Abstract
Phosphatidylglycerol (PG) is a ubiquitous phospholipid in thylakoid membranes of cyanobacteria and chloroplasts and plays an important role in the structure and function of photosynthetic membranes. The last step of the PG biosynthesis is dephosphorylation of phosphatidylglycerophosphate (PGP) catalyzed by PGP phosphatase. However, the gene-encoding PGP phosphatase has not been identified and cloned from cyanobacteria or higher plants. In this study, we constructed a PG-deficient mutant from cyanobacterium Anabaena sp. PCC7120 with a disrupted gene (alr1715, a gene for Alr1715 protein, GenBank accession no. BAB78081) encoding a putative PGP phosphatase. The obtained mutant showed an approximately 30% reduction in the cellular content of PG. Following the reduction in the PG content, the photoautotrophical growth of the mutant was restrained, and the cellular content of chlorophyll was decreased. The decreases in net photosynthetic and photosystem II (PSII) activities on a cell basis also occurred in this mutant. Simultaneously, the photochemical efficiency of PSII was considerably declined, and less excitation energy was transferred toward PSII. These findings demonstrate that the alr1715 gene of Anabaena sp. PCC7120 is involved in the biosynthesis of PG and essential for photosynthesis.
Thylakoid membranes of cyanobacteria and chloroplasts of plants and algae are the sites of the oxygenic photosynthesis, where highly organized pigment-protein complexes are embedded in the lipid matrix. Thylakoid membranes are characterized by its unique lipid composition, which is in contrast with animal, bacterial, and nonchloroplastic cell membranes dominated by phospholipids. The major lipids of thylakoid membranes are the uncharged galactolipids, namely, monogalacotsyl diacylglycerol (MGDG) and digalactosyl diacylglycerol (DGDG). These galactolipids account for at least 50 mol% of the total thylakoid lipids. In addition, thylakoid membranes are specific in that there is a negatively charged sulfolipid, sulfoquinovosyl diacylglycerol (SQDG), which seems not to be present in nonphotosynthetic organisms. The only significant phospholipid is phosphatidylglycerol (PG), rather than phosphatidylcholine or phosphatidylethnolamine (Murata and Siegenthaler, 1998).
PG is an anionic phospholipid that is also extensively distributed in the functional domains of thylakoid membranes, suggesting that PG is critical for the structure and function of thylakoid membrane. Indeed, increasing biochemical and genetic studies provided evidence that PG plays an important role in photosynthesis of thylakoid membranes. It has shown that PG is indispensable for the dimerization of PSII (Kruse et al., 2000) and trimerization of the light-harvesting complex of PSII (Nußberger et al., 1993; Hobe et al., 1994; Liu et al., 2004). PG is also essential for the oligomerization of PSI reaction center of cyanobacterium (Domonkos et al., 2004), consistent with the fact that PG is an integral component of PSI (Jordan et al., 2001). It was found that PG is required for the biogenesis of PSII core complex in Chlamydomonas reinhardtii mutants mf1 and mf2 with the decreased rate of translation of the D1 and CP47 owing to PG deficiency (Pineau et al., 2004), compatible with a previous suggestion that PG might bind to the D1 protein, where PG could sustain the optimal conformation of D1 polypeptide (Kruse and Schmid, 1995). The PGP1 mutants of Arabidopsis (Arabidopsis thaliana) defective in biosynthesis of PG obtained through chemical or T-DNA insertional mutagenesis disrupted in the PGP1 gene for phosphatidylglycerophosphate (PGP) synthase in chloroplasts showed an impairment of photosynthesis and inhibition of growth and chloroplast development, indicating that PG is essential for growth and chloroplast development (Hagio et al., 2002; Xu et al., 2002; Babiychuk et al., 2003). The function of PG has also been studied with Synechocystis sp. PCC6803 mutants with the disruption of the pgsA or the cdsA gene, encoding a PGP synthase and a CDP-diacylglycerol synthase, respectively (Hagio et al., 2000; Sato et al., 2000), which have severe defects in growth and photosynthesis. The growth and concomitant photosynthetic activity of these mutants have an absolute requirement for PG supplementation and vary with the level of PG that can be manipulated in the mutant cells by exogenous addition of PG, indicating the specific dependence of the function of photosynthesis on PG concentration. Deprivation of PG induced the impairment of the electron transfer from the primary electron-accepting plastoquinone of PSII to the secondary electron-accepting plastoquinone of PSII (QB) within PSII and the conformational change at and/or in the environment of the QB-binding site (Gombos et al., 2002), consistent with the in vitro observations that the PSII activity decreased with depletion of PG by phospholipase A2 (remove the β-positioned fatty acyl chains of the phospholipid molecules) treatment (Jordan et al., 1983; Siegenthaler et al., 1987; Kruse et al., 1994). These findings demonstrate that PG is indispensable for the functioning of PSII.
In all organisms, PG is synthesized from phosphatidic acid (PA) in three steps. CDP-diacylglycerol (CDP-DG) is first synthesized by transfer of CMP from CTP to PA with the action of CTP: phosphatidate cytidylyltransferase (EC 2.7.7.41; Carter and Kennedy, 1966). CDP-DG is then converted to PGP catalyzed by PGP synthase (EC 2.7.8.5; Hirabayashi et al., 1976; Bleasdale and Johnston, 1982), which transfers the phosphatidyl group from CDP-DG to sn-glycerol-3-P. The last step is the rapid dephosphorylation of PGP by PGP phosphatase (EC 3.1.3.27; Chang and Kennedy, 1967; Moore, 1982; Andrews and Mudd, 1986) to produce PG. The genes encoding CDP-DG synthase and PGP synthase have been identified for cyanobacteria and higher plants (Kopka et al., 1997; Hagio et al., 2000, 2002; Sato et al., 2000; Xu et al., 2002; Babiychuk et al., 2003), but the gene for PGP phosphatase has not been cloned from photosynthetic organisms.
Although biochemical and genetic results have established that PG is indispensable for photosynthetic organisms and is necessary for photosynthesis process, it has not as yet been unambiguously demonstrated. There still exists the possibility that either the precursor of PG biosynthesis, PGP, or the metabolite of PG biosynthesis, cardiolipin, which remains to be clarified in thylakoid membranes, serves as an essential component for photosynthetic organisms.
It is thus of particular interest to clone the gene involved in the biosynthesis of PGP. Two genes encoding for PGP phosphatase, pgpA and pgpB, have been characterized in the nonphotosynthetic bacterial strain Escherichia coli (Icho, 1988a, 1988b). Through a similarity search of the DNA sequence of the genome of a filamentous, nitrogen-fixing cyanobacterium, Anabaena sp. PCC7120 (Kaneko et al., 2001), we found that two open reading frames, alr1715 and all7623, show a significant sequence similarity to pgpB of E. coli and probably encode the PGP phosphatase in Anabaena sp. PCC7120. In this study, we produced a mutant by inactivation of the gene alr1715 through homologous recombination and have determined the photosynthetic characteristics of this mutant.
RESULTS
Disruption of the Gene alr1715 in Anabaena sp. PCC7120
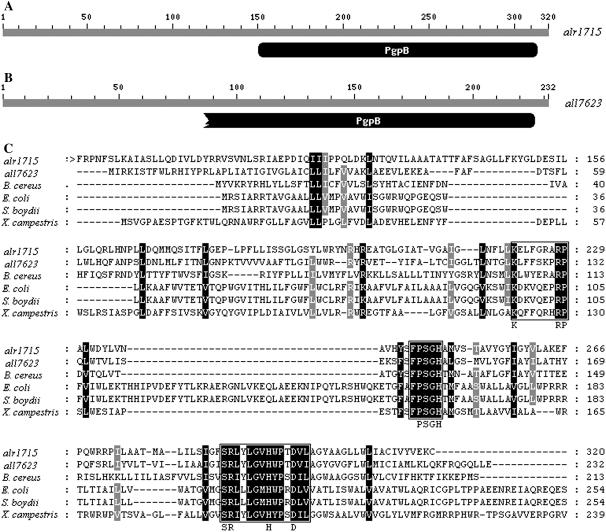
It was demonstrated that there are at least two genes, pgpA and pgpB, encoding for membrane-associated phospholipid phosphatases in E. coli (Icho, 1988a, 1988b). The pgpA gene product is specific for PGP, whereas the pgpB gene product has the ability to dephosphorylate PGP, PA, and lysophosphatidic acid (Icho and Raetz, 1983). The product of E. coli pgpB gene also exhibits diacylglycerol pyrophosphate phosphatase activity (Dillon et al., 1996). Using the gene products of pgpA and pgpB from E. coli in a similarity search for Anabaena sp. PCC7120 genomic sequences, we observed two proteins encoded, respectively, by the genes alr1715 and all7623 showing the remarkable similarities to that of pgpB in E. coli. The sequence alignment indicated that the amino acid sequence deduced from the gene alr1715 exhibits about 32% identity and 46% similarity over 320 amino acids with that of E. coli pgpB gene, while the gene all7623 has 40% identity and 56% similarity over 232 amino acids to the E. coli pgpB gene product. alr1715 and all7623 have conserved domains from the 151st to 314th and from the 65th to 226th amino acid residue, respectively, at their C terminus with phosphatidylglycerophosphatase B (PgpB), as revealed by the result of a conserved domain search in the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/structure/cdd/wrpsb.cgi; Fig. 1, A and B). The polypeptide encoded by the gene alr1715 or all7623 was also found to be homologous to PGP phosphatases of other nonphotosynthetic bacterial strains such as Bacillus cereus (Ivanova et al., 2003), Shigella boydii (Yang et al., 2005), and Xanthomonas campestris (da Silva et al., 2002; Fig. 1C). Several regions of the deduced amino acids of alr1715 and all7623 were observed to be conserved to the nonphotosynthetic bacterial PGP phosphatases, as indicated in Figure 1C. The conserved regions contain a larger sequence motif (boxed in Fig. 1C) that is shared by a superfamily of phosphatase enzymes. This phosphatase signature motif has three conserved domains with the consensus sequences KXXXXXXRP (domain 1, the sequence nearest the N terminus), PSGH (domain 2, the centrally located sequence), and SRXXXXXHXXXD (domain 3, the sequence nearest the N terminus), where X is any amino acid (Hemrika et al., 1997; Neuwald, 1997; Stukey and Carman, 1997). Separating each of the three domains are protein segments ranging from 12 to 54 amino acids in length. Amino acids within this larger motif are believed to be essential for catalytic activity (Brindley and Waggoner, 1998; Sigal et al., 2005). Indeed, we found that the product of the gene alr1715 or all7623 belongs to the superfamily of type 2 PA phosphatase, including the enzyme PAP2, Glc-6-P (EC 3.1.3.9), PgpB (EC 3.1.3.27), and bacterial acid phosphatase (EC 3.1.3.2).
Figure 1.
A and B, The National Center for Biotechnology Information conserved domain database-identified domains for a putative phosphatidylglycerophosphatase in Anabaena sp. PCC7120. The gray line represents the amino acid sequence of alr1715 or all7623 gene of Anabaena sp. PCC7120. Domains are represented in black. PgpB represents the domain of phosphatidylglycerophosphatase B, a membrane-associated phospholipid phosphatase involved in phospholipid metabolism. C, Sequence alignments of alr1715 and all7623 of Anabaena sp. PCC7120 with pgpB genes of nonphotosynthetic bacteria. The amino acid residues are numbered on the right. Identical residues are highlighted in black. Residues conserved in 80% are shown in gray. Gaps (indicated by dashes) were introduced to optimize sequence alignment. The symbol > indicates that parts of the protein of alr1715 have been omitted because they show no similarity to pgpB. The amino acid sequence alr1715 gene exhibits 37% identity with that of all7623 and 32% to 34% identity with the sequences of other nonphotosynthetic bacterial pgpB genes: E. coli (32%), B. cereus (33%), S. boydii (32%), and X. campestris (34%). Conserved phosphatase domains (1, 2, and 3) are boxed. Amino acids within those domains indicated below the box are believed to be essential for catalytic activity (Hemrika et al., 1997; Neuwald, 1997; Stukey and Carman, 1997).
In this work, we have investigated the mutation of the gene alr1715 of Anabaena sp. PCC7120 and characterized the photosynthetic properties of this mutant. Owing to remarkable sequence similarity to the pgpB gene of E. coli, this gene alr1715 is therefore also designated pgpB in this study. The gene alr1715 was disrupted by insertion of a kanamycin-resistant cassette into the coding region of alr1715, as described in “Materials and Methods.” The transformation was conducted with the construct pPGP3 that was transformed into the wild type of Anabaena sp. PCC7120 through homologous recombination using the triparental mating method (Elhai and Wolk, 1988).
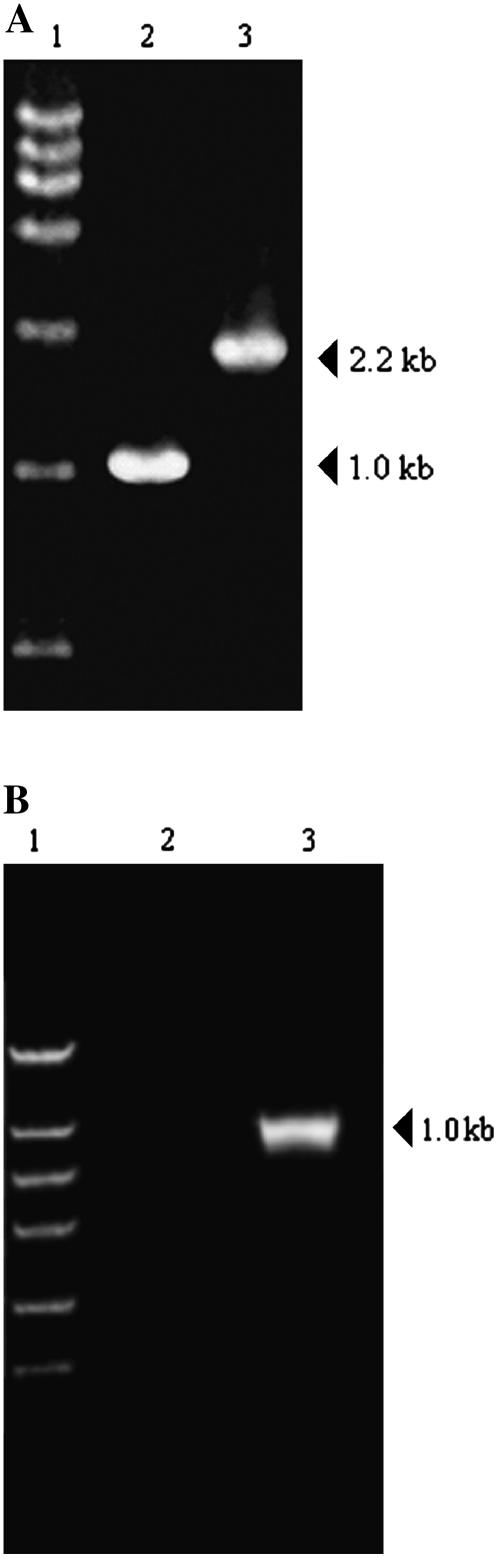
To check whether or not all copies of this gene were disrupted in the genome of the mutant with the neomycin-resistant phenotype, we carried out PCR analysis of the wild type and the mutant, with primers for the 5′- and 3′-terminal regions of the putative pgpB gene. For the wild type, an approximately 1.0-kb DNA fragment corresponding to the native pgpB gene was amplified. In the case of the mutant, a 2.2-kb DNA fragment corresponding to the pgpB∷Kanr gene owing to insertion of the 1.2-kb cassette was observed, but a 1.0-kb DNA fragment corresponding to the native pgpB gene was not amplified (Fig. 2A). To determine if any transcript for the gene is still present in this mutant, reverse transcription (RT)-PCR was performed to examine pgpB expression in the wild type and pgpB mutant. Figure 2B shows RT-PCR results using total RNA isolated from the wild-type and pgpB mutant cells. No pgpB transcription product was detected in the pgpB mutant cells (lane 2). This was in contrast to the wild-type cells, where an approximately 1.0-kb transcription product was observed (lane 3). Thus, the pgpB gene was successfully knocked out.
Figure 2.
Confirmation of a disruptant of alr1715 from Anabaena sp. PCC7120. A, PCR analysis of alr1715 in the wild type (lane 2) and the mutant of Anabaena sp. PCC7120 (lane 3). Lane 1 is the DNA marker DL15 000 (TaKaRa Bio), with sizes of 15, 10, 7.5, 5, 2.5, 1, and 0.25 kb from top to bottom. The sizes of amplified fragments are indicated on the right. B, Confirmation of the pgpB gene mutation by RT-PCR. Lane 1, maker DL 2 000, with sizes of 2, 1, 0.75, 0.5, 0.25, and 0.1 kb from top to bottom; lane 2, the pgpB mutant; lane 3, the wild type.
Growth Phenotype of the Mutant
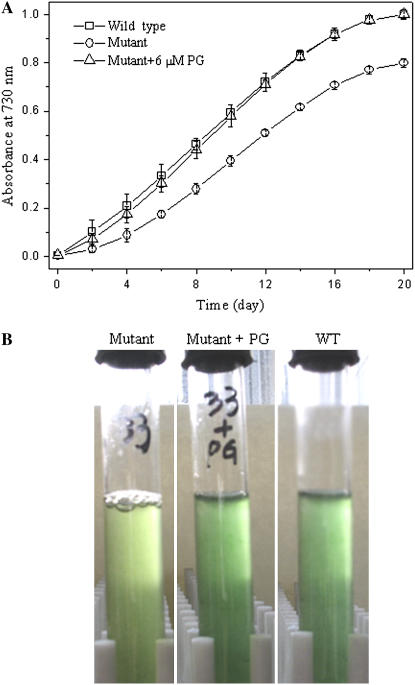
To investigate the role of the putative pgpB gene in growth, the growth profiles of the wild type and the pgpB mutant under photoautotrophic growth condition were investigated. Figure 3 shows the growth of the wild-type and pgpB mutant cells in the liquid growth medium BG11. It can be clearly seen that although both the wild type and pgpB mutant could grow, a different growth existed between the wild type and the mutant. The mutant showed a slower growth than the wild type (Fig. 3A). Even after lengthy growth, the mutant was pale green in appearance, while the wild type remained normal blue green (Fig. 3B). However, similar growth curves were observed for the wild type and the pgpB mutant when the growth medium was supplemented with exogenous PG at concentrations above 2 μm (Fig. 3A). In this situation, the mutant appeared blue green (Fig. 3B).
Figure 3.
Growth curves of the wild-type strain and the pgpB mutant cultured in liquid medium BG11 with and without the addition of PG at 30°C. A, Samples (1 mL) were taken and their optical density at 730 nm was measured. The data are presented as the means for four independent measurements. B, Cells of the wild type (right), mutant (left), and mutant with the addition of exogenous PG (middle) were grown in liquid medium BG11 for 10 d.
Lipid Phenotype of the Mutant
To explore the effect of the pgpB gene on the lipid composition of cyanobacterial cells, lipids from both the wild type and the mutant were extracted and analyzed. Table I shows the composition of lipid classes from intact cells of the wild type and the pgpB mutant cultured in the liquid medium BG11. In the wild type, MGDG was the most abundant lipid, making up 34% of the total lipids. DGDG and PG were second, each accounting for about 23% to approximately 24%. The amount of SQDG was only 20%. A major finding from the data is the clear changes in the contents of two acidic lipids, SQDG and PG. The relative amount of PG was reduced from 24 mol% in wild-type cells to 16% in the pgpB mutant. Concomitantly, the content of SQDG was increased in the mutant (29%) compared with the wild type (20%).The relative amount of MGDG and DGDG were slightly changed in the mutant with respect to the wild type, indicating that the pgpB mutant is little affected as to galactolipid metabolism in Anabaena sp. PCC7120.
Table I.
Cellar lipid composition of the wild type and the pgpB mutant of Anabaena sp. PCC7120
Each value represents the means of two independent experiments.
| Strain | Lipid
|
||||
|---|---|---|---|---|---|
| PG | MGDG | DGDG | SGDG | PG | |
| mol% | |||||
| Wild type | +a | 37.0 | 19.8 | 17.0 | 26.2 |
| −b | 34.0 | 22.5 | 19.5 | 24.0 | |
| Mutant | + | 36.6 | 21.0 | 20.8 | 21.6 |
| − | 31.9 | 23.6 | 28.5 | 16.0 | |
Cells were grown in the presence of 6 μm PG for 10 d.
Cells were grown without exogenous PG.
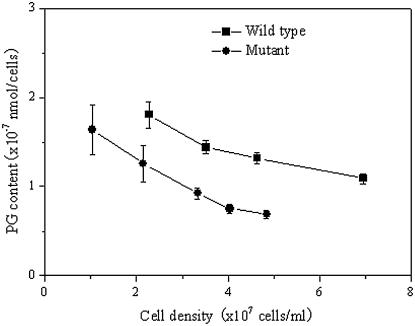
The changes in the content of PG in the pgpB mutant cells with growth were also determined. Figure 4 shows changes in the content of PG in intact cells of the wild type and the pgpB mutant. During incubation with the culture medium BG11 for 10 d, the content of PG in the wild-type cells slightly decreased from 1.8 to 1.1 × 10−16 mol cells−1. By contrast, the content of PG in the pgpB mutant cells markedly decreased from 1.6 to 0.6 × 10−16 mol cells−1. However, the pgpB mutant cells with the addition of exogenous PG showed a near recovery of PG content to the level of PG in the wild type (Table I). These results suggest that the synthesis of PG in the pgpB mutant is affected and the pgpB gene is involved in the biosynthesis of PG.
Figure 4.
Changes in PG content of the pgpB mutant during the growth in liquid medium BG11. The data are presented as the means for three measurements.
Characteristics of Photosynthetic Pigments in the Mutant
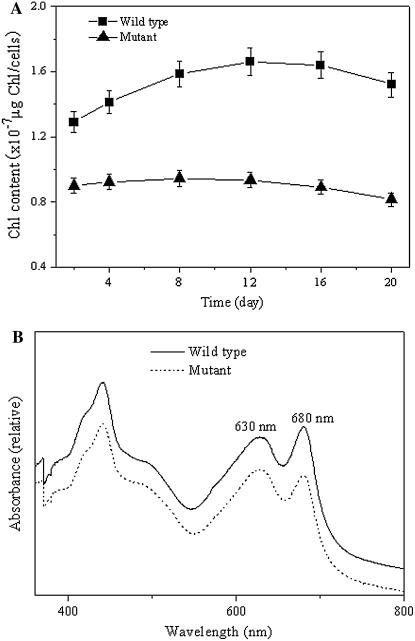
To check the effect of the pgpB mutation on the photosynthetic pigments, the cellular content of chlorophyll (Chl) a and phycobilisome pigments were investigated for the wild type and pgpB mutant. A reduction in Chl a levels was observed in the mutant. During the growth phase, the Chl a content on a per-cell basis increased slightly for the wild type, whereas it changed little for the pgpB mutant, being only about 54% of the level in the wild type (Fig. 5A). In the meantime, the phycobilisome pigments such as phycocyanin and allophycocyanin in the mutant were little affected throughout growth and maintained at almost the same levels, similar to the wild type (data not shown). These findings indicate that the contents of Chl-protein complexes were reduced and the levels of phycobilisomes were nearly constant for the pgpB mutant cells following the decrease in the content of PG.
Figure 5.
Chl content and absorption spectra of the wild type and the pgpB mutant. A, The values of Chl content are on a per-cell basis. The data are presented as the means for four independent measurements. B, The spectra were measured using whole cells of the wild type and mutant. Cells were grown in BG11 for 10 d. The spectra of cells are normalized at A730.
In accordance with the above results, the pgpB mutant cells showed the lower Chl absorbance, with a decrease in the ratio of Chl absorption at 680 nm to phycocyanin absorption at 630 nm, as compared to the wild type (Fig. 5B).
Photosynthetic Characteristics of the Mutant
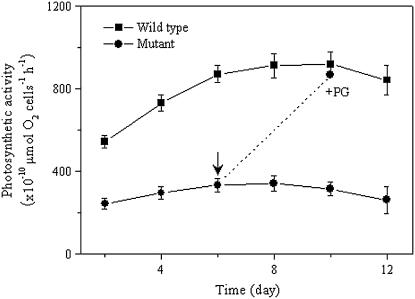
To characterize the effect of a decrease in the PG content on the oxygenic photosynthesis, the photosynthetic oxygen evolution activity (net photosynthesis, water to carbon dioxide) was determined for the pgpB mutant (Fig. 6). During the growth in liquid culture medium BG11, the photosynthetic activity on a per-cell basis of intact cells of the wild type increased gradually. By contrast, the activity of intact cells of the pgpB mutant remained almost constant. However, the CO2-dependent photosynthesis activity in the wild type was always higher than that in the pgpB mutant at the same culture time (Fig. 6A). The reduced photosynthetic activity of the mutant cells could, however, reach the similar level of the activity of the wild-type cells following addition of 6 μm PG to the growth medium for a further 4 d. These results indicate that the decrease in PG content brought about reduction in the cellular content of Chl a and, consequently, in the photosynthetic activity of a cell.
Figure 6.
Changes in the photosynthetic activities of the wild-type and pgpB mutant cells. The wild-type and pgpB mutant cells were washed with BG11 and resuspended in fresh BG11 medium for measurement of photosynthetic activity with 10 mm NaHCO3. The values are the means of three independent measurements. The arrow indicates the time of addition of 6 μm PG.
The effect of the reduced content of PG on PSII activity with an artificial electron acceptor, 2,6-dichloro-p-benzoquinone (DCBQ), was also determined (Table II). As shown in Table II, the PSII activity in intact cells of the wild type and the pgpB mutant was essentially the same in the presence of PG. However, the pgpB mutant, in comparison with the wild type, showed about 73% of decrease in PSII activity when the cells were grown without the added PG. It is known that DCBQ shows an affinity for accepting electrons from the primary electron-accepting plastoquinone of PSII site of the D2 protein in the PSII reaction center (Schneider et al., 2004). These data suggest that the changes in the conformation of the PSII reaction center are induced by the decrease of the PG content in the pgpB mutant and the affinity of DCBQ to QB site is thereby changed. Consistent with the above results, the pgpB mutant showed decreased values in dark-level fluorescence yield (F0), the maximum fluorescence yield (Fm), and therefore a lower variable fluorescence yield (Fv). Consequently, the ratio of Fv/Fm, which is a measure of the photochemical efficiency of PSII in the dark-adapted state, was decreased in the mutant (Table III).
Table II.
Net photosynthetic and PSII activities in intact cells of the wild type and the pgpB mutant of Anabaena sp. PCC7120
Net photosynthetic activity was measured for cells in BG11 medium containing 10 mm NaHCO3, while PSII activity was assayed in BG11 medium containing 0.25 mm DCBQ and 0.25 mm ferricyanide at 30°C. All values represent the means (±se) of three independent measurements.
| Strain | PG | Net Photosynthetic Activity | PSII Activity |
|---|---|---|---|
| μmol O2 10−10cells−1 h−1 | μmol O2 10−10cells−1 h−1 | ||
| Wild type | +a | 895 ± 40 | 1,432 ± 60 |
| −b | 918 ± 50 | 1,515 ± 80 | |
| Mutant | + | 868 ± 55 | 1,142 ± 60 |
| − | 314 ± 15 | 414 ± 20 |
Cells were grown in the presence of 6 μm PG.
Cells were grown without exogenous PG.
Table III.
Chl fluorescence parameters obtained with intact cells of the wild type and the pgpB mutant of Anabaena sp. PCC7120
Samples in BG11 medium were adjusted to equal numbers of cells that corresponded to 5 μg Chl/mL for wild-type cells. (Fv = Fm − F0). All values represent the means of four independent measurements.
| Parameters | Wild Type | Mutant |
|---|---|---|
| F0 | 400 ± 4 | 312 ± 5 |
| Fm | 632 ± 10 | 395 ± 6 |
| Fv | 232 ± 3 | 83 ± 3 |
| Fv/Fm | 0.37 ± 0.05 | 0.21 ± 0.04 |
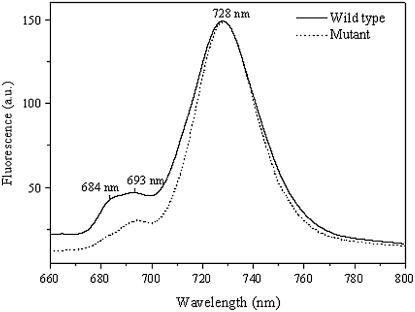
To investigate further alterations of the pigment-protein complexes and the excitation energy transfer to the reaction centers, low temperature fluorescence spectra were measured for the wild type and the pgpB mutant. The 77 K fluorescence spectra of wild-type and mutant strains obtained after Chl a excitation at 436 nm are shown in Figure 7. There are three distinct emission maxima at 684, 693, and 728 nm. The large emission peak at 728 nm is attributed to emanate primarily from PSI antenna, whereas the two peaks at 684 and 693 nm can be ascribed to emit mainly from the PSII core antenna complexes CP43 and CP47, respectively (Satoh et al., 1995). No position shift in the Chl fluorescence emission maxima was observed for both strains, indicating the complete assembly of PSII and PSI complexes in both wild type and mutant (Hartel et al., 1998). However, the ratio of PSII-associated fluorescence to PSI fluorescence was markedly reduced in the mutant cells as compared to the wild type. It reflects that there is a decrease in the number of PSII units, while the number of PSI units remains almost the same in the mutant (Krause and Weis, 1991). In addition, the magnitude of fluorescence emission at 684 nm slightly decreased relative to the 693 nm fluorescence emission in the pgpB mutant. These findings demonstrated that the molecular environment of the PSII core complexes and PSI is almost not affected, but more excitation energy is redistributed toward PSI in the mutant.
Figure 7.
The 77 K fluorescence emission spectra of Anabaena sp. PCC7120 wild type and pgpB mutant following Chl a excitation at 436 nm. Spectra were normalized to the large emission maximum at 728 nm.
DISCUSSION
In this study, we isolated a mutant of Anabaena sp. PCC7120 through the disruption of a gene, alr1715. This gene shares considerable sequence identity with the pgpB genes of several nonphotosynthetical bacterial organisms (e.g. E. coli and B. cereus). A 320-amino acid sequence of this gene has a conserved domain at its C terminus with PgpB. In addition, there is very little similarity between the amino acid sequence of this gene and that of the PGP synthase (all4063) or the phosphatidate cytidylyltransferase (all3875) of Anabaena sp. PCC7120 (data not shown). Furthermore, inactivation of this gene could bring about a 30% reduction in PG content of intact cells, and supplementation of the alr1715 mutant with exogenous PG could restore a near wild-type state. Taken together, we believe that the alr1715 gene is involved in the pathway of PG biosynthesis in Anabaena sp. PCC7120 and most likely encodes a PGP phosphatase that catalyzes the formation of PGP.
The alr1715 mutant has the PG content reduced to only about 70% of that found in the wild type; the majority of PG was still present in this mutant. In addition, there is no transcript for this gene in the mutant as confirmed by RT-PCR, revealing that it is unlikely that the disrupted gene is still partially functional. These facts indicate that there is at least one other gene that encodes PGP phosphatase and thus accounts for the presence of residual PG in the pgpB mutant of Anabaena sp. PCC7120. Indeed, we found that one open reading frame, all7623, is highly homologous to the putative pgpB gene alr1715 through a similarity search of the genome of Anabaena sp. PCC7120 (37% identity and 53% similarity over 320 amino acids). The question of whether there is gene superfamily for PGP phosphatase in Anabaena sp. PCC7120 needs to be clarified and is currently under investigation in our group. In E. coli, at least two genes, pgpA and pgpB, and gene products responsible for the in vivo conversion of PGP to PG were identified (Icho, 1988a, 1988b). A third gene locus for PGP phosphatase was proposed to exist (Funk et al., 1992). These results suggested that dephosphorylation of PGP has more than two independent genes associated with a single metabolic activity in E. coli.
Although only 30% of PG content in the pgpB mutant was reduced in comparison with the wild type, the visible phenotype and photosynthetic activity were considerably affected. The pgpB mutant showed a reduced photoautotrophic growth and a pale blue-green appearance. This is similar to the null PG-deficient strains of Synechocystis sp. PCC6803 (Hagio et al., 2000; Sato et al., 2000), where PG supplementation was required for the growth. In the presence of a relatively lower concentration of exogenous PG or shift to PG-free medium upon preincubation with PG supplementation, these mutants of Synechocystis sp. PCC6803 exhibited a slow growth and even stopped growing.
The alteration of visible phenotype in the pgpB mutant could be related to the changes in pigment composition and photosynthetic light utilization. Indeed, as compared to the wild type, the pgpB mutant had a reduction in the content of Chl a on a per-cell basis, the sole Chl species of PSI and PSII complexes, with phycobilisome pigments being less affected, suggesting the decreased levels of Chl-protein complexes in mutant cells with decreasing PG content.
Low temperature fluorescence measurements suggest that the PG deficiency in the mutant led to the structural modifications of the reaction center/core antenna complex of PSII. The mutant showed no change in the PSI and PSII fluorescence maxima positions following Chl a excitation at 436 nm, suggesting that the molecular environment or binding environment of the Chl a emitting from the core antenna proteins CP43 and CP47 of PSII core antenna complexes and the Chl a from PSI core antenna complexes are not affected by the PG reduction in the mutant. However, the mutant spectrum is characterized by a decrease in the magnitude of the fluorescence emission of PSII. This effect reflects that PG deficiency causes the structural changes (reduction) of the PSII antenna and/or redistribution of excitation energy more in favor of PSI.
The decrease in the low temperature Chl fluorescence ratio of PSII to PSI together with the decreased Chl content in the pgpB mutant indicated a decreased number of PSII (Vermaas et al., 1994; Wilde et al., 1995). The reduction in the CO2-dependent photosynthesis activity and oxygen evolution activity of PSII under saturating light was also observed for the mutant. Together, these results may explain the decreased variable Chl fluorescence yield of PSII in the mutant (Chl fluorescence at room temperature; Table II), because the decrease in the variable Chl fluorescence yield could arise from either a decreased activity of PSII reaction centers or a decreased number of PSII as well as an increased number of PSI reaction centers (Vermaas et al., 1994; Wilde et al., 1995). Corresponding to the decrease in Fv/Fm that reflects a decrease in the quantum yield of photochemistry of PSII, the function of the PSII reaction center was also impaired by PG reduction in the mutant, as the QB-binding site of PSII was affected, which may be caused by depletion of PG from PSII subunits such as of D1 protein in the pgpB mutant, further indicating the modifications in the conformation of the PSII complex.
The pgpB mutant exhibited a decrease in PG content, which was accompanied concomitantly by an increase in the content of SQDG, the other anionic lipid of cyanobacteria and chloroplasts. It has been thought that photosynthetic organisms require constant anionic lipid content by keeping a certain total level of PG and SQDG; if necessary, the two anionic lipids could substitute for each other to some extent, such that the photosynthetic apparatus can function properly and normally. It was found that several photosynthetic organisms, depending on available sources of nutrition, could reciprocally adjust the PG and SQDG contents and maintain the level of the sum of these anionic lipids (Benning et al., 1993; Sato et al., 1995; Guler et al., 1996; Essigmann et al., 1998; Yu et al., 2002; Aoki et al., 2004). However, the increased SQDG in this pgpB mutant could not recover its decreased CO2-dependent photosynthetic activity and DCBQ-dependent PSII activity, suggesting the different role for PG and SQDG in maintenance of thylakoid properties in Anabaena sp. PCC7120, as suggested by Aoki et al. (2004).
In summary, we generated and characterized a PG-deficient mutant from Anabaena sp. PCC7120 with the disrupted alr1715 gene. The inactivation resulted in a reduction by 30% in the PG content and had a marked effect on growth, Chl levels, photosynthetic oxygen evolution, and photochemistry, particularly associated with PSII activity. The phenotype of the mutant was similar but less severe than those for other Synechocystis PG biosynthesis mutants that have reductions in PG by as much as 80% to 90% and require exogenous PG supplements for growth. These findings indicate that the alr1715 gene of Anabaena sp. PCC7120 is involved in the biosynthesis of PG and is essential for photosynthesis. Therefore, this mutant provides a promising opportunity toward understanding the biosynthesis and function of PG in photosynthetic organisms.
MATERIALS AND METHODS
Cell Culture
Wild-type and mutant cells of Anabaena (Nostoc) sp. strain PCC 7120 (in this study referred to as Anabaena sp. PCC 7120) were grown photoautotrophically in BG11 medium (Allen, 1968) at 30°C under continuous illumination by white fluorescent light at mean light intensity of 35 μmol photon m−2 s−1. For growth on plates, the medium was solidified with separately autoclaved 1.5% (w/v) agar. When appropriate, antibiotics were added to plates at the following final concentrations: erythromycin, 30 μg/mL; and neomycin, 25 μg/mL. For growth in liquid cultures, wild-type and mutant cells were shaken in flasks, but mutants were grown in the presence of 100 to 150 μg neomycin/mL.
Isolation of DNA
Cells of Anabaena sp. PCC7120 grown in the mid-log phase of growth were harvested by centrifugation (5,000g, 10 min, 4°C) and suspended in a buffer with 10 mm Tris and 0.1 mm EDTA, pH 7.5. DNA was isolated from these cells as described in Cai and Wolk (1990) and Goyal (1992).
Construction of Mutant Strains with Disruption of the Putative pgpB Gene
Two primers were designed for PCR to clone the putative pgpB gene on the genome of Anabaena sp. PCC7120, with upper primer CCGCCATGGTTTTACTCTTGTTTATCCTCTATC (NcoI) and lower primer CGTCTGCAGTTAGAGGCACAAATTCTAGAGGTA (PstI). Both primers have three extra bases and one NcoI or PstI restriction site (underlined) at the 5′ end, respectively. PCR was performed according to standard procedures using Taq DNA polymerase with a DNA Thermal Cycler (Perkin-Elmer) under the following thermocycle conditions: 5 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 55°C, and 60 s at 72°C, and 10 min at 72°C at end to allow complete elongation of all product DNA.
A 4,822-bp DNA fragment containing the gene alr1715 with a 962-bp length was amplified by PCR with the genomic DNA of Anabaena sp. PCC7120 as a template and then ligated into pMD18-T vector at its EcoRV restriction site for production of the plasmid pPGP1. At one restriction site, XbaI of the alr1715 gene of the pPGP1 plasmid, we inserted the kanamycin phosphotransferase gene (1.2 kb) from pUC4K by digestion with SalI to yield a plasmid, pPGP2. The pPGP2 plasmid containing the disrupted putative pgpB gene was then cloned to the NcoI/SphI-digested conjugative plasmid pRL271, a cloning vector for sacB-mediated positive selection for targeted gene replacement and double recombinants in cyanobacteria (Black et al., 1993). The resultant plasmid was designated as pPGP3 and conjugated into Anabaena PCC 7120 as previously described (Cai and Wolk, 1990). The initial conjugants were selected by screening for resistance to 25 μg/mL of neomycin and were then cultured on BG11 plates containing 5% (w/v) Suc to select for double recombinants lacking the sacB gene (Black et al., 1993). Double recombinants were identified by the Suc- and neomycin-resistant and erythromycin-sensitive phenotype. In all cases, the genomic DNA of the resultant mutant strain was checked by PCR and Southern analysis.
RNA Isolation and RT-PCR
RNA from Anabaena sp. PCC 7120 was prepared as described previously (Golden et al., 1987). The resulting RNA preparations were digested with RNase-free DNase to eliminate residual DNA. RT-PCR was performed using the RT-PCR kit (Promega).
Lipid Analysis
The total lipids were extracted from the cells of the wild type and the mutant according to the method of Sato and Murata (1988). Individual lipid classes were separated from the total lipid extract by two-dimensional thin-layer chromatography, with a solvent system consisting of chloroform/methanol/water (65:25:4, v/v/v) for the first dimension and chloroform/methanol/ammonia (65:35:5, v/v/v) for the second dimension. Individual lipids were used to prepare fatty acid methyl esters with 5% (v/v) H2SO4 in anhydrous methanol. The methyl esters were analyzed by gas chromatography, and the lipid contents were calculated with heptadecanoic acid as an internal standard.
Determination of Pigment Contents
Chl was extracted with 100% methanol from cell pellets obtained from 1 mL of the wild-type and mutant cell cultures, and the Chl content was determined according to Porra et al. (1989). The concentrations of phycocyanin, allophycocyanin, and phycoerythrin were determined using the method of Tandeau de Marsac and Houmard (1988). All absorption spectroscopic determinations were performed on a UV8500 UV-VIS spectrophotometer (Scientific Instrument Network of China).
Measurement of Photosynthetic Activity
Photosynthetic O2 evolution coupled with CO2 fixation (net photosynthesis) was measured for intact cells equivalent to 2.5 μg Chl/mL in culture medium containing 10 mm NaHCO3 as the electron acceptor with a Clark-type oxygen electrode (DW 1 Liquid Clark electrode, Hansatech Institute). PSII activity was measured in BG11 medium containing intact cells equivalent to 2.5 μg Chl/mL, 0.25 mm DCBQ was used as the electron acceptor, and 0.25 mm ferricyanide added to keep the quinones in an oxidized form. For each measurement, an aliquot of cell culture was spun down and resuspended in 1 mL of fresh BG11 medium. The reaction mixture was kept at 30°C and illuminated under saturating white light (approximately 1,000 μmol photon m−2 s−1).
Chl Fluorescence Measurements
Low temperature (77 K) Chl emission fluorescence spectra were recorded in a spectrofluorimeter (F-4500, Hitachi) with 5 nm/2.5 nm (excitation/emission) slit widths. For fluorescence measurements, cells were diluted in BG11 medium containing 20% glycerol to a final Chl concentration of 5 μg/mL and frozen in liquid nitrogen (Schneider et al., 2001).
Room temperature Chl fluorescence was determined with a pulse amplitude modulation fluorometer (PAM101, Walz) according to the procedure of Clarke et al. (1993) and Schneider et al. (2004).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers BAB78081/Q8YWA0.
Acknowledgments
We are grateful to Professor C. Peter Wolk (Michigan State University-U.S. Department of Energy Plant Research Laboratory, Michigan State University) for the generous gift of plasmid pRL271. We thank Dr. Zeneng Wang (Department of Cell Biology, Lerner Research Institute, Cleveland Clinic Foundation, Ohio) for critically reading the manuscript and giving useful suggestions. We also thank Professor Ding-Ji Shi (Institute of Botany, the Chinese Academy of Sciences, China) for his kind help throughout the experiments.
This work was supported by the Innovation Program of the Chinese Academy of Sciences.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Zhenle Yang (yangzhl@ibcas.ac.cn).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.083451.
References
- Allen MM (1968) Simple conditions for growth of unicellular blue-green algae on plates. J Phycol 4: 1–4 [DOI] [PubMed] [Google Scholar]
- Andrews J, Mudd JB (1986) Phosphatidylglycerol synthesis in pea chloroplasts. Plant Physiol 79: 259–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki M, Sato N, Meguro A, Tsuzuki M (2004) Differing involvement of sulfoquinovosyl diacylglycerol in photosystem II in two species of unicellular cyanobacteria. Eur J Biochem 271: 685–693 [DOI] [PubMed] [Google Scholar]
- Babiychuk E, Muller F, Eubel H, Braun HP, Frentzen M, Kushnir S (2003) Arabidopsis phosphatidylglycerophosphate synthase 1 is essential for chloroplast differentiation, but is dispensable for mitochondrial function. Plant J 33: 899–909 [DOI] [PubMed] [Google Scholar]
- Benning C, Beatty JT, Prince RC, Somerville CR (1993) The sulfolipid sulfoquinovosyldiacylglycerol is not required for photosynthetic electron transport in Rhodobacter sphaeroides but enhances growth under phosphate limitation. Proc Natl Acad Sci USA 90: 1561–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black TA, Cai Y, Wolk CP (1993) Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol Microbiol 9: 77–84 [DOI] [PubMed] [Google Scholar]
- Bleasdale JE, Johnston JM (1982) CMP-dependent incorporation of [14C] glycerol 3-phosphate into phosphatidylglycerol and phosphatidylglycerol phosphate by rabbit lung microsomes. Biochim Biophys Acta 710: 377–390 [DOI] [PubMed] [Google Scholar]
- Brindley DN, Waggoner DW (1998) Mammalian lipid phosphate phosphohydrolases. J Biol Chem 3: 24281–24284 [DOI] [PubMed] [Google Scholar]
- Cai Y, Wolk CP (1990) Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J Bacteriol 172: 3138–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter JR, Kennedy EP (1966) Enzymatic synthesis of cytidine diphosphate diglyceride. J Lipid Res 7: 678–683 [PubMed] [Google Scholar]
- Chang YY, Kennedy EP (1967) Phosphatidyl glycerophosphate phosphatase. J Lipid Res 8: 456–462 [PubMed] [Google Scholar]
- Clarke AK, Hurry VM, Gustafsson P, Oquist G (1993) Two functionally distinct forms of the photosystem II reaction center D1 protein in the cyanobacterium Synechococcus sp. PCC7942. Proc Natl Acad Sci USA 90: 11985–11989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva ACR, Ferro JA, Reinach FC, Farah CS, Furlan LR, Quaggio RB, Monteiro-Vitorello CB, Van Sluys MA, Almeida NF Jr, Alves LMC, et al (2002) Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417: 459–463 [DOI] [PubMed] [Google Scholar]
- Dillon DA, Wu WI, Riedel B, Wissing JB, Dowhan W, Carman GM (1996) The Escherichia coli pgpB gene encodes for a diacylglycerol pyrophosphate phosphatase activity. J Biol Chem 271: 30548–30553 [DOI] [PubMed] [Google Scholar]
- Domonkos I, Malec P, Sallai A, Kovacs L, Itoh K, Shen G, Ughy B, Bogos B, Sakurai I, Kis M, et al (2004) Phosphatidylglycerol is essential for oligomerization of photosystem I reaction center. Plant Physiol 134: 1471–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhai J, Wolk CP (1988) Conjugal transfer of DNA to cyanobacteria. Methods Enzymol 167: 747–755 [DOI] [PubMed] [Google Scholar]
- Essigmann B, Guler S, Narang RA, Linke D, Benning C (1998) Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 95: 1950–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk CR, Zimniak L, Dowhan W (1992) The pgpA and pgpB genes of Escherichia coli are not essential: evidence for a third phosphatidylglycerophosphate phosphatase. J Bacteriol 174: 205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden SS, Brusslan J, Haselkorn R (1987) Genetic engineering of the cyanobacterial chromosome. Methods Enzymol 153: 215–231 [DOI] [PubMed] [Google Scholar]
- Gombos Z, Varkonyi Z, Hagio M, Iwaki M, Kovacs L, Masamoto K, Itoh S, Wada H (2002) Phosphatidylglycerol requirement for the function of electron acceptor plastoquinone Q(B) in the photosystem II reaction center. Biochemistry 41: 3796–3802 [DOI] [PubMed] [Google Scholar]
- Goyal D (1992) A simplified method for screening and characterization of plasmid DNA in cyanobacteria. J Microbiol Methods 15: 7–15 [Google Scholar]
- Guler S, Seeliger A, Hartel H, Renger G, Benning C (1996) A null mutant of Synechococcus sp. PCC7942 deficient in the sulfolipid sulfoquinovosyl diacylglycerol. J Biol Chem 271: 7501–7507 [DOI] [PubMed] [Google Scholar]
- Hagio M, Gombos Z, Varkonyi Z, Masamoto K, Sato N, Tsuzuki M, Wada H (2000) Direct evidence for requirement of phosphatidylglycerol in photosystem II of photosynthesis. Plant Physiol 124: 795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagio M, Sakurai I, Sato S, Kato T, Tabata S, Wada H (2002) Phosphatidylglycerol is essential for the development of thylakoid membranes in Arabidopsis thaliana. Plant Cell Physiol 43: 1456–1464 [DOI] [PubMed] [Google Scholar]
- Hartel H, Essigmann B, Lokstein H, Hoffmann-Benning S, Peters-Kottig M, Benning C (1998) The phospholipid-deficient pho1 mutant of Arabidopsis thaliana is affected in the organization, but not in the light acclimation, of the thylakoid membrane. Biochim Biophys Acta 1415: 205–218 [DOI] [PubMed] [Google Scholar]
- Hemrika W, Renirie R, Dekker HL, Barnett P, Wever R (1997) From phosphatases to vanadium peroxidases: a similar architecture of the active site. Proc Natl Acad Sci USA 94: 2145–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi T, Larson TJ, Dowhan W (1976) Membrane-associated phosphatidyl-glycerophosphate synthetase from Escherichia coli: purification by substrate affinity chromatography on cytidine 5′-diphospho-1,2-diacyl-sn-glycerol sepharose. Biochemistry 15: 5205–5211 [DOI] [PubMed] [Google Scholar]
- Hobe S, Prytulla S, Kuhlbrandt W, Paulsen H (1994) Trimerization and crystallization of reconstituted light-harvesting chlorophyll a/b complex. EMBO J 13: 3423–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icho T (1988. a) Membrane-bound phosphatases in Escherichia coli: sequence of the pgpA gene. J Bacteriol 170: 5110–5116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icho T (1988. b) Membrane-bound phosphatases in Escherichia coli: sequence of the pgpB gene and dual subcellular localization of the pgpB product. J Bacteriol 170: 5117–5124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icho T, Raetz CR (1983) Multiple genes for membrane-bound phosphatases in Escherichia coli and their action on phospholipid precursors. J Bacteriol 153: 722–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Sorokin A, Anderson I, Galleron N, Candelon B, Kapatral V, Bhattacharyya A, Reznik G, Mikhailova N, Lapidus A, et al (2003) Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423: 87–91 [DOI] [PubMed] [Google Scholar]
- Jordan BR, Chow WS, Baker AJ (1983) The role of phospholipids in the molecular organization of pea chloroplast membranes: effect of phospholipid depletion on photosynthetic activities. Biochim Biophys Acta 725: 77–86 [Google Scholar]
- Jordan P, Fromme P, Witt HT, Klukas O, Saenger W, Krauss N (2001) Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature 411: 909–917 [DOI] [PubMed] [Google Scholar]
- Kaneko T, Nakamura Y, Wolk CP, Kuritz T, Sasamoto S, Watanabe A, Iriguchi M, Ishikawa A, Kawashima K, Kimura T, et al (2001) Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res 8: 205–213 [DOI] [PubMed] [Google Scholar]
- Kopka J, Ludewig M, Muller-Rober B (1997) Complementary DNAs encoding eukaryotic-type cytidine-5′-diphosphate-diacylglycerol synthases of two plant species. Plant Physiol 113: 997–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause GH, Weis E (1991) Chlorophyll fluorescence and photosynthesis: the basics. Annu Rev Plant Physiol Plant Mol Biol 42: 313–349 [Google Scholar]
- Kruse O, Hankamer B, Konczak C, Gerle C, Morris E, Radunz A, Schmid GH, Barber J (2000) Phosphatidylglycerol is involved in the dimerization of photosystem II. J Biol Chem 275: 6509–6514 [DOI] [PubMed] [Google Scholar]
- Kruse O, Radunz A, Schmid GH (1994) Phosphatidylglycerol and β-carotene bound onto the D1-core peptide of photosystem II in the filamentous cyanobacterium Oscillatoria chalybea. Z Naturforsch 49c: 115–124 [Google Scholar]
- Kruse O, Schmid GH (1995) The role of phosphatidylglycerol as a functional effector and membrane anchor of the D1-core peptide from photosystem II particles of the cyanobacterium Oscillatoria chalybea. Z Naturforsch 50c: 380–390 [PubMed] [Google Scholar]
- Liu Z, Yan H, Wang K, Kuang T, Zhang J, Gui L, An X, Chang W (2004) Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature 4282: 87–92 [DOI] [PubMed] [Google Scholar]
- Moore TS (1982) Phospholipid biosynthesis. Annu Rev Plant Physiol 33: 235–259 [Google Scholar]
- Murata N, Siegenthaler PA (1998) Lipids in photosynthesis: an overview. In PA Siegenthaler, N Murata, eds, Lipids in Photosynthesis: Structure, Function and Genetics. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–20
- Neuwald AF (1997) An unexpected structural relationship between integral membrane phosphatases and soluble haloperoxidases. Protein Sci 6: 1764–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nußberger S, Dörr K, Wang DN, Kuhlbrandt W (1993) Lipid-protein interaction in crystals of plant light-harvesting complex. J Mol Biol 224: 347–356 [DOI] [PubMed] [Google Scholar]
- Pineau B, Girard-Bascou J, Eberhard S, Choquet Y, Tremolieres A, Gerard-Hirne C, Bennardo-Connan A, Decottignies P, Gillet S, Wollman FA (2004) A single mutation that causes phosphatidylglycerol deficiency impairs synthesis of photosystem II cores in Chlamydomonas reinhardtii. Eur J Biochem 271: 329–338 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Sato N, Hagio M, Wada H, Tsuzuki M (2000) Requirement of phosphatidylglycerol for photosynthetic function in thylakoid membranes. Proc Natl Acad Sci USA 97: 10655–10660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Murata N (1988) Membrane lipids. Methods Enzymol 167: 251–259 [Google Scholar]
- Sato N, Tsuzuki M, Matsuda Y, Ehara T, Osafune T, Kawaguchi A (1995) Isolation and characterization of mutants affected in lipid metabolism of Chlamydomonas reinhardtii. Eur J Biochem 230: 987–993 [DOI] [PubMed] [Google Scholar]
- Satoh K, Oh-hashi M, Kashino Y, Koike H (1995) Mechanism of electron flow through the QB site in photosystem II. I. Kinetics of the reduction of electron acceptors at the QB and plastoquinone sites in photosystem II particles from the cyanobacterium Synechococcus vulcanus. Plant Cell Physiol 36: 597–605 [Google Scholar]
- Schneider D, Berry S, Rich P, Seidler A, Rögner M (2001) A regulatory role of the PetM subunit in a cyanobacterial cytochrome b6f complex. J Biol Chem 276: 16780–16785 [DOI] [PubMed] [Google Scholar]
- Schneider D, Berry S, Volkmer T, Seidler A, Rögner M (2004) PetC1 is the major rieske iron-sulfur protein in the cytochrome b6f complex of Synechocystis sp. PCC 6803. J Biol Chem 279: 39383–39388 [DOI] [PubMed] [Google Scholar]
- Siegenthaler PA, Smutny J, Rawler A (1987) Involvement of distinct populations of phosphatidylglycerol and phosphatidylcholine molecules in photosynthetic electron-flow activities. Biochim Biophys Acta 891: 85–93 [DOI] [PubMed] [Google Scholar]
- Sigal YJ, McDermott MI, Morris AJ (2005) Integral membrane lipid phosphatases/phosphotransferases: common structure and diverse functions. Biochem J 387: 281–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukey J, Carman GM (1997) Identification of a novel phosphatase sequence motif. Protein Sci 6: 469–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandeau de Marsac N, Houmard J (1988) Complementary chromatic adaptation: physiological conditions and action spectra. Methods Enzymol 43: 318–328 [Google Scholar]
- Vermaas WF, Shen G, Styring S (1994) Electrons generated by photosystem II are utilized by an oxidase in the absence of photosystem I in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett 337: 103–108 [DOI] [PubMed] [Google Scholar]
- Wilde A, Hartel H, Hubschmann T, Hoffmann P, Shestakov SV, Borner T (1995) Inactivation of a Synechocystis sp strain PCC 6803 gene with homology to conserved chloroplast open reading frame 184 increases the photosystem II-to-photosystem I ratio. Plant Cell 7: 649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Hartel H, Wada H, Hagio M, Yu B, Eakin C, Benning C (2002) The pgp1 mutant locus of Arabidopsis encodes a phosphatidylglycerolphosphate synthase with impaired activity. Plant Physiol 129: 594–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Yang J, Zhang X, Chen L, Jiang Y, Yan Y, Tang X, Wang J, Xiong Z, Dong J, et al (2005) Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res 33: 6445–6458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Xu C, Benning C (2002) Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc Natl Acad Sci USA 99: 5732–5737 [DOI] [PMC free article] [PubMed] [Google Scholar]