of RT-PCR results from normal pancreatic tissue, PDAC (upper panel), and cell lines (lower panel). GAPDH and G6PD expression analysis served as control. White background: not determined; green: no PCR signal (-), cT value > 35; yellow: PCR signal visible (-/+); orange: clear PCR signal (+), cT value > 27< 35; red: strong PCR signal (++), cT value > 20 < 27; dark red: very strong RT-PCR signal (+++), cT value < 20.
In parallel to RT-PCR, we performed Western blot analysis with three corresponding normal/tumor tissue pairs to investigate the expression pattern of CCNB1 (cyclin B1) and CDC25B. Although the expression intensity between the tested samples varied, an upregulation of both proteins in tumor tissues was verifiable, except in one sample with comparable expression (data not shown).
Discussion
Using the Affymetrix U133 GeneChip set, we performed a whole genome gene expression analysis of microdissected cells of PDAC and microdissected normal ductal epithelia of the pancreas. Because the stromal portion in PDAC often exceeds that of neoplastic cells, we carefully microdissected the tumor tissue, employing manual microdissection, and obtained highly homogeneous cell compartments. The same holds true for the dissection of normal pancreatic duct cells, which were selectively removed from large-sized and medium-sized ducts. The fact that there is a need to distinguish neoplastic from non-neoplastic tissues is shown by studies on prostate carcinomas comparing the differential gene expression in dissected and nondissected tissues. Of the differential genes identified in microdissected tissues, only one third was also found in bulk tissues [27]. Generally, microdissection results in smaller amounts of poly A+ RNA. We therefore applied a linear amplification protocol with minimal bias as already described by us and other groups [20,28].
We compared the obtained gene expression profiles with each other as well as with the expression profiles of pancreatic cancer cell lines, and we identified 616 differentially expressed genes. We found 412 to be upregulated and 204 genes to be downregulated in PDAC. A number of these genes were already reported by other groups, which conducted global gene expression studies in PDAC [12,14,16,29–36]. However, we also identified additional genes that have not yet been found to be differentially expressed in pancreatic cancer but were already identified within other malignancies. The third and largest group of genes consists of genes that have not been associated with any type of carcinoma so far.
Comparing our results with other studies employing largescale gene expression analysis of PDAC using DNA microarrays [13–16,31,32,37–39], we found that only a few genes were shown to be differentially expressed in more than one study (Table 3). There are several potential reasons for the low concordance of these studies. First, the type, histology, and number of samples used (i.e., established cell lines or primary tissue) differed. This could be even more important for the type of normal tissue used (commercially available RNA, normal tissue from resected pancreatic tumors, or donor organs). Second, microdissection for large-scale gene expression analysis was applied only by two groups [15,17]. Third, different arrays and array technologies may lead to different gene expression results [40]. Fourth, there is no common standard to assign differential expression within a gene expression profiling experiment. Furthermore, there are different implementations of standard techniques leading to different results. For these reasons, the results of expression profiling studies are intrinsic, subjective, and not easily comparable. However, all these studies lead to sets of candidate genes for further investigations as potential markers and/or therapeutic targets.
From the genes that were overexpressed in PDAC, we chose CCNB1, CCNB2, CDC25B, CENPF, CFLAR, MCM2, MCM7, PLAG1, ECT2, PTTG1, osf-2, and RAMP for validation. CDC25B, CCNB1, CCNB2, CENPF, MCM2, and MCM7 are involved in cell cycle regulation. CCNB1 (cyclin B1) complexes with CDC2 (cell division cycle 2) and is important during G2/M phase transition. CCNB1 overexpression was significantly correlated with tumor size, stage, and survival of patients with laryngeal squamous cell carcinoma [41]. An overexpression of cyclins B1 and B2, which may play a key role in transforming growth factor β-mediated cell cycle control, was also found in colorectal cancers and other tumors [42]. The cell division cycle 25B protein (CDC25B) belongs to phosphatases and activates CDC2 by removing phosphate groups. This step is necessary for entry into mitosis. Recently, it could be demonstrated by others that CDC25B is overexpressed in pancreatic carcinomas and their metastases. Interestingly, treatment with CDC25B inhibitors caused a growth reduction of pancreatic cancer cell lines [43]. MCM2 and MCM7 are both involved in initiation replication. Evidence suggests that MCM7 acts as a cofactor for oncogenic transformation [44], whereas MCM2 is a biomarker of proliferating cells independently of the p53 status [45]. These differentially expressed genes underline the fact that pancreatic cancer is highlighted by an intense proliferation. However, we identified also other genes involved in the signal transduction of several pathways.
For the first time, we report here the overexpression of the pituitary tumor transforming gene 1 (PTTG1) in pancreatic cancer. PTTG1 is expressed at very low levels in normal tissues, except for few cell types such as spermatocytes and spermatids. Overexpression of PTTG1 has been demonstrated in breast and other tumors [46–48]. It is a potent oncogene because of its ability to complex with p53 and thus to prevent p53 from binding to DNA and inducing cell death [49]. It further regulates the secretion of basic fibroblast growth factor (bFGF), which promotes angiogenesis and mitogenesis [48]. A role in cell migration was proposed for the osteoblast-specific factor 2 (osf-2) [50]. Its upregulation has been demonstrated in ovarian tumors [50], neuroblastomas [51], and head and neck squamous cell carcinomas [52]. For RAMP, an increase in cell proliferation of NT2 cells has been demonstrated, and a role in increasing the proliferation rate of human embryonal carcinoma cells was suggested [53].
The proto-oncogene pleiomorphic adenoma gene 1 (PLAG1), which is deregulated in pleiomorphic adenomas of the salivary glands, belongs to the family of zinc finger proteins. It has been demonstrated to be overexpressed in hepatoblastomas, too [54]. Interestingly, overexpression of PLAG1 in HEK 293 cells leads to an overexpression of cancer-related genes such as IGF-II, VEGF, and BCL2 [55].
Except for some variabilities in the gene expression pattern of the abovementioned genes in individual tissues, we found strong support for our microarray data by the validation techniques used.
We also found overexpression of the protein kinase IRAK1 within PDAC. IRAK1 acts as a activator of NF-κB presumably through TRAF6 [56]. Activation of NF-κB has been reported for several pancreatic cancer cell lines that also secrete IL-1α [57]. Therefore, the overexpression of IRAK1 might contribute to the activation NF-κB presumably through an autocrine activation loop involving IL1-α. NF-κB is one of the most important transcription factors implied in cancer formation [58]. Interestingly, NF-κB regulates the expression of proteins of the SMAD family through interaction with the SMAD7 promoter, which leads to an abrogation of the antiproliferative effects of an activated TGF-β signalling pathway [59]. Therefore, as to pancreatic cancer development, the overexpression of IRAK1 might mimic the loss of the SMAD4 tumor suppressor.
In summary, the use of microarray analysis, in combination with tissue microdissection, is a powerful tool for identifying the changes in gene expression associated with tumor development and progression in PDAC. Because of the reduced heterogeneity of microdissected tissues, small changes in gene expression can be observed and used to generate new molecular markers and therapeutic targets.
Acknowledgements
The authors thank K. Dege for critical reading of the manuscript, as well as Alfred E. Neumann whose comments were always appreciated.
Footnotes
The data set can be obtained through http://vtg.uniklinikum-dresden.de/Pankreaslabor.
Robert Grützmann, Christian Pilarsky, and Ole Ammerpohl contributed equally.
This paper was supported by Deutsche Krebshilfe (70-2937-SaI).
References
- 1.Jemal A, Murray T, Samuels A, Ghafoor A, Ward E, Thun MJ. Cancer statistics, 2003. CA Cancer J Clin. 2003;53:5–26. doi: 10.3322/canjclin.53.1.5. [DOI] [PubMed] [Google Scholar]
- 2.Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549–554. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- 3.Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, Kern SE. DPC4, a candidate tumor suppressor gene at human chromosome 18q21. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 4.Hruban RH, Offerhaus GJ, Kern SE, Goggins M, Wilentz RE, Yeo CJ. Tumor-suppressor genes in pancreatic cancer. J Hepatobiliary Pancr Surg. 1998;5:383–391. doi: 10.1007/s005340050062. [DOI] [PubMed] [Google Scholar]
- 5.Slebos RJ, Hoppin JA, Tolbert PE, Holly EA, Brock JW, Zhang RH, Bracci PM, Foley J, Stockton P, McGregor LM, Flake GP, Taylor JA. K-ras and p53 in pancreatic cancer: association with medical history, histopathology, and environmental exposures in a population-based study. Cancer Epidemiol Biomark Prev. 2000;9:1223–1232. [PubMed] [Google Scholar]
- 6.Bardeesy N, DePinho RA. Pancreatic cancer biology and genetics. Nat Rev Cancer. 2002;2:897–909. doi: 10.1038/nrc949. [DOI] [PubMed] [Google Scholar]
- 7.van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 8.Notterman DA, Alon U, Sierk AJ, Levine AJ. Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res. 2001;61:3124–3130. [PubMed] [Google Scholar]
- 9.Luo J, Duggan DJ, Chen Y, Sauvageot J, Ewing CM, Bittner ML, Trent JM, Isaacs WB. Human prostate cancer and benign prostatic hyperplasia: molecular dissection by gene expression profiling. Cancer Res. 2001;61:4683–4688. [PubMed] [Google Scholar]
- 10.Selaru FM, Zou T, Xu Y, Shustova V, Yin J, Mori Y, Sato F, Wang S, Olaru A, Shibata D, Greenwald BD, Krasna MJ, Abraham JM, Meltzer SJ. Global gene expression profiling in Barrett's esophagus and esophageal cancer: a comparative analysis using cDNA microarrays. Oncogene. 2002;21:475–478. doi: 10.1038/sj.onc.1205111. [DOI] [PubMed] [Google Scholar]
- 11.Hippo Y, Taniguchi H, Tsutsumi S, Machida N, Chong JM, Fukayama M, Kodama T, Aburatani H. Global gene expression analysis of gastric cancer by oligonucleotide microarrays. Cancer Res. 2002;62:233–240. [PubMed] [Google Scholar]
- 12.Friess H, Ding J, Kleeff J, Liao Q, Berberat PO, Hammer J, Buchler MW. Identification of disease-specific genes in chronic pancreatitis using DNA array technology. Ann Surg. 2001;234:769–778. doi: 10.1097/00000658-200112000-00008. discussion 778-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crnogorac-Jurcevic T, Missiaglia E, Blaveri E, Gangeswaran R, Jones M, Terris B, Costello E, Neoptolemos JP, Lemoine NR. Molecular alterations in pancreatic carcinoma: expression profiling shows that dysregulated expression of S100 genes is highly prevalent. J Pathol. 2003;201:63–74. doi: 10.1002/path.1418. [DOI] [PubMed] [Google Scholar]
- 14.Iacobuzio-Donahue CA, Maitra A, Olsen M, Lowe AW, Van Heek NT, Rosty C, Walter K, Sato N, Parker A, Ashfaq R, Jaffee E, Ryu B, Jones J, Eshleman JR, Yeo CJ, Cameron JL, Kern SE, Hruban RH, Brown PO, Goggins M. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am J Pathol. 2003;162:1151–1162. doi: 10.1016/S0002-9440(10)63911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grutzmann R, Foerder M, Alldinger I, Staub E, Brummendorf T, Ropcke S, Li X, Kristiansen G, Jesnowski R, Sipos B, Lohr M, Luttges J, Ockert D, Kloppel G, Saeger HD, Pilarsky C. Gene expression profiles of microdissected pancreatic ductal adenocarcinoma. Virchows Arch. 2003;443:508–517. doi: 10.1007/s00428-003-0884-1. [DOI] [PubMed] [Google Scholar]
- 16.Iacobuzio-Donahue CA, Ashfaq R, Maitra A, Adsay NV, Shen-Ong GL, Berg K, Hollingsworth MA, Cameron JL, Yeo CJ, Kern SE, Goggins M, Hruban RH. Highly expressed genes in pancreatic ductal adenocarcinomas: a comprehensive characterization and comparison of the transcription profiles obtained from three major technologies. Cancer Res. 2003;63:8614–8622. [PubMed] [Google Scholar]
- 17.Nakamura T, Furukawa Y, Nakagawa H, Tsunoda T, Ohigashi H, Murata K, Ishikawa O, Ohgaki K, Kashimura N, Miyamoto M, Hirano S, Kondo S, Katoh H, Nakamura Y, Katagiri T. Genomewide cDNA microarray analysis of gene expression profiles in pancreatic cancers using populations of tumor cells and normal ductal epithelial cells selected for purity by laser microdissection. Oncogene. 2004;23:2385–2400. doi: 10.1038/sj.onc.1207392. [DOI] [PubMed] [Google Scholar]
- 18.Sager R. Expression genetics in cancer: shifting the focus from DNA to RNA. Proc Natl Acad Sci USA. 1997;94:952–955. doi: 10.1073/pnas.94.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugiyama Y, Sugiyama K, Hirai Y, Akiyama F, Hasumi K. Microdissection is essential for gene expression profiling of clinically resected cancer tissues. Am J Clin Pathol. 2002;117:109–116. doi: 10.1309/G1C8-39MF-99UF-GT2K. [DOI] [PubMed] [Google Scholar]
- 20.Pilarsky CP, Schmitt AO, Dahl E, Rosenthal A. Microarrays: chances and challenges. Curr Opin Mol Ther. 1999;1:727–739. [PubMed] [Google Scholar]
- 21.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 24.Luttges J, Stigge C, Pacena M, Kloppel G. Rare ductal adenocarcinoma of the pancreas in patients younger than age 40 years. Cancer. 2004;100:173–182. doi: 10.1002/cncr.11860. [DOI] [PubMed] [Google Scholar]
- 25.Li D, Zhu J, Firozi PF, Abbruzzese JL, Evans DB, Cleary K, Friess H, Sen S. Overexpression of oncogenic STK15/BTAK/Aurora A kinase in human pancreatic cancer. Clin Cancer Res. 2003;9:991–997. [PubMed] [Google Scholar]
- 26.Jensen LE, Muzio M, Mantovani A, Whitehead AS. IL-1 signaling cascade in liver cells and the involvement of a soluble form of the IL-1 receptor accessory protein. J Immunol. 2000;164:5277–5286. doi: 10.4049/jimmunol.164.10.5277. [DOI] [PubMed] [Google Scholar]
- 27.Ernst T, Hergenhahn M, Kenzelmann M, Cohen CD, Bonrouhi M, Weninger A, Klaren R, Grone EF, Wiesel M, Gudemann C, Kuster J, Schott W, Staehler G, Kretzler M, Hollstein M, Grone HJ. Decrease and gain of gene expression are equally discriminatory markers for prostate carcinoma: a gene expression analysis on total and microdissected prostate tissue. Am J Pathol. 2002;160:2169–2180. doi: 10.1016/S0002-9440(10)61165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang E, Miller LD, Ohnmacht GA, Liu ET, Marincola FM. High-fidelity mRNA amplification for gene profiling. Nat Biotechnol. 2000;18:457–459. doi: 10.1038/74546. [DOI] [PubMed] [Google Scholar]
- 29.Grutzmann R, Pilarsky C, Staub E, Schmitt AO, Foerder M, Specht T, Hinzmann B, Dahl E, Alldinger I, Rosenthal A, Ockert D, Saeger HD. Systematic isolation of genes differentially expressed in normal and cancerous tissue of the pancreas. Pancreatology. 2003;3:169–178. doi: 10.1159/000070087. [DOI] [PubMed] [Google Scholar]
- 30.Ryu B, Jones J, Blades NJ, Parmigiani G, Hollingsworth MA, Hruban RH, Kern SE. Relationships and differentially expressed genes among pancreatic cancers examined by large-scale serial analysis of gene expression. Cancer Res. 2002;62:819–826. [PubMed] [Google Scholar]
- 31.Iacobuzio-Donahue CA, Maitra A, Shen-Ong GL, van Heek T, Ashfaq R, Meyer R, Walter K, Berg K, Hollingsworth MA, Cameron JL, Yeo CJ, Kern SE, Goggins M, Hruban RH. Discovery of novel tumor markers of pancreatic cancer using global gene expression technology. Am J Pathol. 2002;160:1239–1249. doi: 10.1016/S0002-9440(10)62551-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han H, Bearss DJ, Browne LW, Calaluce R, Nagle RB, Von Hoff DD. Identification of differentially expressed genes in pancreatic cancer cells using cDNA microarray. Cancer Res. 2002;62:2890–2896. [PubMed] [Google Scholar]
- 33.Crnogorac-Jurcevic T, Efthimiou E, Nielsen T, Loader J, Terris B, Stamp G, Baron A, Scarpa A, Lemoine NR. Expression profiling of microdissected pancreatic adenocarcinomas. Oncogene. 2002;21:4587–4594. doi: 10.1038/sj.onc.1205570. [DOI] [PubMed] [Google Scholar]
- 34.Crnogorac-Jurcevic T, Efthimiou E, Capelli P, Blaveri E, Baron A, Terris B, Jones M, Tyson K, Bassi C, Scarpa A, Lemoine NR. Gene expression profiles of pancreatic cancer and stromal desmoplasia. Oncogene. 2001;20:7437–7446. doi: 10.1038/sj.onc.1204935. [DOI] [PubMed] [Google Scholar]
- 35.Argani P, Rosty C, Reiter RE, Wilentz RE, Murugesan SR, Leach SD, Ryu B, Skinner HG, Goggins M, Jaffee EM, Yeo CJ, Cameron JL, Kern SE, Hruban RH. Discovery of new markers of cancer through serial analysis of gene expression: prostate stem cell antigen is overexpressed in pancreatic adenocarcinoma. Cancer Res. 2001;61:4320–4324. [PubMed] [Google Scholar]
- 36.Gress TM, Wallrapp C, Frohme M, Muller-Pillasch F, Lacher U, Friess H, Buchler M, Adler G, Hoheisel JD. Identification of genes with specific expression in pancreatic cancer by cDNA representational difference analysis. Genes Chromosomes Cancer. 1997;19:97–103. doi: 10.1002/(sici)1098-2264(199706)19:2<97::aid-gcc5>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 37.Friess H, Ding J, Kleeff J, Fenkell L, Rosinski JA, Guweidhi A, Reidhaar-Olson JF, Korc M, Hammer J, Buchler MW. Microarray-based identification of differentially expressed growth- and metastasis-associated genes in pancreatic cancer. Cell Mol Life Sci. 2003;60:1180–1199. doi: 10.1007/s00018-003-3036-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, Misek DE, Hanash S. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003;63:2649–2657. [PubMed] [Google Scholar]
- 39.Tan ZJ, Hu XG, Cao GS, Tang Y. Analysis of gene expression profile of pancreatic carcinoma using cDNA microarray. World J Gastroenterol. 2003;9:818–823. doi: 10.3748/wjg.v9.i4.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuo WP, Jenssen TK, Butte AJ, Ohno-Machado L, Kohane IS. Analysis of matched mRNA measurements from two different microarray technologies. Bioinformatics. 2002;18:405–412. doi: 10.1093/bioinformatics/18.3.405. [DOI] [PubMed] [Google Scholar]
- 41.Dong Y, Sui L, Watanabe Y, Sugimoto K, Tokuda M. Clinical relevance of cyclin B1 overexpression in laryngeal squamous cell carcinoma. Cancer Lett. 2002;177:13–19. doi: 10.1016/s0304-3835(01)00770-4. [DOI] [PubMed] [Google Scholar]
- 42.Sarafan-Vasseur N, Lamy A, Bourguignon J, Pessot FL, Hieter P, Sesboue R, Bastard C, Frebourg T, Flaman JM. Overexpression of B-type cyclins alters chromosomal segregation. Oncogene. 2002;21:2051–2057. doi: 10.1038/sj.onc.1205257. [DOI] [PubMed] [Google Scholar]
- 43.Guo J, Kleeff J, Li J, Ding J, Hammer J, Zhao Y, Giese T, Korc M, Buchler MW, Friess H. Expression and functional significance of CDC25B in human pancreatic ductal adenocarcinoma. Oncogene. 2004;23:71–81. doi: 10.1038/sj.onc.1206926. [DOI] [PubMed] [Google Scholar]
- 44.Yoshida K, Inoue I. Conditional expression of MCM7 increases tumor growth without altering DNA replication activity. FEBS Lett. 2003;553:213–217. doi: 10.1016/s0014-5793(03)01018-4. [DOI] [PubMed] [Google Scholar]
- 45.Kodani I, Shomori K, Osaki M, Kuratate I, Ryoke K, Ito H. Expression of minichromosome maintenance 2, (MCM2), Ki-67, and cell-cycle-related molecules, and apoptosis in the normal-dysplasia-carcinoma sequence of the oral mucosa. Pathobiology. 2001;69:150–158. doi: 10.1159/000048770. [DOI] [PubMed] [Google Scholar]
- 46.Shibata Y, Haruki N, Kuwabara Y, Nishiwaki T, Kato J, Shinoda N, Sato A, Kimura M, Koyama H, Toyama T, Ishiguro H, Kudo J, Terashita Y, Konishi S, Fujii Y. Expression of PTTG (pituitary tumor transforming gene) in esophageal cancer. Jpn J Clin Oncol. 2002;32:233–237. doi: 10.1093/jjco/hyf058. [DOI] [PubMed] [Google Scholar]
- 47.Puri R, Tousson A, Chen L, Kakar SS. Molecular cloning of pituitary tumor transforming gene 1 from ovarian tumors and its expression in tumors. Cancer Lett. 2001;163:131–139. doi: 10.1016/s0304-3835(00)00688-1. [DOI] [PubMed] [Google Scholar]
- 48.Heaney AP, Singson R, McCabe CJ, Nelson V, Nakashima M, Melmed S. Expression of pituitary-tumour transforming gene in colorectal tumours. Lancet. 2000;355:716–719. doi: 10.1016/S0140-6736(99)10238-1. [DOI] [PubMed] [Google Scholar]
- 49.Bernal JA, Luna R, Espina A, Lazaro I, Ramos-Morales F, Romero F, Arias C, Silva A, Tortolero M, Pintor-Toro JA. Human securin interacts with p53 and modulates p53-mediated transcriptional activity and apoptosis. Nat Genet. 2002;32:306–311. doi: 10.1038/ng997. [DOI] [PubMed] [Google Scholar]
- 50.Gillan L, Matei D, Fishman DA, Gerbin CS, Karlan BY, Chang DD. Periostin secreted by epithelial ovarian carcinoma is a ligand for alpha(V)beta(3) and alpha(V)beta(5) integrins and promotes cell motility. Cancer Res. 2002;62:5358–5364. [PubMed] [Google Scholar]
- 51.Sasaki H, Sato Y, Kondo S, Fukai I, Kiriyama M, Yamakawa Y, Fuji Y. Expression of the periostin mRNA level in neuroblastoma. J Pediatr Surg. 2002;37:1293–1297. doi: 10.1053/jpsu.2002.34985. [DOI] [PubMed] [Google Scholar]
- 52.Gonzalez HE, Gujrati M, Frederick M, Henderson Y, Arumugam J, Spring PW, Mitsudo K, Kim HW, Clayman GL. Identification of 9 genes differentially expressed in head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2003;129:754–759. doi: 10.1001/archotol.129.7.754. [DOI] [PubMed] [Google Scholar]
- 53.Cheung WM, Chu AH, Chu PW, Ip NY. Cloning and expression of a novel nuclear matrix-associated protein that is regulated during the retinoic acid-induced neuronal differentiation. J Biol Chem. 2001;276:17083–17091. doi: 10.1074/jbc.M010802200. [DOI] [PubMed] [Google Scholar]
- 54.Zatkova A, Rouillard JM, Hartmann W, Lamb BJ, Kuick R, Eckart M, Von Schweinitz D, Koch A, Fonatsch C, Pietsch T, Hanash SM, Wimmer K. Amplification and overexpression of the IGF2 regulator PLAG1 in hepatoblastoma. Genes Chromosomes Cancer. 2004;39:126–137. doi: 10.1002/gcc.10307. [DOI] [PubMed] [Google Scholar]
- 55.Voz ML, Mathys J, Hensen K, Pendeville H, Van Valckenborgh I, Van Huffel C, Chavez M, Van Damme B, De Moor B, Moreau Y, Van de Ven WJ. Microarray screening for target genes of the protooncogene PLAG1. Oncogene. 2004;23:179–191. doi: 10.1038/sj.onc.1207013. [DOI] [PubMed] [Google Scholar]
- 56.Luftig M, Prinarakis E, Yasui T, Tsichritzis T, Cahir-McFarland E, Inoue JI, Nakano H, Mak TW, Yeh WC, Li X, Akira S, Suzuki N, Suzuki S, Mosialos G, Kieff E. Epstein-Barr virus latent membrane protein 1 activation of NF-kappaB through IRAK1 and TRAF6. Proc Natl Acad Sci USA. 2003;100:15595–15600. doi: 10.1073/pnas.2136756100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niu J, Li Z, Peng B, Chiao PJ. Identification of an autoregulatory feedback pathway involving IL-1alpha in induction of constitutive NF-kappaB activation in pancreatic cancer cells. J Biol Chem. 2003;279:16452–16462. doi: 10.1074/jbc.M309789200. [DOI] [PubMed] [Google Scholar]
- 58.Garg A, Aggarwal BB. Nuclear transcription factor-kappaB as a target for cancer drug development. Leukemia. 2002;16:1053–1068. doi: 10.1038/sj.leu.2402482. [DOI] [PubMed] [Google Scholar]
- 59.Nagarajan RP, Chen F, Li W, Vig E, Harrington MA, Nakshatri H, Chen Y. Repression of transforming-growth-factor-beta-mediated transcription by nuclear factor kappaB. Biochem J. 2000;348(Part 3):591–596. [PMC free article] [PubMed] [Google Scholar]