Abstract
In most genome-wide linkage studies, implication of a causative disease gene often requires years of expanding the study to more families and finer mapping of the initially described region. Even after such efforts, unobtainable sample sizes can be required to make statistically meaningful conclusions about a single gene. Here we demonstrate that by adding a layer of functional biology to statistical genetic results, this process can be accelerated. The diabetes susceptibility locus (chromosome 18p11) was systematically dissected by using a cell-based secretion assay and RNA interference, and we identified laminin α1 to have a role in pancreatic β cell secretion. The screen was extended to identify laminin receptor 1 as a functional partner in regards to β cell function. Our approach can potentially be widely used in the setting of high-throughput cellular screening of other loci to identify candidate genes.
Keywords: diabetes, functional genomics, insulin, laminin, RNA interference
The risks that contribute toward the development and progression of type 2 diabetes include both environmental and inherited factors. As such, determining the underlying genetic basis of diabetes is critical for diagnosing and preventing the disease. As with many diseases with a demonstrated genetic component, identification of susceptibility genes by whole-genome linkage studies is widely used (1). In such studies, the entire genome of an individual is mapped by using genetic markers. Statistical tests are applied to link specific genomic regions to disease risk. Outcomes from such studies can lead to the isolation of a single causative gene but often leave investigators confronting broad genomic regions containing hundreds of genes. Factors such as the impracticality of dramatically increasing the number of families tested, low allelic frequency, and incomplete penetrance of the effect often prevent the statistical narrowing down of these large regions to identify the involved gene.
In regard to studies of diabetes risk, successes in this approach have largely been limited to monogenic forms of the disease. Cases in point are the Maturity-Onset Diabetes of the Young (MODY) genes, which typically exert a strong phenotype mainly by interfering with the normal release of insulin. Altogether, the six forms of MODY can account for only 1–2% of all diabetes patients (2). More common for many diseases, and in particular type 2 diabetes, is the involvement of multiple genes (3), which individually have small phenotypic effects. This characteristic is the defining feature of so-called “complex trait” diseases (4), a feature that greatly complicates the statistical narrowing of genetic loci into causative genes.
In this study, we have designed a cell biology-based approach to accelerate the dissection of disease susceptibility loci. Our strategy utilizes RNA interference (RNAi), the process where double-stranded RNA induces the homology-dependent degradation of cognate mRNA (5). RNAi is well suited for screening numerous target genes, as demonstrated by other successful functional genomic studies (6, 7). By combining the available genome data with prediction algorithms for efficient RNAi agents, we have designed a panel of RNAi agents against both known and predicted genes encoded within a defined chromosome region associated with diabetes risk. These RNAi agents were screened by using a cell-based assay to functionally dissect a diabetes-susceptibility locus and isolate a causative gene.
Results
Functional Mapping of a Susceptibility Locus, a Four-Step Method.
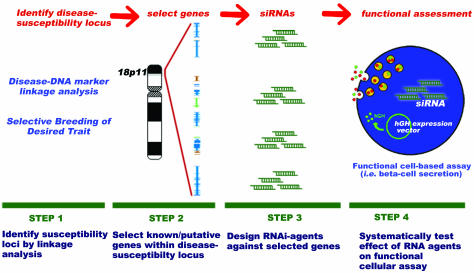
To dissect a susceptibility locus into individual testable genes, we developed a four-step strategy described as follows: (i) identify the disease-susceptibility region, (ii) select known and/or predicted genes from this region, (iii) design RNAi agents against these selected genes, and (iv) evaluate the effects of the RNAi agents against a functional cell-based assay. Such a functionally mapping of disease genes is diagrammed in Fig. 1.
Fig. 1.
Functional mapping of disease genes using cellular biology. Step 1: Susceptibility loci are identified by several strategies, which can include disease–DNA marker analysis using SNP, restriction fragment length polymorphisms (RFLP), variable number of tandem repeats (VNTR), simple sequence repeats (SSR), and simple sequence length polymorphisms (SSLP). Alternatively, selectively breeding a desired trait against a constant genetic background can isolate a trait-containing locus in animal disease models. Step 2: Flanking linkage markers identified in Step 1 delineate the chromosomal region, which is subsequently analyzed for known and predicted genes. Step 3: Multiple siRNAs are designed and synthesized against the panel of selected genes. Step 4: Each siRNA is functionally assessed in a cell-based assay. In regard to this study, the screen is for effects on pancreatic β cell secretion.
Step 1: Identification of diabetes-susceptibility region.
To select an appropriate susceptibility locus for functional analysis, we prioritized previously identified type 2 diabetes-susceptibility loci based upon the following four criteria: (i) Replicates significant linkage in independent studies; (ii) replicates significant linkage across independent populations, (iii) contains <20 genes within the identified locus, and (iv) does not contain a previously described diabetes-associated gene. Criterion iii reduced the complexity of this initial proof-of-principle study but can be relaxed when libraries of small interfering (si)RNAs (8, 9) are screened by using high-throughput methods. Criterion iv fostered an unbiased analysis of the results.
One such type 2 diabetes-susceptibility locus that fulfils each of these criteria is located within human chromosome 18p11. Parker et al. (10) performed a genome-wide type 2 diabetes linkage analysis of Finnish and Swedish families and identified this region flanked by DNA markers D18S452 and D18S843. They calculated a logarithm of odds (lod) of 0.66, and after stratification of their data set by obesity (defined as the 20% most obese subjects), the multipoint logarithm of odds increased to 4.22. Identification of this genomic region was replicated in a Dutch population (11, 12) and in American Caucasians (13). The convergence of each of these independent studies on the same region facilitated a further narrowing of the region to the chromosome segment chosen for gene selection and siRNA design (Fig. 5, which is published as supporting information on the PNAS web site; see also Figs. 6–8 and Tables 1–3, which are published as supporting information on the PNAS web site).
Step 2: Selection of known or predicted genes to include in the functional screen.
Eighteen gene models, as characterized in National Center for Biotechnology Information build 35 of the human genome, are within the region flanked by the outermost markers used to delineate the susceptibility locus (Fig. 5). Of these 18, 6 are confirmed gene models (14) with functional evidence of the gene product described in the scientific literature. The functional assay used in our primary screen utilizes a rat insulinoma (see Step 4). Because RNAi is highly sequence-dependent, variations in the sequences between the human and rat genes necessitate siRNA design derived from rat sequences. Thus, an additional step to map the human diabetes-susceptibility locus against the rat genome was required for effective siRNA design. All six of the functionally described human genes mapped on the rat genome within rat chromosome 9q37–38 (105–110 megabases) and maintained an identical sequential order (Fig. 5). Of the 12 remaining human gene models, four had significant similarity to the syntenic region of rat chromosome 9, as determined by blast sequence alignments (15). Based on this analysis, we prioritized our gene list and selected 10 genes for siRNA design (Table 1).
Step 3: Design and synthesis of the collection of RNAi agents to target genes within susceptibility locus.
Using previously described selection rules (16, 17), four siRNAs were designed against each target. The use of multiple siRNAs increases the probability of an effective siRNA against the target (18). Additionally, confidence in the interpretation of the screen is enhanced when similar functional consequences are observed with siRNAs against the same target but with different sequences (19).
Step 4: Screen the collection of RNAi agents against cell-based assay for β cell function.
We developed a general screening assay that would be capable of detecting perturbations of β cell function and would be sensitive to a broad range of cellular activities. The assay, glucose-stimulated hormone release, can be affected by fuel metabolism, tissue-specific transcription factors, calcium homeostasis, and components of the exocytotic machinery. If the transfection efficiency of the RNAi agents into the β cell approach 100%, insulin release could be measured. However, this is not the case in purified β cells, most β cell lines, and the INS1E rat insulinoma cell line (20) used in this study. To correct for the shortcomings of low transfection efficiency, the human growth hormone (hGH) cotransfection technique is used to screen β cell function (21). Briefly, a plasmid encoding hGH is cotransfected with the siRNA. Once expressed, hGH is known to colocalize with insulin and is secreted in a glucose-dependent manner mirroring insulin secretion (22). With such a strategy, hormone release is measured from only the transfected population of cells receiving the RNAi agent and is not masked by secretion from the untransfected cells (23).
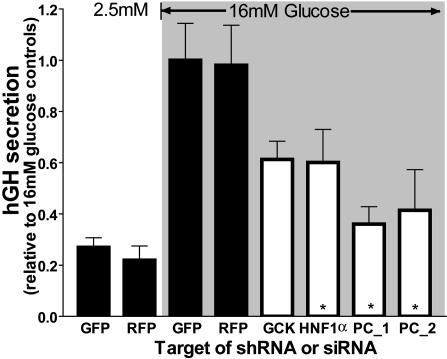
Glucose-stimulated hGH release is demonstrated in Fig. 2. Typically in the INS1E cell line, a 30-min stimulation of 16 mM glucose elicits a 3- to 5-fold increase of insulin secretion over the basal secretion observed at 2.5 mM glucose (24). As demonstrated in Fig. 2, hGH secretion parallels this fold induction in control groups with siRNAs against GFP and red fluorescent protein (RFP) and is suppressed by an siRNA against glucokinase (the known disease gene in MODY2 patients) and a short hairpin RNA (shRNA) vector against hepatocyte nuclear factor-1α, the known disease gene in MODY3 patients (25). The suppression of secretion by knockdown of these two MODY genes confirms the ability of the rat β cell-screening assay to detect such genes involved in human diabetes. It should be noted that, during the identification of these MODY genes, substantially higher logarithm of odds scores were obtained as compared with the majority of diabetes-susceptibility loci, including the region screened in this study. To determine whether the hGH assay could identify genes from regions of similar statistical strength to 18p11, we next targeted a gene recently identified from a diabetes-susceptibility locus on chromosome 11. This particular locus was identified in the insulin resistance atherosclerosis family study [logarithm of odds = 3.2 (26)], and further studies identified pyruvate carboxylase as a potential disease gene (47). We developed two shRNA vectors against pyruvate carboxylase. As shown in Fig. 2, each significantly impairs hormone secretion elicited by 16 mM glucose (by 65% and 60%).
Fig. 2.
The hGH-based functional assay of β cell secretion can assess perturbations of glucose-stimulated secretion and is sensitive to known diabetogenes. Control siRNAs against GFP and RFP (black bars) do not affect glucose-stimulated secretion in hGH cotransfected INS1E cells. Secretion is increased 4-fold at 16 mM glucose relative to 2.5 mM glucose. Impaired secretion is observed with an siRNA against glucokinase (GCK, MODY2 gene), an shRNA vector against hepatocyte nuclear factor 1-α (HNF1α, MODY3 gene), and two shRNA vectors against pyruvate carboxylase (PC, susceptibility loci chromosome 11). Data are means ± SEM of three to four individual experiments performed in triplicate.
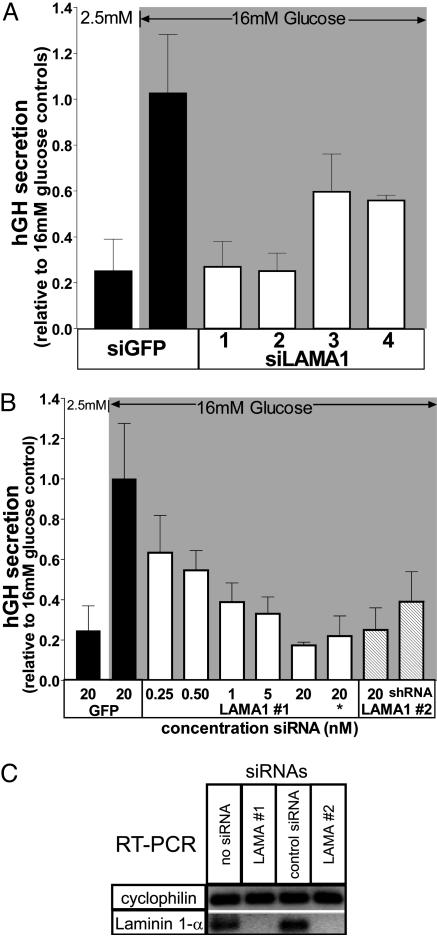
Knowing that both strong causal and weaker susceptibility genes can be detected with the β cell assay, we initiated a functional screen across the susceptibility locus on human chromosome 18. Using the 38 siRNAs designed and synthesized in Step 3, each siRNA was screened by using the hGH secretion assay (Fig. 5). Of the 10 gene models examined, one in particular had a significant impact on β cell secretion, which is summarized in Fig. 3A. Four siRNAs were synthesized against the rat gene corresponding to the human laminin α1 (LAMA1), the α subunit of the heterotrimeric laminin complex (see note in Fig. 4C). Each of the four siRNAs reduced glucose-stimulated secretion, albeit to varying degrees (73%, 75%, 40%, and 45%) (Fig. 4A). The two most potent siRNAs (siLAMA1 #1 and #2) were further characterized as described in Fig. 3B. Inhibition of glucose-stimulated secretion was dose-dependent over the range of 0.25–20 nM with siLAMA1 #1. Using the sequence of siLAMA1 #2, a DNA-based shRNA construct was prepared (shLAMA1 #2) and confirmed the observed decrease in glucose-stimulated secretion (by 60%).
Fig. 3.
Transfected siRNAs designed against LAMA1 impair glucose-stimulated secretion. (A) Four different siRNAs against LAMA1 (siLAMA1) decrease glucose-stimulated secretion relative to the siGFP control. Data are means ± SEM of three experiments performed in triplicate. (B) The dose dependency of siLAMA #1 is demonstrated between 250 pM and 20 nM of the siRNA. The 20 nM siRNA concentration marked with an asterisk is from a second RNA synthesis, from a separate vendor. A DNA-based shRNA construct was designed against the sequence targeted by siLAMA #2. Data are means ± SD, performed in triplicate. (C) LAMA1 mRNA is reduced in siLAMA1-treated cells. INS1E cells were cotransfected with various siRNAs and a GFP expression vector. Transfected cells were enriched by fluorescence-activated cell sorting. From these cells, mRNA was prepared, reverse-transcribed, and amplified by PCR with specific primers for cyclophilin (invariant control) and LAMA1. Note that LAMA1 mRNA was undetectable (up to 40 cycles) in both siLAMA1 #1- and #2-treated cells compared with controls.
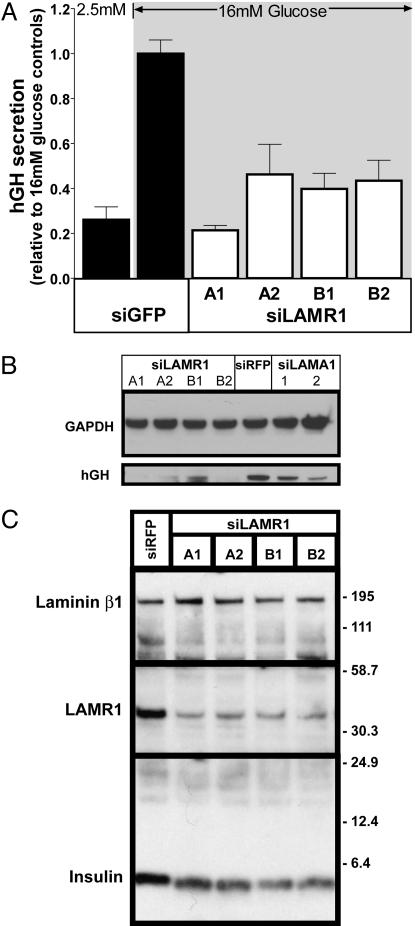
Fig. 4.
Effects of LAMR1 siRNAs on β cell secretion, hgH, and insulin expression. (A) Transfected siRNAs designed against LAMR1 impair glucose-stimulated secretion. Four different siRNAs against LAMR1 decrease glucose-stimulated secretion relative to a control group transfected with siGFP. Data are means ± SEM of three individual experiments performed in triplicate. (B) hGH protein levels are decreased in siLAMA1- and siLAMR1-treated cells. INS1E cells were cotransfected with an hGH expression vector and siRNAs as indicated. A reduction of hGH protein levels was observed with all siRNAs against LAMR1 and less severely with siRNAs against LAMA1. GAPDH was used as a protein-loading control. (C) Insulin expression is decreased in siLAMR1-treated cells. INS1E cells were cotransfected with the indicated siRNAs and a vector for the expression of the puromycin resistance gene. Transfected cells were selected by puromycin treatment for 36 h, and cell lysates were analyzed by Western blot. Note that each siRNA against LAMR1 specifically reduced both LAMR1 and insulin expression compared with the loading control, laminin β1 (LAMB1). Note that laminins are heterotrimeric proteins consisting of α, β, and γ subunits. LAMA1 is a component of laminin-1 and -3, which differ by the β subunit present, β1 and β2, respectively. In INS1E cells, the β1 isoform was detected by immunoblot, but not the β2 isoform.
To validate that the designed siRNAs specifically reduced LAMA1 mRNA, the cells receiving siRNA were enriched from the untransfected population by using a cotransfected GFP expression plasmid and fluorescence-activated cell sorting. RT-PCR was carried out after the enrichment of transfected cells. As shown in Fig. 3C, LAMA1 mRNA from INS1E cells transfected with siLAMA1 #1 and #2 was undetectable compared with cells transfected with a control siRNA or no siRNA.
Secondary Screen of Laminin-Binding Proteins.
Laminins are glycoproteins of the extracellular matrix (ECM) and function through binding various protein ligands to trigger signaling cascades (27). To delineate which of these pathways could be involved in laminin signaling in the context of β cell function, a second screen was performed on selected members of three laminin receptor types (integrins α6, β1; dystroglycan; and laminin receptor 1, Fig. 6). Four siRNAs were designed for each gene and screened by the hGH secretion assay (see Table 2 for details). Of these four genes, a dramatic impairment of secretion was observed only with siRNAs against laminin receptor 1 (LAMR1). All four siRNAs designed against the LAMR1 gene severely reduced hGH secretion (80%, 55%, 60%, and 57%) as shown in Fig. 4A.
Role of Laminin–LAMR1 Interaction.
LAMR1 has been described as a bifunctional protein based on physical binding assays (28) and sequence comparisons. The domain required for laminin binding resides within the C terminus of LAMR1 (28, 29). The N-terminal domain has sequence similarity to ribosomal protein S2 (30). Accordingly, the N-terminal domain has been proposed to be involved in ribosomal function; however, no such activity has been experimentally demonstrated. Although our functional screen has been optimized to detect perturbations of glucose-stimulated secretion, it also remains susceptible to pathways that could alter the biosynthesis of hGH. Because of the possibility that LAMR1 may be involved in ribosome function and thus translation of proteins, we next investigated whether knockdown of LAMA1 and LAMR1 genes have a direct effect on hGH protein levels.
An immunoblot against hGH was performed on cells treated with siRNAs against LAMA1 and LAMR1. The two most potent siRNAs against LAMA1 reduced hGH protein levels by 30% and 50%, respectively, compared with an siRFP control and normalized to GAPDH (Fig. 4B). The severity of this effect was greater for siRNAs against LAMR1, where hGH protein levels were decreased by >80%. The knockdown of both LAMA1 and LAMR1 genes specifically reduces hGH levels, and not endogenous genes, such as GAPDH (Fig. 4B) or other ectopically expressed genes such as GFP (Fig. 7). It has been well described that biosynthesis of secreted hormones, such as insulin in the β cell, is highly regulated at the level of translation (31). In the case of insulin, this process utilizes specific recognition sequences contained within the 5′ and 3′ untranslated ends of specific genes, including insulin (32). Although the transcription of hGH mRNA in our reporter construct is driven by the CMV promoter, the expected transcript retains portions of the native 5′ and 3′ untranslated ends of the hGH gene and may therefore be available for gene specific translational control.
Based on the specific effect of LAMA1 and LAMR1 siRNAs on hGH levels, we hypothesized a similar regulation of biosynthesis may occur with the endogenous insulin. To determine whether insulin levels are affected by LAMR1 knockdown, it was necessary to first enrich the transfected population of cells. INS1E cells were transfected with siRNAs against LAMR1 and RFP. To exclude untransfected cells from the analysis, the siRNAs were cotransfected with the puromycin resistance gene followed by 2 days of selection with the puromycin antibiotic. The enriched transfected population of cells was collected and processed for immunoblot analysis (Fig. 4C). Each of the four LAMR1 siRNAs reduced both LAMR1 and insulin protein levels compared with the siRFP control, whereas laminin β1 (LAMB1) was relatively unchanged across the samples. The observed decrease of insulin suggests that the biosynthesis of insulin is highly dependent on LAMR1 expression.
Discussion
By using a functional screen across genes in susceptibility loci 18p11, we have isolated LAMA1 as a strong candidate for associated diabetes risk. To further understand the role of laminin-1 in the β cell, we conducted a secondary screen of laminin effectors, and identified LAMR1. Further analysis of LAMR1 knockdown provided previously undescribed functional evidence of LAMR1 exerting effects on the translational machinery.
Considering other observations of laminin function on β cells, we can begin to assemble a model of how laminin–LAMR1 interactions can influence β cell function. Several studies have observed the beneficial effects of laminin-1 and extracellular matrix (ECM) preparations on pancreatic islet development and function. Purified laminin-1 or Matrigel (a commercial preparation of ECM, which is highly enriched with laminin-1) promotes differentiation of cultured rat β cells (33, 34). Insulin content of human adult pancreatic cell cultures increases when grown on Matrigel (35, 36). These beneficial effects are inhibited by antibodies against laminin-1 (37). A role for LAMR1 has been proposed in angiogenesis (38), and the pancreatic islets are vascularized with a highly fenestrated capillary network (39). These observations, coupled with the demonstrated presence of laminin-1 in intraislet capillaries (40), lead to the intriguing possibility that the laminin–LAMR1 interaction may facilitate a functional crosstalk between insulin-secreting cells and their associated vasculature. This would ensure that the highly metabolically active β cell receives adequate oxygen, and these secretory cells maintain appropriate levels of insulin.
Replenishment of secreted insulin is critical in the maintenance of insulin stores and as such, the β cell has adapted a highly regulated translation control of insulin (31). Because the knockdown of LAMR1 decreases insulin protein levels and, due to the homology of the intracellular C terminus (29) of LAMR1 to the S2 ribosomal protein, we propose that LAMR1 modulates translational control of insulin. In Fig. 8, we diagram three models of the numerous scenarios by which LAMR1 can enhance translational activation. Each model takes into account previous observations describing translational control of insulin biosynthesis and shared elements of hGH biosynthesis, such as a regulatory UTR and a signal peptide. In model one, LAMR1 modulates translational activity via the UTR of insulin mRNA (32). LAMR1 exerts its effects via preproinsulin prebound to the ribosome and signal recognition particle in model two (41) and via cotranslocation translation of the secreted protein in model three (42).
In addition to effects on the pancreatic β cell, altered LAMR1 activity may also affect insulin-sensitive tissues such as liver, skeletal muscle, and adipose tissue. Additional studies to determine the role of LAMR1 in these tissues will be required to fully understand the role of the laminin–LAMR1 interaction in the context of diabetes. Expanding our screening strategy toward functional assays for these tissues, such as insulin sensitivity and glucose metabolism, will further improve the odds of identifying genes involved in diabetes.
Our findings from this functional screen exemplify the promise of using population genetic studies to guide functional genomic screens, whereas the process of identifying a causative gene from the susceptibility locus was greatly accelerated. Reciprocally, functional screens can focus the search for causal sequence variations on smaller genomic regions, which will in turn accelerate discoveries in these studies. For instance, high-density SNP mapping could initially be restricted to the LAMA1 gene, rather than across the entire susceptibility locus. Likewise, the functional identification of LAMR1 in this study directs future genetic studies to analyze this gene in regards to diabetes.
In addition to identifying disease-risk genes, the unbiased nature of this approach allows the identification of previously unknown pathways and interactions and should assist in understanding the functional consequences among multiple genes in complex trait diseases such as diabetes. Methods such as congenic (43) and chromosome substitution strains of animals (44) and expanded human genetic studies become far less feasible as the number of identified genes increase and various combinations of genes are required to elicit a phenotype. However, a readily scalable high-throughput cell-based functional assay can accommodate such a combinatorial discovery approach and thus help construct a more complete network of disease-risk genes and their functional interrelationships. Describing such genetic relationships to disease will assist the interpretation of an individual’s genetic profile toward diagnosis and personalized treatment programs.
Materials and Methods
Cell Culture and hGH Secretion.
INS1E cells were cultured as described by Asfari et al. (45) and seeded at a density of 2.5 × 105 cells per well in a 24-well tissue culture plate (Falcon). Cells were washed three times with modified Krebs–Ringer bicarbonate buffer (KRBH) containing 135 mM NaCl, 3.6 mM KCl, 10 mM Hepes (pH 7.4), 5 mM NaHCO3, 0.5 mM NaH2PO4, 0.5 mM MgCl, and 1.5 mM CaCl2 and preincubated for 30 min in KRBH with 2.5 mM glucose. Cells were washed an additional two times and incubated with KRBH supplemented with either 2.5 or 16 mM glucose. After 30 min of incubation at 37°C, medium was collected for hGH ELISA and measured according to vendor protocol (Roche Diagnostics).
siRNA Design and Synthesis.
siRNAs were designed by using publicly available algorithms (16, 17). siRNAs were either synthesized at the Memorial Sloan–Kettering Cancer Center’s siRNA core facility or purchased from a commercial vendor (Integrated DNA Technologies, Coralville, IA). DNA-based shRNA plasmids were constructed by annealing two complementary oligonucleotides, followed by ligation to pSilencer 1.0 (Ambion, Austin, TX) digested with ApaI/EcoRI per vendor-supplied protocol. See Supporting Text, which is published as supporting information on the PNAS web site, for the oligonucleotide sequences used.
Transfections.
On day 1, INS1E cells were seeded on Falcon 24-well plates, 2.5 × 105 cells per well. On day 2, cells were transfected [0.25–0.5 μg of DNA, siRNA (0.1–40 nM), 4 μl of DMRIE-C-transfection reagent (Invitrogen) in 500 μl of RPMI medium 1640 without FCS and without antibiotics]. Three hours later, the transfection complexes were removed and replaced with complete media. For larger-scale transfections (i.e., 10-cm2 dishes), 5 × 106 cell were plated and transfected in 3 ml using scaled-up conditions as described above.
Enrichment of Transfected Cells by FACS and Puromycin Selection.
Cells were cotransfected with GFP expression vector, pEGFPN3 (Clontech), and either no siRNA or siRNAs designed against the target gene. After 2 days, to allow for sufficient GFP expression, transfected cells were enriched by fluorescence-activated cell sorting by GFP fluorescence (FACSAria; Becton Dickinson). Sorted cells were centrifuged (6 min at 3,800 × g) and immediately processed for mRNA purification.
Alternatively, cells were cotransfected with the vector pSofA (see Supporting Text for vector details), which includes the CMV promoter followed by a puromycin resistance gene, an internal ribosome entry site, and the EGFP gene. After 24 h, the media were supplemented 1.5 μg/ml puromycin for 2 additional days and then for 8 h without puromycin. Cells were washed two times with 5 ml of PBS media, then collected in 1.5 ml of PBS (without calcium and magnesium) and centrifuged (6 min at 3,800 × g).
Western Blotting.
Collected cells were lysed in 1% Nonidet P-40 supplemented with protease inhibitor mixture (Roche Diagnostics), one tablet per 50 ml. Total protein was quantified by a modified Lowry method (Bio-Rad), and 20 μg of protein was loaded on a precast 10% Bis-Tris gel (Invitrogen). Proteins were transferred to 0.45 μM poly(vinylidene difluoride) membrane, which was then blocked with 1% BSA in TBS-T buffer (25 mM Tris/0.15 M NaCl/0.2%Tween 20) for 1 h. After five washes with TBS-T (5 min each), the membrane was incubated with the primary antibody diluted in TBS-T for 1 h. This was followed by five washes with TBS-T (5 min each) and incubation with the secondary antibody (if required) for 1 h and then five washes with TBS-T. Antibodies and their dilutions are described in Supporting Text. Chemiluminescent detection was performed with the West Pico Supersignal Kit (Pierce) according to the vendor-provided protocol. The chemiluminescent reaction was captured with Kodak BioMax XAR film, which was exposed at multiple time intervals. The developed film was scanned on a high-resolution flat-bed scanner, and protein bands were quantitated with image j (46) image analysis software.
RT-PCR.
RNA and first-strand DNA synthesis were prepared from FACS cells by using the PROSTAR First Strand RT-PCR Kit (Stratagene), according to vendor instructions. PCR of the desired targets was performed with specific oligonucleotide primers (see Supporting Text for additional details).
Supplementary Material
Acknowledgments
We greatly appreciate the assistance of the following individuals in this study: Bernd Jagla for siRNA design; Nathalie Aulner for INCELL 3000 confocal analyzer expertise; Kristie Gordon for FACS; Ouatek Ourfelli for siRNA synthesis; and Donald Bowden, Fabienne Paumet, Tom Melia, Hong Ji, and Lynn Caporale for helpful discussions. This work was supported by grants from the American Diabetes Association (to P.A.A.), the Juvenile Diabetes Research Foundation (toP.A.A.), and the Mathers Foundation (to J.E.R.).
Abbreviations
- RNAi
RNA interference
- hGH
human growth hormone
- shRNA
short hairpin RNA
- LAMA1
laminin α1
- LAMR1
laminin receptor 1
- si
small interfering
- RFP
red fluorescent protein.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Hanis C. L., Boerwinkle E., Chakraborty R., Ellsworth D. L., Concannon P., Stirling B., Morrison V. A., Wapelhorst B., Spielman R. S., Gogolin-Ewens, et al. Nat. Genet. 1996;13:161–166. doi: 10.1038/ng0696-161. [DOI] [PubMed] [Google Scholar]
- 2.Almind K., Doria A., Kahn C. R. Nat. Med. 2001;7:277–279. doi: 10.1038/85405. [DOI] [PubMed] [Google Scholar]
- 3.Florez J. C., Hirschhorn J., Altshuler D. Annu. Rev. Genomics Hum Genet. 2003;4:257–291. doi: 10.1146/annurev.genom.4.070802.110436. [DOI] [PubMed] [Google Scholar]
- 4.Phillips T. J., Belknap J. K. Nat. Rev. Neurosci. 2002;3:478–485. doi: 10.1038/nrn847. [DOI] [PubMed] [Google Scholar]
- 5.Elbashir S. M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 6.DasGupta R., Kaykas A., Moon R. T., Perrimon N. Science. 2005;308:826–833. doi: 10.1126/science.1109374. [DOI] [PubMed] [Google Scholar]
- 7.Kim J. K., Gabel H. W., Kamath R. S., Tewari M., Pasquinelli A., Rual J.-F., Kennedy S., Dybbs M., Bertin N., Kaplan J. M., et al. Science. 2005;308:1164–1167. doi: 10.1126/science.1109267. [DOI] [PubMed] [Google Scholar]
- 8.Kaykas A., Moon R. T. BMC Cell Biol. 2004;5:16. doi: 10.1186/1471-2121-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng L., Liu J., Batalov S., Zhou D., Orth A., Ding S., Schultz P. G. Proc. Natl. Acad. Sci. USA. 2004;101:135–140. doi: 10.1073/pnas.2136685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parker A., Meyer J., Lewitzky S., Rennich J. S., Chan G., Thomas J. D., Orho-Melander M., Lehtovirta M., Forsblom C., Hyrkkö A., et al. Diabetes. 2001;50:675–680. doi: 10.2337/diabetes.50.3.675. [DOI] [PubMed] [Google Scholar]
- 11.van Tilburg J. H. O., Sandkuijl L. A., Strengman E., van Someren H., Rigters-Aris C. A. E., Pearson P. L., van Haeften T. W., Wijmenga C. J. Clin. Endocrinol Metab. 2003;88:2223–2230. doi: 10.1210/jc.2002-021252. [DOI] [PubMed] [Google Scholar]
- 12.Aulchenko Y. S., Vaessen N., Heutink P., Pullen J., Snijders P. J. L. M., Hofman A., Sandkuijl L. A., Houwing-Duistermaat J. J., Edwards M., Bennett S., et al. Diabetes. 2003;52:3001–3004. doi: 10.2337/diabetes.52.12.3001. [DOI] [PubMed] [Google Scholar]
- 13.Elbein S. C., Hoffman M. D., Teng K., Leppert M. F., Hasstedt S. J. Diabetes. 1999;48:1175–1182. doi: 10.2337/diabetes.48.5.1175. [DOI] [PubMed] [Google Scholar]
- 14.Yeh R. F., Lim L. P., Burge C. B. Genome Res. 2001;11:803–816. doi: 10.1101/gr.175701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 16.Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W. S., Khvorova A. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 17.Jagla B., Aulner N., Kelly P. D., Song D., Volchuk A., Zatorski A., Shum D., Mayer T., Angelis D. A. D., Ouerfelli O., et al. RNA. 2005;11:864–872. doi: 10.1261/rna.7275905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huppi K., Martin S. E., Caplen N. J. Mol. Cell. 2005;17:1–10. doi: 10.1016/j.molcel.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 19.Nat. Cell Biol. 2003;5:489–490. doi: 10.1038/ncb0603-490. Anonymous. [DOI] [PubMed] [Google Scholar]
- 20.Merglen A., Theander S., Rubi B., Chaffard G., Wollheim C. B., Maechler P. Endocrinology. 2004;145:667–678. doi: 10.1210/en.2003-1099. [DOI] [PubMed] [Google Scholar]
- 21.Wick P. F., Senter R. A., Parsels L. A., Uhler M. D., Holz R. W. J. Biol. Chem. 1993;268:10983–10989. [PubMed] [Google Scholar]
- 22.Maechler P., Antinozzi P. A., Wollheim C. B. IUBMB Life. 2000;50:27–31. doi: 10.1080/15216540050176557. [DOI] [PubMed] [Google Scholar]
- 23.Iezzi M., Kouri G., Fukuda M., Wollheim C. B. J. Cell Sci. 2004;117:3119–3127. doi: 10.1242/jcs.01179. [DOI] [PubMed] [Google Scholar]
- 24.Antinozzi P. A., Ishihara H., Newgard C. B., Wollheim C. B. J. Biol. Chem. 2002;277:11746–11755. doi: 10.1074/jbc.M108462200. [DOI] [PubMed] [Google Scholar]
- 25.Yamagata K., Oda N., Kaisaki P. J., Menzel S., Furuta H., Vaxillaire M., Southam L., Cox R. D., Lathrop G. M., Boriraj V. V., et al. Nature. 1996;384:455–458. doi: 10.1038/384455a0. [DOI] [PubMed] [Google Scholar]
- 26.Rich S. S., Bowden D. W., Haffner S. M., Norris J. M., Saad M. F., Mitchell B. D., Rotter J. I., Langefeld C. D., Wagenknecht L. E., Bergman R. N. Diabetes. 2004;53:1866–1875. doi: 10.2337/diabetes.53.7.1866. [DOI] [PubMed] [Google Scholar]
- 27.Miner J. H., Yurchenco P. D. Annu. Rev. Cell Dev. Biol. 2004;20:255–284. doi: 10.1146/annurev.cellbio.20.010403.094555. [DOI] [PubMed] [Google Scholar]
- 28.Wewer U. M., Liotta L. A., Jaye M., Ricca G. A., Drohan W. N., Claysmith A. P., Rao C. N., Wirth P., Coligan J. E., Albrechtsen R. Proc. Natl. Acad. Sci. USA. 1986;83:7137–7141. doi: 10.1073/pnas.83.19.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castronovo V., Taraboletti G., Sobel M. E. J. Biol. Chem. 1991;266:20440–20446. [PubMed] [Google Scholar]
- 30.Kazmin D. A., Chinenov Y., Larson E., Starkey J. R. Biochem. Biophys. Res. Commun. 2003;300:161–166. doi: 10.1016/s0006-291x(02)02772-9. [DOI] [PubMed] [Google Scholar]
- 31.Goodge K. A., Hutton J. C. Semin. Cell Dev. Biol. 2000;11:235–242. doi: 10.1006/scdb.2000.0172. [DOI] [PubMed] [Google Scholar]
- 32.Knight S. W., Docherty K. J. Mol. Endocrinol. 1992;8:225–234. doi: 10.1677/jme.0.0080225. [DOI] [PubMed] [Google Scholar]
- 33.Jiang F.-X., Harrison L. C. Differentiation. 2005;73:45–49. doi: 10.1111/j.1432-0436.2005.07301002.x. [DOI] [PubMed] [Google Scholar]
- 34.Vasir B., Aiello L. P., Yoon K. H., Quickel R. R., Bonner-Weir S., Weir G. C. Diabetes. 1998;47:1894–1903. doi: 10.2337/diabetes.47.12.1894. [DOI] [PubMed] [Google Scholar]
- 35.Bonner-Weir S., Taneja M., Weir G. C., Tatarkiewicz K., Song K. H., Sharma A., O’Neil J. J. Proc. Natl. Acad. Sci. USA. 2000;97:7999–8004. doi: 10.1073/pnas.97.14.7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao R., Ustinov J., Pulkkinen M.-A., Lundin K., Korsgren O., Otonkoski T. Diabetes. 2003;52:2007–2015. doi: 10.2337/diabetes.52.8.2007. [DOI] [PubMed] [Google Scholar]
- 37.Jiang F. X., Cram D. S., DeAizpurua H. J., Harrison L. C. Diabetes. 1999;48:722–730. doi: 10.2337/diabetes.48.4.722. [DOI] [PubMed] [Google Scholar]
- 38.Hu C., Oliver J. A., Goldberg M. R., Al-Awqati Q. Am J. Physiol. Renal Physiol. 2001;281:F739–F750. doi: 10.1152/ajprenal.2001.281.4.F739. [DOI] [PubMed] [Google Scholar]
- 39.Lifson N., Lassa C. V., Dixit P. K. Am J. Physiol. 1985;249:E43–E48. doi: 10.1152/ajpendo.1985.249.1.E43. [DOI] [PubMed] [Google Scholar]
- 40.Geutskens S. B., Homo-Delarche F., Pleau J.-M., Durant S., Drexhage H. A., Savino W. Cell Tissue Res. 2004;318:579–589. doi: 10.1007/s00441-004-0989-0. [DOI] [PubMed] [Google Scholar]
- 41.Wiedmann M., Huth A., Rapoport T. A. FEBS Lett. 1986;194:139–145. doi: 10.1016/0014-5793(86)80065-5. [DOI] [PubMed] [Google Scholar]
- 42.Kalies K. U., Görlich D., Rapoport T. A. J. Cell Biol. 1994;126:925–934. doi: 10.1083/jcb.126.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aitman T. J., Glazier A. M., Wallace C. A., Cooper L. D., Norsworthy P. J., Wahid F. N., Al-Majali K. M., Trembling P. M., Mann C. J., Shoulders C. C., et al. Nat. Genet. 1999;21:76–83. doi: 10.1038/5013. [DOI] [PubMed] [Google Scholar]
- 44.Singer J. B., Hill A. E., Burrage L. C., Olszens K. R., Song J., Justice M., O’Brien W. E., Conti D. V., Witte J. S., Lander E. S., et al. Science. 2004;304:445–448. doi: 10.1126/science.1093139. [DOI] [PubMed] [Google Scholar]
- 45.Asfari M., Janjic D., Meda P., Li G., Halban P. A., Wollheim C. B. Endocrinology. 1992;130:167–178. doi: 10.1210/endo.130.1.1370150. [DOI] [PubMed] [Google Scholar]
- 46.Rasband W. S. image j. Bethesda: National Institutes of Health; 19972006. [Google Scholar]
- 47.Palmer N. D., Langefeld C. D., Campbell J. K., Williams A. H., Saad M. F., Norris J. M., Haffner S. M., Rotter J. I., Wagenknecht L. E., Bergman R. N., et al. Diabetes. 2006 in press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.