Abstract
Besides being a metabolic fuel, carbohydrates play important roles in plant growth and development, in stress responses, and as signal molecules. We exploited natural variation in Arabidopsis (Arabidopsis thaliana) to decipher the genetic architecture determining carbohydrate content. A quantitative trait locus (QTL) approach in the Bay-0 × Shahdara progeny grown in two contrasting nitrogen environments led to the identification of 39 QTLs for starch, glucose, fructose, and sucrose contents representing at least 14 distinct polymorphic loci. A major QTL for fructose content (FR3.4) and a QTL for starch content (ST3.4) were confirmed in heterogeneous inbred families. Several genes associated with carbon (C) metabolism colocalize with the identified QTL. QTLs for senescence-related traits, and for flowering time, water status, and nitrogen-related traits, previously detected with the same genetic material, colocalize with C-related QTLs. These colocalizations reflect the complex interactions of C metabolism with other physiological processes. QTL fine-mapping and cloning could thus lead soon to the identification of genes potentially involved in the control of different connected physiological processes.
Sugars and starch are important plant products used for human diet and as raw material in industry (Roeper, 2002). The level and quality of carbohydrates depends on the species, the organ, growth conditions, and many other parameters, but globally reflects the balance between photosynthesis and growth. In addition, total plant biomass and, therefore, yield in the case of many crops largely depend on photosynthetic carbon (C) fixation and carbohydrate metabolism.
During photosynthesis, CO2 is fixed in the chloroplast into triose-phosphates, which are mainly used to regenerate ribulose-1,5- bisphosphate. Only the surplus is exported in the cytosol in counter exchange with inorganic orthophosphate (Edwards and Walker, 1983). In the cytosol triose-phosphates are converted to end products, including Suc, by releasing phosphate. Suc is the main transport form of C, which is translocated via the phloem to the sink organs. Suc phosphate synthase and Suc synthase (Susy) catalyze the formation of Suc in the cytosol, either with the substrates Fru-6-P and UDP-Glc or from Fru and UDP-Glc, respectively. Susy, a reversible enzyme, is more important for Suc degradation than for Suc synthesis. The catalytic reaction of Suc phosphate synthase results in Suc-6-P, and Suc synthesis is completed by the action of Suc-6-P phosphatase.
In the chloroplast, some of the photosynthate is directly converted to starch, which represents a transient storage, and is remobilized during the night to maintain leaf metabolism and Suc export to sink organs (Geiger and Servaites, 1994). ADP-Glc pyrophosphorylase catalyzes the first committed step of starch synthesis, producing ADP-Glc, the substrate for soluble and granule-bound starch synthases. The first product, amylose, is then further processed by starch branching enzymes I and II (Dennis and Blakeley, 2000).
Free hexoses are not direct end products of photosynthesis, but arise from Suc and starch degradation. Glc and Fru are produced by the action of invertases on Suc. There are several compartment-specific invertase isoforms (vacuolar, apoplastic, cytosolic) having different biological roles (for review, see Koch, 2004). Suc can be also degraded by Susy, producing Fru and UDP-Glc. Another source of free hexoses is the degradation of starch, which can either occur by phosphorolytic degradation or by hydrolysis via amylases to yield Glc. Free hexoses reenter metabolism via phosphorylation by hexokinases or fructokinases.
Starch, which is transitorily stored in leaves, is degraded mainly during the night, thus providing C for Suc synthesis and for growth and maintenance. Starch degradation in cereal endosperm has been studied in great depth at biochemical and molecular levels (Ritchie et al., 2002). Nevertheless, relatively little is known about the regulation of starch degradation in chloroplasts of photosynthetic tissue, where this pathway must be regulated in a manner that reflects its integration with many metabolic pathways, in contrary to the situation in endosperm, a cellular tissue at the time of starch degradation. Multiple forms of several starch-degrading enzymes, like a and b amylases, glucosidases, debranching enzyme, starch phosphorylase, and disproportionating enzyme (Trethewey and Smith, 2000), have been described for Arabidopsis (Arabidopsis thaliana) leaves, which suggests that several possible pathways of starch degradation exist. The coexistence of hydrolytic and phosphorolytic activities in chloroplast might directly direct the degradation products, in the first case free Glc and in the second case phosphorylated intermediates, into different metabolic pathways (Stitt et al., 1985). Nevertheless, results on mutants missing chloroplastic starch phosphorylase activity suggest that this enzyme has no major impact in general but a specific role under some circumstances, which needs to be defined (Smith et al., 2003). To this rather complicated picture is added the recent identification of a maltose exporter, which plays a major role in the export of C during starch breakdown (Niittyla et al., 2004).
Starch synthesis and starch degradation need to be well regulated to make sure that the transient starch in leaf supplies enough energy and C skeletons for sink demand during the night. Plants defective in starch synthesis do not survive in short-day conditions, and in the same way mutants in the degradation of the transitory starch grow more slowly than wild type (Zeeman et al., 1998).
Photosynthetic production of sugars and starch is regulated by the sink demand, which acts via sugars (Krapp and Stitt, 1994). During the last decade, the role of sugars not only as products of photosynthesis and the “fuel” for plants but also as important signal molecules has been discovered. Soluble sugars are implicated in the regulation of developmental processes, such as seed and embryo development (Hills, 2004), the formation of adult structures (Gibson, 2005), timing of flowering (Bernier et al., 1993), and the development of senescence (Rolland et al., 2002). They are supposed to play important roles as signals to regulate metabolic pathways, for example, photosynthesis, anthocyanin biosynthesis, β-oxidation, and nitrogen (N) metabolism (for review, see Gibson, 2005).
Indeed, one major sink for C skeletons is the production of organic nitrogenous compounds, like amino acids, proteins, and many secondary compounds. The fixation and reduction of CO2 during photosynthesis and its reoxidation during respiration are crucial to provide both energy and C skeletons for the incorporation of inorganic N into amino acids by the N assimilation pathway. Vice versa, N availability and assimilation are required to sustain the use of C skeletons. In situations when N becomes limited, carbohydrates, starch, and soluble sugars accumulate in photosynthetic-active organs (Ono et al., 1996; Sun et al., 2002) and cannot be used for further synthesis of amino acids and their derivates. As a consequence of the high C status, photosynthesis is then down-regulated. The fine tuning of this complicated network is crucial to plant development and productivity, and over the last decades multiple levels of regulation between C and N metabolism have been revealed (Stitt and Hurry, 2002). The pattern of C and N regulation observed in global gene expression studies validates the combined CN signaling hypothesis (Palenchar et al., 2004; Price et al., 2004). Four distinct N- and/or C-sensing systems have been identified in higher plants today (for review, see Foyer et al., 2003). However, we do not have a complete picture of this well-orchestrated network.
Despite all the knowledge acquired on these central metabolic pathways in plants, genome sequencing revealed that we are far from understanding the structure and regulation of the basic metabolic pathways (Benning and Stitt, 2004). Multiple isoforms of enzymes exist and posttranslational modifications add to the complexity. In most instances there is no single form of an enzyme that catalyzes a given step. Instead, multiple isoforms provide the regulatory framework that is needed to adjust plant metabolism during development and in response to the environment. High-throughput functional genomics will speed up our knowledge on these pathways. However, due to the complexity and the multilevel regulation of metabolic networks, these approaches might reach their limits. Exploitation of natural variation can help to overcome these limitations by giving useful information on complex polygenic, quantitative traits such as, for instance, carbohydrate content (Koornneef et al., 2004). Most natural variation segregating between Arabidopsis accessions is of continuous nature and hence controlled by more than one gene. Quantitative trait locus (QTL) mapping is a means to identify the individual genetic factors, i.e. the QTLs, controlling quantitative traits. Interest for QTL analysis approaches only recently grew in Arabidopsis (Alonso-Blanco and Koornneef, 2000; Maloof, 2003). QTL studies do not directly identify the genes controlling a quantitative trait, but they yield valuable information on their minimal number and their approximate genomic position. The availability of the complete genomic sequence of Arabidopsis (Arabidopsis Genome Initiative, 2000) allows a preliminary identification of candidate genes and greatly facilitates the map-based cloning of QTLs.
Here, we describe a QTL analysis for soluble sugar and starch content in the rosettes of Arabidopsis plants growing in short-day condition under two contrasting N environments, one that limits growth (N−) and the other that is not limiting (N+). The Bay-0 × Shahdara recombinant inbred line (RIL) population studied here displayed large variations for all traits measured, and 39 QTLs corresponding to at least 14 distinct loci were detected. Taking advantage of the residual heterozygosity and the large size of this RIL population, we used a heterogeneous inbred family (HIF) strategy (Tuinstra et al., 1997) and were able to confirm two interesting QTLs in near-isogenic lines (NILs).
RESULTS
Statistical Analyses of the Variation
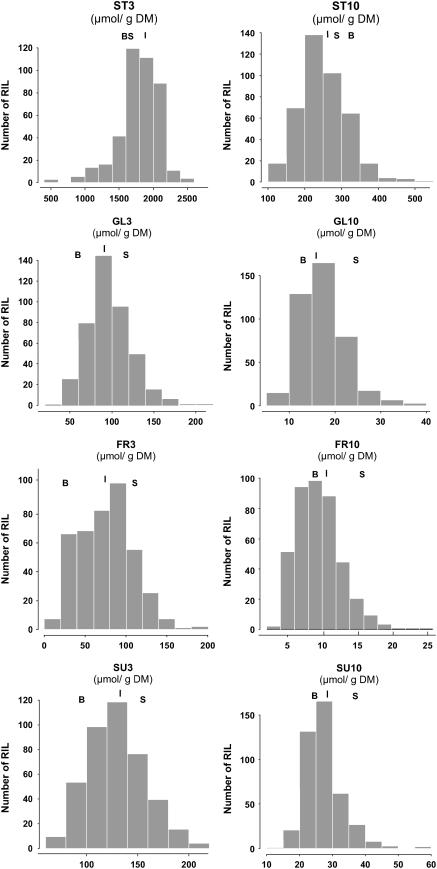
Analyses of variance revealed a significant genotypic effect for each trait measured (P < 0.0001). A significant effect of the nitrate environment was also found for each trait (P < 0.0001). Cultivation repetitions also had significant effects on phenotypic variations for all traits (P < 0.0001) except ST3 and ST10, thus revealing the probable influence of uncontrolled environmental factors, although repetitions were performed in the same growth chamber. No significant effects of genotype × repetition interactions were found. All subsequent QTL analyses were performed on unadjusted mean values across the different repetitions, which should represent a good estimation of the average behavior of a genotype in a specific N environment. Distributions of the phenotypic values among RILs (Fig. 1) are relatively normal, with the population mean at or between the parental values. Frequency distributions (Fig. 1) of the traits measured show that transgressive individuals are found in the population for each trait, although Bay-0 and Shahdara phenotypes are very similar for most traits. Starch content, for instance, was very similar in Bay-0 and Shahdara both on 10 mm and 3 mm nitrate, but a large variability was nevertheless observed in the progeny. Frequency distributions of the traits measured are in accordance with a polygenic determinism, i.e. clearly quantitative. Broad-sense heritability (Table I) was very high only for Fru content on 3 mm nitrate (82%), very low for Suc content on 10 mm nitrate (14%), and intermediate for the other traits (34%–49%) measured.
Figure 1.
Histograms of repartition of the phenotypic values in the Bay-0 × Shahdara population. For trait meaning, refer to Table I. B and S positions indicate values obtained for the parental accessions Bay-0 and Shahdara, respectively. The position of the vertical line above bars indicates the population mean value.
Table I.
Means, ranges, and broad-sense heritabilities of sugars and starch content in Bay-0, Shahdara, and the Bay-0 × Shahdara RILs, according to the nitrate environment (N+: 10 mm; N−: 3 mm)
| Trait | Code | Means
|
RIL Range | Heritability | ||
|---|---|---|---|---|---|---|
| Bay-0 | Shahdara | RIL | ||||
| N+ | ||||||
| Starch content (μmol/g DM) | ST10 | 294.5 | 257.8 | 250.6 | 105.9–529.8 | 0.49 |
| Glu content (μmol/g DM) | GL10 | 16.3 | 25.6 | 17.4 | 6.9–39.6 | 0.34 |
| Fru content (μmol/g DM) | FR10 | 8.2 | 14.7 | 9.5 | 3.7–24.4 | 0.47 |
| Suc content (μmol/g DM) | SU10 | 27.5 | 34.0 | 27.1 | 14.5–57.5 | 0.14 |
| N− | ||||||
| Starch content (μmol/g DM) | ST3 | 1,732.8 | 1,738.4 | 1,776.8 | 406.9–2,583.2 | 0.37 |
| Glu content (μmol/g DM) | GL3 | 67.0 | 121.7 | 96.5 | 30.5–200.2 | 0.48 |
| Fru content (μmol/g DM) | FR3 | 15.5 | 109.0 | 74.9 | 13.9–197.5 | 0.82 |
| Suc content (μmol/g DM) | SU3 | 90.0 | 148.5 | 130.0 | 41.8–130.0 | 0.36 |
The strongest correlations between the traits measured (Table II) were found between Suc and Glc contents (0.86 and 0.92) and Suc and Fru contents (0.69 and 0.49) on both N+ and N−, respectively. A stronger positive correlation was found in N+ between Glc and Fru (0.71) than in N− (0.46). A rather high negative correlation on both N+ and N− was found between total N content and starch content (−0.65 and −0.73, respectively). Interestingly, a strong correlation was found between amino acid content and starch content in N− (−0.75), whereas it was not significant in N+.
Table II.
Phenotypic correlations among N- and sugar-related traits in the Bay-0 × Shahdara RIL progeny (Pearson correlation coefficients)
N-related trait (total N content [NP], free amino acid concentration [AA], or nitrate concentration [NO]), flowering time under long days (LD), and DM values were obtained in previous studies (Loudet et al., 2002, 2003a). Sugar-related trait (starch [ST], Glu [GL], Fru [FR], or Suc [SU]) values were obtained in this study. Values in bold correspond to N− environment correlations; values in italic correspond to N+ environment correlations; and values in normal type correspond to correlations across N environments (N+/N−). ns, Correlation not significant at P < 0.01; ***, **, and *, correlation significant at P < 0.001, P < 0.01, and P < 0.05; NA, not available. N+/N−, Plants grown on 10 mm/3 mm nitrate.
| N−/N+ | DM | NP | AA | NO | LD | ST | GL | FR | SU |
|---|---|---|---|---|---|---|---|---|---|
| DM | +0.53*** | −0.34*** | ns | 0.23*** | 0.21*** | 0.47*** | 0.17*** | ns | ns |
| NP | −0.27*** | +0.12*** | +0.10* | 0.73*** | −0.21*** | −0.65*** | −0.31*** | −0.14** | −0.27*** |
| AA | −0.33*** | 0.84*** | +0.22*** | ns | 0.16** | ns | ns | ns | 0.16*** |
| NO | NA | NA | NA | NA | ns | −0.47*** | −0.22*** | ns | −0.20*** |
| LD | NA | NA | NA | NA | NA | 0.18*** | ns | ns | ns |
| ST | 0.39*** | −0.73*** | −0.75*** | −0.48*** | NA | 0.20*** | 0.38*** | 0.15** | 0.33*** |
| GL | 0.22*** | −0.10** | ns | ns | NA | ns | 0.36*** | 0.71*** | 0.86*** |
| FR | ns | ns | 0.17*** | ns | NA | ns | 0.46*** | 0.56*** | 0.69*** |
| SU | 0.21*** | −0.18*** | ns | −0.16*** | NA | 0.15* | 0.92*** | 0.49*** | 0.24*** |
QTL Analyses
QTL mapping results are shown in Table III and Figure 2. QTLs are named with the trait name suffixed with an ordering number starting with the first chromosome. A total of 39 independent QTLs distributed over all chromosomes and two QTL × QTL interactions were identified for the five traits measured on two contrasting nitrate environments (N+/N−). QTL effects were rather weak, with contributions to phenotypic variation (R2) ranging from 3% to 40% and with only four QTLs with R2 higher than 10%. When considering QTL likelihood peak positions and direction of allelic effects, it appears that nine QTLs were identified in both limiting (N−) and nonlimiting (N+) N environments, so we consider that 30 independent loci (and not 39) were actually identified. For each of these nine QTLs, significant QTL × N environment interaction effects were identified (P < 0.0001). Each trait measured is controlled by three to eight QTLs. Positive allelic effects on trait derive from Shahdara for 26 out of the 39 QTLs detected (negative “2a” value).
Table III.
Results of QTL analyses for starch (ST), Glc (GL), Fru (FR), and Suc (SU) contents in the Bay-0 × Shahdara RIL population
| Trait | Cofactorsa | Chromosome | Positionb | LOD Score | 2ac | R2d | h2e |
|---|---|---|---|---|---|---|---|
| ST10.1 | NGA248 | 1 | 24.6 | 10.2 | 43.7 | 8 | |
| ST10.2 | MSAT1-13 | 1 | 70.4 | 4.9 | −27.6 | 4 | |
| ST10.3 | NGA172 | 3 | 0.3 | 6.7 | 32.3 | 6 | |
| ST10.4 | MSAT4-8 | 4 | 5.7 | 7.0 | −37.5 | 7 | |
| ST10 complete model | 25% | 49% | |||||
| ST3.1 | MSAT1-13 | 1 | 67.1 | 6.1 | −156.3 | 4 | |
| ST3.2 | MSAT2-38 | 2 | 13.0 | 6.6 | −156.2 | 5 | |
| ST3.3 | NGA172 | 3 | 0.0 | 3.7 | 115.0 | 5 | |
| ST3.4 | MSAT3-21 | 3 | 49.8 | 14.3 | 234.5 | 14 | |
| ST3.5 | MSAT4-35 | 4 | 29.4 | 4.3 | −134.5 | 3 | |
| ST3.2 × ST3.4 | 2 × 3 | 3 | |||||
| ST3.4 × ST3.5 | 3 × 4 | 2 | |||||
| ST3 complete model | 36% | 37% | |||||
| GL10.1 | MSAT2-36 | 2 | 24.6 | 2.9 | 1.8 | 5 | |
| GL10.2 | MSAT4-9 | 4 | 53.2 | 6.1 | −2.7 | 6 | |
| GL10.3 | MSAT5-22 | 5 | 40.9 | 4.0 | −2.2 | 4 | |
| GL10.4 | MSAT5-19 | 5 | 75.2 | 6.4 | −2.6 | 4 | |
| GL10 complete model | 19% | 34% | |||||
| GL3.1 | MSAT1-10 | 1 | 23.1 | 2.5 | 8.4 | 3 | |
| GL3.2 | NGA128 | 1 | 56.0 | 5.5 | 11.1 | 5 | |
| GL3.3 | MSAT2-22 | 2 | 62.5 | 3.5 | −8.7 | 3 | |
| GL3.4 | MSAT4-8 | 4 | 1.8 | 8.1 | 14.0 | 6 | |
| GL3.5 | MSAT4-9 | 4 | 55.8 | 2.3 | −7.1 | 3 | |
| GL3.6 | NGA225 | 5 | 0.0 | 2.5 | −7.4 | 3 | |
| GL3.7 | MSAT5-9 | 5 | 41.2 | 10.2 | −17.3 | 10 | |
| GL3.8 | MSAT5-19 | 5 | 72.4 | 3.7 | −11.8 | 3 | |
| GL3 complete model | 36% | 48% | |||||
| FR10.1 | MSAT2-36 | 2 | 20.8 | 9.0 | 1.8 | 8 | |
| FR10.2 | MSAT4-15 | 4 | 30.5 | 16.4 | −2.5 | 16 | |
| FR10.3 | MSAT5-22 | 5 | 42.4 | 7.9 | −1.7 | 6 | |
| FR10.4 | MSAT5-19 | 5 | 71.8 | 17.1 | −2.5 | 12 | |
| FR10 complete model | 42% | 47% | |||||
| FR3.1 | MSAT2-38 | 2 | 18.0 | 5.7 | 11.4 | 5 | |
| FR3.2 | NGA172 | 3 | 3.7 | 13.3 | −17.1 | 4 | |
| FR3.3 | MSAT3-21 | 3 | 47.5 | 14.2 | −18.1 | 4 | |
| FR3.4 | MSAT4-15 | 4 | 33.2 | 57.7 | −40.2 | 40 | |
| FR3.5 | MSAT5-22 | 5 | 42.9 | 12.3 | −18.5 | 7 | |
| FR3.6 | MSAT5-19 | 5 | 73.2 | 10.0 | −15.7 | 5 | |
| FR3 complete model | 65% | 82% | |||||
| SU10.1 | MSAT2-36 | 2 | 22.1 | 5.2 | 2.7 | 5 | |
| SU10.2 | MSAT5-22 | 5 | 36.4 | 2.4 | −1.7 | 3 | |
| SU10.3 | MSAT5-19 | 5 | 73.2 | 6.4 | −3.0 | 5 | |
| SU10 complete model | 13% | 14% | |||||
| SU3.1 | NGA128 | 1 | 53.1 | 4.0 | 13.3 | 4 | |
| SU3.2 | MSAT2-22 | 2 | 62.5 | 3.5 | −11.4 | 3 | |
| SU3.3 | MSAT4-8 | 4 | 2.0 | 6.2 | 15.6 | 6 | |
| SU3.4 | MSAT4-9 | 4 | 54.7 | 3.1 | −11.6 | 4 | |
| SU3.5 | MSAT5-9 | 5 | 43.5 | 14.1 | −26.6 | 10 | |
| SU3 complete model | 27% | 36% |
Cofactors used in CIM model 6, as well as in ANOVA analysis.
Position expressed in centiMorgans from the first marker of the chromosome.
The mean effect of the replacement of both Shahdara alleles by Bay-0 alleles at the QTL.
Percentage of variance explained by the QTL or by QTL × QTL interactions when significant.
Broad-sense heritability.
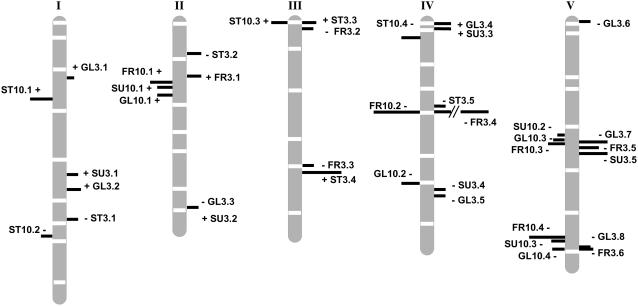
Figure 2.
QTLs detected for starch (ST), Glc (GL), Fru (FR), and Suc (SU) contents in the Bay-0 × Shahdara population. Each QTL is represented by a bar located at its most probable position (likelihood peak). QTLs detected on N+ (respectively, N−) are represented on the left (resp. right) side of chromosomes. The length of the bar is proportional to the QTL contribution to the phenotypic variation in the population (R2). The sign of the allelic effect is indicated for each QTL. The framework genetic map (indicating positions of the microsatellites markers) is from Loudet et al. (2002).
For starch content, four minor QTLs with R2 ranging from 4% to 8% were identified in N+ and five QTLs with R2 ranging from 3% to 14% in N−. One starch QTL (ST10.3/ST3.3) was common to both nitrate conditions with similar effects, on the top of chromosome 3 (marker NGA172). Two QTL × QTL interactions with weak R2, both involving QTL ST3.4, were found significantly associated with starch variation in the RILs (Table III). Four minor QTLs for Glc contents were identified in N+ conditions with R2 ranging from 4% to 6%, and eight QTLs were identified in N− conditions with effects ranging from 3% to 10%. Three QTLs (GL10.2/GL3.5, GL10.3/GL3.7, and GL10.4/GL3.8) were identified in both environments with similar effects, on chromosomes 4 (marker MSAT4-9) and 5 (MSAT5-19 and MSAT5-22). Four QTLs for Fru content were identified in N+ with R2 from 6% to 16% and six QTLs in N− with R2 from 4% up to 40%. Four Fru QTLs (FR10.1/FR3.1, FR10.2/FR3.4, FR10.3/FR3.5, and FR10.4/FR3.6) were identified in both conditions. The major QTL FR10.2/FR3.4 on chromosome 4 displayed different R2 according to the N environment: 16% in N+ and 40% in N−. In N−, as much as 40.2 μmol/g dry matter (DM) of Fru content variation could be explained by this locus. Conversely, QTL FR10.4/FR3.6 on chromosome 5 had higher effects (12%) in N+ compared to N− (5%). Only one QTL for Fru content shows positive 2a effects (in both environments). Three minor QTLs (R2 < 6%) for Suc content were identified in N+ and five QTLs in N− with R2 from 3% to 10%. One Suc QTL (SU3.5/SU10.2) was common to both N conditions on chromosome 5.
QTL Confirmation in NILs
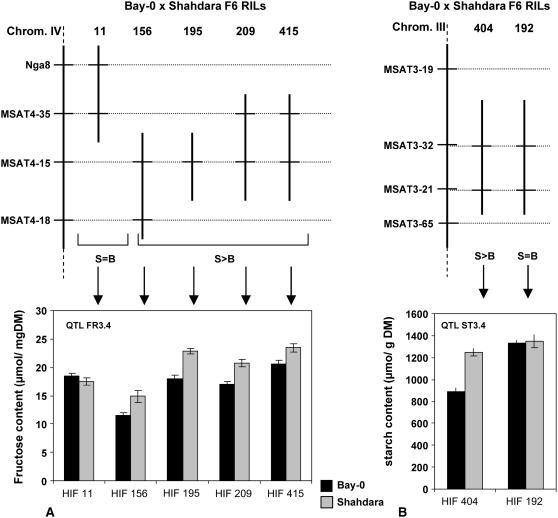
Two QTLs were chosen for confirmation in NILs: FR3.4 and ST3.4. Our NIL strategy (HIF) relies on the existence of RILs still segregating for the region of interest. Five F6 RILs with a residual heterozygosity around FR3.4 position (i.e. MSAT4.15) were found, whereas two such RILs were found for ST3.4 with a residual heterozygosity around MSAT3.21 (Fig. 3). Each of the distinct candidate RILs for a QTL displays a different genetic background, which is useful when QTLs are involved in epistatic interactions with other loci. Four F7 plants derived from each candidate RIL and fixed for either the Bay-0 or the Shahdara allele at the region of interest were selfed to compare F8 phenotypes. As displayed on Figure 3, FR3.4 was confirmed in N− conditions in four of the five HIFs tested (HIF156, HIF195, HIF209, and HIF415). These HIFs segregate around MSAT4-15, which is close to the estimated QTL position. Fru content was higher in the plants fixed for the Shahdara allele at MSAT4-15, which is consistent with the QTL analysis results (Table III). Conversely, Fru content was not significantly affected by the allele present at the region segregating in HIF11 (which does not include MSAT4-15). ST3.4 was confirmed in N− conditions with HIF404 (starch content being higher in Bay-0 fixed plants) but not with HIF192, although both HIFs are segregating at MSAT3-21, which is close to the estimated QTL position.
Figure 3.
Confirmation of QTLs FR3.4 and ST3.4 through NIL comparisons. A, HIFs 209, 415, 195, and 156 segregate around marker MSAT4-15. Comparison of NILs derived from these HIFs and fixed for either the Bay-0 (B) or the Shahdara (S) genotype confirms FR3.4. HIF11 segregates around markers NGA8 and MSAT4.35, and comparison of B/S-fixed NIL derived from this HIF does not confirm this QTL. B, HIF404 segregates around markers MSAT3-32 and MSAT3-21, and B/S-fixed NILs from this HIF confirm QTL ST3.4. Vertical bars indicate ses. S > B (S = B), Phenotypic values of NILs with the Shahdara genotype at the segregating region are (are not) significantly different from NILs with the Bay-0 genotype at the 5% probability threshold. Recombination breakpoints delimiting heterozygous regions are arbitrarily depicted in the middle of the marker interval.
Candidate Genes for Sugar-Related Traits
When taking into account 10-cM intervals around the QTL positions, several colocalizations between some of the 118 C metabolism-related candidate genes and QTLs were found (Table IV). Ten-centiMorgan intervals in Arabidopsis contain, on average, 450 genes, so such colocalizations cannot be interpreted without excessive speculation. Nevertheless, the colocalization of APL2 (large subunit of ADP-Glc pyrophosphorylase) with ST10.1 and the colocalization of a Suc transporter-like protein with SU3.5 are striking.
Table IV.
Colocalizations occurring between the QTLs for C-related traits detected and the 118 candidate genes chosen with a priori for their involvement in C metabolism
Ten-centiMorgan confidence intervals around the most probable position of QTLs (i.e. position of the LOD score maximum value) were taken into account to look for colocalizations. Chr., Chromosome; CHO, carbohydrate.
| Chr. | QTL | Gene | Gene Description | Function |
|---|---|---|---|---|
| I | ST10.1 | At1g27680 | ADP-Glc pyrophosphorylase large subunit 2, APL2 | Major CHO metabolism, starch synthesis |
| ST3.1 | At1g66430 | b2 pfkB-type CHO kinase family protein | Major CHO metabolism, Suc degradation | |
| SU3.1 | At1g32900 | β-Granule-bound starch synthase, GBS1 | Major CHO metabolism, starch synthesis | |
| II | SU3.2 | At1g47840 | Putative G hexokinase, similar to hexokinase 1 | Major CHO metabolism, Suc degradation |
| GL3.3 | At2g45880 | β-Amylase 7, BAM7 | Major CHO metabolism, starch degradation | |
| III | FR3.3 | At3g45940 | α-Glucosidase-like 3, AGL3 | Major CHO metabolism, starch degradation |
| FR3.2 | At3g08580 | Adenylate translocator identical to GB:X65549 | Major CHO metabolism, starch synthesis | |
| ST3.4 | At3g46970 | Glucan phosphorylase (cytosolic), PHS2 | Major CHO metabolism, starch degradation | |
| ST10.3 | At3g03250 | Putative UTP Glc 1 phosphate uridylyltransferase | Major CHO metabolism | |
| ST3.3 | ||||
| IV | GL10.2 | At4g32400 | ADP-Glc translocator, AGT1 | Major CHO metabolism, starch synthesis |
| FR3.4 | At4g15210 | β-Amylase 5, BAM5 RAM1 | Major CHO metabolism, starch degradation | |
| SU3.4 | At4g00490 | β-Amylase 2, BAM2 | Major CHO metabolism, starch degradation | |
| ST10.4 | At4g02280 | Susy/arab3/Susy4 (Suc synthase) | Major CHO metabolism, Suc degradation | |
| ST3.5 | At4g10260 | b2 pfkB-type CHO kinase family protein | Major CHO metabolism, Suc degradation | |
| V | GL10.4 | At5g64380 | Fru-1,6-bisphosphatase family protein | Photosynthesis, Calvin cycle |
| FR3.8 | ||||
| GL3.8 | ||||
| SU3.5 | At5g43610 | Suc transporter related | Major CHO metabolism | |
| FR3.5 | At5g45300 | β-Amylase 8, BAM8 | Major CHO metabolism, starch degradation | |
| FR10.3 | At5g39790 | Putative 5-AMP-activated protein kinase, β-1 subunit | Regulation |
We focused our candidate gene approach more particularly on FR3.4 and ST3.4. By genotyping the HIFs heterozygous around FR3.4 with new markers (data not shown), the candidate region surrounding this QTL was narrowed to 1,080 kb. This region contains 268 genes, including several putative candidate genes coding for enzymes involved in C metabolism. None of the known fructokinases, Suc synthases, invertase, or invertase inhibitors is located near FR3.4. This interval contains a cytosolic β-amylase (At4g15210), which is expressed in shoots at the vegetative stage and is inducible by sugar. The candidate region for ST3.4, if referring to HIF404, covers 10,175 kb. Although this region is still much too large to designate a candidate gene, it is nevertheless remarkable that a starch phosphatase (At3g46970) is encoded in this region. This region also contains a gene encoding Asn synthetase 1 (At3g47340), a major enzyme for amino acid synthesis.
DISCUSSION
The identification of new loci controlling quantitative traits related to metabolism should contribute to a better understanding of metabolic pathways and their regulation. QTL mapping in Arabidopsis has been only used a few times to study physiological traits (Mitchell-Olds and Pedersen, 1998; Bentsink et al., 2000, 2003; Juenger et al., 2000; Loudet et al., 2003a, 2003b; Sergeeva et al., 2003). This study focused on traits related to carbohydrate metabolism in interaction with N availability, taking advantage of the large Bay-0 × Shahdara RIL population (Loudet et al., 2002). DM, N metabolite content, root architecture, flowering time, water and ion homeostasis, and trichome density have been studied previously in this progeny (Loudet et al., 2002, 2003a,2003b, 2005; Symonds et al., 2005). We used strictly the same genetic material, which allowed us not only to detect QTLs for C traits but also to determine major interactions with other traits through QTL colocalizations. Besides being the metabolic fuel in plants, sugars have multiple roles during growth and development and in stress responses. In addition, their importance as signal molecules was realized during the last decade. Therefore, we will discuss in the following paragraphs not only the colocalization of QTLs for different C-related traits, but also compare the localization of C-related QTLs with other physiological traits, as, for example, N traits. Furthermore, we discuss possible interaction with stress response and developmental QTLs. All colocalizations have to be considered with great care since it is not possible to distinguish easily between pleiotropy and linkage at this time. Nevertheless, this analysis is always interesting for the identification of new physiological interactions.
Thirty-nine C metabolism-related QTLs have been detected in total, which correspond to at least 14 distinct polymorphic loci, each representing one to six colocalizing QTLs. The genetic dissection revealed at least three QTLs for each trait and up to eight QTLs for Glc content in N-limiting conditions. This is coherent with previous QTL studies in Arabidopsis, detecting usually two to seven QTLs for a given trait (Alonso-Blanco et al., 1998, 1999; Juenger et al., 2000; Loudet et al., 2003a, 2003b). Significant epistatic interactions were observed only for trait ST3, with two remarkable interactions both involving the ST3.4 locus. More epistatic interactions could be expected, knowing the complexity of the C metabolism pathways, but these remained undetected despite the high number of lines studied. With the notable exception of QTL FR3.4, mostly minor- and medium-effect QTLs have been detected (R2 = 3%–16%). The transgression observed for each trait among RILs is well genetically explained by the presence of both positive and negative allelic effect loci. Transgression for FR3 is not as strong as for other traits, which can be attributed to the major effect of QTL FR3.4.
QTLs for C-Related Traits Interact with N Availability
The variation observed for sugar-related traits in the Bay-0 × Shahdara progeny is wide (Fig. 1). All measured traits are strongly influenced by the level of nitrate provided to the plants (Fig. 1; Table I). In addition, many QTLs are specific to an N environment. Such an interaction of the QTLs for sugar-related traits with the N environment was expected, knowing the strong interrelation between C and N metabolisms in plants. Expression of C metabolism-related genes varies according to N status (Scheible et al., 1997; Dian et al., 2003). In addition, interaction between sugar and N sensing has been largely described (Coruzzi and Zhou, 2001). In recent studies, it appeared that the regulation exerted by C on gene expressions was usually rather modulated by N (Palenchar et al., 2004; Price et al., 2004).
We propose that loci common to both N+ and N− environments encode key enzymes or control elements for C metabolism, which are essential independently of the N status, whereas loci that are not stable across environments reflect adaptation of metabolism due to the constraint, encoding, for example, specific isoforms or regulatory factors. However, it is also possible that QTLs for sugar and starch are more difficult to detect in N+ condition due to lower carbohydrate levels in plant materials. Two out of four ST10 loci colocalize with a ST3 locus, and three out of four GL10 loci colocalize with a GL3 locus, which might indicate the general importance of these loci for starch and Glc accumulation. Interestingly, almost all QTLs for Suc are dependent on the environment, whereas, in contrast, all Fru QTLs detected in N+ also are detected in N−. The major Fru QTL found in this study (FR3.4), accounting for 40% of the variation in Fru content in N−, colocalizes with a Fru locus in N+ (FR10.2), which is only responsible for 16% of the variation. The importance of the FR10.2 effect (R2) is higher if compared to the genetic variation (estimated via the broad-sense heritability, h2): R2/h2 = 0.35 for FR10.2 (N+) versus 0.49 for FR3.4 (N−). This might indicate a greater influence of environmental variations in N+, combined with a possibly reduced power in QTL detection. Nevertheless, this locus (FR3.4/FR10.2) is clearly interacting with the N environment (P < 0.0001). The underlying gene might be especially important in conditions when high levels of Fru accumulate due to the misbalance in the C and N supply in N−, or the expression of this gene might be induced by N limitation.
QTLs for Different C Traits Colocalize
In this study, QTLs for different soluble sugars are often found clustered (Fig. 2). Interestingly, inside all such clusters, the allelic effects are in the same direction, which strengthens the possibility that one gene accounts for all the colocalized QTLs. For one region in the middle of chromosome 5, six QTLs for all possible C-related traits were detected. If there is only one gene underlying these QTLs, it must be of most concern a step leading to general carbohydrate accumulation, but not starch accumulation. It is often found that Fru, Glc, and Suc levels rise (or fall) in parallel in leaves. An initial accumulation of Suc, one end product of photosynthesis, is followed by an increase in free hexose levels generated by the cleavage of Suc. At two positions, on chromosome 2 and at the south end of chromosome 5, GL10, FR10, and SU10 QTLs (N+) colocalize, but in N− a QTL for Fru only is found. On the other hand, three times we observed either the combination of SU3 with GL3 or SU3 with FR3. Different metabolic pathways and specific regulations are certainly underlying these quantitative traits. Interestingly, the strongest QTL found in this study, FR3.4, is not accompanied by other sugar- or starch-content QTLs. It colocalizes with a FR10 locus, which might correspond to the same gene.
C QTLs Colocalize with QTLs for Other Physiological Traits
DM and N
The population used here has been studied previously for N-related traits (Loudet et al., 2003b). Five CN loci, including QTLs for different compositions of C and N traits, can be described (Krapp et al., 2005). CN1 (chromosome I) is specific for N+, CN2 (chromosome II) and CN3 (chromosome III) include QTLs from both N+ and N−, and CN4 (chromosome III) and CN5 (chromosome IV) are specific for N−. Colocalizations between starch and DM QTLs with positive allelic effects occur in N+. The increase in starch might be responsible for the higher DM, which itself could be due to an increase in C and energy supply for plant growth, especially during the night, when transitory starch becomes the only C and energy source. In N−, although transitory starch is unlikely limiting growth, since it represents 25% of total shoot dry weight, colocalizations are found between DM and starch loci, in accordance with the positive correlation between these two traits (+0.39). In both N+ and N−, colocalizations between starch- and N-content QTLs are found with opposite allelic effect signs, which is in line with the data showing an inverse relationship between N content and starch or sugar accumulation (Scheible et al., 1997). Two clusters involve an amino acid-content QTL with a sign opposite to starch-content QTL, which might reflect a role for amino acid level in the regulatory network between low N and high starch levels. Amino acids have for a long time been discussed as major signal molecules in N assimilation, and the finding of putative Glu sensors in plants (Lam et al., 1998) supports this idea. Interestingly, there are very few colocalizations of QTLs for soluble sugars contents and DM QTLs, which is reflected by the low correlation coefficient between these parameters. Starch content seems to have more impact on biomass accumulation than the level of free sugars. However, the carbohydrate levels studied were collected at the beginning of day (2 h light), and it would be interesting to perform the same study at the end of the day.
Water and Anion Content
Sugar accumulation is implicated in osmotic regulation. Sugars act either as osmotica or protect specific macromolecules during dehydration (Larcher, 1995; Danyluk et al., 1998). Indeed, one medium-effect QTL for water content, HU3.5 (Loudet et al., 2003a), colocalizes with SU3.3 with the same allelic effect sign, which could be due to the fact that an increase in sugar level osmotically increases water uptake. Anions such as chloride (CL) and phosphate (PO) also function as osmotica, mainly when replacing nitrate in nitrate-limiting conditions (Lamaze et al., 1984; White and Broadley, 2001). Soluble sugars and starch, as well as anions (mainly CL and PO), increase during N limitation, which might explain the colocalizations found between QTLs for these traits. One starch QTL (ST3.1) colocalizes with CL3.2 and PO3.2 (Loudet et al., 2003a). Another QTL cluster involves CL3.6, FR3.6, FR10.4, SU10.3, and GL10.4. Two phosphate QTLs are located close to loci for free sugars: PO3.5 near SU3.4, and PO3.6 near GL3.7, FR3.5, and SU3.5. If each of these loci corresponds to only one gene, it could be a factor involved in control of the osmotic balance control. More generally, the colocalizations observed might also be due to the fact that phosphate is closely linked to sugar metabolism. Inorganic phosphate plays a key role in the coupling of light and dark reactions in photosynthesis and in the export of triose phosphates from the chloroplasts. Depletion of free phosphate by acquisition of phosphate as sugar-phosphates (Leegood and Furbank, 1986) can then be overcome by the synthesis of starch and Suc to release again free phosphate. Inorganic phosphate also is a substrate or a product in many of the reactions of sugar metabolism and an effector of key enzymes in starch and Suc synthesis. In addition, several interactions have been observed on the gene expression level. Several Suc-responsive genes also are induced by phosphate starvation (Nielsen et al., 1998), and the expression of the high-affinity phosphate transporter (Lejay et al., 2003) and of many other phosphate-regulated genes is regulated by sugars (Franco-Zorrilla et al., 2005).
Flowering Time
Soluble sugar content affects flowering time in many plants (Bernier et al., 1993). In Arabidopsis, sugar export from the leaf correlates with flower induction (Corbesier et al., 1998) and supply of carbohydrates on the apical parts induces flowering in darkness (Roldan et al., 1999). One important QTL for flowering time in short day (SD1; Loudet et al., 2002) colocalizes with opposite allelic effect with GL3.4 and SU3.3 on the top of chromosome IV. Although the data analyzed were collected at 35 d, i.e. before flowering time in short-day conditions, this result would be in accordance with an earlier flowering matching to higher sugar content. SD1 very likely corresponds to FRIGIDA (Loudet et al., 2002), which is a direct component of the flowering pathway. On this locus maps as well the water-content QTL HU3.5 (see above). A tempting, but rather speculative hypothesis would be the regulation of flowering time via osmotic pressure changes by FRIGIDA.
Senescence
Senescence-associated yellowing and stress-related redness (anthocyanins) have recently been analyzed in the Bay-0 × Shahdara population in N-limiting condition (Diaz et al., 2006). A sugar accumulation is observed during senescence in leaves (Wingler et al., 1998; Stessman et al., 2002), which might even be involved in the induction of senescence (Yoshida, 2003). However, some contradictory data indicate that senescence in petals can be induced by sugar starvation as external application of sugar can delay senescence (van Doorn, 2004), and the reduction of photosynthetic efficiency, resulting in sugar starvation in the leaf, has been supposed to be a signal for induction of senescence (Hensel et al., 1993). This is in addition supported by the observation that the dark-induced expression of many senescence-enhanced genes is repressed in the presence of Suc (Chung et al., 1997). Obviously, the relationship between sugar levels and senescence is therefore not yet clear and might depend on the combination with other developmental signals (Ono et al., 2001). Roitsch and colleagues have recently shown that extracellular invertase is an essential component of cytokinin-mediated delay of senescence in tobacco (Nicotiana tabacum; Balibrea Lara et al., 2004). Cytokinins induce the expression of extracellular invertase, resulting in increased sugar utilization, reduced Glc levels, and delayed senescence. Furthermore, hexokinase, postulated as Glc sensor (Rolland et al., 2002), has been discussed as a player in regulating senescence, as transgenic tomato (Solanum lycopersicum) plants overexpressing hexokinase exhibit premature senescence (Dai et al., 1999) and Arabidopsis hexokinase 1 mutant (gin2-1) shows delayed senescence (Moore et al., 2003), which can either be due to the implication in sugar signaling or to its role in metabolism.
All of the loci for yellowness (YP) and redness (RV) are located close to C QTLs found in this study. For example, YP3.2 and YP3.4 colocalize with inverse effect with ST3.2 and ST3.4. In the same manner, RV3.6 colocalizes with GL3.7, FR3.5, and SU3.5. Thus, the inverse relationship between carbohydrates and senescence-related traits found in our analysis rather points to the link between sugar starvation and the onset of senescence.
Candidate Genes for C QTLs
Candidate gene approaches have already been undertaken on starch or soluble sugar content in many crop species. For instance, Chen et al. (2001) conducted a large analysis for candidate genes controlling potato tuber starch content. Beside several loci encoding enzymes involved in starch synthesis and degradation, such as ADP-Glc pyrophosphorylase, starch synthase and starch-degrading enzyme STP23, genes coding for enzymes involved in the production of the substrates for starch synthesis (including Rubisco, Rubisco activase, Susy, pyruvate kinase, and malic enzyme), also colocalized with QTLs for starch tuber content. In tomato, it was shown after the fine-mapping of a major QTL that fruit sugar content is controlled by molecular variants of the apoplastic invertase gene (Fridman et al., 2000). Several interesting colocalizations were found in this study (Table IV). The most remarkable of them involve QTL FR3.4 with a cytosolic β-amylase, QTL ST3.4 with a starch phosphorylase, QTL ST10.1 with APL2, and QTL SU3.5 with a Suc transporter.
We focused our candidate gene approach more finely on FR3.4 and ST3.4. We looked at all the genes included in the genomic regions in which these QTLs were validated by exploiting HIFs. Fru release arises by the cleavage of Suc by invertase, but none of the known invertase or invertase inhibitors is located near FR3.4. Susy also produces Fru by Suc cleavage, yielding Fru and UDP-Glc, which can reenter metabolism without any further ATP cost, but none of the annotated Susy genes maps close to FR3.4. However, Susy is highly regulated (Koch, 2004), and one of the potential regulatory genes might be underlying this major locus. An interesting candidate gene could be a cytosolic β-amylase (At4g15210), which is expressed in shoots at the vegetative stage and is inducible by sugar. However, Ram1 mutants defective in this gene contain wild-type levels of soluble sugars and starch (Laby et al., 2001). It is remarkable that a starch phosphatase (At3g46970) is encoded in the candidate region for ST3.4. Digital northern analysis (https://www.genevestigator.ethz.ch) reveals expression in leaves and differential expression during stress treatments. The gene encoding Asn synthetase 1 (At3g47340), a major enzyme for amino acid synthesis, is located on this part of chromosome 3 as well and also is a potential candidate. An enzyme involved in amino acid biosynthesis, whose expression, in addition, responds to the cell sugar level, could very well underlie this starch QTL, if considering that C and N traits colocalize at this locus (Krapp et al., 2005) and keeping in mind the close interaction between C and N metabolisms.
Many genes with putative regulatory functions, such as transcription factors and protein kinases, are located in the large confidence intervals delimiting the QTLs detected. Some of our QTLs likely represent new regulatory loci. In addition, we are aware that the restriction to major carbohydrate metabolism is rather limiting, and enzymes in pathways like photosynthesis, respiration, photorespiration, and many others would be possible candidate underlying QTLs for carbohydrate content. Research in recent years has hinted that the importance of the respiratory pathway in photosynthesis metabolism is greater than once imagined (Raghavendra and Padmasree, 2003). Clearly, due to the large confidence intervals delimiting QTLs and to the huge number of potential candidate genes for C-related traits, it seems absolutely necessary to precisely determine the QTL positions by a fine-mapping approach, which in Arabidopsis could potentially lead to the gene(s) underlying the QTL.
Confirmation of Two QTLs Using HIFs
QTL confirmation and fine-mapping requires the construction of NILs that differ only at a small region around the QTL of interest (Glazier et al., 2002). We used NILs derived from HIFs (Tuinstra et al., 1997). HIFs are derived from the selfing of lines displaying a specific residual heterozygosity and, therefore, segregating at a region of interest. The Bay-0 × Shahdara F6 progeny displays approximately 3% of residual heterozygosity (Loudet et al., 2002) and is large enough to successfully identify one or more heterozygous lines around each QTL of interest (Loudet et al., 2005). We chose to focus on two different loci. FR3.4 is the largest-effect QTL among all carbohydrate-related QTLs detected here (R2 = 40% in N−) and seems to interact with the N environment (significant QTL × N environment interaction, P < 0.0001). In addition, we chose to confirm ST3.4, the largest-effect QTL for ST3 trait, which colocalizes with a hot spot for N-related traits, L4 (Loudet et al., 2003b). The presence of FR3.4 could be confirmed in four of five different HIFs segregating in the region of this QTL (Fig. 3). HIF11, which is heterozygous in the northern half of the confidence interval of FR3.4, did not segregate for Fru content, which limits the QTL to the south of marker NGA8. ST3.4 was confirmed in HIF404 but not in HIF192 (Fig. 3). The segregating regions of these HIFs are potentially of different lengths in the south of MSAT3-21, so that, under the hypothesis that the QTL is located between MSAT3-21 and MSAT3-65, one HIF could segregate for the QTL while the other could be fixed at this locus. Epistatic interactions with other loci could also explain the different phenotypes of these two HIFs. HIF404 and HIF192 display different genotypes at marker MSAT4-35, which was detected as interacting with QTL ST3.4 (Table III), so that the allele at MSAT4-35 in HIF192 (Bay-0) might mask the effect of QTL ST3.4 in this HIF, provided it is actually segregating.
CONCLUSION
This work dissects for the first time, to our knowledge, the genetic architecture behind carbohydrate accumulation in Arabidopsis grown in two contrasting N environments. Beside the discovery of one major locus for Fru accumulation, which responds to N constraints, many small- or medium-effect QTLs are observed. Interesting colocalizations with traits related to N metabolism, water status, flowering, root growth, and senescence add together to a global picture of the multiple roles of sugars in metabolism and signaling, during stress responses, as well as in development and plant growth. The cloning of the underlying genes will provide a basis to optimize plant growth efficiency and thereby plant yield.
MATERIALS AND METHODS
Plant Material
The material used in this study was developed in our laboratory and has been deposited in public Arabidopsis (Arabidopsis thaliana) stock centers. The Bay-0 × Shahdara RIL population has been fully described previously (Loudet et al., 2002; http://www.inra.fr/qtlat). F7 seeds of the 415 RILs of the population obtained from the last generation of single-seed descent were used. They were harvested from plants grown once for all lines, thus minimizing the maternal environment effect. Plants were cultivated for 35 d in the growth room under short-day conditions and controlled temperature, light, and hygrometry as described by Loudet et al. (2003a). The RIL were completely and independently randomized in each cultivation repetition (performed successively in the same growth chamber). Two N environments were studied: the first one (10 mm nitrate, N+) is not limiting for plant growth, whereas the second one (3 mm nitrate, N−) strongly limits shoot growth. The data from three cultivation repetitions of the N+ environment and two repetitions of the N− environment have been collected and analyzed in detail.
NILs were developed as HIFs following the idea published by Tuinstra et al. (1997). At the F6 stage, lines 11, 209, 415, 195, and 156 were all heterozygous for two to four microsatellite markers surrounding QTL FR3.4 on chromosome 4 and homozygous for the other markers (Fig. 3). Lines 192 and 404 were heterozygous at two microsatellite markers close to QTL ST3.4 on chromosome 3 and homozygous for the other markers (Fig. 3). F7 plants derived from these lines were selected: two plants fixed with the Bay-0 allele and two other plants fixed with the Shahdara allele at segregating markers were chosen. F8 plants were used for phenotyping.
Phenotyping
Plants were previously harvested 2 h after the beginning of day, freeze-dried, weighted (DM), and extracted with a two-step ethanol-water procedure (Loudet et al., 2003a). The extracts obtained had been used previously to measure N-related traits: nitrate concentration, free amino acid concentration, and total N content (Loudet et al., 2003b). Aliquots of the same extracts were used in this study to measure starch, Glc, Fru, and Suc contents. (Table I).
For the determination of Glc and Fru contents, 140 μL of water and 60 μL of NADP/ATP were added to 20 μL of each extract. A first measure of optical density (OD) at 340 nm was made after 10 min of agitation at 30°C. A second measure (OD2) was made after adding 0.3 units of hexokinase and 0.15 units of Glc-6-P dehydrogenase and a second series of agitation at 30°C. A last measure of OD (OD3) was made after adding 0.7 units of phosphoglucoisomerase per sample and a third series of agitation at 30°C. OD2-OD1 and OD3-OD2 are proportional to the Glc and Fru concentrations, respectively. For the determination of the Suc content, 100 μL of extract plus 10 μL of invertase were agitated for 20 min at 30°C. OD1 was measured after adding 60 μL of NADP-ATP buffer and agitation for 10 min at 30°C. OD2 was measured after adding 0.3 units of hexokinase and 0.15 units of Glc-6-P dehydrogenase and agitation for 10 min at 30°C. OD2-OD1 is proportional to the Suc content. For the calculation of the results (in μmol/g DM), a calibration curve with known Glc and Fru concentrations was built. For starch content determination, the residual DMs from ethanolic extractions were dried at 50°C, mixed with 1 mL of water per sample, and incubated at 100°C for 2 h. After adding β-amyloglucosidase, α-amylase, and sodium acetate buffer (0.2 m, pH 4.8), samples were incubated overnight at 50°C. Starch content, which is proportional to Glc content, was then calculated by measuring Glc content in the obtained extracts.
Statistical Analyses and QTL Mapping
Data were collected from two (N−) or three (N+) independent replications. The complete set of data was included in an ANOVA model to determine the specific effects of the “genotype” and “cultivation repetition” factors. This ANOVA enabled us to quantify the broad-sense heritability (genetic variance/total phenotypic variance). Subsequent analyses involved mean values over cultivation repetitions for each line. Phenotypic correlations were calculated for all combinations of traits in both nitrate conditions studied (Table II). Correlations between sugar-related traits measured in this study and N-related traits measured previously (Loudet et al., 2003b) were also calculated (Table II). ANOVA estimations were obtained using the aov() function of the S-PLUS Version 3.4 statistical package (Statistical Sciences).
The original set of markers (38 microsatellite markers) and the genetic map obtained with MAPMAKER 3.0, as described previously (Loudet et al., 2002; http://www.inra.fr/qtlat), were used in this study. All QTL analyses were performed using the Unix version of QTL Cartographer 1.14 (Basten et al., 1994, 2000). We used standard methods as described previously (Loudet et al., 2002), interval mapping, and composite interval mapping (CIM). First, interval mapping (Lander and Botstein, 1989) was used to determine putative QTLs involved in the variation of the trait. CIM model 6 of QTL Cartographer was then performed on the same data; the closest marker to each local logarithm of odds ratio (LOD) score peak (putative QTL) was used as a cofactor to control the genetic background while testing at a position of the genome. When a cofactor was also a flanking marker of the tested region, it was excluded from the model. The number of cofactors involved in our models varied between four and seven. The walking speed chosen for all QTL analyses was 0.1 cM. The LOD significance threshold (2.3 LOD) was estimated from several permutation test analyses, as suggested by Churchill and Doerge (1994).
Additive effects of detected QTLs were estimated from CIM results; 2a represents the mean effect of the replacement of the Shahdara alleles by Bay-0 alleles at the studied locus (i.e. the difference between Bay-0 and Shahdara phenotypic effects associated with each allele at this locus). The contribution of each identified QTL to the total variance (R2) was estimated by variance component analysis. For each trait, the model involved the genotype at the closest marker to the corresponding detected QTLs as random factors in ANOVA. Only homozygous genotypes were included in the ANOVA analysis. Significant QTL × QTL interactions were also added to the linear model via the corresponding marker × marker interactions, and their contribution to the total variance also was estimated (Table II). QTL × N environment interaction was assessed by a two-factor ANOVA, with the corresponding marker genotype and N environment as classifying factors.
Candidate Genes for Sugar-Related Traits
Potential candidate genes involved in sugar metabolism were searched in silico at two distinct QTLs: QTL FR3.4 and QTL ST3.4. Large genomic regions were taken into account to mine for putative positional candidates on the Web site of The Arabidopsis Information Resource (http://www.arabidopsis.org/). On the other hand, genomic positions of genes coding for key enzymes of the carbohydrate metabolism, chosen a priori for their function, were searched to look for potential colocalizations with the other QTLs detected at the genome level. A huge number of genes involved in C metabolism are known in Arabidopsis. We limited our analysis to 118 genes annotated as involved in major carbohydrate metabolism. We also included known regulatory genes recently described for their involvement in the regulation of C and N metabolisms, or their participation to sugar and N sensing, as well as to the regulation of the C to N ratio.
This work was supported by the European project NATURAL (grant no. QLRT–2000–01097, 2001–2005 fellowship to V.S.-C.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Anne Krapp (krapp@versailles.inra.fr).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.082396.
References
- Alonso-Blanco C, Blankestijn-de Vries H, Hanhart CJ, Koornneef M (1999) Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 4710–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, El-Assal SE, Coupland G, Koornneef M (1998) Analysis of natural allelic variation at flowering time loci in the Landsberg erecta and Cape Verde Islands ecotypes of Arabidopsis thaliana. Genetics 149: 749–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Koornneef M (2000) Naturally occurring variation in Arabidopsis: an underexploited resource for plant genetics. Trends Plant Sci 5: 22–29 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Balibrea Lara ME, Gonzalez Garcia MC, Fatima T, Ehness R, Lee TK, Proels R, Tanner W, Roitsch T (2004) Extracellular invertase is an essential component of cytokinin-mediated delay of senescence. Plant Cell 16: 1276–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basten CJ, Weir BS, Zeng Z-B (1994) Zmap: a QTL cartographer. In C Smith, JS Gavora, B Benkel, J Chesnais, W Fairfull, JP Gibson, BW Kennedy, EB Burnside, eds, Proceedings of the 5th World Congress on Genetics Applied to Livestock Production: Computing Strategies and Software, Vol 22. University of Guelph Press, Guelph, Ontario, Canada, pp 65–66
- Basten CJ, Weir BS, Zeng Z-B (2000) QTL Cartographer, Version 1.14. Department of Statistics, North Carolina State University, Raleigh, NC
- Benning C, Stitt M (2004) Physiology and metabolism reacting to the full complexity of metabolic pathways in a post-genomic era. Curr Opin Plant Biol 7: 231–234 [Google Scholar]
- Bentsink L, Alonso-Blanco C, Vreugdenhil D, Tesnier K, Groot SP, Koornneef M (2000) Genetic analysis of seed-soluble oligosaccharides in relation to seed storability of Arabidopsis. Plant Physiol 124: 1595–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Yuan K, Koornneef M, Vreugdenhil D (2003) The genetics of phytate and phosphate accumulation in seeds and leaves of Arabidopsis thaliana, using natural variation. Theor Appl Genet 106: 1234–1243 [DOI] [PubMed] [Google Scholar]
- Bernier G, Havelange A, Houssa C, Petitjean A, Lejeune P (1993) Physiological signals that induce flowering. Plant Cell 5: 1147–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Salamini F, Gebhardt C (2001) A potato molecular-function map for carbohydrate metabolism and transport. Theor Appl Genet 102: 284–295 [Google Scholar]
- Chung BC, Lee SY, Oh SA, Rhew TH, Nam HG, Lee CH (1997) The promoter activity of sen1, a senescence-associated gene of Arabidopsis, is repressed by sugars. J Plant Physiol 151: 339–345 [Google Scholar]
- Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L, Lejeune P, Bernier G (1998) The role of carbohydrates in the induction of flowering in Arabidopsis thaliana: comparison between the wild type and a starchless mutant. Planta 206: 131–137 [DOI] [PubMed] [Google Scholar]
- Coruzzi GM, Zhou L (2001) Carbon and nitrogen sensing and signaling in plants: emerging ‘matrix effects’. Curr Opin Plant Biol 4: 247–253 [DOI] [PubMed] [Google Scholar]
- Dai N, Schaffer A, Petreikov M, Shahak Y, Giller Y, Ratner K, Levine A, Granot D (1999) Overexpression of Arabidopsis hexokinase in tomato plants inhibits growth, reduces photosynthesis, and induces rapid senescence. Plant Cell 11: 1253–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danyluk J, Perron A, Houde M, Limin A, Fowler B, Benhamou N, Sarhan F (1998) Accumulation of an acidic dehydrin in the vicinity of the plasma membrane during cold acclimation of wheat. Plant Cell 10: 623–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis DT, Blakeley SD (2000). Carbohydrate metabolism. In B Buchanan, W Gruissem, R Jones, eds, Biochemistry and Molecular Biology of Plants, Ed 1. American Society of Plant Physiologists, Rockville, MD, pp 630–675
- Dian W, Jiang H, Chen Q, Liu F, Wu P (2003) Cloning and characterization of the granule-bound starch synthase II gene in rice: gene expression is regulated by the nitrogen level, sugar and circadian rhythm. Planta 218: 261–268 [DOI] [PubMed] [Google Scholar]
- Diaz C, Saliba-Colombani V, Loudet O, Belluomo P, Moreau L, Daniel-Vedele F, Morot-Gaudry J-F, Masclaux-Daubresse C (2006) Leaf yellowing and anthocyanin accumulation are two genetically independent strategies in response to nitrogen limitation in Arabidopsis thaliana. Plant Cell Physiol 47: 74–83 [DOI] [PubMed] [Google Scholar]
- Edwards G, Walker DA (1983) C3, C4: Mechanisms and Cellular and Environmental Regulation of Photosynthesis. Blackwell Scientific Publications, London
- Foyer CH, Parry M, Noctor G (2003) Markers and signals associated with nitrogen assimilation in higher plants. J Exp Bot 54: 585–593 [DOI] [PubMed] [Google Scholar]
- Geiger DR, Servaites JC (1994) Diurnal regulation of photosynthetic carbon metabolism in C3 plants. Annu Rev Plant Physiol Plant Mol Biol 45: 235–256 [Google Scholar]
- Hensel LL, Grbic V, Baumgarten DA, Bleecker AB (1993) Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in Arabidopsis. Plant Cell 5: 553–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hills MJ (2004) Control of storage-product synthesis in seeds. Curr Opin Plant Biol 7: 302–308 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Martin AC, Leyva A, Paz-Ares J (2005) Interaction between phosphate-starvation, sugar, and cytokinin signaling in Arabidopsis and the roles of cytokinin receptors CRE1/AHK4 and AHK3. Plant Physiol 138: 847–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman E, Pleban T, Zamir D (2000) A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc Natl Acad Sci USA 97: 4718–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SI (2005) Control of plant development and gene expression by sugar signalling. Curr Opin Plant Biol 8: 93–102 [DOI] [PubMed] [Google Scholar]
- Glazier AM, Nadeau JH, Aitman TJ (2002) Finding genes that underlie complex traits. Science 298: 2345–2349 [DOI] [PubMed] [Google Scholar]
- Juenger T, Purugganan M, Mackay TF (2000) Quantitative trait loci for floral morphology in Arabidopsis thaliana. Genetics 156: 1379–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch K (2004) Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr Opin Plant Biol 7: 235–246 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D (2004) Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol 55: 141–172 [DOI] [PubMed] [Google Scholar]
- Krapp A, Saliba-Colombani V, Daniel-Vedele F (2005) Analysis of C and N metabolisms and of C/N interactions using quantitative genetics. Photosynth Res 83: 251–263 [DOI] [PubMed] [Google Scholar]
- Krapp A, Stitt M (1994) An evaluation of direct and indirect mechanisms for the “sink regulation” of photosynthesis in spinach: changes in gas exchange, carbohydrates, metabolites, enzyme activities and steady state transcript levels after cold girdling source leaves. Planta 195: 313–323 [Google Scholar]
- Laby RJ, Kim D, Gibson SI (2001) The ram1 mutant of Arabidopsis exhibits severely decreased beta-amylase activity. Plant Physiol 127: 1798–1807 [PMC free article] [PubMed] [Google Scholar]
- Lam HM, Chiu J, Hsieh MH, Meisel L, Oliveira IC, Shin M, Coruzzi G (1998) Glutamate-receptor genes in plants. Nature 396: 125–126 [DOI] [PubMed] [Google Scholar]
- Lamaze T, Sentenac H, Grignon C (1984) Effects of nitrate on phosphate accumulation and transport by corn roots. Physiol Veg 22: 155–161 [Google Scholar]
- Lander ES, Botstein D (1989) Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larcher W (1995) Physiological Plant Ecology, Ed 3. Springer, Berlin
- Leegood RC, Furbank RT (1986) Stimulation of photosynthesis by O2 at low temperature is restored by phosphate. Planta 168: 84–93 [DOI] [PubMed] [Google Scholar]
- Lejay L, Gansel X, Cerezo M, Tillard P, Muller C, Krapp A, von Wiren N, Daniel-Vedele F, Gojon A (2003) Regulation of root ion transporters by photosynthesis: functional importance and relation with hexokinase. Plant Cell 15: 2218–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F (2002) Bay-0 x Shadara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor Appl Genet 104: 1173–1184 [DOI] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Krapp A, Daniel-Vedele F (2003. a) Quantitative trait loci analysis of water and anion content in interaction with nitrogen availability in Arabidopsis thaliana. Genetics 163: 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Merigout P, Talbotec J, Daniel-Vedele F (2003. b) Quantitative trait loci analysis of nitrogen use efficiency in Arabidopsis. Plant Physiol 131: 345–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Gaudon V, Trubuil A, Daniel-Vedele F (2005) Quantitative trait loci controlling root growth and architecture in Arabidopsis thaliana confirmed by heterogeneous inbred family. Theor Appl Genet 110: 742–753 [DOI] [PubMed] [Google Scholar]
- Maloof JN (2003) Genomic approaches to analyzing natural variation in Arabidopsis thaliana. Curr Opin Genet Dev 13: 576–582 [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T, Pedersen D (1998) The molecular basis of quantitative genetic variation in central and secondary metabolism in Arabidopsis. Genetics 149: 739–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 11: 332–336 [DOI] [PubMed] [Google Scholar]
- Nielsen TH, Krapp A, Roeper-Schwarz U, Stitt M (1998) The sugar-mediated regulation of genes encoding the small subunit of Rubisco and the regulatory subunit of ADP glucose pyrophosphorylase is modified by phosphate and nitrogen. Plant Cell Environ 21: 443–454 [Google Scholar]
- Niittyla T, Messerli G, Trevisan M, Chen J, Smith AM, Zeeman SC (2004) A previously unknown maltose transporter essential for starch degradation in leaves. Science 303: 87–89 [DOI] [PubMed] [Google Scholar]
- Ono K, Nishi Y, Watanabe A, Terashima I (2001) Possible mechanisms of adaptative leaf senescence. Curr Opin Plant Biol 3: 234–243 [Google Scholar]
- Ono K, Terashima I, Watanabe A (1996) Interaction between nitrogen deficit of a plant and nitrogen content in the old leaves. Plant Cell Physiol 37: 1083–1089 [Google Scholar]
- Palenchar PM, Kouranov A, Lejay LV, Coruzzi GM (2004) Genome-wide patterns of carbon and nitrogen regulation of gene expression validate the combined carbon and nitrogen (CN)-signaling hypothesis in plants. Genome Biol 5: R91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J, Laxmi A, St Martin SK, Jang J-C (2004) Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16: 2128–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra AS, Padmasree K (2003) Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci 8: 546–553 [DOI] [PubMed] [Google Scholar]
- Ritchie SM, Swanson SJ, Gilroy S (2002) From common signalling components to cell specific responses: insights from the cereal aleurone. Physiol Plant 115: 342–351 [DOI] [PubMed] [Google Scholar]
- Roeper H (2002) Renewable raw material in Europe—industrial utilisation of starch and sugar. Starch Staerke 54: 89–99 [Google Scholar]
- Roldan M, Gomez-Mena C, Ruiz-Garcia L, Salinas J, Martinez-Zapater JM (1999) Sucrose availability on the aerial part of the plant promotes morphogenesis and flowering of Arabidopsis in the dark. Plant J 20: 581–590 [DOI] [PubMed] [Google Scholar]
- Rolland F, Moore B, Sheen J (2002) Sugar sensing and signaling in plants. Plant Cell 14: 185–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible WR, Gonzalez-Fontes A, Lauerer M, Muller-Rober B, Caboche M, Stitt M (1997) Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell 9: 783–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeeva LI, Vonk J, Keurentjes JJB, van der Plas LHW, Koornneef M, Vreugdenhil D (2003) Histochemical analysis reveals organ-specific quantitative trait loci for enzyme activities in Arabidopsis. Plant Physiol 134: 237–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC, Thorneycroft D, Smith SM (2003) Starch mobilization in leaves. J Exp Bot 54: 577–583 [DOI] [PubMed] [Google Scholar]
- Stessman D, Miller A, Spalding M, Rodermel S (2002) Regulation of photosynthesis during Arabidopsis leaf development in continuous light. Photosynth Res 72: 27–37 [DOI] [PubMed] [Google Scholar]
- Stitt M, Bulpin PV, ap Rees T (1985) Pathway of starch breakdown in photosynthetic tissues of Pisum sativum. Biochim Biophys Acta 544: 200–214 [DOI] [PubMed] [Google Scholar]
- Stitt M, Hurry V (2002) A plant for all seasons: alterations in photosynthetic carbon metabolism during cold acclimation in Arabidopsis. Curr Opin Plant Biol 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Sun J, Gibson KM, Kiirats O, Okita TW, Edwards GE (2002) Interactions of nitrate and CO2 enrichment on growth, carbohydrates, and Rubisco in Arabidopsis starch mutants. Significance of starch and hexose. Plant Physiol 130: 1573–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds VV, Godoy AV, Alconada T, Botto JF, Juenger TE, Casal JJ, Lloyd AM (2005) Mapping quantitative trait loci in multiple populations of Arabidopsis thaliana identifies natural allelic variation for trichome density. Genetics 169: 1649–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trethewey RN, Smith AM (2000) Starch metabolism in leaves. In RC Leegood, TD Sharkey, S von Caemmerer, eds, Advances in Photosynthesis: Physiology and Metabolism, Vol 9. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 205–231
- Tuinstra MR, Ejeta G, Goldsbrough PB (1997) Heterogeneous inbred family (HIF) analysis: an approach for developing near-isogenic lines that differ at quantitative trait loci. Theor Appl Genet 95: 1005–1011 [Google Scholar]
- van Doorn WG (2004) Is petal senescence due to sugar starvation? Plant Physiol 34: 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PJ, Broadley MR (2001) Chloride in soils and its uptake and movement within the plant. Ann Bot (Lond) 88: 967–988 [Google Scholar]
- Wingler A, von Schaewen A, Leegood RC, Lea PJ, Quick WP (1998) Regulation of leaf senescence by cytokinin, sugars, and light. Effects on NADH-dependent hydroxypyruvate reductase. Plant Physiol 116: 329–335 [Google Scholar]
- Yoshida S (2003) Molecular regulation of leaf senescence. Curr Opin Plant Biol 6: 79–84 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Umemoto T, Lue WL, Au-Yeung P, Martin C, Smith AM, Chen J (1998) A mutant of Arabidopsis lacking a chloroplastic isoamylase accumulates both starch and phytoglycogen. Plant Cell 10: 1699–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]