Abstract
The exocyst is an essential multiprotein complex mediating polarized secretion in yeast. Here we describe a gene, SEM1, that can multicopy-suppress exocyst mutants sec3–2, sec8–9, sec10–2, and sec15–1. SEM1 is highly conserved among eukaryotic species. Its human homologue, DSS1, has been suggested as a candidate gene for the split hand/split foot malformation disorder. SEM1 is not an essential gene. However, its deletion rescued growth of the temperature-sensitive exocyst mutants sec3–2, sec8–9, sec10–1, and sec15–1 at the restrictive temperature. Cell fractionation showed that Sem1p is mainly cytosolic but also associates with the microsomal fraction. In linear sucrose gradients, Sem1p cosedimented with the exocyst component Sec8p. In diploid cells that normally do not form pseudohyphae (S288C background), deletion of SEM1 triggered pseudohyphal growth. This phenotype was abolished after reintroduction of either SEM1 or the mouse homologue Dss1 into the cells. In diploids that have normal capacity for pseudohyphal growth (Σ1278b background), deletion of SEM1 enhanced filamentous growth. The functionality of both SEM1 and Dss1 in a differentiation process in yeast suggests that Dss1 indeed could be the gene affected in the split hand/split foot malformation disorder. These results characterize SEM1 as a regulator of both exocyst function and pseudohyphal differentiation and suggest a unique link between these two cellular functions in yeast.
The secretory pathway in eukaryotic cells is composed of series of membrane-bound compartments that communicate with each other using transport vesicles. Targeting and fusion of vesicles is regulated by transport step-specific, homologous sets of proteins and general factors shared by multiple, if not all, targeting and fusion events. The general factors in yeast include Sec18p (NSF) and Sec17p (α-SNAP) (1, 2). The synaptobrevins/cellubrevins and syntaxins, present on transport vesicles and target membranes, respectively, act as receptors for NSF and SNAP, forming the so called SNARE (SNAP receptor) complexes (3) that participate in targeting and fusion of vesicles with the target membrane (4). The Sec1p-related proteins and small GTPases of the Rab/Ypt subfamily represent additional levels of regulation for each vesicle fusion event (5, 6).
Fusion of the vesicle with the plasma membrane appears to require yet another additional level of regulation. A protein complex, the exocyst, composed of at least Sec3p, -5p, -6p, -8p, -10p, and -15p and Exo70p (7, 8) is indispensable for vesicle fusion at the plasma membrane and for cell viability in yeast. The presence of Sec8p in the bud tip and random budding observed in certain sec3 mutants suggested that the complex is involved in the bud site selection (9–11). Recently, it was shown that Sec3p localizes stably at the sites of exocytosis independent of ongoing membrane traffic or actin and septin cytoskeleton or other components of the exocyst (12). This indicates that Sec3p could mediate the association of the exocyst with the plasma membrane and have a role as a spatial landmark for polarized secretion (12). However, the molecular interactions of the exocyst subcomponents and the precise role of the complex in vesicle transport remain to be revealed. In mammalian cells, a similar multiprotein complex has been characterized, indicating that this level of regulation in the secretory process also is conserved (13, 14).
A central feature of Saccharomyces cerevisiae is a polarized mode of growth (15). Haploid cells bud axially, i.e., the new bud site on the mother cell forms adjacent to the former bud. In diploid cells, budding follows a bipolar mode in which the new bud forms alternately, either adjacent to the former bud site or at the opposite pole of the cell. The ability of S. cerevisiae to grow in polarized fashion is clearly manifested in cells grown on solid low-nitrogen medium. Under these conditions, diploid cells normally undergo a differentiation process causing a change from bipolar budding to unipolar budding. This change is connected to elongation of individual cells, formation of pseudofilaments, and a capability of cells to invade the growth substratum (16). Here we report isolation and characterization of a gene of S. cerevisiae, SEM1, which is highly conserved in evolution and appears to act as a regulator both for the exocyst complex function and pseudohyphal differentiation.
MATERIALS AND METHODS
Yeast Strains and Culture Conditions.
The yeast strains used are shown in Table 1. Meiosis and sporulation were induced on agar plates (0.1% Difco yeast extract, 1% potassium acetate, 0.05% glucose, and 2% Difco Bacto agar). The nitrogen starvation medium (SLAD) contained 132 mg/l (NH4)2SO4, Difco yeast nitrogen base with no amino acids, 2% washed Bacto agar (16), and (when appropriate) uracil.
Table 1.
Yeast strains
| Strain | Genotype | Source |
|---|---|---|
| NY179 | a leu2-3,112 ura3-52 | P. Novick |
| NY15 | α ura3-52 his4-619 | P. Novick |
| NY3 | a sec1-1 ura3-52 | P. Novick |
| NY24 | a sec1-11 ura3-52 | P. Novick |
| NY770 | α sec2-41 leu2-3,112 ura3-52 | P. Novick |
| NY772 | a sec3-2 leu2-3,112 ura3-52 | P. Novick |
| NY774 | α sec4-8 leu2-3,112 ura3-52 | P. Novick |
| NY776 | α sec5-24 leu2-3,112 ura3-52 | P. Novick |
| NY778 | a sec6-4 leu2-3,112 ura3-52 | P. Novick |
| sf821-8A | a sec7-1 his4-580 leu2-3,112 trp1-289 ura3-52 | R. Schekman |
| NY780 | a sec8-9 leu2-3,112 ura3-52 | P. Novick |
| NY782 | a sec9-4 leu2-3,112 ura3-52 | P. Novick |
| NY784 | a sec10-2 leu2-3,112 ura3-52 | P. Novick |
| NY786 | a sec15-1 leu2-3,112 ura3-52 | P. Novick |
| BY54 | a sec17-1 ura3-52 | P. Brennwald |
| mBY12-6D | α sec18-1 his− leu2-3,112 trpl-289 ura3-52 | R. Schekman |
| NY1213 | α sec19-1 leu2-3,112 ura3-52 | P. Novick |
| NY1427 | α ura3-52 sec8Δ∷URA3 leu2-3,112∷(LEU2, SEC8-3X-c-myc) L-A-o | P. Novick |
| H615 | a leu2-3,112 ura3-52 mso1∷LEU2 | Ref. 22 |
| H1286 | a ura3-52 leu2-3, 112 sem1∷URA3 | This study |
| H1352 | α ura3-52 leu2-3, 112 sem1∷URA3 | This study |
| H1557 | a ura3-52 leu2-3, 112 sem1Δ∷kanr | This study |
| H1908 | a sec1-1 ura3-52 sem1∷URA3 | This study |
| H1900 | a sec3-2 leu2-3,112 ura3-52 sem1∷URA3 | This study |
| H1906 | α sec4-8 leu2-3,112 ura3-52 sem1∷URA3 | This study |
| H2034 | α sec5-24 leu2-3,112 ura3-52 sem1∷URA3 | This study |
| H2035 | a sec6-4 leu2-3,112 ura3-52 sem1∷URA3 | This study |
| H2036 | a sec8-9 leu2-3,112 ura3-52 sem1∷URA3 | This study |
| H1902 | a sec10-2 leu2-3,112 ura3-52 sem1∷URA3 | This study |
| H1904 | a sec15-1 leu2-3,112 ura3-52 sem1∷URA3 | This study |
| H1558 | a sec3-2 leu2-3,112 ura3-52 sem1Δ∷kanr | This study |
| H1561 | α sec5-24 leu2-3,112 ura3-52 sem1Δ∷kanr | This study |
| H1562 | a sec8-9 leu2-3,112 ura3-52 sem1Δ∷ kanr | This study |
| H1559 | a sec10-2 leu2-3,112 ura3-52 sem1Δ∷kanr | This study |
| H1560 | a sec15-1 leu2-3,112 ura3-52 sem1Δ∷kanr | This study |
| HS33-1 | α sec15-1 leu2-3,112 ura3-52 his4-260 trp l ade2-1 | Ref. 26 |
| 12T7c* | a ura3 | E. Dubois |
| O2933b* | α ura3 | E. Dubois |
| H1927* | a/α ura3/ura3 | This study |
| SKY24* | a/α ura3/ura3 sem1∷URA3/sem1∷URA3 | This study |
| H1700 | a/α ura3-52/ura3-52 leu2-3,112/LEU2 HIS4/his4-619 | This study |
| H1699 | a/α ura3-52/ura3-52 leu2-3,112/LEU2 HIS4/his4-619 sem1∷URA3/sem1∷URA3 | This study |
| H1809 | a/α ura3-52/ura3-52 leu2-3,112/LEU2 HIS4/his4-619 sem1Δ∷kanr/sem1Δ∷kanr | This study |
Strains congenic to Σ 1278b.
Plasmids.
The original SEM1 clone (YEpSEM1T) was isolated from a cDNA library (17) in HS33–1. SEM1 ORF was cloned by using PCR from genomic DNA (S288C background) with BamHI/XhoI sites added to Bluescript (SK)− to yield pBSEM1 and sequenced. YEpSEM1U is pVT102U (18) (from T. Vernet, Biotechnology Research Institute, Montreal, Canada) containing SEM1 ORF as a BamHI fragment. The plasmid pGEXSEM1∷URA3 was created by inserting the URA3 gene in SEM1 between XbaI and NdeI sites. His6-Sem1p was expressed from pGAT-4 (from J. Peränen, Institute of Biotechnology, University of Helsinki, Finland). The Dss1 was cloned by using PCR from mouse kidney cDNA to the BamHI/XhoI sites in Bluescript SK (−), resulting in pBDss1. The product was sequenced and cloned further to pVT102U to yield YEpDss1U. FLO8 was expressed from pHL135 (from G. Fink, Massachusetts Institute of Technology, Cambridge, MA) (19).
Disruption of the Chromosomal SEM1.
Strains were transformed with the sem1∷URA3 cassette, and the Ura+ phenotype was selected. The deletion of the entire SEM1 ORF was done by using the kanMX4 module (20). The sem1∷kanr transformants were grown on yeast extract/peptone/dextrose/(YPD) plates overnight and then replicated on YPD containing 200 mg/l G418 (Geneticin, GIBCO/BRL). The disruptions were verified either by Southern blotting or by PCR and Western blotting.
Antibodies.
Polyclonal antibodies against the purified His6–Sem1p were generated in New Zealand White rabbits. The antibodies against the Sso2p have been described (21). The anti-myc mAb was produced as ascites fluid in BALB/c mice inoculated with the hybridoma 9E10 [European Collection of Animal and Cell Cultures (ECACC) 85102202].
Genetic Interactions.
For multicopy suppression analysis, the sec mutants (sec1–1, sec1–11, sec2–4, sec3–2, sec4–8, sec5–24, sec6–4, sec7–1, sec8–9, sec9–4, sec10–2, sec15–1, sec17–1, sec18–1, and sec19–1) were transformed with YEpSEM1U or pVT102U at the permissive temperature. The growth of four individual transformants was monitored for three days at 38.5, 37, 36, 35, 30, and 24°C. For synthetic lethalities the a- and α-sem1∷URA3-disruptant strains were crossed with the sec mutants above and with a MSO1 disruptant (22). Diploids were sporulated and tetrads were dissected, and their ability to grow on SC-Ura medium at 24°C and on YPD at 37°C was monitored.
Cell Fractionation and Sucrose Velocity Gradients.
Cell lysate preparation, cell fractionation, and analysis of Sem1p membrane association was carried out as described (22). For sucrose gradient analysis, lysis buffer contained 0.2 M sorbitol. The cell homogenate was centrifuged for 10 min at 10,000 rpm in a Sorvall SS-34 rotor, and the supernatant was either frozen at −70°C or used directly. Samples were layered on top of linear 10–30% (wt/vol) sucrose gradients containing 0.1% Triton X-100 and 1 mM DTT. Gradients were spun at 40,000 rpm in a Beckman SW-41 rotor for 8 hr, after which 500-μl fractions were collected from the top. Sedimentation standards were BSA (4.5 S), catalase (11.5 S), and thyroglobulin (19.3 S) (Pharmacia).
Other Methods.
Proteins were separated by using either the Laemmli (23) or the Schägger and Jagov (24) system and detected in Western blots with specific antibodies followed either by 35S-labeled protein A (Amersham) or enhanced chemiluminescence (Amersham). To remove nonspecific background in the Western blots, Sem1p antiserum was incubated for 1 hr at 4°C with 1% (wt/vol) acetone powder prepared from the sem1-Δ1 strain (25), centrifuged for 10 min at 10,000 × g, and used for labeling. Sequence homologies were analyzed with gap and pileup programs of the Genetics Computer Group (Madison, WI).
RESULTS
SEM1 Encodes a Small Acidic Protein.
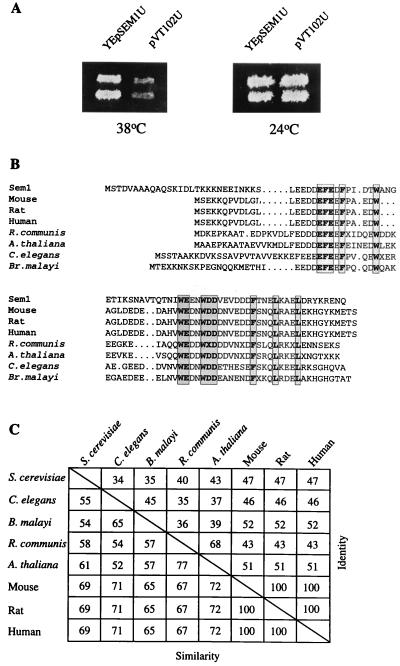
The complex genetic interactions with several of the late-acting SEC genes suggest a central role for SEC15 in the regulation of vesicle fusion at the plasma membrane (26, 27). We therefore reasoned that additional components might be uncovered through genetic interactions with SEC15. The SEM1 (Suppressor of Exocyst Mutations) gene was cloned as a multicopy suppressor of the temperature-sensitive mutation sec15–1 (Fig. 1A). DNA sequencing revealed an 89-aa ORF. The encoded protein has a predicted molecular mass of 10,386 daltons, is hydrophilic, and has an acidic pI (4.2). SEM1 is located in chromosome 4 adjacent to CDC40, but because of its small size, Sem1p does not appear as a putative ORF in the yeast genome database. The only known protein motifs in Sem1p detected in the prosite database are two putative protein kinase C and 2 casein kinase II phosphorylation sites.
Figure 1.
(A) A multicopy plasmid YEpSem1U, but not the vector plasmid alone, suppresses the growth of sec15–1 at 38°C. Two independent sec15–1 transformants are shown. (B) SEM1 is highly conserved in evolution. Comparison of several Sem1p homologues translated from dbEST database sequences. The fully conserved amino acids are shown in shaded boxes. R. communis, Ricinus communis; A. thaliana, Arabidopsis thaliana; C. elegans, Caenorhabditis elegans; B. malayi, Brugia malayi. (C) The degree of similarity and identity of different Sem1p homologues. The GenBank accession numbers for the homologues are R. communis, T14813; A. thaliana, T41856; C. elegans, D76210; B. malayi, R88405; mouse, X853589; rat, H35331; and human, U41515.
Sem1p Is Conserved in Evolution.
Close homologues for SEM1 were found in the dbEST database in a wide spectrum of eukaryotes (Fig. 1 B and C). The human homologue, DSS1 (together with two other genes) has been mapped to the locus affected in the autosomal-dominant form of the split hand/split foot developmental disorder (28). The Sem1p homologues share a high level of sequence similarity, with the amino terminus being least conserved (Fig. 1B). Thirteen amino acids (15–19%) are completely conserved throughout the species, suggesting a crucial role for those residues. Remarkably, the mouse, rat, and human homologues are 100% identical (Fig. 3 B and C), and a partial cDNA clone of the zebrafish (Danio rerio) homologue in dbEST (AA566308) shares within the 54 amino-terminal amino acids >90% identity with the human protein.
Figure 3.
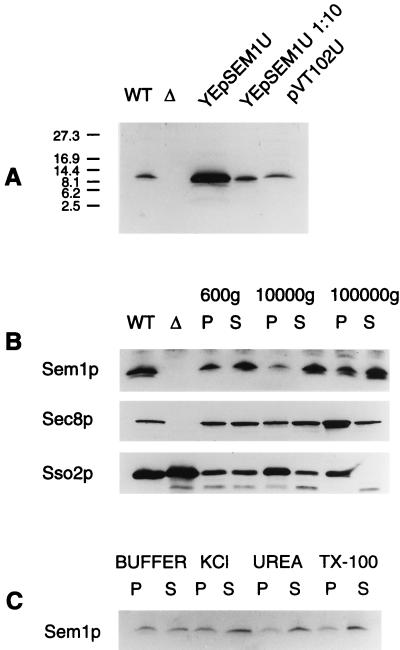
The Sem1 protein. (A) Polyclonal antibodies to Sem1p recognize a 12-kDa protein in wt cells. This protein is absent in sem1-Δ1 and present in increased amounts in SEM1-overexpressing cells. (B) Cell fractionation of NY1427. Sem1p is mainly soluble but also is found associated with the 100,000 × g pellet in wt cells. WT, total-cell lysate prepared from NY1427; Δ, total-cell lysate from H1286 (sem1-Δ1). (C) A majority of Sem1p is removed from the pellet material by 1 M KCl, 2.5 M urea, or 1% (wt/vol) Triton X-100 in 10 mM Hepes (pH 7.2). Roughly 50% of Sem1p can be extracted from the 100,000 × g pellet with 10 mM Hepes (pH 7.2) alone.
SEM1 Is Not Essential for Growth or Invertase Secretion.
Southern analysis suggested that SEM1 is a single-copy gene (data not shown), and indeed no significantly related sequences were found in the yeast genomic database. To test whether SEM1 is an essential gene, it was disrupted in diploid or haploid wild-type (wt) cells with URA3 or deleted entirely with the kanr cassette, producing sem1-Δ1 and sem1-Δ2, respectively. Disruption or deletion of SEM1 did not cause any obvious growth defects at different temperatures on synthetic or rich media containing different carbon sources. We conclude that SEM1 is a single-copy gene nonessential for growth.
Possible effects of SEM1 deletion on invertase secretion or on cellular morphology were studied in haploid sem1-Δ1 at 30°C. No significant difference in the amount of secreted invertase was observed between the wt and the sem1-Δ1 strain (data not shown). SEM1 deletion or overexpression in haploid strains at 24°C or 37°C, as examinated in the electron microscope, did not cause morphological changes, e.g. transport vesicle accumulation (data not shown).
Genetic Interactions Suggest a Function for SEM1 in Association with the Exocyst Complex.
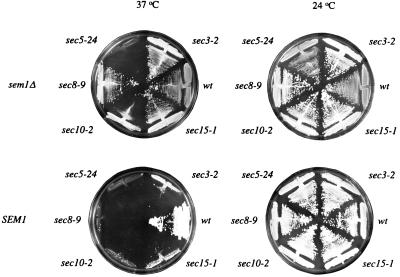
Cloning of SEM1 as a suppressor of sec15–1 suggested that SEM1 is involved in a late step of the secretory process. We therefore tested whether SEM1 could suppress other late-acting sec mutants. The mutant strains were transformed with a multicopy plasmid (YEpSEM1U) at the permissive temperature and screened for growth at elevated temperatures. SEM1 overexpression rescued also the growth of sec3–2, sec8–9, and sec10–2 at 37–38°C but had no effect on growth of other sec strains (data not shown). To gain further insight into the function of SEM1, we tested whether SEM1 deletion creates synthetic phenotypes with the sec mutants defective in Golgi to plasma membrane transport. sem1-Δ1 (Ura+) was crossed with the sec mutants (Ura−), sporulated, dissected, and the phenotypes of the spores were analyzed. An unexpected type of genetic interaction was detected with a subset of exocyst genes. Among the spore colonies derived from 20 tedrads of each cross, no Ura+ temperature-sensitive strains were recovered from crosses of sem1-Δ1 with sec3–2, sec8–9, sec10–2, or sec15–1. Interestingly, in most of the tetrads more than two spores grew at 37°C, whereas the Ura+/Ura− phenotype always segregated 2:2. This suggested that disruption of SEM1 rescued the growth of these mutants at 37°C. Rescue was confirmed by direct deletion of SEM1 in the haploid mutants. At 37°C, SEM1 deletion rescued the growth of sec3–2, sec8–9, sec10–2, and sec15–1 but not that of sec5–24 (Fig. 2), sec1–1, sec4–2, or sec6–4 (data not shown). The results suggest a negative regulatory role for SEM1 in connection with the exocyst complex.
Figure 2.
SEM1 is implicated as a negative regulator of the exocyst complex. Wt cells, exocyst mutants sec3–2, sec5–24, sec8–9, sec10–2, and sec15–1, and double mutants sem1-Δ2 sec3–2, sem1-Δ2 sec5–24, sem1-Δ2 sec8–9, sem1-Δ2 sec10–2, and sem1-Δ2 sec15–1 were grown on YPD plates at 24°C or 37°C. Deletion of SEM1 suppresses the temperature-sensitive phenotype of sec3–2, sec8–9, sec10–2, and sec15–1, in contrast to sec5–24.
The Sem1 Protein.
To study the Sem1 protein, polyclonal antibodies were produced in rabbits against Sem1p. In Western blots of wt cells, the antiserum detected a 12-kDa protein that was absent in the disruptant strain but was increased in cells overexpressing SEM1 (Fig. 3A). These data indicate that the 12-kDa band represents the Sem1p. The intracellular localization of Sem1p was studied by using cell fractionation. To simultanously analyze the intracellular distribution of the exocyst component Sec8p, the fractionation was performed in a strain in which the endogenous Sec8p had been replaced by a myc-tagged, functional Sec8p (8). Sem1p was found, to a large extent, in the soluble fraction after the high-speed centrifugation (Fig. 3B). However, a fraction of the protein associated with the 100,000 × g microsomal pellet. Under these conditions, Sec8p mainly associated with the 100,000 × g microsomal pellet, whereas a small amount of the protein was found in the soluble fraction. Sso2p was found entirely in the microsomal pellet—as expected for an integral membrane protein. Treatment of the microsomal pellet with 10 mM Hepes (pH 7.4) or with the buffer supplemented with 1 M KCl, 1% Triton X-100, or 2.5 M urea, followed by 100,000 × g pelleting, released most of the Sem1p to the supernatant (Fig. 3C). These results show that Sem1p exists as a soluble, loosely microsome-associated protein and that it partially cofractionates with Sec8p.
Sem1p Cosediments with Sec8p in Sucrose Gradients.
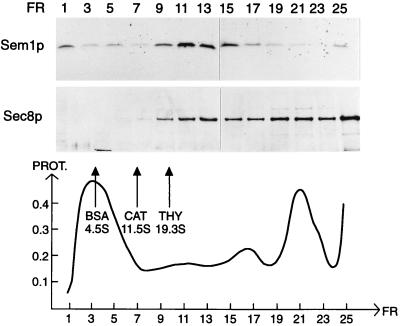
The genetic interactions between SEM1 and the exocyst genes suggested that Sem1p may functionally interact with the complex. To study possible cosedimentation of Sem1p with the exocyst complex or with its subcomponents, we analyzed sedimentation of Sem1p in linear sucrose gradients. The exocyst originally was identified as a complex sedimenting at 19.5 S in sucrose gradients (7). Recently, many of the exocyst components also were shown to be present in a significantly larger complex (9). Sedimentation of Sem1p and Sec8p-myc was analyzed in 10–30% linear sucrose gradients in the presence of 0.1% Triton X-100 and 1 mM DTT. Under these conditions, Sem1p cosedimented with Sec8p-myc at 20 S (Fig. 4). In accordance with the previous results (9), a significant portion of the Sec8p sedimented with a much higher sedimentation value than 20 S. Sem1p was not found in this region of the gradient.
Figure 4.
Sem1p cosediments with Sec8-myc at 20 S in linear sucrose gradients. Yeast-cell lysate (NY1427) was centrifuged in a linear 10–30% (wt/vol) sucrose gradient containing 0.1% Triton X-100 for 8 hr. Sedimentation of Sem1p and Sec8p was analyzed from fractions by using SDS/PAGE and Western blotting. The sedimentation markers are BSA (4.5 S), catalase (11.5 S), and thyroglobulin (19.3S). The amount of total protein is presented in arbitrary units.
SEM1 Deletion Induces Pseudohyphal Growth.
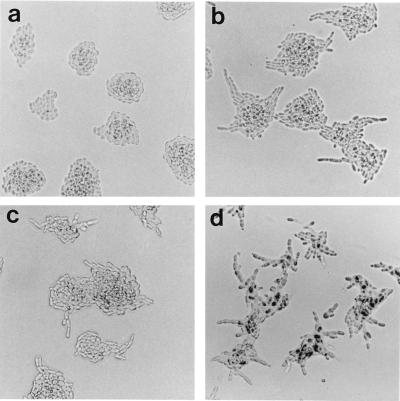
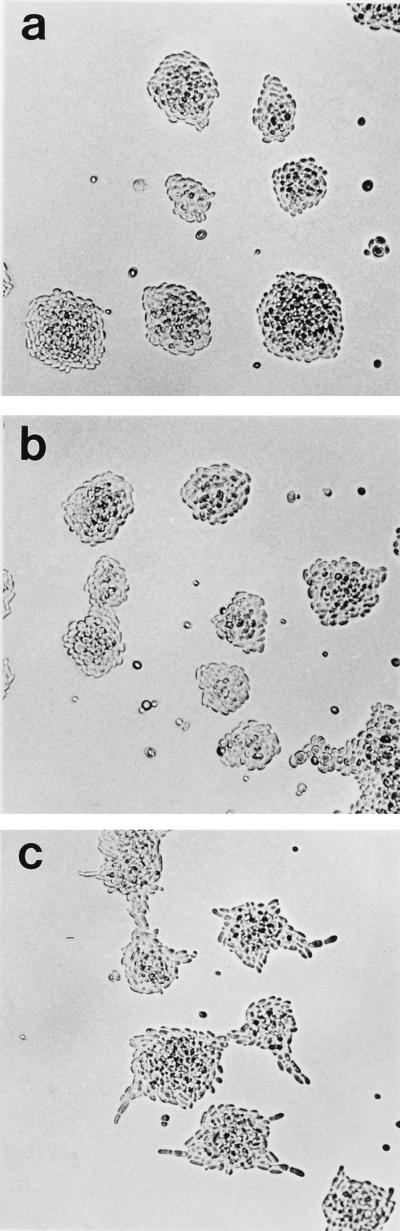
Because SEM1 deletion did not cause any phenotype under normal growth conditions, we proceeded to test possible effects in more specialized situations. Under nitrogen starvation, diploid S. cerevisiae normally undergoes a differentiation process leading to growth of pseudohyphae (16). The diploid S. cerevisiae strain used in these studies (S288C background) does not form pseudohyphae on SLAD low-nitrogen growth medium, presumably because of the flo8 mutation (19). Interestingly, pseudohyphal differentiation was induced in sem1-Δ1/sem1-Δ1 diploids grown overnight at 30°C on SLAD. These cells formed highly irregular colonies with chains of clearly elongated cells radiating outwards (Fig. 5b). At the same time, wt SEM1/SEM1 colonies were round and symmetrical (Fig. 5a). To test whether the lack of pseudohyphae formation in the wt strain was caused by the flo8 mutation, the wt diploid was transformed with FLO8. FLO8 expression allowed pseudohyphae formation on solid low-nitrogen medium (data not shown), thus proving that the only defect in pseudohyphae formation in the wt cells was the result of flo8 mutation. In sem1Δ/sem1Δ (S288C) cells, FLO8 expression further enhanced pseudohyphal growth (data not shown). The effect of SEM1 deletion (sem1-Δ1/sem1-Δ1) also was studied in the Σ1278b background, which is capable of forming pseudohyphae on SLAD. The wt Σ1278b showed a similar pseudohyphal phenotype (Fig. 5c), as did sem1/sem1 in S288C (Fig. 5b). Deletion of SEM1 appeared to enhance the pseudohyphal phenotype in Σ1278b (Fig. 5d). These cells formed many filaments from each colony and during longer growth periods on SLAD, they formed longer filaments that the wt Σ1278b (data not shown). The conserved nature of Sem1p tempted us to test the ability of a mammalian homologue to functionally replace Sem1p in yeast. For this purpose the mouse homologue, Dss1, was cloned from a mouse kidney cDNA. SEM1 and Dss1 were reintroduced in multicopy plasmids to sem1-Δ2/sem1-Δ2 diploids (S288C). Cells transformed with YEpSEM1U (Fig. 6a) or YEpDss1U (Fig. 6b) could not be distinguished from the wt cells (S288C) on SLAD plates, whereas cells transformed with the control plasmid retained pseudohyphal morphology (Fig. 6c). This result demontrates that the observed phenotype was caused by the deletion of SEM1 and that Dss1 can functionally replace SEM1 in the pseudohyphal differentation process in yeast.
Figure 5.
Deletion of SEM1 in diploid S288C cells triggers pseudohyphal differentiation. Homozygous SEM1 deletants (sem1-Δ1/sem1-Δ1) were grown in low-nitrogen SLAD medium (16). After overnight growth, SEM1 deletants form asterisk-shaped colonies (b). These colonies contain clearly elongated cells forming short filaments in contrast to wt diploids in which the cells are round and the colonies are smooth (a). wt Σ1278b diploids form pseudofilaments on SLAD (c). In Σ1278b deleted for SEM1 (sem1-Δ1/sem1-Δ1) (d), pseudohyphae formation is enhanced compared with the wt cells (c).
Figure 6.
The mammalian Dss1 is functional in a differentiation process in yeast. Transformation of sem1-Δ2/sem1-Δ2 diploids with YEpSEM1U (a) or YEpDss1U (b) multicopy plasmids represses the filament formation on SLAD plates, in contrast to control cells containing only the vector plasmid pVT102U (c). The colonies were grown overnight.
DISCUSSION
Sem1p Is Implicated in the Regulation of the Exocyst Complex.
Several lines of evidence suggest that Sem1p function is associated with the exocyst complex in yeast. SEM1 multicopy suppresses the exocyst mutants sec3–2, sec8–9, sec10–2, and sec15–1. Interestingly, deletion of SEM1 rescued (at the restrictive temperature) the growth of the same exocyst mutants (sec3–2, sec8–9, sec10–2, and sec15–1) but showed no genetic interaction with any other sec genes functioning in the Golgi to plasma membrane transport. Because removal of SEM1 from the exocyst mutants was needed to reestablish the essential function of the complex at the restrictive temperature, it appears that SEM1 acts as a negative regulator for the exocyst function. The possible role of Sem1p in the exocyst-complex regulation was studied further by using biochemical experiments. In linear sucrose gradients, Sem1p sedimented at 20 S. In the same gradients, Sec8p sedimented in two distinct positions, at 20 S and at a much higher S value. The existence of a form of exocyst complex with a sedimentation value over 20 S has been reported previously (9). Interestingly, Sem1p and Sec8p cosedimented only at 20 S. Such a partial cosedimentation is in accordance with observed partial cofractionation of Sem1p and Sec8p in cell fractionation.
Most exocyst mutants show either decreased stability or reduced amounts of the properly assembled exocyst complex (9). In such a situation, removal of Sem1p (a putative negative regulator) could allow sufficient complex formation to support cell growth. In this case, Sem1p would only associate with the 20S exocyst complex;it could be imagined that removal of Sem1p from this complex could allow formation of a larger, possibly functional exocyst complex. Another possibility is that deletion of SEM1 creates or activates an exocyst-bypass pathway. The fact that not only removal but also overexpression of Sem1p could suppress a subset of exocyst mutants can be explained in several ways. Overproduction of Sem1p could sequester a possible cofactor needed for Sem1p function, or the overproduced Sem1p may become nonfunctional because of misfolding, mislocalization, or lack of a posttranslational modification.
SEM1 and the Mammalian Homologue Dss1 Are Functionally Conserved.
The finding that SEM1 is highly conserved in evolution and is present in a wide spectrum of eukaryotes suggests that Sem1p performs an important cellular function in yeast. Close SEM1 homologues could be found from lower eukaryotic parasites to plants and to mammals. The presence of homologues in the dbEST database containing cDNA sequences indicates that SEM1 homologues are also widely expressed. Interestingly, the yeast and the mammalian proteins are not only homologous by protein sequence but also are closely related functionally. The mammalian homologue, Dss1, was able to restore the wt pseudohyphal phenotype of S288C identically to SEM1. This result suggests that Dss1 performs in mammalian cells a closely related or identical activity to that of SEM1 in yeast.
Possible Role of SEM1 in Cell Differentiation.
A striking observation was that removal of Sem1p in diploid cells resulted in induction of pseudohyphal growth. Many commonly used laboratory strains of S. cerevisiae (e.g., S288C) carry a mutation in FLO8 gene (19), which makes these strains unable to undergo transition to filamentous growth or to flocculate. The onset of pseudohyphal differentiation did not occur in haploid or in heterozygous diploid S288C cells and was detected only during nitrogen starvation in sem1Δ/sem1Δ. Interestingly, deletion of SEM1 in Σ1278b background enhanced the pseudohyphal phenotype. Sem1p thus seems to play a negative regulatory role in this differentiation process.
Recently, DSS1 and Dss1, the human and mouse homologues of SEM1, respectively, were identified as possible candidate genes for the autosomal dominant form of split hand/split foot malformation disorder (28, 29). In addition to DSS1, the affected locus contains two other genes that could be involved in the observed phenotype in mammals. The results showing that expression of Dss1 in yeast was able to complement the loss of SEM1 in a cellular differentiation process suggests that Dss1 could be involved in a differentiation process in mammals and could be the key determinant affected in the split hand/split foot malformation disorder.
Currently, no direct evidence exists to explain why deletion of SEM1 in yeast and (possibly) altered expression level of Dss1 in mammalian cells could lead to pseudohyphal growth and to split hand/split foot phenotypes. It is, however, tempting to speculate how deletion of a putative vesicle transport protein could cause such effects. In diploid S. cerevisiae, the onset of filamentous growth requires a shift from bipolar budding to unipolar budding (16). Because in S. cerevisiae the budding process as such is tightly linked to directed secretion toward the growing bud, it is likely that a change in the budding mode requires regulation of the secretory pathway. The exocyst complex has been suggested to play an important role in bud-site selection (10, 11), and in fact recent results by Novick and colleagues (12) position one of the exocyst components, the Sec3p, as an important factor defining the site of exocytosis. It could be that SEM1, by directly or indirectly regulating the life cycle of the exocyst complex, affects bud-site selection. In mammalian cells, Dss1 has been suggested to be involved in the development of limb bud, craniofacial primordia, and skin (28). Although the role of membrane traffic in the developmental stages is poorly understood, it is likely that formation of new cellular structures requires efficient, polarized membrane transport.
A large body of data on the regulation of pseudohyphal growth has been obtained during the past few years, and a growing number of genes involved in the mitogen-activated protein kinase-dependent and -independent signal transduction pathways regulating the pseudohyphal growth have been characterized (30, 31). The easily scorable pseudohyphal phenotype can now be employed for further analysis on the role and interactions of Sem1p and its mammalian homologue with other components of the induction process. The connection of the secretory pathway to pseudohyphal differentiation through Sem1p may reveal an interesting new level of regulation of these processes.
Acknowledgments
We are grateful to Peter Novick, Evelyne Dubois, Gerald Fink, Thierry Vernet, and Johan Peränen for reagents and to Rainer Duden and Hans Ronne for comments on the manuscript. Esa Kuismanen and members of the Keränen lab are acknowledged for helpful discussions. Technical help of Riitta Lampinen, Aili Grundström, and Hanna Karmakka also is gratefully acknowledged. We thank Oili Lappalainen for help in preparing figures. This study was supported by The Academy of Finland (Grants 29520 and 42160) and The Human Frontiers Science Program Grant RG63/95 (S.K.).
ABBREVIATIONS
- wt
wild type
- SEM
suppressor of exocyst mutations. Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF059310)
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Clary D O, Griff I C, Rothman J E. Cell. 1990;61:709–721. doi: 10.1016/0092-8674(90)90482-t. [DOI] [PubMed] [Google Scholar]
- 2.Malhotra V, Orci L, Glick B S, Block M R, Rothman J E. Cell. 1988;15:221–227. doi: 10.1016/0092-8674(88)90554-5. [DOI] [PubMed] [Google Scholar]
- 3.Söllner T, S, Whiteheart W, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman J E. Nature (London) 1993;632:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 4.Rothman J E, Wieland F T. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- 5.Halachmi N, Lev Z. J Neurochem. 1996;66:889–897. doi: 10.1046/j.1471-4159.1996.66030889.x. [DOI] [PubMed] [Google Scholar]
- 6.Olkkonen V M, Stenmark H. Int Rev Cytol. 1997;176:1–85. doi: 10.1016/s0074-7696(08)61608-3. [DOI] [PubMed] [Google Scholar]
- 7.Bowser R, Muller H, Govindan B, Novick P. J Cell Biol. 1992;118:1041–1056. doi: 10.1083/jcb.118.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.TerBush D R, Maurice T, Roth D, Novick P. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- 9.TerBush D R, Novick P. J Cell Biol. 1995;130:299–312. doi: 10.1083/jcb.130.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haarer B K, Corbett A, Kweon Y, Petzold A S, Brown S S. Genetics. 1996;144:495–510. doi: 10.1093/genetics/144.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finger F P, Novick P. Mol Biol Cell. 1997;8:647–662. doi: 10.1091/mbc.8.4.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finger F P, Hughes T E, Novick P. Cell. 1998;92:559–571. doi: 10.1016/s0092-8674(00)80948-4. [DOI] [PubMed] [Google Scholar]
- 13.Hsu S-C, Ting A E, Hazuka C D, Davenger S, Kenny J W, Kee Y, Scheller R H. Neuron. 1996;17:1209–1219. doi: 10.1016/s0896-6273(00)80251-2. [DOI] [PubMed] [Google Scholar]
- 14.Kee Y, Yoo J-S, Hazuka C D, Peterson K E, Hsu S-C, Scheller R H. Proc Natl Acad Sci USA. 1997;94:14438–14443. doi: 10.1073/pnas.94.26.14438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chant J. Curr Opin Cell Biol. 1996;8:557–565. doi: 10.1016/s0955-0674(96)80035-4. [DOI] [PubMed] [Google Scholar]
- 16.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 17.McKnight G L, McConaughy B L. Proc Natl Acad Sci USA. 1983;80:4412–4416. doi: 10.1073/pnas.80.14.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vernet T, Dignard D, Thomas D Y. Gene. 1987;52:225–233. doi: 10.1016/0378-1119(87)90049-7. [DOI] [PubMed] [Google Scholar]
- 19.Liu H, Styles C A, Fink G R. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wach A, Brachat A, Pohlmann R, Philippsen P. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 21.Jäntti J, Keränen S, Toikkanen J, Kuismanen E, Ehnholm C, Söderlund H, Olkkonen V M. J Cell Sci. 1994;107:3623–3633. doi: 10.1242/jcs.107.12.3623. [DOI] [PubMed] [Google Scholar]
- 22.Aalto M, Jäntti J, Östling J, Keränen S, Ronne H. Proc Natl Acad Sci USA. 1997;94:7331–7336. doi: 10.1073/pnas.94.14.7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Schägger H, von Jagow G. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 25.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 26.Aalto M, Ronne H, Keränen S. EMBO J. 1993;12:4095–4104. doi: 10.1002/j.1460-2075.1993.tb06093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brennwald P, Kearns B, Champion K, Keränen S, Bankaitis V, Novick P. Cell. 1994;79:245–258. doi: 10.1016/0092-8674(94)90194-5. [DOI] [PubMed] [Google Scholar]
- 28.Crackower M A, Schere S W, Rommens J M, Hui C C, Parvoneh P, Soder S, Cobben J M, Hudgins L, Evans J P, Tsui L-C. Hum Mol Genet. 1996;5:571–579. doi: 10.1093/hmg/5.5.571. [DOI] [PubMed] [Google Scholar]
- 29.Temtamy S A, McKusick V A. In: The Genetics of Hand Malformations. Alan R, editor. New York: Liss; 1978. pp. 53–71. [PubMed] [Google Scholar]
- 30.Herskowitz I. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 31.Lorentz M C, Heitman J. EMBO J. 1998;17:1236–1247. doi: 10.1093/emboj/17.5.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]