Abstract
The identification of genes associated with colonization and persistence of Helicobacter pylori in the gastric mucosa has been limited by the lack of robust animal models that support infection by strains whose genomes have been completely sequenced. Here we report that an interleukin-12 (IL-12)-deficient mouse (IL-12−/− p40 subunit knockout in C57BL/6 mouse) is permissive for infection by a motile variant (KE88-3887) of The Institute For Genomic Research-sequenced strain (KE26695) of H. pylori. The IL-12-deficient mouse was also more permissive for colonization by the mouse-colonizing Sydney 1 strain of H. pylori than were wild-type C57BL/6 mice. Differences in colonization efficiency were demonstrated by mouse challenge with SS1 strains containing loss-of-function mutations in two genes (hspR and hrcA), whose products negatively regulate several heat shock genes. At 5 weeks postinfection, double-knockout mutants (SS1 hspR hrcA) efficiently colonized IL-12-deficient mice (5 of 5 animals compared to 4 of 10 for C57BL6 mice) and bacterial counts were higher in stomachs of IL-12-deficient mice (106 versus 105 CFU/g of stomach, respectively). IL-12-deficient mice were efficiently colonized by KE88-3887 (29 of 30), but not by nonmotile KE26695, and bacterial numbers (104 to 105 CFU/g of stomach) were unchanged over an 8-week period postinfection. In contrast, C57BL/6 mice were inefficiently colonized by KE88-3887 (8 of 20 animals with bacterial loads at the limit of detection, ∼103 CFU/g), and infection did not persist much beyond 5 weeks. Cytokine responses (tumor necrosis factor alpha and gamma interferon), pathology, and antral-predominant infection were indistinguishable between IL-12-deficient and C57BL/6 mice. The increased permissiveness of the IL-12-deficient mouse for infection with H. pylori should facilitate whole-genome-based strategies to study genes associated with virulence and immune modulation.
Helicobacter pylori colonizes the gastric mucosa of humans, producing a chronic gastritis that may remain asymptomatic for many years. In about 10% of individuals, more severe disease manifestations will occur such as duodenal and gastric ulcers, atrophic gastritis, and intestinal metaplasia, all risk factors for gastric cancer (20, 40, 42). The remarkable ability of H. pylori to establish lifelong infections is not well understood but likely involves evasion or modulation of host immune responses as well as adaptation (through mutation and selection) to the unique physiology of each individual host (3). In addition, different disease pathologies seem to correlate with particular H. pylori genotypes, with strains containing the cag-associated pathogenicity island and cagA gene (cytotoxin-associated gene) and vacA (vacuolating cytotoxin) correlating with more severe disease (11, 14, 39). There is much genetic diversity in these genes and in others such as the restriction modification genes (9) among strains from different geographic regions and people of different ethnicities (1, 10, 13). The evaluation of genetic diversity among strains and identification of genes associated with severity of infection have generally been hampered by the lack of good animal models (9, 25, 36).
Robust animal models of infection are also a necessary component in the discovery process for new therapeutics or the evaluation of vaccine candidates (12, 23, 47). While several animal models have been developed, these models are limited to a few animal-adapted strains (9, 22, 36) or support only transient infection (25). Mouse-adapted strains such as the Sydney 1 strain (SS1), Hp1, and a few others are difficult to manipulate genetically (37) and usually require high infectious doses in order to establish infection (36). In such mouse models requiring high colonization thresholds, many genes scored as necessary for colonization may be dispensable in a more permissive animal. Another limitation of existing mouse-colonizing strains is that systematic whole-genome approaches to the study of virulence determinants cannot be performed, as the genomes of mouse-colonizing strains have not been sequenced and strains for which genome information is available do not colonize mice (22, 25). Recent reports also indicate that several genes of the cag pathogenicity island of SS1 may not be functional, raising the possibility that this strain is attenuated for virulence (15). In this regard, Philpott et al. have shown that the mouse-adapted SS1 strain produces less inflammation in the mucosa of colonized mice than that produced by more recent clinical isolates that can colonize mice (43). However, the burgeoning number of H. pylori strains of unknown genomic sequence being used as model systems together with the wide variation in animal infection models has made comparisons of findings difficult.
The inability of most H. pylori strains to colonize mice might in part be due to development of a type 1 immune response in the gastric mucosa of these animals (17, 28, 41). Interleukin-12 (IL-12) is a critical inducer of type 1 cytokine responses that plays a key role in both innate and acquired immune responses. While IL-12 is a heterodimer of a p40 and a p35 subunit, most studies are performed with p40-deficient mouse strains (2, 34). IL-12-deficient mice have previously been demonstrated to diminish immune responses to a variety of pathogens (19, 34), and products of H. pylori elicit IL-12 production (41).
In this study we report that IL-12-deficient C57BL/6 mice (IL-12−/−) are more permissive for colonization by SS1 than are parental C57BL/6 mice. Using SS1 strains with defined mutations in genes whose products control the level of expression of Hsp60 and Hsp70 (hspR hrcA double mutant), we demonstrate that these mutants more efficiently colonize IL-12-deficient mice, suggesting that the threshold for colonization is lower in this mouse. The lower colonization threshold of IL-12-deficient mice permitted colonization and persistence by a motile variant (KE88-3887) of the Institute for Genomic Research (TIGR)-sequenced strain of H. pylori, providing a potentially new model system for the study of genes associated with pathogenesis in a strain for which the entire genome has been sequenced.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
H. pylori strains SS1 (36) and KE26695 (22, 51) and the latter's motile variant KE88-3887 (piglet-passaged motile strain obtained from K. Eaton, Department of Veterinary Biosciences, Ohio State University) were grown under humid microaerobic conditions with 5% CO2 at 37°C on brucella-based medium supplemented with 7.5% newborn calf serum (Gibco Laboratories) as previously described (24). Bacterial cultures were observed microscopically for purity and to ensure that they were devoid of coccoid forms and if necessary were confirmed as H. pylori by biochemical tests. Escherichia coli strains were cultured in Luria-Bertani medium and supplemented as required with appropriate antibiotics.
Generation of mutants.
The gene encoding HspR (HP1025), a regulator of heat shock genes, was cloned by PCR amplification with flanking DNA primers designed on the basis of published genomic sequences (51). To amplify hspR, primers HSPRF (5′ CCATCGATGGAGGAATCGTGTGCGATTATGA) and HSPRR (5′ CGGAATTCTCCCTACTAAATGCTCTTGGCCT) were used in a PCR containing chromosomal DNA obtained from KE26695 by protocols described previously (6, 24). The resulting 0.46-kb amplicon was TA cloned into pBluescript-T vector (Stratagene, La Jolla, Calif.). A chloramphenicol resistance cassette (cam) (54) was restricted with XbaI from shuttle vector pDH26 (27) and after treatment with Klenow fragment of DNA polymerase I (to create blunt ends) was inserted into the unique HindIII site (also treated with Klenow fragment) located in hspR. Following PCR-based validation of the construct, pBSK-hspR::cam DNA was introduced into KE26695 by natural transformation (53) and Cmr colonies were picked and verified by PCR.
The gene encoding another heat shock regulatory protein (HrcA) was inactivated by insertion of a kanamycin cassette (aph-3) by the same strategy as that described for hspR above. DNA containing the hrcA gene (HP0111) was PCR amplified from H. pylori strain KE26695-obtained DNA with primer pair hrcAF (5′ GGAATTCCATATGGTGATTGACGAGATT) and hrcAR (5′ CCGCTCGAGTTCCTCCTCAGAAATCGT). The 0.84-kb PCR fragment was ligated into pBSK-T (Stratagene) vector, resulting in recombinant plasmid pBSK-hrcA. The unique restriction site for a blunt-end cutter, BalI (MlsI), was used to insert the kanamycin cassette, which was recovered from plasmid pDH37 (27) as a 1.4-kb XbaI fragment (insert was blunt ended with Klenow fragment before the ligation). The resulting mutant had the HrcA coding sequence interrupted by a Kmr cassette and was designated pBSK-hrcA::aph-3. Allelic-exchange mutagenesis following natural transformation of H. pylori with pBSK-hrcA::aph-3 DNA and selection for kanamycin resistance produced HrcA-knockout mutants that were verified by PCR with the same primer pair.
The same strategy and procedures were used to construct the double-knockout (DKO) mutants as described above for each of the genes hspR and hrcA. DKO mutants of H. pylori were selected on brucella-based medium containing chloramphenicol and kanamycin. hspR hrcA DKO mutants were verified by PCR with two primer pairs (in the same PCR mixture), hspRF-hspRR and hrcAF-hrcAR.
DNA techniques.
DNA manipulations were carried out by general techniques as described by Sambrook et al. (45). Miniscale plasmid preparations were carried out with Wizard Plus SV Minipreps (Promega). DNA fragments or PCR amplification products for cloning purposes were purified from agarose gels with the Geneclean II kit (Bio 101). H. pylori genomic DNA was isolated by the cetyltrimethylammonium bromide method (5).
PCR.
The PCR mixtures used to analyze knockout mutants routinely contained (in 25 μl) 10 ng of H. pylori genomic DNA, 10 pmol of each primer, 2.5 mM (each) deoxynucleoside triphosphates, and 1 U of Taq DNA polymerase in standard PCR buffer (MBI Fermentas). Thirty amplification cycles were performed (94°C for 30 s, 50°C for 30 s, and 72°C for 3 min) in a Perkin-Elmer 2400 thermal cycler.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting.
Crude extracts of H. pylori cell cultures prepared as described previously (30) and equal amounts of protein (25 μg each) were separated on an SDS-10 to 20% polyacrylamide gel and transferred onto an 0.45-μm-pore-size nitrocellulose membrane (Protran; Schleicher & Schuell) for 18 h at 50 V as outlined previously (26). After transfer, the nitrocellulose membrane was immersed in 1× Ponceau S to check transfer efficiency. The membrane was blocked in 1× TBST (Tris-buffered saline with 0.05% Tween 20) containing 5% skim milk at room temperature for 2 h and processed as described by Helsel et al. (26). Polyclonal antibodies used in these studies have been previously described (31, 32): Hsp60 serum was diluted 1:4,000, anti-Hsp70 was diluted 1:1,000, and the developing antibody was used at 1:5,000 (goat anti-rabbit antibody ([immunoglobulin G {heavy plus light}]) conjugated to alkaline phosphatase (Cedarlane Laboratories Ltd.). Immunoreactive proteins were visualized by using 0.01% nitroblue tetrazolium and 0.005% 5-bromo-4-chloro-3-indolylphosphate in alkaline phosphatase buffer as a substrate.
Animals and housing.
Female pathogen-free C57BL/6 mice (approximately 3 to 5 weeks old) were obtained from Charles River Canada. IL-12-deficient mice (C57BL/6 p40−/−) (obtained from Jean Magram, Hoffmann-La Roche, Nutley, N.J.) were bred in-house at the Carleton Animal Care Facility, Dalhousie University, Halifax, Nova Scotia, Canada. Animals were bred under isolator conditions and placed in conventional housing for at least 4 weeks prior to experimentation. All animals were given access to water and commercial chow throughout the course of the experiments. Animals were sacrificed by CO2 asphyxiation. Animal protocols were approved by the animals ethics committee of the Dalhousie Medical School animal care facility.
Mouse infection protocols.
Mice (groups of five) were infected with 0.1 ml of saline (controls) or 0.1 ml of bacteria grown for 48 h (approximately 109 CFU) as previously described (35). Animals were infected by oral gavage three times over a 5-day period, unless otherwise indicated. Blood was collected from each mouse by the saphenous vein method before infection, at 3 weeks postinfection, and on the day of sacrifice (serum samples were stored at −20°C). The collected serum was tested for antibody levels by cytofluorometric analysis. Animals were allowed to fast overnight prior to sacrifice, and animal cages were cleaned on the day before sacrifice to reduce transient microbial contamination of stomachs (particularly to reduce the incidence of Proteus mirabilis). At the desired times postinfection, animals were sacrificed by intraperitoneal injection of chloral hydrate and then necropsied. Stomachs were removed aseptically and opened. The contents were rinsed with phosphate-buffered saline (PBS), and then the splayed stomach was cut into two sections, with one section being weighed, homogenized, and plated onto Columbia blood agar plates with Dent's antibiotics for microbiological counts (16) and the other being prepared for histology. In some cases biopsy punch samples (0.3-cm diameter) were taken from stomachs for cytokine measurements.
Histology and gastritis score.
One half of each stomach was placed into 10% buffered formalin and processed in paraffin, and 4-μm sections were stained with a modified Steiner silver stain. Slides were coded and examined blind. Colonization was assessed on a five-point scale: 0, no bacteria; 1, less than one-third of crypts colonized with 1 to 10 bacteria; 2, one-third to two-thirds of crypts colonized with 10 to 20 bacteria; 3, two-thirds of the crypts colonized with >20 bacteria; 4, all crypts colonized with >20 bacteria. Colonization was assessed in three regions of the stomach: antrum, body, and the transition zone between these regions (35). The antrum-body transitional zone was defined as the area where parietal cells of the body-type glands disappear and antral-type glands become dominant. Evaluation of colonization within the antrum-body transitional zone was performed by identifying the center point of the transitional zone and counting bacteria within a five-gland width on either side.
Severity of inflammation was assessed for infiltration of the mucosa by mononuclear and polymorphonuclear cells with the Sydney histology grading system (35). This consisted of a four-point scale ranging from none to mild to moderate to severe.
Cytokine determinations.
Punch biopsy samples were obtained from different regions of the washed stomach tissue and then further washed in sterile PBS and placed in culture for 24 h in RPMI 1640 (Gibco BRL) medium containing 10% fetal calf serum, 1% penicillin-streptomycin, and 100 μg of soybean trypsin inhibitor (Sigma)/ml. After this time the biopsy samples and cell debris were removed by centrifugation and the harvested supernatants were stored in aliquots at −80°C until assay. Supernatant levels of tumor necrosis factor alpha (TNF-α), IL-1β, and gamma interferon (IFN-γ) were determined by enzyme-linked immunosorbent assay (ELISA) with matched antibody pairs. Briefly, 96-well ELISA plates (Maxisorb; Nunc) were coated with anticytokine antibodies (TNF-α, goat anti-mouse TNF-α [R&D Systems]; IL-1β, anti-murine IL-1β monoclonal antibody 410 [R&D Systems]; IFN-γ, rat anti-mouse IFN-γ 554431 [PharMingen]). Remaining binding sites were blocked by incubation of the plates with 1% bovine serum albumin in PBS, and samples and/or standards were then added. Cytokine bound to the wells was detected with biotinylated specific antibodies (TNF-α, anti-murine TNF-α [MM35000-B Endogen]; IL-1β, goat anti-mouse IL-1β BAF 401 [R&D Systems]; IFN-γ, rat anti-mouse IFN-γ 554410 [PharMingen]) followed by streptavidin-alkaline phosphatase and a commercial ELISA amplification-development system (Life Technologies). The sensitivities of the ELISAs were as follows: TNF-α, 30 pg/ml; IL-1β, 20 pg/ml; and IFN-γ, 20 pg/ml.
Statistics.
Cytokine ELISA results were compared by analysis of variance followed by t tests with the Bonferroni correction.
RESULTS
IL-12-deficient mouse model.
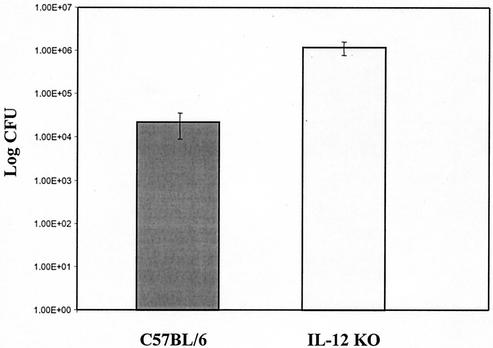
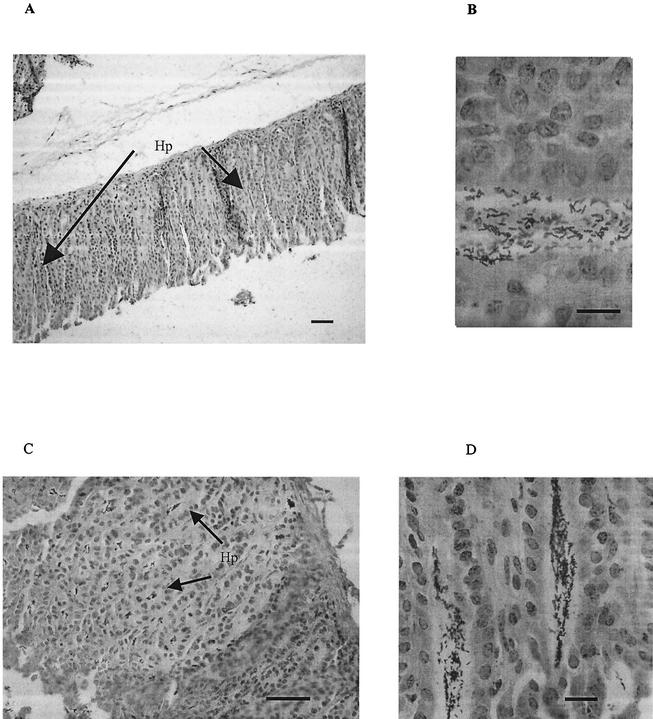
IL-12 (p40)-deficient and wild-type mice were identically infected by single orogastric gavage with the SS1 strain of H. pylori and scored for colonization efficiency at 3 weeks postinfection. As shown in Fig. 1, IL-12-deficient mice were consistently infected (five of five versus four of five for C57BL/6 mice) and to a higher colonization density (>106 CFU/g of stomach versus 2 × 104 CFU/g of stomach for C57BL/6 mice). The higher colonization density was not accompanied by increases in inflammation or location of infection, which was antral predominant (Fig. 2A and B). These findings suggested that, in the absence of IL-12 production, mice were more permissive for infection by H. pylori.
FIG. 1.
Colonization density comparison of mice infected with H. pylori strain SS1. Mice were infected once by oral gavage, and at 20 days postinfection, mouse stomachs were removed and bacterial counts were determined as described in the text. Bacterial density is CFU per gram of stomach.
FIG. 2.
Histological examination of H. pylori-infected stomach tissue. The figure shows silver-stained gastric tissue from the stomachs of C57BL/6 (A and B) and IL-12-deficient (C and D) mice infected with the SS1 strain of H. pylori. (A) Antral-predominant infection; (B) expanded region and bacterial load; (C) antral region of an IL-12-deficient mouse stomach colonized by SS1; (D) expanded region. Bars, ∼100 μm (A and C) and ∼10 μm (B and D).
Characterization of hrcA and hspR mutants.
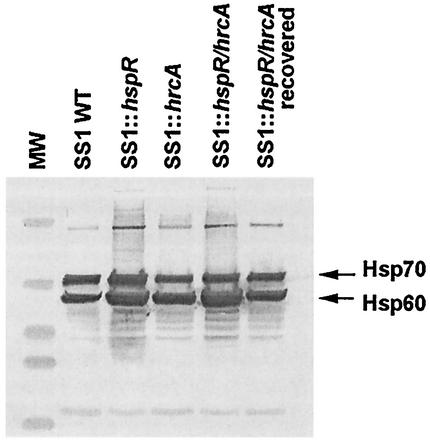
To test whether IL-12-deficient mice were more permissive for infection by SS1 strains, we generated mutations in two regulatory genes that putatively control heat shock or stress-induced genes. The hspR gene encodes a negative transcription regulator of several heat shock operons including those for hspAB (Hsp60) and dnaK (Hsp70) (18, 48). We had chosen these genes because heat shock proteins in general potentiate inflammation (29, 49) and have been implicated previously in gastric inflammation (52). Functional inactivation of hspR leads to constitutive overexpression of Hsp60 and Hsp70 as depicted in Fig. 3 for the SS1 strain (immunoblot with antisera specific for Hsp60 and Hsp70) and consistent with findings reported by others (18, 49). Comparable results were obtained with strains KE26695 and Hp1 (data not presented). Functional inactivation of hrcA, a putative regulator of heat shock genes (38), appears to have little or no effect on relative levels of Hsp60 and Hsp70 (wild-type SS1-equivalent levels) (Fig. 3). The results for mutational inactivation of both hspR and hrcA (DKO mutant in Fig. 3) were indistinguishable from those obtained for the hspR mutant.
FIG. 3.
Immunoblot of Hsp60 and Hsp70 of H. pylori strains. Bacterial extracts (equal concentrations of protein) from H. pylori strain SS1 and mutants grown on agar medium were subjected to SDS-PAGE and transferred to nitrocellulose as described in the text. The blot was developed with antiserum to Hsp70 and Hsp60 as detailed in the text. WT, wild type; MW, molecular weight markers.
Efficiency of colonization of SS1- and mutant-challenged C57BL/6 and IL-12-deficient mice.
Preliminary challenge experiments with C57BL/6 mice had shown that the SS1-hrcA::aph-3 mutant was unaltered in colonization efficiency, whereas the SS1-hspR::cam mutant and the SS1-hspR::cam hrcA::aph-3 DKO mutant (DKO) were equally enfeebled for colonization at 5 weeks postinfection (20 to 40% decrease in number of animals infected), and bacterial counts from infected animals were 1 to 2 logs lower than those for mice similarly infected with SS1 (data not presented). To determine whether a more permissive mouse model might be more efficiently colonized by these mutants, C57BL/6 and IL-12-deficient mice were identically infected with the DKO mutant and SS1 strains, and at 5 weeks postinfection stomachs were removed and bacterial counts were determined. The results of these challenges are summarized in Table 1. The DKO mutant poorly colonized C57BL/6 mice (4 of 10 animals colonized), and bacterial counts were variable with an average of ∼7.2 × 105 CFU/g of stomach. In contrast to the colonization efficiencies noted with C57BL/6 mice, five of five IL-12-deficient mice at 5 weeks postinfection were colonized by the SS1 and DKO strains. Moreover, the bacterial loads from the stomachs of the IL-12-deficient animals colonized by the DKO mutant were only slightly higher (2.3 × 106) than for similarly infected C57BL/6 animals. These results show that colonization efficiency of H. pylori mutants, illustrated here with the DKO mutant, can vary depending on the permissiveness of the mouse strain used, suggesting that the colonization threshold may be lower in the absence of IL-12.
TABLE 1.
Analysis of mutants in IL-12-deficient and C57BL/6 mice at 5 weeks postinfectiona
| Strain | CFU/g of stomach tissue for mouse group:
|
|
|---|---|---|
| IL-12-deficient | C57BL/6 | |
| SSI | (3.8 ± 4) × 107 | (2.7 ± 1.7) × 106 |
| DKO | (2.3 ± 1.7) × 106 | (7.2 ± 5.2) × 105 (4/10) |
| RDKO | (7.9 ± 4.5) × 107 | (2.4 ± 2) × 106 (7/10) |
Numbers in parentheses after the CFU denote the number of animals infected of the total number challenged and are included only where the number of animals infected was less than the number challenged, which in most experiments was five. Strain designations: SSI, wild-type strain; DKO, hspR hrcA double mutant in SSI; RDKO, mouse-recovered revertant of DKO expressing wild-type levels of Hsp60.
Recovered mouse-passaged SS1 DKO mutant.
In our preliminary screenings of SS1 mutants in mice, we noticed that one mouse colonized by the DKO mutant exhibited near-wild-type SS1 bacterial numbers (i.e., 106 CFU/g of stomach), suggesting that it might have reverted to wild-type infectivity during mouse passage. This variant (named RDKO for mouse-recovered DKO mutant) was resistant to both chloramphenicol and kanamycin, and PCR analysis confirmed that hrcA and hspR genes were still disrupted by antibiotic resistance markers. To investigate this mutant further, protein extracts were examined by SDS-PAGE and immunoblotting, and the analysis revealed near-wild-type levels of Hsp60 and Hsp70 (Fig. 3). Similar genetic analysis of DKO mutants recovered from other infected mice (exhibiting decreased colonization efficiency) showed that they retained the increased Hsp60 and Hsp70 expression phenotype (data not presented), suggesting that the RDKO mutant likely contained a second-site mutation, which affected the relative level of production of Hsp60. The RDKO variant was somewhat enfeebled for colonization in the C57BL/6 mouse model (7 of 10 animals colonized) but not in the IL-12-deficient mouse model (Table 1). These results suggest that increased production of Hsp60, rather than loss of regulatory functions of HspR, is associated with the colonization deficiency phenotype of hspR mutants.
Colonization of IL-12-deficient mice by KE26695.
We then examined the possibility that IL-12-deficient mice might be permissive for colonization by the KE26695 strain of H. pylori, one of two non-mouse-colonizing strains for which the whole genome sequence is available (4, 51). Since the KE26695 strain sequenced by TIGR was nonmotile and avirulent (33), we obtained a piglet-passaged motile variant of KE26695 (KE88-3887) from K. Eaton. Challenge of either the C57BL/6 or IL-12-deficient mice with the nonmotile KE26695 strain resulted in no colonization, and no bacteria were recovered from mouse stomach material. The results depicted in Table 2 show that KE88-3887 and a once-mouse-passaged strain (KE88-3887R) colonized C57BL6 and IL-12-deficient mice. However, C57BL/6 mice were poorly colonized (8 of 20 total animals at 3 and 5 weeks postinfection), and bacterial loads were ∼103 CFU/g of stomach (at the limit of detection). By 8 weeks postinfection, none of the C57BL/6 mice were colonized by KE88-3887 as indicated by culture (Table 2). In contrast, 10 of 10 IL-12-deficient mice were colonized by KE88-3887 at 3 weeks postinfection and 9 of 10 mice were colonized at 5 weeks postinfection. Bacterial loads in the stomachs were also around 1 to 2 logs higher than those seen with the C57BL/6 animals, reflecting an increased permissiveness of this animal for colonization by H. pylori. In addition, 10 of 10 of the IL-12-deficient mice remained colonized at 8 weeks, and bacterial numbers were decreased only slightly from numbers obtained earlier. The once-mouse-passaged KE88-3887R strain yielded slightly higher bacterial counts, consistent with the general theme of adaptation and selection of more fit variants. While further studies are required to determine the duration of colonization beyond 8 weeks, our findings indicate that IL-12-deficient mice can support infection by motile variants of the KE26695 strain of H. pylori.
TABLE 2.
Colonization of IL-12-knockout and C57BL/6 mice with KE88-3887, KE88-3887R (once mouse passaged) and the avirulent TIGR strain of 26695 (26695-Lab)a
| Mouse and H. pylori strain | CFU/g of stomach at wk:
|
||
|---|---|---|---|
| 3 | 5 | 8 | |
| C57BL/6 | |||
| Uninfected control | 0 | 0 | 0 |
| 26695-Lab | 0 | 0 | 0 |
| KE88-3887/R | (1.3 ± 0.47) × 103 (3/10) | (6.0 ± 4) × 103 (5/10) | 0 |
| IL-12-knockout mouse | |||
| Uninfected control | 0 | 0 | 0 |
| 26695-Lab | 0 | 0 | 0 |
| KE88-3887 | (6.6 ± 5) × 104 | (4.4 ± 3) × 104 (4/5) | (4.14 ± 2.1) × 104 |
| KE88-3887R | (1.0 ± 2.8) × 105 | (1.07 ± 0.9) × 105 | (5.76 ± 3) × 104 |
For each experiment, five animals were infected as per the protocol in text. Except where indicated, all infections represented five of five animals infected. For the C57BL/6 mice, data from KE88-3887 and KE88-3887R were combined due to low infectivity by these strains.
Cytokine comparisons.
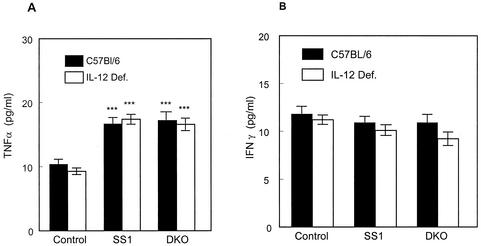
To investigate whether the activation of type 1 cytokines, such as IFN-γ, might be altered in the infected IL-12-deficient mouse and contribute to the increased permissiveness for infection, we examined stomach biopsy samples for TNF-α and IFN-γ production following short-term culture of SS1- and DKO-infected animals. Figure 4 shows that TNF-α production increased by 3 weeks postinfection, and while the values were significant over controls (the triple asterisk denotes P = 0.001), no differences in TNF-α levels were noted between wild-type and IL-12-deficient animals. In contrast, no differences in production of IFN-γ were noted from controls for SS1- or DKO-infected animals. These results suggest that levels of IFN-γ production in the stomach tissue of mice are not primarily regulated by IL-12 in either the presence or the absence of H. pylori infection.
FIG. 4.
Cytokine profiles of mice infected with H. pylori. TNF-α and IFN-γ levels were measured from punch biopsy material obtained from the stomachs of infected mice 20 days postinfection with H. pylori SS1 and DKO mutant (hrcA hspR). The means and standard deviations were determined, and the triple asterisks indicate statistically significant differences over uninfected controls (P = 0.001). Def., deficient.
DISCUSSION
Gastric species of Helicobacter efficiently colonize the gastric mucosa of mammals, where they often persist for the life of the host. Since nearly all mammalian species are colonized by a unique, genetically distinct species of Helicobacter (8), development of robust animal models for study of the human gastric species H. pylori must overcome natural barriers to infection, which may differ for each animal model. Here we demonstrate that mice deficient in the production of IL-12 are highly permissive for colonization by H. pylori. In comparison with parental C57BL/6 mice, the IL-12-deficient mice were (i) colonized to a higher bacterial density by the SS1 mouse-adapted strain (>1 log CFU/g of stomach), (ii) were more efficiently colonized by mutant strains deficient in regulation of heat shock genes, and (iii) were permissive for colonization by the motile variant (KE88-3887) of KE26695. Previously, pathogenesis studies with the KE88-3887 strain were limited to gnotobiotic piglets (21, 22) and beagle dogs (44). Our studies would suggest that, in mice, IL-12 production and its downstream immunomodulating activities might contribute to the natural resistance of mice to infection by human strains of H. pylori. Alternatively, human strains might express IL-12, promoting antigens that are not produced by Helicobacter species that naturally colonize mice (e.g., Helicobacter felis).
The piglet-passaged KE88-3887 strain of H. pylori is a motile variant of the nonmotile strain whose genome was completely sequenced by TIGR (51); presumably the two variants are isogenic and differ only in the DNA sequence of the fliP gene (33). Functional FliP is required for flagellum synthesis and therefore motility, and genetic analysis demonstrated that expression of this gene is controlled by slipped-strand mispairing mutagenesis in an 8-base repeat sequence of cytosines (8-nucleotide C tracks) in the fliP gene (33). The nonmotile KE26695 strain did not colonize either mouse model, and analysis of the proteomes of KE26695 and KE88-3887 by two-dimensional gel electrophoresis confirmed that the nonmotile KE26695 strain was deficient in production of FlaA and FlaB (data not presented), as had been previously reported (33), but was otherwise indistinguishable.
While the piglet-passaged KE88-3887 strain reportedly does not colonize mice, we found that this strain transiently colonized C57BL/6 mice, albeit poorly, and with bacterial loads at the limit of detection (∼103 CFU/g of stomach). We suggest that the production of IL-12 by the C57BL/6 mice ultimately leads to clearance of the infection by 8 weeks postinfection.
The KE26695 strain has been tested recently in two other animal models (25, 50). In a suckling mouse model KE26695 was one of several strains of H. pylori shown to very transiently colonize this mouse (25). In another study, Lex and Ley mutants constructed in KE26695 (motile) were shown to colonize C3H/HeJ lipopolysaccharide (LPS)-nonresponder mice (point mutation in Toll-like receptor 4 [TLR4]) (50). The near-equivalence of this mouse model for KE26695 (KE88-3887) suggests that LPS-mediated activation of inflammatory cytokines through the action of TLR4 might also lead to deficiency in IL-12 production. While the issue was not directly addressed in either study, molecular studies show that TLR4 activates the promoter for human p40 of IL-12 (55). Limited studies with H. pylori further establish that LPS from type I strains (cagA+) stimulates cytokine production via TLR4 (46). In addition to LPS, our studies with the DKO mutant suggest that the overproduction of Hsp60 and to a lesser extent Hsp70 might contribute to host resistance. In support of overexpression of heat shock proteins contributing to host resistance, a spontaneous mutant of DKO (RDKO) that produced normal levels of Hsp60 (Fig. 3) was nearly wild type for colonization of mice. This would suggest that it is the increased production of Hsp60 and immune modulation by this antigen (innate or acquired), rather than other genes whose expression is controlled by HspR, that are responsible for the lower colonization efficiency observed for C57BL/6 mice. Taken together, these studies suggest a role for both LPS and heat shock proteins in promoting a Th1-type response in the gastric mucosa to H. pylori infection by pathways linking TLR4 and IL-12.
IL-12-deficient mice infected with H. pylori produced IFN-γ from stomach tissue at levels indistinguishable from that produced by C57BL/6 mice, suggesting that other routes of IFN-γ induction, such as by IL-18-regulated pathways, might be involved. Our studies suggest that IFN-γ alone may not contribute to eradication of gastric H. pylori. While no differences were noted in IFN-γ production between uninfected control animals and SS1-infected mice, gastric biopsy material from infected mice (C57BL/6 and IL-12-deficient mice) exhibited significant increases in levels of TNF-α. These observations are in keeping with the development of a local inflammatory response as a result of transient or sustained infection. However, again, the absence of IL-12 did not blunt the degree of TNF-α production observed. Elevations of IL-12 have been previously demonstrated to be associated with H. pylori infection in both mouse models (35) and human disease (7, 28). Moreover, recent studies have shown that IL-12 production is required for development of protective immunity in a mouse infection model (2). Our findings confirm a pivotal role for this cytokine in host defense against H. pylori, while demonstrating that neither the local TNF-α nor IFN-γ production in the stomach of infected or uninfected animals is IL-12 dependent. Both the lower threshold of infection and the increased colonization density noted with IL-12-deficient mice support a role for IL-12 in early stages of colonization (innate immunity) rather than in persistence (adaptive immunity).
In summary, we have characterized an IL-12-deficient mouse model that is more permissive than its parent C57BL/6 mouse for colonization by H. pylori. This study also confirms an important role for IL-12 in host defense against H. pylori (2, 17, 41). Using this model, we were able to establish infection for at least 8 weeks with KE88-3887, a motile variant of KE26695 for which the complete genomic DNA sequence is available. The ability to work with an H. pylori strain for which complete genomic information is available, as well as one containing a functional cag pathogenicity island (15), should greatly facilitate studies of these genes and others in mechanisms of pathogenesis. The model has advantages in that genes associated with persistence can be distinguished from genes associated with colonization. When combined with high-throughput vector-free allelic replacement mutagenesis, developed for use in KE26695 (12), it should be possible to rapidly screen nonessential genes for their roles in pathogenesis.
Acknowledgments
We thank Ye Song Wei and Linda Best for technical assistance, Pat Cole for histological support, Kathryn A. Eaton for kindly providing the piglet-passaged KE88-3887 strains, and Douglas E. Berg for helpful suggestions.
This work was supported by grants from the Canadian Institutes for Health Research (ROP 357514) and Astrazeneca to P.S.H., S.J.O.V.V., and J.S.M.
Editor: B. B. Finlay
REFERENCES
- 1.Achtman, M., T. Azuma, D. E. Berg, Y. Ito, G. Morelli, Z. J. Pan, S. Suerbaum, S. A. Thompson, A. van der Ende, and L. J. van Doorn. 1999. Recombination and clonal groupings within Helicobacter pylori from different geographic regions. Mol. Microbiol. 32:459-470. [DOI] [PubMed] [Google Scholar]
- 2.Akhiani, A. A., J. Pappo, Z. Kabok, K. Schon, W. Gao, L. E. Franzen, and N. Lycke. 2002. Protection against Helicobacter pylori infection following immunization is IL-12-dependent and mediated by Th1 cells. J. Immunol. 169:6977-6984. [DOI] [PubMed] [Google Scholar]
- 3.Akopyants, N. S., K. A. Eaton, and D. E. Berg. 1995. Adaptive mutation and cocolonization during Helicobacter pylori infection of gnotobiotic piglets. Infect. Immun. 63:116-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alm, R. A., L.-S. L. Ling, D. T. Moir, N. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelson, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel, F. M., et al. (ed.). 1998. Current protocols in molecular biology, vol. 1, p. 1.1.1. Wiley Interscience, New York, N.Y.
- 6.Baker, L. M., A. Raudonikiene, P. S. Hoffman, and L. B. Poole. 2001. Essential thioredoxin-dependent peroxiredoxin system from Helicobacter pylori: genetic and kinetic characterization. J. Bacteriol. 183:1961-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauditz, J., M. Ortner, M. Bierbaum, G. Niedobitek, H. Lochs, and S. Schreiber. 1999. Production of IL-12 in gastritis relates to infection with Helicobacter pylori. Clin. Exp. Immunol. 117:316-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergin, I. L., and J. G. Fox. 2002. Animal models of Helicobacter pylori infection, p. 215-251. In Y. Yamamoto, H. Friedman, and P. S. Hoffman (ed.), Helicobacter pylori infection and immunity. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 9.Bjorkholm, B. M., J. L. Guruge, J. D. Oh, A. J. Syder, N. Salama, K. Guillemin, S. Falkow, C. Nilsson, P. G. Falk, L. Engstrand, and J. I. Gordon. 2002. Colonization of germ-free transgenic mice with genotyped Helicobacter pylori strains from a case-control study of gastric cancer reveals a correlation between host responses and HsdS components of type I restriction-modification systems. J. Biol. Chem. 277:34191-34197. [DOI] [PubMed] [Google Scholar]
- 10.Blaser, M. J., and D. Kirschner. 1999. Dynamics of Helicobacter pylori colonization in relation to the host response. Proc. Natl. Acad. Sci. USA 96:8359-8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chalker, A. F., H. W. Minehart, N. J. Hughes, K. K. Koretke, M. A. Lonetto, K. K. Brinkman, P. V. Warren, A. Lupas, M. J. Stanhope, J. R. Brown, and P. S. Hoffman. 2001. Systematic identification of selective essential genes in Helicobacter pylori by genome prioritization and allelic replacement mutagenesis. J. Bacteriol. 183:1259-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Covacci, A., J. L. Telford, G. Del Guidice, J. Parsonnet, and R. Rappuoli. 1999. Helicobacter pylori virulence and genetic geography. Science 284:1328-1333. [DOI] [PubMed] [Google Scholar]
- 14.Cover, T. L., and H. J. Blaser. 1999. Helicobacter pylori factors associated with disease. Gastroenterology 117:257-261. [DOI] [PubMed] [Google Scholar]
- 15.Crabtree, J. E., R. L. Ferrero, and J. G. Kusters. 2002. The mouse colonizing Helicobacter pylori strain SS1 may lack a functional cag pathogenicity island. Helicobacter 7:139-141. [DOI] [PubMed] [Google Scholar]
- 16.Debets-Ossenkopp, Y. J., R. G. Pot, D. J. van Westerloo, A. Goodwin, C. M. Vandenbroucke-Grauls, D. E. Berg, P. S. Hoffman, and J. G. Kusters. 1999. Insertion of mini-IS605 and deletion of adjacent sequences in the nitroreductase (rdxA) gene cause metronidazole resistance in Helicobacter pylori NCTC11637. Antimicrob. Agents Chemother. 43:2657-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Jonge, R., J. G. Kusters, M. S. Timmer, V. Gimmel, B. J. Appelmelk, S. Bereswill, A. H. van Vliet, S. G. Meuwissen, M. Kist, C. M. Vandenbroucke-Grauls, and E. J. Kuipers. 2001. The role of Helicobacter pylori virulence factors in interleukin production by monocytic cells. FEMS Microbiol. Lett. 196:235-238. [DOI] [PubMed] [Google Scholar]
- 18.Delany, I., G. Spohn, R. Rappuoli, and V. Scarlato. 2002. In vitro selection of high affinity HspR-binding sites within the genome of Helicobacter pylori. Gene 283:63-69. [DOI] [PubMed] [Google Scholar]
- 19.Derrico, C. A., and K. J. Goodrum. 1996. Interleukin-12 and tumor necrosis factor alpha mediate innate production of gamma interferon by group B Streptococcus-treated splenocytes of severe combined immunodeficiency mice. Infect. Immun. 64:1314-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eaton, K. A., D. Kersulyte, M. Mefford, S. J. Danon, S. Krakowka, and D. E. Berg. 2001. Role of Helicobacter pylori cag region genes in colonization and gastritis in two animal models. Infect. Immun. 69:2902-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eaton, K. A., D. R. Morgan, and S. Krakowka. 1989. Campylobacter pylori virulence factors in gnotobiotic piglets. Infect. Immun. 57:1119-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garhart, C. A., R. W. Redline, J. G. Nedrud, and S. J. Czinn. 2002. Clearance of Helicobacter pylori infection and resolution of postimmunization gastritis in a kinetic study of prophylactically immunized mice. Infect. Immun. 70:3529-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodwin, A., D. Kersulyte, G. Sisson, S. J. Veldhuyzen van Zanten, D. E. Berg, and P. S. Hoffman. 1998. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol. Microbiol. 28:383-393. [DOI] [PubMed] [Google Scholar]
- 25.Guo, B. P., and J. J. Mekalanos. 2002. Rapid genetic analysis of Helicobacter pylori gastric mucosal colonization in suckling mice. Proc. Natl. Acad. Sci. USA 99:8355-8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helsel, L. O., W. F. Bibb, C. A. Butler, P. S. Hoffman, and R. M. McKinney. 1987. Recognition of a genus-wide antigen of Legionella by a monoclonal antibody. Curr. Microbiol. 16:201-208. [Google Scholar]
- 27.Heuermann, D., and R. Haas. 1998. A stable shuttle vector system for efficient genetic complementation of Helicobacter pylori strains by transformation and conjugation. Mol. Gen. Genet. 257:519-528. [DOI] [PubMed] [Google Scholar]
- 28.Hida, N., T. Shimoyama, Jr., P. Neville, M. F. Dixon, A. T. Axon, T. Shimoyama, Sr., and J. E. Crabtree. 1999. Increased expression of IL-10 and IL-12 (p40) mRNA in Helicobacter pylori infected gastric mucosa: relation to bacterial cag status and peptic ulceration. J. Clin. Pathol. 52:658-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffman, P. S., and R. A. Garduno. 1999. Surface-associated heat shock proteins of Legionella pneumophila and Helicobacter pylori: roles in pathogenesis and immunity. Infect. Dis. Obstet. Gynecol. 7:58-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffman, P. S., A. Goodwin, J. Johnsen, K. Magee, and S. J. Veldhuyzen van Zanten. 1996. Metabolic activities of metronidazole-sensitive and -resistant strains of Helicobacter pylori: repression of pyruvate oxidoreductase and expression of isocitrate lyase activity correlate with resistance. J. Bacteriol. 178:4822-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huesca, M., S. Borgia, P. S. Hoffman, and C. Lingwood. 1996. Acidic pH changes receptor binding specificity of Helicobacter pylori: a binary adhesion model in which surface heat shock (stress) proteins mediate sulfatide recognition in gastric colonization. Infect. Immun. 64:2643-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huesca, M., A. Goodwin, A. Bhagwansingh, P. S. Hoffman, and C. Lingwood. 1998. Characterization of an acidic-pH-inducible stress protein (hsp70), a putative sulfatide binding adhesin, from Helicobacter pylori. Infect. Immun. 66:4061-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Josenhans, C., K. A. Eaton, T. Thevenot, and S. Suerbaum. 2000. Switching of flagellar motility in Helicobacter pylori by reversible length variation of a short homopolymeric sequence repeat in fliP, a gene encoding a basal body protein. Infect. Immun. 68:4598-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kishikawa, H., R. Song, and D. A. Lawrence. 1997. Interleukin-12 promotes enhanced resistance to Listeria monocytogenes infection of lead-exposed mice. Toxicol. Appl. Pharmacol. 147:180-189. [DOI] [PubMed] [Google Scholar]
- 35.Konturek, P. C., T. Brzozowski, S. J. Konturek, J. Stanchura, E. Karczewska, R. Pajdo, P. Ghiara, and E. G. Hahn. 1999. Mouse model of Helicobacter pylori infection: studies of gastric function and ulcer healing. Aliment. Pharmacol. Ther. 13:333-346. [DOI] [PubMed] [Google Scholar]
- 36.Lee, A., J. O'Rourke, M. C. De Ungria, B. Robertson, G. Daskalopoulos, and M. F. Dixon. 1997. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology 112:1386-1397. [DOI] [PubMed] [Google Scholar]
- 37.Logan, S. M., J. W. Conlan, M. A. Monteiro, W. W. Wakarchuk, and E. Altman. 2000. Functional genomics of Helicobacter pylori: identification of a beta-1,4 galactosyltransferase and generation of mutants with altered lipopolysaccharide. Mol. Microbiol. 35:1156-1167. [DOI] [PubMed] [Google Scholar]
- 38.Lund, P. A. 2001. Microbial molecular chaperones. Adv. Microb. Physiol. 44:93-140. [DOI] [PubMed] [Google Scholar]
- 39.Marchetti, M., and R. Rappuoli. 2002. Isogenic mutants of the cag pathogenicity island of Helicobacter pylori in the mouse model of infection: effects on colonization efficiency. Microbiology 148:1447-1456. [DOI] [PubMed] [Google Scholar]
- 40.Marshall, B. J., and J. R. Warren. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet i:1311-1315. [DOI] [PubMed]
- 41.Meyer, F., K. T. Wilson, and S. P. James. 2000. Modulation of innate cytokine responses by products of Helicobacter pylori. Infect. Immun. 68:6265-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parsonnet, J., G. D. Friedman, D. P. Vandersteen, Y. Chang, J. H. Vogelman, N. Orentreich, and R. K. Sibley. 1991. Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med. 325:1127-1131. [DOI] [PubMed] [Google Scholar]
- 43.Philpott, D. J., D. Belaid, P. Troubadour, J. M. Thiberge, J. Tankovic, A. Labigne, and R. L. Ferrero. 2002. Reduced activation of inflammatory responses in host cells by mouse-adapted Helicobacter pylori isolates. Cell. Microbiol. 4:285-296. [DOI] [PubMed] [Google Scholar]
- 44.Radin, M. J., K. A. Eaton, S. Krakowka, D. R. Morgan, A. Lee, G. Otto, and J. Fox. 1990. Helicobacter pylori gastric infection in gnotobiotic beagle dogs. Infect. Immun. 58:2606-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 46.Shimoyama, T., and J. E. Crabtree. 1997. Mucosal chemokines in Helicobacter pylori infection. J. Physiol. Pharmacol. 48:315-323. [PubMed] [Google Scholar]
- 47.Sizemore, C. F., J. D. Quispe, K. M. Amsler, T. C. Modzelewski, J. J. Merrill, D. A. Stevenson, L. A. Foster, and A. M. Slee. 2002. Effects of metronidazole, tetracycline, and bismuth-metronidazole-tetracycline triple therapy in the Helicobacter pylori SS1 mouse model after 1 day of dosing: development of an H. pylori lead selection model. Antimicrob. Agents Chemother. 46:1435-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spohn, G., and V. Scarlato. 1999. The autoregulatory HspR repressor protein governs chaperone gene transcription in Helicobacter pylori. Mol. Microbiol. 34:663-674. [DOI] [PubMed] [Google Scholar]
- 49.Suerbaum, S., J.-M. Thiberge, I. Kansau, R. L. Ferrero, and A. Labigne. 1994. Helicobacter pylori hspA-hspB heat shock gene cluster: nucleotide sequence, expression, putative function and immunogenicity. Mol. Microbiol. 14:959-974. [DOI] [PubMed] [Google Scholar]
- 50.Takata, T., E. El-Omar, M. Camorlinga, S. A. Thompson, Y. Minohara, P. B. Ernst, and M. J. Blaser. 2002. Helicobacter pylori does not require Lewis X or Lewis Y expression to colonize C3H/HeJ mice. Infect. Immun. 70:3073-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomb, J.-F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 52.Vorobjova, T., O. Ananieva, H. Maaroos, P. Sipponen, K. Villako, M. Utt, I. Nilsson, T. Wadstrom, and R. Uibo. 2001. Seropositivity to Helicobacter pylori heat shock protein 60 is strongly associated with intensity of chronic inflammation, particularly in antrum mucosa: an extension of an 18-year follow-up study of chronic gastritis in Saaremaa, Estonia. FEMS Immunol. Med. Microbiol. 30:143-149. [DOI] [PubMed] [Google Scholar]
- 53.Wang, Y., P. Roos, and D. E. Taylor. 1993. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. J. Gen. Microbiol. 139:2485-2493. [DOI] [PubMed] [Google Scholar]
- 54.Wang, Y., and D. E. Taylor. 1990. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene 94:23-28. [DOI] [PubMed] [Google Scholar]
- 55.Weinmann, A. S., D. M. Mitchell, S. Sanjabi, M. N. Bradley, A. Hoffmann, H.-C. Liou, and S. T. Smale. 2001. Nucleosome remodeling at the IL-12 p40 promoter is a TLR-dependent, Rel-independent event. Nat. Immunol. 2:51-57. [DOI] [PubMed] [Google Scholar]