Abstract
In the environment, multiple microbial taxa typically coexist as communities, competing for resources and, often, physically associated within biofilms. A dual-species cocultivation model has been developed by using two ubiquitous and well studied microbes Pseudomonas aeruginosa (P.a.) and Agrobacterium tumefaciens (A.t.) as a tractable system to identify molecular mechanisms that underlie multispecies microbial associations. Several factors were found to influence coculture interactions. P.a. had a distinct growth-rate advantage in cocultures, increasing its relative abundance during planktonic and biofilm growth. P.a. also demonstrated a slight quorum-sensing-dependent increase in growth yield in liquid cocultures. P.a. dominated coculture biofilms, “blanketing” or burying immature A.t. microcolonies. P.a. flagellar and type IV pili mutant strains exhibited deficient blanketing and impaired competition in coculture biofilms, whereas, in planktonic coculture, these mutations had no effect on competition. In contrast, A.t. used motility to emigrate from coculture biofilms. In both planktonic and biofilm cocultures, A.t. remained viable for extended periods of time, coexisting with its more numerous competitor. These findings reveal that quorum-sensing-regulated functions and surface motility are important microbial competition factors for P.a. and that the outcome of competition and the relative contribution of different factors to competition are strongly influenced by the environment in which they occur.
Keywords: flagella, interspecies interactions
In theoretical and empirical ecology, competition between species plays a central role in defining community structure and activity. Stable coexistence of diverse organisms in communities is thought to be fostered by individual tradeoffs and optimization of competitive strategies along resource gradients (1). Outside of the laboratory, microorganisms usually coexist in multicellular communities, governed by competition for common nutritional resources with other community members (2). Competitive fitness can be realized simply by occupying a suitable or specialized nutritional niche. Motility provides a mechanism by which microbes continually reposition themselves, adapting to changing nutritional and physical conditions. Another effective competitive strategy is to secrete antimicrobial compounds that kill or impair other species that occupy the same niche. Competitive interactions are most likely to occur when local microbial population densities are high, such as in biofilm communities. At high population densities, the process of quorum sensing is an important mechanism that coordinates and reinforces community behaviors in many bacterial species. Biofilm formation and quorum sensing are two bacterial community behaviors that clearly have significant potential to influence multispecies interactions (3–5). We hypothesize that motility, quorum sensing, and biofilm formation are among the mechanisms by which bacteria compete and persist within microbial communities.
To examine this hypothesis, we have developed a dual-species model system, composed of Pseudomonas aeruginosa (P.a.) and Agrobacterium tumefaciens (A.t.). P.a. is a Gram-negative γ-proteobacterium ubiquitous in soil and aquatic environments. It is also an opportunistic pathogen that causes many nosocomial infections and is frequently associated with the chronic lung infections that plague people suffering from cystic fibrosis. P.a. is a paradigm for the study of acyl-HSL-based quorum sensing and the formation of surface-associated communities called biofilms (6, 7). A.t. is a Gram-negative α-proteobacterium that causes crown gall disease in plants. This microbe has served as a model for horizontal gene transfer, host–microbe interactions, pathogenesis, and acyl-HSL-based signaling for many years. P.a. and A.t. have been isolated from the same environment, where they coexist as common residents of freshwater, bulk soil, and the rhizosphere (8–10).
We report here, that P.a. manifests a significant competitive advantage over A.t., simply through its rapid growth rate in laboratory culture. We show that other functions can influence competition independently of growth rate. Our studies reveal important roles for quorum sensing and surface motility in the competitive interaction of P.a. with A.t. in cocultures. Examination of an A.t. flagellar-motility mutant suggests a very different role for A.t. swimming in biofilm coculture interactions.
Results
P.a. Dominates Planktonic Coculture Interactions.
Because growth rate is a key variable for interpreting subsequent experiments, the doubling times of P.a. and A.t. were determined in a defined growth medium supplemented with three distinct carbon sources: glucose, succinate, and glutamate. For all three carbon sources tested, P.a. grew at least twice as quickly as A.t. (see Table 2, which is published as supporting information on the PNAS web site). The carbon source succinate was used in subsequent experiments, where the doubling times of P.a. and A.t. were estimated to be 29 and 59 min, respectively.
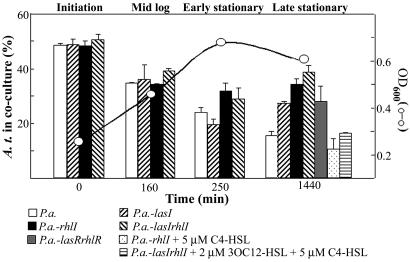
We initially examined the population dynamics of wild-type P.a. and A.t. planktonic cocultures. Cocultures were inoculated with different ratios (ranging from 10:1 to 1:10) of wild-type P.a. and A.t. The relative percentage of A.t. was determined in the coculture at three points during the growth curve. In all tested cocultures, the percentage of A.t. decreased over time (Fig. 1 and data not shown). In a coculture inoculated with a 1:1 ratio, A.t. dropped from 50% of total cells to ≈16% in late stationary phase. To determine whether our laboratory strain of P.a. (originally a clinical isolate) exhibited coculture trends generally representative of other P.a. strains, four environmental isolates (see Table 2) were also examined in coculture with A.t. Both pure culture growth rates and coculture trends with A.t. were similar to PAO1 (data not shown).
Fig. 1.
Planktonic cocultures. Population dynamics of wild-type, mutant, and complemented mutant cocultures inoculated at a 1:1 ratio. Displayed on the y axis (at the left) is the percentage of A.t. present in the culture at four different points of the growth curve. Standard deviation of three replicates is indicated. Open circles indicate the OD600 (at the right) of a representative growth curve at which a coculture sample was assayed. All coculture growth curves were nearly identical (data not shown). Quorum-sensing mutant strains were complemented by the exogenous addition of the indicated amount of purified acyl-HSLs.
The relative percentage of P.a. present in the coculture increased during logarithmic growth, most likely because of its faster doubling time. However, the P.a. present in the coculture continued to increase in stationary phase, long after growth had ceased, as measured by OD600 (compare percentage of A.t. present at 250 min with that at 1,440 min in Fig. 1). Because differences in exponential growth rate cannot account for this observation, it was further investigated.
P.a. Quorum-Sensing Mutants Show Reduced Growth Yields in Planktonic Cocultures.
P.a. quorum sensing controls the expression of several secreted factors related to antimicrobial activity and nutrient acquisition; thus, we hypothesized that such functions might contribute to the observed increase in P.a. percentage between early and late stationary-phase cultures. Acyl-HSL signal-production mutants defective in the synthesis of butyryl-HSL (P.a.-rhlI), 3-oxododecanoyl-HSL (P.a.-lasI), or both signals (P.a.-lasIrhlI) were examined in liquid cocultures with A.t. as was PAO1, the relative amount of A.t. decreased throughout log phase in all three cocultures (Fig. 1). This decrease was not surprising, because the mutant strains had a doubling time similar to that of PAO1 (see Table 3, which is published as supporting information on the PNAS web site). However, unlike wild-type P.a. cocultures, A.t. percentages for all three quorum-sensing mutant cocultures increased or plateaued between early and late stationary phase (Fig. 1). Exogenous addition of purified acyl-HSL signals to the growth medium restored the wild-type coculture phenotype of the P.a. acyl-HSL-synthase mutants (Fig. 1). A P.a.-lasRrhlR double mutant showed coculture trends similar to those of the quorum-sensing signal-synthase mutant strains (Fig. 1).
Previous studies have indicated that acyl-HSLs produced by one species may influence quorum sensing in another species, such as in modulation of the amount and type of signal produced (4, 11). To examine this possibility, acyl-HSL signal profiles present in pure and cocultures of wild-type A.t. and P.a. were examined by thin-layer chromatography and acyl-HSL-responsive bioassays. The cocultivation of P.a. and A.t. did not significantly affect acyl-HSL levels produced by either species (see Fig. 7, which is published as supporting information on the PNAS web site). It is important to note that, although A.t. utilizes an acyl-HSL-based quorum-sensing system and does produce low levels of 3OC8-HSL in standard culture, this system remains nonfunctional in the absence of specific compounds called opines, produced only by Agrobacterium-infected plant tissue (12).
We hypothesized that P.a. quorum-sensing-regulated functions, or the acyl-HSL signals themselves, might impair or kill A.t. Purified C4-HSL and 3OC12-HSL were added to stationary-phase A.t. cultures and found to have no effect on A.t. viability (data not shown). Three quorum-sensing-controlled toxins are rhamnolipid, cyanide, and pyocyanin (13–16). We found that a triple mutant unable to make any of these compounds (AHP4C) had a coculture phenotype with A.t. identical to wild-type (data not shown). Finally, filtrates from P.a. stationary-phase cultures and P.a./A.t. cocultures were examined for A.t.-inhibitory activity. A.t. suffered no observable effects in the presence of these filtrates (data not shown). Thus, it appeared that inhibition or killing of A.t. by P.a. quorum-sensing-regulated function(s) did not explain the dominance of P.a. in coculture.
The hypothesis that quorum sensing imparts a growth advantage to P.a. in stationary-phase cocultures was tested next. Total colony-forming units (CFUs) present in the culture were determined in early vs. late stationary-phase liquid cultures. In A.t. and P.a. pure cultures, total CFUs did not significantly change between late and early stationary-phase time points (Table 1). However, in coculture, P.a. numbers continued to increase between early and late stationary phase, whereas A.t. CFUs remain constant. Although the coculture OD600 did not change significantly between early and late stationary phase, the total cell number did (compare the open circles on Fig. 1 to Table 1). This apparent inconsistency may, in part, be attributed to a decrease in overall cell size for P.a. in late stationary phase. On the other hand, total P.a. CFUs do not increase in stationary-phase cocultures for the quorum-sensing mutants as they did for wild-type (Table 1). These data suggest that quorum-sensing-regulated functions allow P.a. to achieve a slightly higher growth yield in cocultures.
Table 1.
Liquid culture CFU/ml in stationary phase (×107)
|
P.a. |
A.t. |
|||
|---|---|---|---|---|
| Early | Late | Early | Late | |
| P.a | 220 ± 14 | 180 ± 35 | ||
| P.a.-lasIrhlI | 180 ± 49 | 120 ± 47 | ||
| A.t. | 170 ± 12 | 210 ± 10 | ||
| P.a./A.t.* | 30 ± 1 | 83 ± 32 | 87 ± 15 | 110 ± 18 |
| P.a.-lasIrhlI/A.t.* | 19 ± 2 | 20 ± 1 | 150 ± 14 | 140 ± 7 |
*Initial inoculum ratio P.a./A.t. = 1:10.
P.a. Outcompetes A.t. in Biofilm Coculture.
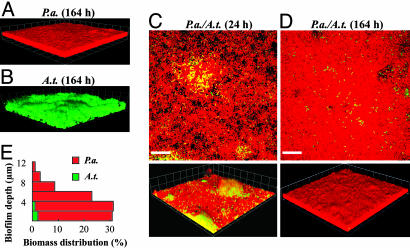
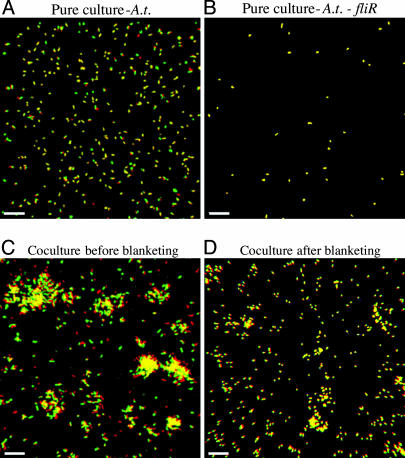
P.a. and A.t. pure cultures and 1:1 cocultures were characterized in flow-cell biofilms. Similar to previous reports, P.a. formed flat, tightly packed biofilms with little heterogeneity in succinate-based minimal medium (Fig. 2A; and see Table 4, which is published as supporting information on the PNAS web site) (17). In contrast, A.t. formed loosely packed biofilms with significant architectural heterogeneity compared with P.a. biofilms (Fig. 2B and Table 4) (18). In biofilm cocultures, P.a. eventually covered A.t., a phenomenon we call “blanketing” (Fig. 2 C and D). The sagittal profile of the coculture displayed in Fig. 2E illustrates that P.a. was predominant throughout the biofilm and that small amounts of A.t. biomass were confined to the glass surface. Before blanketing, the amount of A.t. biofilm biomass was similar to that in pure cultures at equivalent stages of growth (Fig. 3D). However, once blanketing was complete (≈48–72 h), A.t. biomass proceeded to decline to ≈1% of the total coculture biomass, remaining at that level for extended periods (≈10 d) (Fig. 3D; and see Table 5, which is published as supporting information on the PNAS web site; and data not shown). Viability staining of the 164-h coculture biofilm showed that the majority of cells in the biofilm were alive, indicating that the blanketed A.t., although spatially confined, remain viable (data not shown). Complete blanketing was not observed when P.a. was added after A.t. had established a biofilm. The addition of planktonic P.a. to preestablished pure-culture A. t. biofilms (164 h) resulted in significant P.a. colonization, although complete blanketing was never achieved (data not shown).
Fig. 2.
P.a. blankets A.t. in coculture biofilms. (A and B) Three-dimensional views of P.a. and A.t. wild-type pure-culture flow-cell biofilms. (C and D) Coculture biofilms at 24 and 164 h after inoculation. (Top) An x–y slice close to the attachment surface. (Bottom) Three-dimensional views. Red cells, P.a., green cells, A.t. (Scale bars, 20 μm; 1 unit in 3D view = 13.4 μm.) (E) A quantitative determination of biomass distributions along biofilm depth in a coculture biofilm at 96 h.
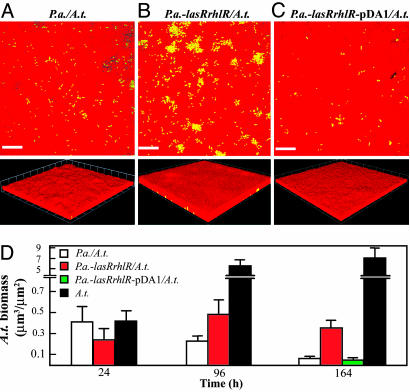
Fig. 3.
Quorum sensing provides an advantage to P.a. in coculture biofilms. (A–C Top) An x–y slice close to the attachment surface. (A–C Bottom) Three-dimensional views. (A) A coculture biofilm of wild-type P.a. and A.t. (B) A coculture biofilm of P.a.-lasRrhlR and wild-type A.t. (C) A coculture biofilm of P.a.-lasRrhlR complemented with pDA1 and wild-type A.t. Red cells, P.a., green cells, A.t. (Scale bars, 20 μm; 1 unit in 3D view = 13.4 μm.) All micrographs were taken at 164 h. (D) comstat determination of the relative amount of A.t. biofilm biomass present in pure and coculture biofilms.
P.a. Quorum Sensing Plays a Role in Older Coculture Biofilms.
We next examined whether P.a. quorum sensing played a role in biofilm cocultures. The P.a.-lasRrhlR mutant strain was used for these studies, because the P.a.-lasIrhlI strain has a known defect in twitching motility unrelated to quorum sensing (19). In pure culture, P.a.-lasRrhlR had a growth rate similar to wild-type and formed biofilms with similar structure (Table 4). In 24-h biofilms, the amount of A.t. biomass present in the coculture with P.a.-lasRrhlR was comparable with the wild-type P.a./A.t. coculture (Fig. 3D). However, at later time points, the amount of A.t. biomass in the P.a.-lasRrhlR coculture biofilm remained constant, in contrast to the wild-type P.a. coculture, where A.t. biomass continued to decrease (compare Fig. 3 A and B; and see Fig. 3D and Table 5). Complementation of the P.a.-lasRrhlR double mutant with pDA1 restored wild-type biofilm coculture phenotypes (Fig. 3C).
Motility Confers a Competitive Advantage to P.a. in Coculture Biofilms.
We hypothesized that blanketing by P.a. was mediated by surface motility. P.a. is capable of moving on a surface using two separate types of motility. Twitching motility is mediated by type-IV pili, whereas swarming motility involves type IV pili, flagella, and secreted surfactants called rhamnolipids (20, 21). Both swarming and twitching motilities have been implicated in P.a. biofilm development (17, 22, 23). A.t. can swim via flagella, but has not been reported to exhibit surface motility. Two P.a. motility mutants were studied in both pure and coculture biofilms. A mutant with a nonfunctional pilA gene, encoding the pilin subunit, is unable to produce a type IV pilus and shows no twitching and reduced swarming motility. The P.a. flgK gene encodes a flagellar hook protein. A flgK mutant is unable to produce flagella and is defective in both swarming and swimming motilities.
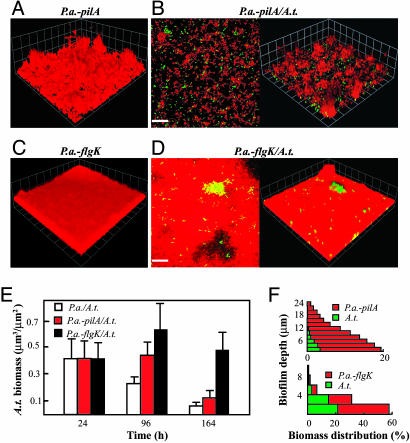
Similar to previous reports, the P.a.-pilA pure-culture biofilm was clearly heterogeneous and thick and had a high degree of surface roughness compared with the wild-type parent (Fig. 4A and Table 4) (17, 24). In coculture biofilms, P.a.-pilA did not exhibit complete blanketing but, instead, colonized discrete patches on top of A.t. microcolonies (Fig. 4B). A.t. biomass present in the P.a.-pilA coculture biofilm was slightly greater than with the wild-type P.a. at 96 h; however, at 164 h, levels of A.t. biomass were similar in both cocultures (Fig. 4E and Table 5). In pure culture, the P.a.-flgK strain formed flat biofilms very similar to wild-type (compare Figs. 2A and 4C; Table 4). In coculture biofilms, P.a.-flgK exhibited reduced blanketing, with much of the A.t. exposed to the overlying bulk liquid (Fig. 4D). A.t. biomass present in the P.a.-flgK coculture biofilm was significantly higher than the P.a. coculture at 164 h (Fig. 4E and Table 5). Analysis of sagittal profiles of the P.a.-flgK/A.t. coculture revealed that A.t. represented a significant portion of the biomass (30–50%) throughout the biofilm (Fig. 4F). The pilA and flgK mutants were successfully complemented in trans with pDA2 (Plac::pilA) and pDA3 (Ptac::flgK), respectively (data not shown). Unlike biofilm cocultures, both flgK and pilA mutant strains were indistinguishable from wild-type P.a. in planktonic coculture (data not shown).
Fig. 4.
P.a. motility is required for blanketing. (A and C) Three-dimensional views of a P.a.-pilA (A) and P.a.-flgK (C) pure-culture biofilms. (B and D) x–y slices close to the attachment surface and 3D views of a P.a.-pilA/A.t. (B) and P.a.-flgK/A.t. (D) Coculture biofilms. Red cells, P.a.; green cells, A.t. (Scale bars, 20 μm; 1 unit in 3D view, 13.4 μm.) All micrographs were taken at 164 h. (E) comstat determination of the relative amount of A.t. biofilm biomass present in cocultures. (F) A quantitative determination of biomass distributions along biofilm depth in P.a.-pilA/A.t. and P.a.-flgK/A.t. coculture biofilms at 96 h.
Influence of A.t. Flagellar Motility on Coculture Biofilms.
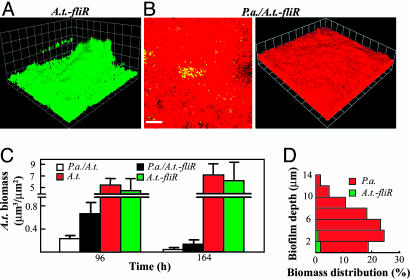
To compare the role of A.t. and P.a. motility functions for liquid and biofilm coculture interactions, A.t. bearing a mutation in the fliR gene (defective in swimming motility) was examined (25). In liquid culture, this strain had an identical growth rate and the same coculture phenotype as the wild-type A.t. strain (data not shown). In pure-culture biofilms, this strain exhibited an attachment-deficient phenotype relative to wild-type A.t., although it ultimately formed biofilms with only slightly less biomass than wild-type at 164 h (compare Fig. 2B with Fig. 5A and C and Fig. 8, which is published as supporting information on the PNAS web site). In coculture, P.a. blanketed the A.t.-fliR mutant and eventually formed a biofilm with similar structure and biomass distribution as with wild-type A.t. (compare Figs. 2D and 5B; Figs. 2E and 5D). Interestingly, at earlier time points, there is more A.t.-fliR biomass in the coculture biofilms than observed for A.t. (Fig. 5C). We hypothesized that this difference might reflect the repulsion or emigration of wild-type A.t. away from surfaces colonized by P.a. in earlier coculture biofilms, for which the A.t.-fliR strain would be deficient.
Fig. 5.
Biofilm coculture phenotypes of an A.t.-fliR mutant strain. (A) A 3D-view of an A.t.-fliR pure-culture biofilm; (B) An x–y slice close to attachment surface and a 3D-view of a P.a./A.t.-fliR coculture biofilm; Red cells, P.a; green cells, A.t. (Scale bars, 20 μm; 1 unit in 3D view, 13.4 μm.) All micrographs were taken at 164 h. (C) comstat determination of the relative amount of A.t. biofilm biomass present in pure cultures and cocultures. (D) A quantitative determination of biomass distributions along biofilm depth in a P.a./A.t.-fliR. coculture biofilm at 96 h.
To test this hypothesis, various experimental approaches were tried. Initially, collection of effluent from the flow-cell system failed to show significant emigration of A.t. from the coculture biofilm (data not shown). However, time-lapse microscopy revealed that, at early time points in pure and coculture biofilms, A.t. has the capacity to detach and leave (Fig. 6 A and C; and see Movies 1 and 2, which are published as supporting information on the PNAS web site). Cells from the bulk liquid were also observed to attach to the surface after the time series was initiated. On the other hand, once A.t.-fliR attached to the surface, no detachment was observed in pure and cocultures (Fig. 6B; and see Movie 3, which is published as supporting information on the PNAS web site; and data not shown). In coculture with P.a., wild-type A.t. was distinctly motile before blanketing and was observed to gradually leave the biofilm. Analysis of time-lapse series revealed an ≈10–15% decrease of A.t. biofilm biomass over a 3-h period before complete blanketing (Fig. 6C and data not shown). It is not clear whether P. a. induces detachment of A.t. in biofilm cocultures. The loss in A.t. biofilm biomass in cocultures may simply be due to normal detachment rates coupled with a decrease or halt in A.t. biofilm growth.
Fig. 6.
Different biofilm time-lapse microscopy series. Yellow cells indicate bacteria that have not moved during the course of the time series. Green cells indicate bacteria that were present in the field of view at the beginning of the time course but not at the end. Red cells are bacteria present at the end of the time series that were not present at the start. (A and B) A 1-h time course of a newly inoculated pure-culture biofilm of wild-type A.t. (A) or A.t.-fliR (B). (Scale bar, 12 μm.) (C and D) A time course of wild-type A.t. in coculture with P.a. (P.a. cells are not visible). (C) A 3-h time course before blanketing has occurred. (D) A 3-h time course after complete blanketing has occurred. (Scale bar, 20.2 μm.)
In older coculture biofilms, after blanketing had occurred, both A.t. and A.t.-fliR were immobilized (Fig. 6D; and see Movie 4, which is published as supporting information on the PNAS web site; and data not shown). Ultimately, at later time points, the amount of A.t.-fliR present in the coculture biofilm was only slightly greater than the wild-type A.t. (Fig. 5C and Table 5).
Discussion
In this study, we have begun to examine the complex interactions between two common environmental microorganisms, P.a. and A.t., in planktonic and biofilm growth modes. During exponential growth in dispersed, liquid culture, P.a. dominated A.t. because of a higher growth rate. Within biofilms grown on glass surfaces in the same defined medium, P.a. was also found to numerically dominate the population and to cover adherent A.t., a process requiring motility via flagella and type IV pili. Quorum-sensing mutants displayed an impaired competition phenotype in both liquid and flow-cell biofilm cultures. Motility was found to be important for both species in coculture biofilms. Although A.t. was outnumbered after the rapid-growth phase in both growth formats, its population remained viable, leading to a period of coexistence of these two microbes.
Quorum sensing appears to allow P.a. to achieve a slightly higher growth yield in liquid cocultures. Several quorum-sensing-regulated secreted functions related to nutrient acquisition might explain this observation. Quorum sensing regulates multiple secreted proteases, which might act to degrade A.t. exoproducts, which could then serve as a nutrient source (26). An alternative possibility is that P.a. may have a lower Ks value than A.t. for key limiting nutrients in coculture. Quorum-sensing-regulated functions may affect the Ks values for these substrates. An example is iron acquisition by the quorum-sensing-controlled siderophore pyoverdine, which is used to secure iron in a form less accessible to other species (although the defined medium used in this study is iron replete).
A P.a. quorum-sensing mutant strain was impaired in biofilm cocultures. The quorum-sensing mutant strain coculture biofilm displayed a modest increase in the amount of A.t. biomass. One potential explanation is that the dense carpet of P.a. cells covering A.t. produces a quorum-sensing-regulated toxic compound(s) that kills or inhibits its growth. A similar role for quorum sensing has been shown for Pseudomonas aureofaciens, which uses quorum-sensing-regulated phenazine antibiotics to compete with the local flora of the wheat rhizosphere (27). Filtered supernatants of P.a. liquid cultures failed to inhibit or kill A.t. However, we cannot rule out the possibility that direct cell contact is necessary for killing/inhibition or the high cell densities of P.a. present in biofilms produced an elevated, lethal concentration of a secreted antimicrobial(s).
In flow-cell biofilms, P.a. formed confluent mats on the biofilm surface that eventually submerged A.t. microcolonies. The blanketing phenotype was impaired in both flgK and pilA mutants, suggesting that both of these surface appendages and, by extension, the motility they provide, are required for this phenomenon. However, the severity of the competitive deficiency was different between the two mutants. The amount of A.t. biofilm biomass remained relatively constant in coculture with the flgK mutant, whereas, in pilA cocultures, the amount of A.t. biomass decreased over time. In the flgK coculture A.t. was present at the surface of the biofilm, whereas, in the pilA coculture, it localized to the biofilm depths. Therefore, the success of A.t. in the flgK coculture may result from its exposure to nutrients present in the overlying liquid medium. The pure culture biofilms of wild-type P.a. and the flgK mutant were structurally indistinguishable, whereas the flgK mutant differed substantially from the wild-type in the coculture biofilms, suggesting that some functions have little impact on the development of pure-culture biofilms, but can have a significant impact in the development of biofilms in a mixed-species environment. Another important point is that flgK and pilA mutants did not exhibit a competitive defect in planktonic coculture. This observation highlights the importance of the microenvironment in dictating key competitive factors.
Surprisingly, disruption of flagellar motility in A.t. did not augment the dominance of P.a. in flow-cell biofilm coculture. In pure culture, such a mutation has a distinct adherence defect (Fig. 8). Instead, in biofilm cocultures, the A.t.-fliR mutant accumulated more initial biomass on the surface (Fig. 5C), although this biomass was eventually subsumed, as with the wild type, by the rapidly moving P.a. cells. Our time-lapse microscopy observations of early stages of surface colonization suggest that a significant fraction of A.t. cells contacting and transiently adhering to the surface is highly motile, and many A.t. cells tend to swim away from the surface. In contrast, once bound to the surface, the fliR mutant is much more stable, with few emigrating cells. It is intriguing to speculate that, in coculture biofilms with P.a., A.t. flagellar motility may be more important as a mechanism of escape than as a factor that enhances surface colonization. These data suggest that there may be two distinct mechanisms that result in the diminished A.t. biomass observed in older coculture biofilms. The first could be the erosion of biomass because of emigration of A.t. from the biofilm before blanketing. The second is the loss of A.t. biomass after blanketing (not attributable to emigration from the biofilm), perhaps because of a quorum-sensing-regulated function (as seen in Fig. 3).
The dual-species format used in this study is a powerful approach for identifying key attributes that allow specific microbes to effectively compete and coexist in different environments. Indeed, other laboratories are increasingly taking advantage of reconstituting simple, defined multispecies laboratory model systems to gain insight into microbial interactions (28–30). Our findings identified P.a. quorum sensing and motility via pili and flagella as functions that contribute to its competitive interactions with A.t. in our binary coculture system. We are currently exploring whether our observations in laboratory coculture represent general features of common competitive interactions that occur in the environment. For example, do other species use quorum sensing to regulate their competitive interactions with cohabiting microbes in dense, polymicrobial environments? If so, the degradation or sequestration of acyl-HSLs within these environments, now well documented for several diverse bacteria, could be particularly influential (31). Ultimately, by using observations extrapolated from simple, tractable coculture model systems, we hope to identify important aspects of microbial interactions in complex systems.
Materials and Methods
Bacterial Strains and Media.
The P.a. and A.t. strains used in this study are described in Table 2 (and see Supporting Text, which is published as supporting information on the PNAS web site). Bacterial cultures were grown in AT minimal medium (32). The carbon sources were 0.5% glucose, 0.4% succinate, or 0.5% glutamate. Where indicated, before coculture assembly, acyl-HSL was added and dried in the culture flask for a working concentration of 5 μM for C4-HSL or 2 μM for 3OC12-HSL. For supernatant killing assays, 5 ml of a 24-h-old stationary-phase culture was passed through 0.22-μm filters (Millex-GV; Millipore, Bedford, MA), and the filtrate was added to washed stationary phase A.t. cells. A.t. viability was measured by plate counts.
Planktonic Pure-Culture and Coculture Growth Curves.
All growth curves were conducted at 30°C and performed in triplicate. Cocultures were inoculated by growing pure cultures to log phase and then diluting the cultures to OD600 = 0.1. Cocultures (50 ml) were initiated by directly combining the pure cultures in the appropriate ratios. At different growth stages of the planktonic cocultures, 3-μl samples were subjected to epifluorescence microscopy. The numbers of the fluorescent A.t. and the total number of cells viewed by light microscopy were counted. The percentages of A.t. were calculated based on >1,000 total cells for each coculture. Pure cultures of A.t. were subjected to epifluorescence microscopy to verify that GFP fluorescence was retained in 100% of the culture. Viable plate counting was also used to quantify the two species in pure and cocultures. The two species were differentiated in coculture on the basis of gentamycin sensitivity (A.t. was resistant, whereas P.a. was sensitive).
Flow-Cell Biofilm Culture.
A flow-cell biofilm system was incubated at 30°C as described in ref. 33. The coculture biofilm inoculum was prepared by combining the diluted pure cultures of P.a. wild type or mutants and A.t. in a 1:1 ratio. P.a. was stained with a 1:30 dilution of SYTO 62 (Molecular Probes), and A.t. was visualized by GFP fluorescence. Viability staining was performed as described in ref. 34. Images were obtained by confocal laser scanning microscopy (CLSM) and processed by using the program volocity (Improvision, Lexington, MA). Every condition tested was run in duplicate channels and in multiple experiments. The image-analysis program comstat (35) was applied to quantify structural aspects of biofilms of 10 randomly chosen representative images. For time-lapse-microscopy experiments, images were acquired of the same field of view for the times indicated in a flow-cell biofilm reactor. During the experiment, flow cells were subjected to continuous flow and were incubated at 30°C on the microscope stage in an environmental chamber. Three replicates were performed for each biofilm condition.
Supplementary Material
Acknowledgments
We thank C. Manoil (University of Washington, Seattle) and G. A. O’Toole (Dartmouth College, Hanover, NH) for provided bacterial strains and P. K. Singh, T. L. Yahr, T. G. Platt, J. D. Bever, and B. E. Ramey for helpful discussion and critical reading of the manuscript. This work was supported by a grant from the National Science Foundation (to M.R.P.) and U.S. Department of Agriculture Grant CRI 2002-35319-12636 (to C.F.).
Abbreviations
- A.t.
Agrobacterium tumefaciens
- CFU
colony-forming unit
- P.a.
Pseudomonas aeruginosa.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Tilman D. Proc. Natl. Acad. Sci. USA. 2004;101:10854–10861. doi: 10.1073/pnas.0403458101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith V. H. Antonie Leeuwenhoek. 2002;81:99–106. doi: 10.1023/a:1020533727307. [DOI] [PubMed] [Google Scholar]
- 3.Taga M. E., Bassler B. L. Proc. Natl. Acad. Sci. USA. 2003;100:14549–14554. doi: 10.1073/pnas.1934514100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riedel K., Hentzer M., Geisenberger O., Huber B., Steidle A., Wu H., Hoiby N., Givskov M., Molin S., Eberl L. Microbiology. 2001;147:3249–3262. doi: 10.1099/00221287-147-12-3249. [DOI] [PubMed] [Google Scholar]
- 5.Moller S., Sternberg C., Andersen J. B., Christensen B. B., Ramos J. L., Givskov M., Molin S. Appl. Environ. Microbiol. 1998;64:721–732. doi: 10.1128/aem.64.2.721-732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller M. B., Bassler B. L. Annu. Rev. Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 7.Costerton J. W., Lewandowski Z., Caldwell D. E., Korber D. R., Lappin-Scott H. M. Annu. Rev. Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 8.Schmeisser C., Stockigt C., Raasch C., Wingender J., Timmis K. N., Wenderoth D. F., Flemming H.-C., Liesegang H., Schmitz R. A., Jaeger K.-E., Streit W. R. Appl. Environ. Microbiol. 2003;69:7298–7309. doi: 10.1128/AEM.69.12.7298-7309.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Troxler J., Azelvandre P., Zala M., Defago G., Haas D. Appl. Environ. Microbiol. 1997;63:213–219. doi: 10.1128/aem.63.1.213-219.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu J. Y., Fan Y., Lin Y.-H., Zhang H.-B., Ong S. L., Dong N., Xu J.-L., Ng W. J., Zhang L.-H. Res. Microbiol. 2003;154:623–629. doi: 10.1016/j.resmic.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 11.McKenney D., Brown K., Allison D. J. Bacteriol. 1995;177:6989–6992. doi: 10.1128/jb.177.23.6989-6992.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuqua W., Winans S. J. Bacteriol. 1994;176:2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuster M., Lostroh C. P., Ogi T., Greenberg E. P. J. Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallagher L. A., Manoil C. J. Bacteriol. 2001;183:6207–6214. doi: 10.1128/JB.183.21.6207-6214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ran H., Hassett D. J., Lau G. W. Proc. Natl. Acad. Sci. USA. 2003;100:14315–14320. doi: 10.1073/pnas.2332354100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kownatzki R., Tummler B., Doring G. Lancet. 1987;329:1026–1027. doi: 10.1016/s0140-6736(87)92286-0. [DOI] [PubMed] [Google Scholar]
- 17.Klausen M., Heydorn A., Ragas P., Lambertsen L., Aaes-Jorgensen A., Molin S., Tolker-Nielsen T. Mol. Microbiol. 2003;48:1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- 18.Ramey B. E., Matthysse A. G., Fuqua C. Mol. Microbiol. 2004;52:1495–1511. doi: 10.1111/j.1365-2958.2004.04079.x. [DOI] [PubMed] [Google Scholar]
- 19.Beatson S. A., Whitchurch C. B., Semmler A. B. T., Mattick J. S. J. Bacteriol. 2002;184:3598–3604. doi: 10.1128/JB.184.13.3598-3604.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohler T., Curty L. K., Barja F., van Delden C., Pechere J.-C. J. Bacteriol. 2000;182:5990–5996. doi: 10.1128/jb.182.21.5990-5996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mattick J. S. Annu. Rev. Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 22.O’Toole G. A., Kolter R. Mol. Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 23.Klausen M., Aaes-Jorgensen A., Molin S., Tolker-Nielsen T. Mol. Microbiol. 2003;50:61–68. doi: 10.1046/j.1365-2958.2003.03677.x. [DOI] [PubMed] [Google Scholar]
- 24.Heydorn A., Ersboll B., Kato J., Hentzer M., Parsek M. R., Tolker-Nielsen T., Givskov M., Molin S. Appl. Environ. Microbiol. 2002;68:2008–2017. doi: 10.1128/AEM.68.4.2008-2017.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramey B. E. PhD Dissertation. Bloomington, IN: Indiana Univ.; 2004. [Google Scholar]
- 26.Parsek M. R., Greenberg E. P. Proc. Natl. Acad. Sci. 2000;97:8789–8793. doi: 10.1073/pnas.97.16.8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morello J. E., Pierson E. A., Pierson L. S., III Appl. Environ. Microbiol. 2004;70:3103–3109. doi: 10.1128/AEM.70.5.3103-3109.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hogan D. A., Vik A., Kolter R. Mol. Microbiol. 2004;54:1212–1223. doi: 10.1111/j.1365-2958.2004.04349.x. [DOI] [PubMed] [Google Scholar]
- 29.Egland P. G., Palmer R. J., Jr, Kolenbrander P. E. Proc. Natl. Acad. Sci. USA. 2004;101:16917–16922. doi: 10.1073/pnas.0407457101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duan K., Dammel C., Stein J., Rabin H., Surette M. G. Mol. Microbiol. 2003;50:1477–1491. doi: 10.1046/j.1365-2958.2003.03803.x. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L. H., Dong Y. H. Mol. Microbiol. 2004;53:1563–1571. doi: 10.1111/j.1365-2958.2004.04234.x. [DOI] [PubMed] [Google Scholar]
- 32.Tempe J., Petit A., Holsters M., Montagu M., Schell J. Proc. Natl. Acad. Sci. USA. 1977;74:2848–2849. doi: 10.1073/pnas.74.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christensen B. B., Sternberg C., Andersen J. B., Palmer R. J., Nielsen A. T., Givskov M., Molin S. Methods Enzymol. 1999;310:20–42. doi: 10.1016/s0076-6879(99)10004-1. [DOI] [PubMed] [Google Scholar]
- 34.Teitzel G. M., Parsek M. R. Appl. Environ. Microbiol. 2003;69:2313–2320. doi: 10.1128/AEM.69.4.2313-2320.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heydorn A., Nielsen A. T., Hentzer M., Sternberg C., Givskov M., Ersboll B. K., Molin S. Microbiology. 2000;146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.