Abstract
Extracellular pathogenesis-related proteins, including glucanases, are expressed at cold temperatures in winter rye (Secale cereale) and display antifreeze activity. We have characterized recombinant cold-induced glucanases from winter rye to further examine their roles and contributions to cold tolerance. Both basic β-1,3-glucanases and an acidic β-1,3;1,4-glucanase were expressed in Escherichia coli, purified, and assayed for their hydrolytic and antifreeze activities in vitro. All were found to be cold active and to retain partial hydrolytic activity at subzero temperatures (e.g. 14%–35% at −4°C). The two types of glucanases had antifreeze activity as measured by their ability to modify the growth of ice crystals. Structural models for the winter rye β-1,3-glucanases were developed on which putative ice-binding surfaces (IBSs) were identified. Residues on the putative IBSs were charge conserved for each of the expressed glucanases, with the exception of one β-1,3-glucanase recovered from nonacclimated winter rye in which a charged amino acid was present on the putative IBS. This protein also had a reduced antifreeze activity relative to the other expressed glucanases. These results support the hypothesis that winter rye glucanases have evolved to inhibit the formation of large, potentially fatal ice crystals, in addition to having enzymatic activity with a potential role in resisting infection by psychrophilic pathogens. Glucanases of winter rye provide an interesting example of protein evolution and adaptation aimed to combat cold and freezing conditions.
The stationary nature of plants has driven the evolution of strategies to cope with environmental stresses such as cold, drought, salinity, and pathogen attacks. The ability to survive temperature fluctuations is particularly critical for plants in temperate and boreal regions where they may be exposed to severe heat in summer and extreme cold in winter. When plants are exposed to freezing temperatures, ice crystals form in the extracellular spaces, in part because the extracellular fluid has a higher freezing point than the cytosol (Sakai and Larcher, 1987; Guy et al., 1992). This difference results in a decrease in the water potential outside the cell and leads to the movement of water from the cytosol into the apoplastic spaces (Sakai and Larcher, 1987). The cellular dehydration caused by extracellular ice crystal formation is one of the most damaging consequences of low temperature in plants.
Freezing-tolerant plants undergo a gradual adaptation to low temperature through a process known as cold acclimation, which allows plants to increase their freezing tolerance in response to cold, but not freezing, temperatures (Browse and Xin, 2001). Cold acclimation alters numerous aspects of cellular metabolism, including hormone levels, sugars, membrane lipid composition, as well as the expression of certain genes (Sakai and Larcher, 1987; Guy et al., 1992; Sung et al., 2003). During cold acclimation, some plants, including many cereals, secrete proteins into the apoplast that inhibit the growth of extracellular ice. These proteins are known as antifreeze proteins (AFPs; for review, see Griffith and Yaish, 2004).
The apoplast of cold-acclimated (CA) winter rye (Secale cereale), an overwintering and freezing-tolerant grass, contains at least eight polypeptides ranging in mass from 9 to 34 kD with varying levels of antifreeze activity (Griffith et al., 1992). N-terminal sequencing, enzymatic activity, and immunoblotting assays of these polypeptides reveal strong similarities to pathogenesis-related (PR) proteins, including β-1,3-glucanases, endochitinases, thaumatins, and lipid transfer proteins (Hon et al., 1994). CA winter rye PR-like proteins have the ability to control the growth of ice crystals in the apoplast (Griffith et al., 1992) where they exist as complexes of different compositions, including two β-1,3-glucanases of 32 and 34 kD (Yu and Griffith, 1999).
Plant β-1,3-glucanases are documented to contribute to several other physiological processes, including pathogen resistance, cell wall synthesis, and pollen development (for review, see Meins et al., 1992; Simmons, 1994; Høj and Fincher, 1995). Despite intensive studies on plant β-1,3-glucanases, their specific role in cold tolerance has not been previously identified and thus this study seeks to address that issue by examining the enzymatic activities of the winter rye glucanases. To achieve this goal, cDNAs encoding β-1,3-glucanases and a β-1,3;1,4-glucanase were isolated from a cDNA library prepared from mRNAs isolated from CA winter rye and expressed in Escherichia coli. The recombinant proteins were purified and their hydrolytic and ice-binding activities were assayed. In addition, a computational structural analysis was employed to identify a putative ice-binding surface (IBS) on these glucanases. Based on our results, we propose that winter rye glucanases provide plants with a protective mechanism against formation of ice crystals in the apoplast in addition to the ability to degrade glucans at low temperature. The latter could provide a potential protective mechanism against psychrophylic pathogenic organisms. β-1,3-Glucanases present an interesting example of PR proteins involved in both biotic and abiotic resistance mechanisms during subzero conditions.
RESULTS
CA Winter Rye Expresses Both β-1,3-Glucanase and β-1,3;1,4-Glucanase Genes
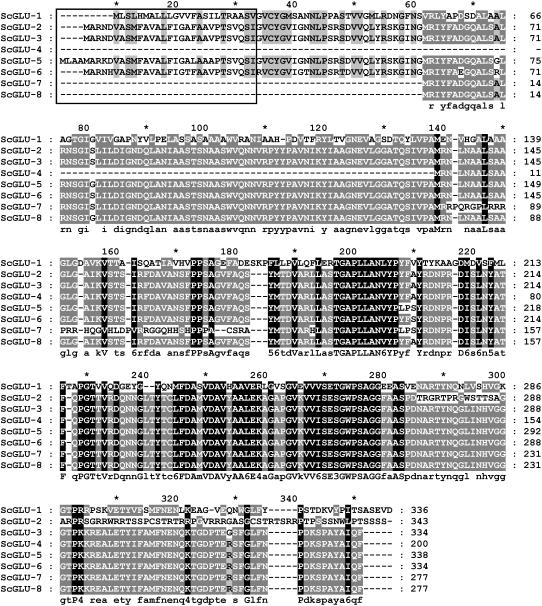
Glucanase sequences were recovered from a mRNA library prepared from CA plants by screening 93,000 colonies from the λ-Zap cDNA library made from poly(A)+ mRNA isolated from CA winter rye leaves (Yeh et al., 2000) with a barley (Hordeum vulgare) β-1,3-glucanase sequence probe (Høj et al., 1989). Forty positive clones were recovered and converted into pBluescript SK (Stratagene). Clones were classified into eight different groups based on their restriction fingerprint and one representative plasmid of each group was sequenced in both directions. The results showed the presence of eight different cDNA sequences that were designated ScGLU-1 to ScGLU-8. The inserts in these clones ranged in length from 947 bp (ScGLU-4) to 1,256 bp (ScGLU-1). All open reading frames from these clones encode full or partial glucanase sequences of 277 to 343 amino acid residues (Fig. 1).
Figure 1.
Alignment of the deduced CA winter rye glucanase amino acid sequences. The sequences were aligned using ClustalX and conserved residues were shaded by GeneDoc (Nicholas et al., 1997). Consensus sequences are shown below the alignment, where conserved groups are individual residue types or one of the following: 1 = DN, 3 = ST (hydroxylated), 4 = KR (basic), 5 = FYW (aromatic), and 6 = LIVM (aliphatic or M). Groups conserved across all sequences are shaded black, those residues conserved 80% to 99% across the sequences are shaded dark gray, those residues conserved 60% to 79% across the sequences are shaded light gray, and nonconserved residues are not shaded. The predicted signal peptide domains are highlighted by a box. Five of the glucanase sequences contain signal peptides targeting them for secretion.
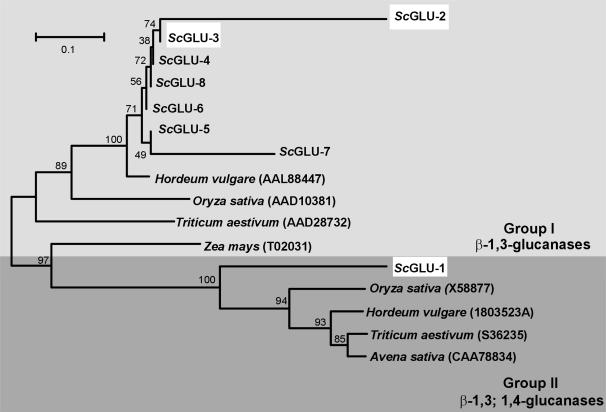
The winter rye sequences partitioned into two different groups (group I and group II) based on phylogenetic analysis with representative glucanases isolated from different cereal species (Fig. 2). Whereas group I contained seven winter rye glucanase sequences (ScGLU-2 to 8), group II contained only one sequence (ScGLU-1). The amino acid sequences within group I were generally conserved, except for a few substitutions and small insertions. One base pair deletion in the open reading frame of ScGLU-2 leads to a frame shift in the C-terminal part of the deduced protein. Group I winter rye sequences have up to 96% amino acid sequence identity to barley, rice (Oryza sativa), and wheat (Triticum aestivum) β-1,3-glucanases. On the other hand, the ScGLU-1 sequence has a maximum of 80% identity with other cereal β-1,3-glucanases and β-1,3;1,4-glucanases. All sequences were predicted to contain a glycosyl hydrolase domain (Marchler-Bauer et al., 2005), consistent with their putative roles as glucanases.
Figure 2.
Phylogenetic tree of glucanases from CA winter rye and other cereal species. Amino acid sequences were aligned using ClustalX and the tree was constructed by neighbor joining on the basis of the Dayhoff 250 matrix using Molecular Evolutionary Genetic Analysis software, version 2.1. Bootstrap values are indicated at nodes as a percentage of 100 replicates. GenBank accession numbers are shown in parentheses. When compared with glucanase sequences from other cereals, the winter rye amino acid sequences partitioned into two distinct groups: one group of seven sequences had high amino acid identities to basic β-1,3-glucanases of other cereals, whereas the sequence of ScGLU-1 was similar to acidic β-1,3-glucanases and β-1,3;1,4-glucanases isolated from barley, oat (Avena sativa), rice, and wheat. Highlighted winter rye sequences were chosen for production of recombinant proteins.
An N-terminal signal peptide directing the protein to the endomembrane system for eventual extracellular secretion was present in ScGLU-1, ScGLU-2, ScGLU-3, ScGLU-5, and ScGLU-6 coding regions, based on their analysis with SignalP (Nielsen et al., 1997). This is consistent with previous work in which immunogold localization of CA β-1,3-glucanases showed accumulation in the extracellular space in winter rye (Pihakaski-Maunsbach et al., 1996). The absence of the signal peptide from the shorter clones (ScGLU-4, ScGLU-7, and ScGLU-8; Fig. 1) is likely a cloning artifact or splicing alteration in the mRNA. Based on the deduced amino acid sequences of the mature polypeptides, the group I sequences code for basic β-1,3-glucanases (predicted pIs 7.3–10.73), whereas the ScGLU-1 sequence of group II encodes an acidic β-1,3;1,4-glucanase (predicted pI of 4.4).
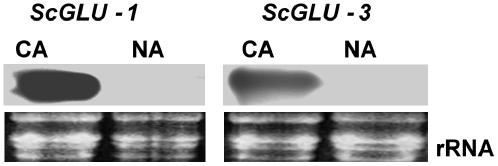
Representatives from the two winter rye glucanase gene families also demonstrate induction in response to cold acclimation. β-1,3;1,4-Glucanase (ScGLU-1) and β-1,3-glucanase (ScGLU-3) transcripts accumulated to high levels in 5-week-old CA leaves, whereas the ScGLU-1 and ScGLU-3 transcripts were not detectable in nonacclimated (NA) leaves (Fig. 3). Because coding regions of the glucanase genes were used as probes in this analysis, cross-reacting hybridizations within the same glucanase gene family members cannot be ruled out.
Figure 3.
Effect of temperature on the transcript accumulation of β-1,3;1,4-glucanase (ScGLU-1) and β-1,3-glucanase (ScGLU-3) in winter rye. Total RNA samples were extracted from NA and CA leaves. RNA was electrophoresed in a 1% denaturing formaldehyde gel, stained with ethidium bromide, and visualized by UV light. RNA was capillary transferred to a nylon membrane and hybridized with DIG-labeled probes for ScGLU-1 and ScGLU-3. Transcripts of ScGLU-1 and ScGLU-3 were detected in CA, but not in NA, leaves.
Production of Winter Rye Glucanases in E. coli
To characterize their activities in vitro, ScGLU-1, ScGLU-2, and ScGLU-3 mature peptides were fused to a glutathione S-transferase (GST) tag, overexpressed in E. coli, purified, and followed by removal of the GST affinity tag. Because it was of interest to compare the enzymatic activities of these CA glucanases with those present in NA plants, two β-1,3-glucanase sequences from NA leaves were recovered by reverse transcriptase (RT)-PCR and cloned as GST fusions for expression in E. coli. These sequences were designated as ScGLUNA-1 and ScGLUNA-2. ClustalX analysis of these sequences, in comparison with ScGLU-3, revealed the presence of three and two nonsynonymous substitutions in ScGLUNA-1 and ScGLUNA-2, respectively (Fig. 4). Relative to ScGLU-3, there are 16 substitutions in the ScGLUNA-1 nucleic acid sequence, resulting in three amino acid changes. These sequence differences confirm that ScGLUNA-1 is encoded by a distinct β-1,3-glucanase gene member and is not an artifact of PCR.
Figure 4.
Alignment of amino acid sequences of a β-1,3-glucanase cloned from CA winter rye (ScGLU-3) and β-1,3-glucanases cloned from NA winter rye (ScGLUNA-1 and ScGLUNA-2). Coding DNA sequences of the mature peptide were amplified by PCR using cDNA prepared from NA RNA leaves, translated, and aligned with the ScGLU-3 mature peptide using ClustalX. Amino acid substitutions are highlighted by boxes.
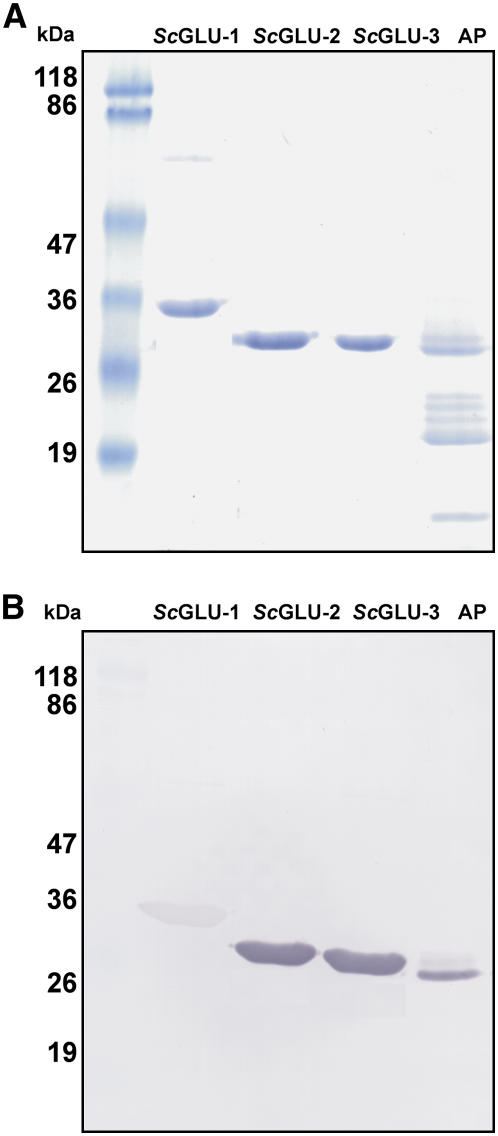
While ScGLU-2 and ScGLU-3 migrated on SDS-PAGE, consistent with their expected masses and the β-1,3-glucanases from the CA winter rye apoplast, ScGLU-1 migrated slower than its calculated molecular mass and had no detectable counterpart in the apoplastic extract (Fig. 5A). A possible explanation for the slow migration of ScGLU-1 is its acidic pI, which often results in overestimation of the protein size in SDS-PAGE (Kaufmann et al., 1984). Mass spectrometry analysis of each recombinant protein showed sharp mass peaks consistent with the virtual translation products of each coding sequence, indicating that these proteins are not posttranslationally modified (Fig. 6). Moreover, in-gel staining of apoplastic proteins from CA and NA winter rye, as well as the recombinant glucanases, did not detect any phosphorylation, glycosylation, or lipidation (data not shown). Although the apoplastic glucanases, ScGLU-2 and ScGLU-3, reacted strongly with the specific antibodies raised previously against the winter rye β-1,3-glucanase (Antikainen et al., 1996), ScGLU-1 bound the same antiserum only weakly (Fig. 5B).
Figure 5.
Expression of recombinant glucanases from CA winter rye. ScGLU-2, ScGLU-3, and ScGLU-1 were expressed in E. coli and the recombinant proteins were purified from the GST tag and examined by SDS-PAGE (A) and immunoblotting (B). A, The indicated recombinant proteins (5 μg/lane) were solubilized, separated on a 12% (w/v) SDS-polyacrylamide gel, and then stained with Coomassie blue. The molecular masses of prestained protein standards are shown on the left and apoplastic proteins (AP) extracted from CA plants are on the right. B, Immunoblot of a duplicate gel treated with polyclonal antibody produced against the purified 34-kD AFP β-1,3-glucanase from winter rye.
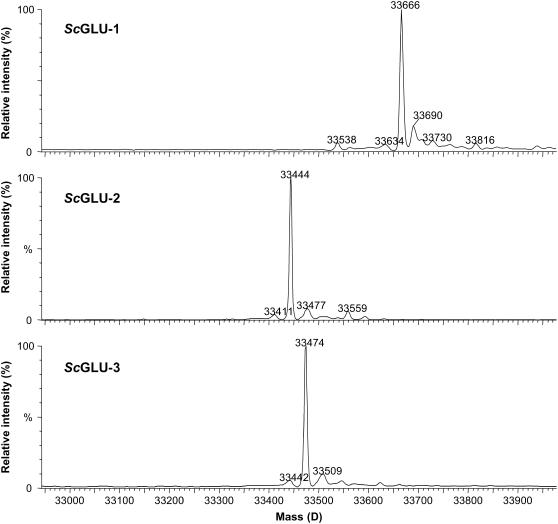
Figure 6.
Molecular masses of recombinant CA winter rye glucanases determined by mass spectrometry. Raw data (mass-to-charge ratio) were processed using MaxEnt1 to yield spectra on a true molecular mass scale. The molecular mass of ScGLU-1 was 33,666 ± 0.7 D, ScGLU-2 was 33,444 ± 0.8 D, and ScGLU-3 was 33,474 ± 0.7 D. The recombinant β-1,3-glucanases and β-1,3;1,4-glucanase were not posttranslationally modified.
Glucanases Expressed in E. coli Exhibit Different Levels of Antifreeze Activity
The antifreeze activity of each recombinant glucanase was assayed based on their ability to modify the morphology of ice crystals grown in solution (Hon et al., 1994). Changes in the ice crystal shapes were noted when the microscope stage was warmed and cooled in slow cycles. Each recombinant protein was serially diluted and tested for its effect on ice crystal morphology. ScGLU-2, ScGLU-3, and ScGLUNA-2 exhibited very similar antifreeze activities at the same protein concentration (2.5 mg mL−1). ScGLU-1 exhibited weaker antifreeze activity at equal or higher protein concentrations. The antifreeze activity of ScGLU-2, ScGLU-3, and ScGLUNA-2 was no longer apparent when the protein concentration was reduced to 1 mg mL−1. Interestingly, twice the concentration of ScGLUNA-1 was required for observable antifreeze activity, with no apparent activity at concentrations below 2.0 mg mL−1 (Fig. 7). Boiling the recombinant glucanase samples, as well as treating with proteinase K, resulted in the elimination of their antifreeze activities.
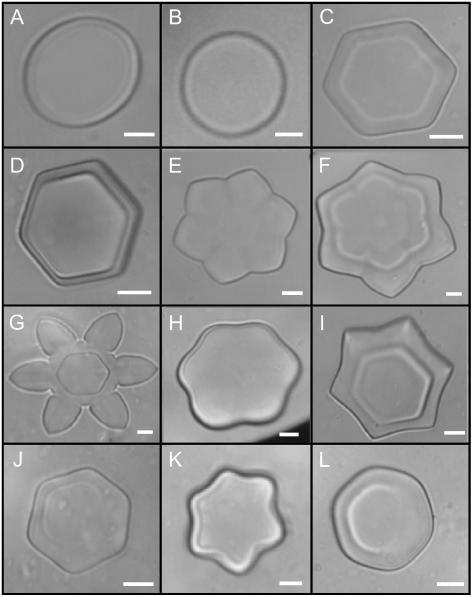
Figure 7.
Antifreeze activity of the purified recombinant glucanases. Solutions were flash frozen on a freezing stage of a microscope, warmed until only one ice crystal was present, and cooled to observe changes in morphology as the ice crystal grew. A, Extraction buffer only (50 mm ammonium acetate and 10 mm EDTA, pH 5.5). B, Buffer containing 2.5 mg mL−1 bovine serum albumin. In A and B, ice grew as a round and flat crystal. Identical crystal morphology was noted when recombinant proteins were boiled for 5 min. C, Buffer containing 2.5 mg mL−1 ScGLU-3, showing hexagonal growth of the ice crystal. D to G, Because the temperature was cooled and warmed in very slow cycles, ice crystals grew into shapes similar to stars and flowers. H and I, Ice crystal grown in the presence of 2.5 mg mL−1 ScGLU-2 showing successive stages of ice crystal development. J and K, Ice grown in the presence of 4 mg mL−1 and 2.5 mg mL−1 of ScGLNA-1 and ScGLNA-2, respectively. Increasing the ScGLNA-1 concentration by 2-fold results in similar antifreeze activity to ScGLU-2, ScGLU-3, and ScGLNA-2. L, Ice crystal grown in the presence of 2.5 mg mL−1 ScGLU-1 demonstrating weak antifreeze activity. The magnification bars represent 10 μm. Purified ScGLU-2, ScGLU-3, ScGLNA-1, and ScGLNA-2 β-1,3-glucanases have observable antifreeze activity; however, ScGLU-1, a β-1,3;1,4-glucanase, has much lower activity.
Recombinant Glucanases Exhibit Hydrolytic Activity at Subzero Temperatures
The hydrolytic activity of each CA recombinant glucanase was measured over a temperature range of −8°C to 72°C by assaying the production of reduced sugars released from polysaccharide substrates. All of the recombinant glucanases were able to catalyze the hydrolysis of laminarin over a wide temperature range (Fig. 8A). The recombinant glucanases varied in their hydrolytic activities and substrate specificities. Whereas ScGLU-2, ScGLU-3, ScGLUNA-1, and ScGLUNA-2 had only β-1,3-glucanase activity (EC 3.2.1.39), ScGLU-1 had both β-1,3-glucanase and β-1,3;1,4-glucanase activities (EC 3.2.1.73), consistent with the phylogenetic analysis (Fig. 2).
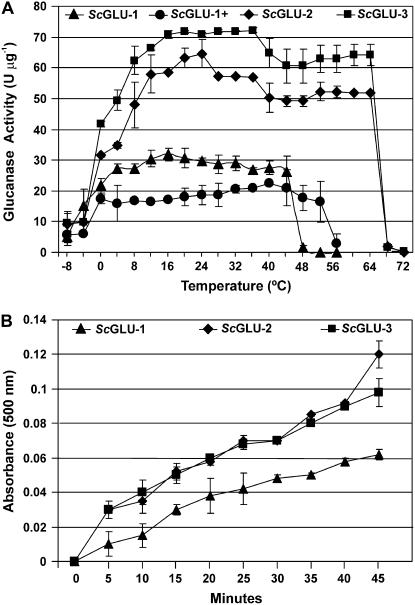
Figure 8.
Hydrolytic activity of recombinant glucanases. A, Specific β-1,3-glucanase activities of ScGLU-1, ScGLU-2, and ScGLU-3 and specific β-1,3;1,4-glucanase activity of ScGLU-1 (ScGLU-1+) measured at different temperatures. β-1,3-Glucanase and β-1,3;1,4-glucanase activities were determined using laminarin from L. digitata and β-1,3;1,4-glucan from barley as substrates, respectively. Specific activities are plotted as means ± sd from three independent replicates. B, Accumulation of reduced sugars as a result of ScGLU-1, ScGLU-2, and ScGLU-3 β-1,3-glucanase activity over time at −4°C.
Surprisingly, these glucanases retained some hydrolytic activity at cold temperatures. For example, ScGLU-1 and ScGLU-3 retained 60% and 65% of their maximal activity at 0°C, respectively. At lower temperatures, their catalytic activities were dramatically reduced (e.g. the β-1,3-glucanase activity of ScGLU-3 was reduced by 86% at −4°C relative to its maximal activity at 36°C). When the incubation time of the assay was extended, it was evident that the recombinant proteins had β-1,3-glucanase activity at −4°C (Fig. 8B).
There is a direct relationship between an enzyme's activity and protein structural stability at cold temperatures (Hochachka and Somero, 1984). Differential scanning calorimetry (DSC) was used to evaluate the effect of temperature variations on the conformational structure of ScGLU-3 when it was subjected to a temperature cycle ranging from 1°C to 100°C. The results showed that ScGLU-3 was structurally stable when exposed to temperatures between 1°C and 64°C, but was irreversibly denatured above 64°C. The protein did not recover its native structure when the temperature dropped again in a second cycle (data not shown). Indeed, when the temperature exceeded 68°C, the ScGLU-3 enzyme had no detectable hydrolytic activity (Fig. 8A).
Structural Modeling Reveals a Putative IBS
X-ray structures of previously characterized glucanases from barley were used as templates to construct three-dimensional models and identify a putative IBS of winter rye glucanases. Structural modeling based on a barley β-1,3-glucanase template (Protein Data Bank [PDB] ID 1GHS_A) revealed a large, relatively flat surface region on ScGLU-2, ScGLU-3, ScGLUNA-1, and ScGLUNA-2. Using ScGLU3 as a representative glucanase, this surface corresponds to residues Arg-23, Gly-26, Asn-28, Asn-45, Gly-47, Pro-82, Asn-85, Lys-87, Gly-118, Ala-121, Gln-305, and Phe-306 (Fig. 9, A and B). These residues form a highly ordered, grid-like arrangement of solvent-accessible groups that match repetitive openings or ice cages (Madura et al., 2000) on the secondary prism plane of ice (2ı̄0). All 12 putative ice-binding residues are charge-conserved amino acids among the glucanases (ScGLU-2, ScGLU-3, ScGLUNA-1, ScGLUNA-2, and template) with the exception of the Asn-28-Asp substitution found in ScGLUNA-1. This substitution introduces a charged residue to the center of the putative IBS (Fig. 9, A and B), which is a probable cause for the observed reduction in antifreeze activity with respect to ScGLU-2, ScGLU-3, and ScGLUNA-2 (Fig. 7). In contrast, these substitutions would be expected to have little effect on the hydrolytic activity. Accordingly, apoplastic proteins extracted from CA and NA winter rye have a similar β-1,3-glucanase hydrolytic activity (Hon et al., 1995). Interestingly, the Gln-305-Pro and Phe-306-Thr substitutions on the putative IBS of the ScGLU-2 did not appear to affect the antifreeze activity of the protein, likely because these amino acids are neutral and do not interrupt the putative IBS net charge. Another key feature of our model is the large protruding region adjacent to, and projecting past, the plane of the putative IBS. In the best AFP ice-docking configuration modeled using ZDOCK (Chen et al., 2003), this region is predicted to hang over the edge of the ice plane (Fig. 9D). When the same approach was used to predict a putative IBS for ScGLU-1, the putative IBS was not as flat or as ordered as for ScGLU-3. Whereas the ScGLU-3, ScGLUNA-1, and ScGLUNA-2 models are based on a template sequence identity of 94%, the ScGLU-1 model is not as accurate because the sequence identity with the template (PDB ID 1A10_A) is approximately 65%.
Figure 9.
Models of glucanase-ice interactions. A, Depiction of the secondary prism face (2ı̄0) of ice displaying a remarkable complimentarity with the putative IBS. One-letter codes for corresponding amino acids on the putative IBS of ScGLU-3 are shown in putative interstitial positions within the ice lattice. B, Molecular surface representation of ScGLU-3 showing the putative IBS (ordered surface groups are highlighted in red). The position of the Asn-28-Asp substitution in ScGLUNA-1 is highlighted in yellow. C, Lowest energy conformation of the ScGLU-3-ice complex as generated by molecular docking (ZDOCK). The hydrolytic domain (highlighted in yellow) is located on the opposite side of the protein relative to its putative IBS. D, Interaction shown in C, rotated to show the planar surface of the putative IBS and the adjacent overhanging region. Water molecules are shown as point locations of oxygen atom centers.
DISCUSSION
Winter Rye Glucanases Retain Partial Hydrolytic Activity at Subzero Temperatures
The recombinant glucanases presented in this study displayed hydrolytic activity at cold and freezing temperatures. Although their maximal activity in vitro occurred at temperatures ranging from 26°C to 36°C, there was clear evidence of activity at zero and subzero temperatures (0°C, −4°C, and −8°C; Fig. 8A). When activity was assayed at −4°C for an extended period, it was evident that all three recombinant glucanases were active (Fig. 8B). The crude apoplastic protein complexes extracted from CA winter rye also exhibit β-1,3-glucanase activity at low temperatures (data not shown); however, it was not possible to unequivocally assign the specific β-1,3-glucanase activity to an individual protein because thaumatin present within the apoplastic extract also possesses β-1,3-glucanase activity (Grenier et al., 1999). The antifungal activity of β-1,3-glucanases characterized from different plant species has been previously assayed in vivo and in vitro against various pathogens (for review, see Meins et al., 1992; Simmons, 1994; Høj and Fincher, 1995); however, their hydrolytic capacity has not been previously tested in cold conditions. It should also be noted that glucanase antifungal activity assessed in vitro and in planta can be quite variable (Meins et al., 1992; Simmons, 1994).
Generally, cold-active enzymes display less temperature dependence, higher catalytic efficiency, and lower activation energies than their thermophilic counterparts (Feller and Gerday, 1997; Taguchi et al., 1998). Besides the winter rye glucanases described in this study, enzymes with cold-active properties have been recovered from various psychrophilic microorganisms (for review, see Marshall, 1997; Lonhienne et al., 2000). Interestingly, several of these enzymes are related to glycosyl hydrolase proteins. These enzymes include an α-amylase (D'Amico et al., 2004) and a xylanase (Collins et al., 2002, 2003, 2005) of the Antarctic bacterium Pseudoalteromonas haloplanktis, a glucanase of Fibrobacter succinogenes S85 (Iyo and Forsberg, 1999), and an α-galactosidase of Carnobacterium piscicola strain BA (Coombs and Brenchley, 2001).
Relatively few physical properties distinguish cold-active enzymes from other proteins. Structural studies have revealed that these enzymes are generally highly fold flexible and therefore less rigid (Georlette et al., 2003, 2004; Collins et al., 2005). Cold-active enzymes are also characterized by stacking of small amino acids around the catalytic residues (Feller et al., 1992; Davail et al., 1994). Indeed, by using Swiss PDB Viewer (Guex and Peitsch, 1997), the calculated loop (random coil) content percentage of the ScGLU-3 template was 48%, which suggests a high degree of fold flexibility. This value is significantly higher than the average loop content (37.6%) of 154 nonprotease proteins reported previously (Stawiski et al., 2000).
Similar to other cold-active enzymes, winter rye glucanases displayed low thermal stability when exposed to high temperatures. This instability was demonstrated by irreversible denaturation as determined by DSC and also by hydrolytic activity assays (Fig. 8A). This is consistent with the relationship between conformational plasticity at low temperatures and instability at high temperatures, which has been described for other cold-active enzymes (Georlette et al., 2004; Collins et al., 2005). Feller and Gerday (1997) concluded that cold-active enzymes probably are perfectly evolved enzymes that can react with the surrounding substrate without the need of high-activation energy.
Glucanase AFPs Modify the Growth of Ice Crystals
Multicomponent protein mixtures secreted to the apoplast of CA winter rye exhibit antifreeze activity. The recombinant β-1,3-glucanases presented in this study, as well as the endochitinases of winter rye, independently exhibit antifreeze activity (Yeh et al., 2000). Preliminary experiments using polypeptides eluted from SDS-PAGE gels provided an initial indication that glucanase-like proteins possessed antifreeze activity (Hon et al., 1995). Those observations are validated in the experiments described here.
Through structural modeling and molecular docking, a putative IBS was identified and matched to the secondary prism plane of ice (2ı̄0); Fig. 9). This ice-binding model is consistent with several of the observations from the antifreeze assays. First, ScGLUNA-1, which exhibited lower antifreeze activity than ScGLU-3, has an amino acid substitution (Asn-28-Asp) in the center of the putative IBS. This substitution may affect ice binding by adding a charge and reducing surface hydrophobicity. Conversely, the putative IBS of ScGLU-1 is significantly more irregular than ScGLU-3, which may account for its reduced antifreeze activity. Because ScGLU-1 has a lower sequence identity (approximately 65%) with the protein template used in homology modeling, this interpretation remains somewhat speculative. Second, the putative IBS model predicts that when a glucanase AFP adsorbs on an ice surface it will most favorably interact with the secondary prism plane, directing growth of the ice crystal to occur by addition to the primary prism face (Figs. 9 and 10). That is, by adsorbing specifically to the secondary prism plane, winter rye ScGLU-3 may preferentially bind to and inhibit growth at the corners of an initial hexagonal crystal, eventually resulting in flower-like crystal morphology. Growth on the basal plane will also be preferentially inhibited at the same vertices, resulting in the appearance of basal plane edges perpendicular to the axes from the initial vertices to the center of the crystal. The result of this is the appearance of a secondary set of hexagonal edges on the basal plane, rotated 30 degrees relative to the initial hexagonal edges (Fig. 10). This unusual morphology, consistent with AFP binding to the edges of the secondary prism plane, is readily apparent in ice crystals grown in the presence of ScGLU-3. This was the best ice-binding configuration as determined by molecular docking and offers an explanation for the observed ice-crystal morphology.
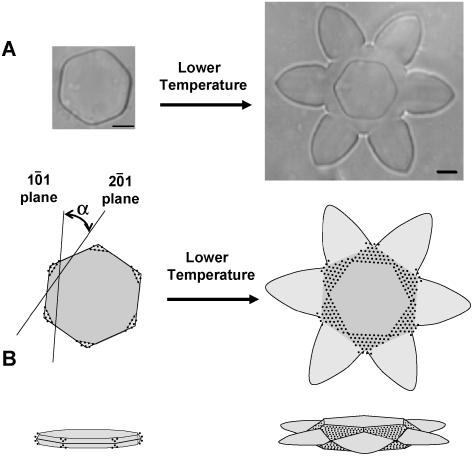
Figure 10.
Proposed β-1,3-glucanse ice-binding mechanism in winter rye. A, Changes in ice crystal morphologies were observed during an antifreeze assay of a sample containing purified ScGLU-3 (2.5 mg mL−1) when the microscope stage was warmed and cooled in slow cycles. B, ScGLU-3 is proposed to bind to secondary prism planes of ice (2ı̄0). As the temperature is lowered, ice crystal growth is preferentially inhibited on the (2ı̄0) faces of the ice crystal. This limits growth of the crystal to the central regions of the primary (1ō0) prism faces, producing a characteristic flower-like morphology. Slow growth on the basal plane of the crystal (parallel to the plane of the page) is likewise inhibited, producing a smaller hexagon with edges formed by the (2ı̄0) prism faces, rotated 30° (α) relative to growth on the primary prism faces (1ō0).
The ice morphologies observed in the presence of the recombinant β-1,3-glucanases compared to those produced by CA winter rye apoplasts are different. Whereas high concentrations of recombinant glucanase modify the growth of the ice crystals to form hexagonal and star-like shapes, apoplast extracts induce the formation of hexagonal and needle-like ice structures. Interestingly, similar star-shaped ice crystals are produced in assays of antifreeze activity of snow mold extract (Snider et al., 2000), although the nature of the proteins producing these crystals is unknown.
The winter rye apoplast extract exhibits higher antifreeze activity than any of its individual protein components. For example, the most active recombinant glucanase studied here required a concentration of 2.5 mg mL−1 to have a comparable antifreeze activity to that of an apoplast extract concentrated to 660 μg mL−1 (Hon et al., 1994). The differences in ice crystal morphology, as well as the antifreeze activity, likely reflect variations in the protein strength of interaction and plane of ice-crystal adsorption. That is, AFP complexes may have synergistic or coordinating effects on the ice crystal inhibition process by binding to various surfaces of ice and therefore efficiently limit their growth at a lower concentration.
How Do Plants React with Pathogens and Ice Crystals during Freezing Conditions?
During cold temperatures, plants are more susceptible to pathogen attack especially if freezing injury has occurred. Whereas the majority of pathogens are less virulent and unable to attack plants at freezing temperatures, some psychrophilic pathogens are able to invade plants under these conditions and cause significant damage (Jamalainen, 1974). Fortunately, exposure to cold provides tolerance to other stresses. Accordingly, winter rye accumulates PR proteins during cold acclimation (Antikainen and Griffith, 1997). The predicted function of these proteins was to provide plants with resistance against pathogens when a plant is covered with a snow blanket. However, it was still unknown whether these PR proteins are really active against pathogens at cold and subzero temperatures or fulfill alternative functions. Fungal cell walls contain β-1,3-glucan chains (Adams, 2004), which act as substrates for glucanase. Hence, the β-1,3-glucanase cold activity mirrors antifungal activity during cold and freezing conditions when the plant is threatened both by psychrophilic pathogens and growing ice crystals. As demonstrated, glucanases from CA winter rye not only possess hydrolytic activity, but also have the ability to adsorb to ice crystal surfaces and thereby modify their growth. Because the substrate-binding and catalytic sites on β-1,3-glucanase (Varghese et al., 1994) are located within a deep hydrophobic cleft approximately 180 degrees from the putative IBS with no overlap (Fig. 9B), they will not be disrupted upon ice binding, and hydrolytic activity will be maintained.
During cold acclimation, various metabolic changes take place in plant cells, particularly with respect to their sugar and protein content (Siminovitch, 1981; Guy et al., 1992; Takagi et al., 2003). As in winter rye (Antikainen and Griffith, 1997; Livingston and Premakumar, 2002), the accumulation of sugar molecules increases plant cold and freezing tolerance (Wanner and Junttila, 1999; Takagi et al., 2003). Generally, accumulation of sugars in the apoplast has an osmotic effect, decreasing ice formation and reducing the impact of freezing-induced dehydration (Sakai and Larcher, 1987). In fact, β-1,3-glucanase production is sometimes induced by sugars (Aziz et al., 2003; Thibaud et al., 2004). Thus, the β-1,3-glucanases presented here may be regulated by sugars and also act by breaking high-Mr substrates into smaller units, a process that may also have an osmoprotective effect.
From the above, we suggest that the cold-induced β-1,3-glucanases in winter rye provide the plant with a dual defense role against biotic and abiotic stresses during subzero conditions. This was demonstrated by the presence of β-1,3-glucan hydrolytic activity and ice-binding activity at low temperatures. Whereas hydrolytic activity was relatively constant for each glucanase studied, there was significant variation in their ability to affect ice growth. We have attributed this variation to specific substitutions on the putative IBS identified through structural modeling.
What Characteristics Distinguish Glucanase AFPs?
The question arises as to whether β-1,3-glucanases act as AFPs in other species and, if so, what characteristics distinguish these from glucanases lacking antifreeze function? For a glucanase to act as an AFP in winter rye, several criteria must be met. First, expression of the glucanase gene should be induced by cold conditions. Second, they must be secreted into the apoplast (Pihakaski-Maunsbach et al., 1996), where they limit ice crystal growth. Third, the glucanase AFP must possess an IBS that is geometrically complementary to an ice surface for strong binding to take place. The CA glucanase genes described in this study meet all three of these criteria: Their transcripts accumulate under cold conditions because they were cloned from a CA cDNA library, and cold-inducible expression was confirmed by northern-blot analysis (Fig. 3); sequence-based prediction (Nielsen et al., 1997) and previous experimental evidence (Pihakaski-Maunsbach et al., 1996) demonstrate localization of β-1,3-glucanases to the apoplast in cold-treated plants, and their protein crystal structures display a unique and specific putative IBS, as proposed here.
In this study, the antifreeze activities for individually sequenced glucanases isolated from CA and NA plants were assayed. It was found that glucanases from NA plants do have some antifreeze activity, although less than glucanases expressed in CA plants. The apparent lack of antifreeze activity in NA apoplast extracts is therefore due primarily to reduced expression of antifreeze-active proteins (Fig. 3), with reduced antifreeze activity of the NA glucanases playing a lesser role. Because small changes in the coding sequence can have a significant effect on antifreeze activity, we conclude that, in this case, sequence similarity does not directly imply similarity of function. Therefore, any putative glucanase AFPs from different plant species should have antifreeze activity confirmed experimentally.
CONCLUSION
Previous research results and those of this study led us to propose that winter rye glucanases may contribute to multiple aspects of plant survival at cold temperatures. It is evident that they exhibit antifreeze activity and control the freezing process in the apoplast by binding to the surface of ice crystals; this limits ice growth, which otherwise could lead to freezing-induced dehydration. These glucanases retain partial hydrolytic activity at subzero temperatures, thereby providing plants with a potential means of defense against some psychrophilic pathogens. Furthermore, they are sugar-related enzymes that may play an important role in the carbohydrate modification processes contributing to osmoprotection and also osmotic inhibition of ice formation in the apoplast. Whereas this work provides experimental evidence on the contribution of glucanase in freezing tolerance in winter rye, further investigation is required to precisely clarify additional roles that glucanase may have in cold tolerance in planta.
MATERIALS AND METHODS
Plant Materials
Winter rye (Secale cereale L. cv Musketeer) seeds were sown at a rate of 5 g seed/15-cm pot of ProMix BX (Premier Horticulture). NA plants were grown at 20°C/16°C (day/night) with a 16-h photoperiod and a photosynthetic photon flux density of 300 μmol m−2 s−1 for 3 weeks. For cold acclimation, plants were transferred after 1 week to 5°C/2°C (day/night) with an 8-h photoperiod and the same irradiance for an additional 7 weeks. CA plants were watered as needed and fertilized weekly with 0.5 g L−1 of 20-20-20 (N-P-K) all-purpose fertilizer (Plant Products).
Apoplastic Protein Extraction and Quantification
Extraction of AFPs from CA and NA winter rye leaves was carried out using the previously described method (Hon et al., 1994). Protein concentrations were determined by the method of Bradford (1976) using bovine serum albumin as the standard.
Assay for β-1,3-Glucanase and β-1,3;1,4-Glucanase Activities
β-1,3-Glucanase hydrolytic activity was measured by determining the release of reducing sugars from laminarin of Laminaria digitata (Sigma) essentially as described by McFeeters (1980). The reaction containing 0.5% reduced laminarin, 20 mm sodium acetate (pH 5.5), and the purified enzyme in a total volume of 100 μL, was incubated at a certain temperature for 10 min. The reaction was stopped after 10 min by the addition of 3,5-dinitrosalycilic acid and boiling for 5 min. β-1,3;1,4-Glucanases were assayed as described previously (Akiyama et al., 1996). The standard reaction mixture contained a 0.25% (w/v) barley (Hordeum vulgare) 1,3;1,4-glucan source, 50 mm sodium acetate (pH 5.5), and the enzyme in a total volume of 100 μL and was incubated at a specific temperature. The reaction was stopped after 10 min by the addition of p-hydroxybenzoic acid hydrazide reagent and boiling for 5 min. The increase in concentration of reducing sugar was determined spectrophotometrically at 500 and 405 nm for the β-1,3-glucanase and β-1,3;1,4-glucanase assays, respectively. For the enzymatic assays carried out at temperatures below 25°C, the reaction components were prechilled and then mixed in a microcentrifuge tube placed in a refrigerated circulating bath of polyethylene glycol. One unit of activity is defined as the release of 1 mmol Glc equivalent min−1.
Antifreeze Activity Assay
Serial dilutions of purified glucanases containing 0.5, 1.0, 1.5, 2.0, 2.5, and 3.0 mg protein mL−1 were assayed for antifreeze activity by examining the morphology of ice crystals grown in solution using the previously described method (Hon et al., 1994).
Screening of cDNA Library and Sequencing
The glucanase cDNA sequences were isolated from a ZAP II library (Stratagene) constructed from poly(A)+ mRNA purified from the leaves of 8-week-old CA winter rye plants (Yeh et al., 2000). The library was screened with a near full-length cDNA encoding the barley β-1,3-glucanase isoenzyme GII (Høj et al., 1989; accession no. M23548). All the positive glucanase clones from the second screening were purified, converted to Bluescript SK plasmids (Stratagene), and the inserts sequenced.
Cloning of β-1,3-Glucanases from NA Winter Rye
Total RNA was extracted from NA 3-week-old plant leaves using the TriPure RNA isolation kit (Roche Diagnostics GmbH). First-strand cDNA was prepared by RT-PCR using 1 μg of the total RNA, the Moloney murine leukemia virus RT kit from Fermentus, and the oligo(dT)18 primer following the manufacturer's instructions. NA glucanases were amplified from the cDNA using PCR. The primer pair was designed to match the conserved regions coding for the N- and C-terminal regions of the ScGLU-3 clone. The cDNA sample was concentrated and used as a template in 50-μL PCRs containing 0.01 μm of each primer: GLB2MATF (5′-TGAATTCTTAGAACTGGATGGCGTAGG-3′) and GLB2ALLR (5′-GGAATTCCCATCGGCGTCTGTTAC-3′) and the High-Fidelity PCR Master mix (Roche Diagnostics GmbH). The PCR program comprised a denaturation step of 95°C for 1 min, followed by 34 cycles of 94°C for 40 s, 65°C for 60 s, and 72°C for 60 s. Because primers were expected to amplify a group of β-1,3-glucanases of different DNA sequences, PCR products were individualized by cloning into the pGEM-T-easy vector (Promega) and classified by restriction enzyme fingerprinting. Two different groups were identified and three representative clones were sequenced.
Northern Blotting
Total RNA (5 μg) was electrophoresed in a 1% denaturing formaldehyde gel and capillary transferred to nylon positive membranes (Roche Diagnostics GmbH). ScGLU-3 and ScGLU-1 cDNA probes were PCR labeled using a PCR digoxigenin (DIG) probe synthetic kit (Roche Diagnostics GmbH) following the manufacturer's instructions and the same PCR cycle used before. Membranes were prehybridized and probed with DIG-labeled glucanase using DIG Easy Hyb granule solution (Roche Diagnostics GmbH) at 45°C. After hybridization, membranes were washed twice with a buffer containing 2× SSC and 0.1% SDS at room temperature and then the membranes were washed twice with the high-stringency buffer (0.1% SSC containing 0.1% SDS) at 68°C. Detection was performed with CDP-Starter ready-to-use substrate (Roche Diagnostics GmbH) and chemiluminescence signals recorded by the Lumi-Imager (Florochem system).
Expression of ScGLU-1, ScGLU-2, ScGLU-3, ScGLUNA-1, and ScGLUNA-2 in Escherichia coli
Primers were designed to amplify by PCR the deduced mature peptide of the glucanase genes and fused in frame with GST of the pGEX-KG vector (Guan and Dixon, 1991). Restriction sites were introduced to the synthetic oligonucleotides to facilitate the cloning process of the sequences. ScGLU-2, ScGLU-3, ScGLUNA-1, and ScGLUNA-2 (cDNA sequences coding for β-1,3-glucanases) were amplified using GLB2MATF and the GLUB2MATR primers. ScGLU-1 (a cDNA sequence coding for β-1,3;1,4-glucanases) was amplified using the MATURA1U (5′-GGAATTCCGGTGGGCGTGTGCTAC-3′) and MATURA1L (5′-CGAATTCCGAACTCAATCAACTTCACTTGC-3′). The recombinant vectors were transformed into the E. coli DH5α strain and the positive colonies were identified. Following this, the recombinant plasmid was amplified and retransformed to the protease-deficient E. coli BL21 (DE3) cells (Novagen). One single colony harboring the pGEX-KG recombinant plasmid was selected and used to inoculate 10 mL of 2× yeast extract tryptone medium. Subsequently, the cultural media was incubated overnight and then used to inoculate 1 L of 2× yeast (Saccharomyces cerevisiae) extract tryptone medium. The culture was grown to obtain an optical density value at 600-nm wavelength of 1 before the addition of isopropyl-β-d-thiogalactopyranoside to 0.2 mm final concentration and then the culture was incubated for an additional 4 h at 37°C.
Purification of the Recombinant Protein
Cells were harvested by centrifugation and the recombinant protein was purified using 1 mL GSTrap FF columns following the manufacturer's instructions (Amersham Biosciences). The GST tag was cleaved out while the fusion protein is bound to the column using thrombin (80 units/mL in phosphate-buffered saline). The columns were incubated at room temperature for 18 h, then the proteins were eluted and the thrombin was removed from the solution using HiTrap Benzamidine FF columns following the manufacturer's instructions (Amersham Biosciences). The purified glucanases were dialyzed for 16 h at 4°C against 1 L of buffer composed of 50 mm ammonium acetate and 10 mm of EDTA, pH 5.5, and subsequently concentrated using the Amicon ultracentrifuge filter device with a Mr cutoff of 10,000 (Millipore).
SDS-PAGE and Immunoblots
Soluble proteins were separated by SDS-PAGE according to the method of Laemmli (1970) and blotted onto a nitrocellulose membrane using a mini-trans-blot electrophoretic transfer cell (Bio-Rad). Blots were blocked overnight at 4°C in 5% (w/v) skim milk powder in Tris-buffered saline, pH 7.5, and then incubated for 2 h at room temperature with the primary polyclonal antibody produced in rabbit against the purified apoplastic 32-kD β-1,3-glucanase (Antikainen et al., 1996) and used at a dilution of 1:7,500 in the blocking buffer, followed by washing with four 15-min washes in Tris-buffered saline plus 0.1% Tween 20. After incubating the blot with goat anti-rabbit IgG (H + L)-alkaline phosphatase conjugate (1:30,000; Sigma), positive immunoreactions were detected using 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium chloride (Amersham Biosciences).
DSC
One milligram of ScGLU-3 diluted in 50 mm sodium acetate buffer, pH 5.5, was analyzed using the Valerian Plotnikov DSC system (VP-DSC; MicroCal LLC). The analysis was carried out for 60 cycles over the temperature range of 1°C to 100°C.
Mass Spectrometry
The purified glucanase proteins were diluted to approximately 9 nmol mL−1 in 1:1 (v/v) acetonitrile and water containing 0.2% formic acid (v/v). The sample was infused at a rate of 10 μL min−1 and positive ion electrospray mass spectra were acquired with a Waters/Micromass Q-Tof Ultima GLOBAL mass spectrometer. Raw data (mass-to-charge ratio) were processed using MaxEnt1 to yield spectra on a true molecular mass scale.
Database Searching and Sequence Analysis
Identification of homologous glucanase sequences was carried out using BLASTN and BLASTP searches (Altschul et al., 1990) of GenBank nucleotide and amino acid sequence databases. A multiple sequence alignment was constructed using ClustalX (Thompson et al., 1997). Phylogenetic analysis was conducted using Molecular Evolutionary Genetic Analysis software, version 3.0 (Kumar et al., 2004). The distance between the amino acid sequences of the glucanases from different plant species were calculated based on the Dayhoff 250 matrix (Dayhoff, 1978) and the phylogenetic tree was constructed using the neighbor-joining method (Saitou and Nei, 1987). Putative signal peptides were identified using SignalP, version 1.1 (Nielsen et al., 1997; Center for Biological Sequence Analysis, Technical University of Denmark [http://www.cbs.dtu.dk/services/SignalP]).
Structural Modeling
Three-dimensional models were generated via the automated SWISS-MODEL server (Schwede et al., 2003). β-1,3-Glucanase from barley (PDB ID 1GHS_A) was the highest BLAST match to structures in the PDB for the winter rye β-1,3-glucanase sequences (ScGLU-2, ScGLU-3, ScGLUNA-1, and ScGLUNA-2), and was chosen as a structural template for homology modeling. Pairwise sequence identities with the template exceeded 94% for these sequences and there were no gaps in the sequence alignments, yielding high-quality structural models. An in-house flatness function algorithm was applied to the structural models to find highly planar surface regions, the largest of which was modeled as the putative IBS. Deepview/Swiss PDB Viewer, version 3.7 (Guex and Peitsch, 1997), was used to identify a regular pattern of surface atoms on the putative IBS. Ice models were obtained from the Web site (http://pout.cwru.edu/approximatelyfrank/afp1) and were previously constructed by Dalal and Sonnichsen (2000). ZDOCK 2.3 (Chen et al., 2003) was used for molecular docking and ranked generated binding configurations of the ScGLU-3 and ice using shape complementarity, desolvation energy, and electrostatics. A structural model for ScGLU-1 (β-1,3;1,4-β-glucanase from winter rye) was generated with the automated SWISS-MODEL server using a barley β-1,3;1,4-glucanase (PDB ID 1A10_A) as a homology modeling template with approximately 65% sequence identity.
Sequence data from this article can be found in the GenBank/EMBL databases under accession numbers AM181305 to AM181314.
Acknowledgments
We would like to thank Dr. G.B. Fincher, Department of Plant Science, University of Adelaide, Australia, for providing the barley β-1,3-glucanase probe used in this study and Dr. Thomas Hsiang, University of Guelph, for comments on the manuscript. We also thank Roy Satmaka, University of Waterloo, for technical assistance.
This work was supported by the Natural Sciences and Engineering Research Council of Canada (Discovery grants to M.G., B.A.M., and B.J.M.).
This paper is dedicated to the memory of Marilyn Griffith.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Mahmoud W.F. Yaish (myaish@uoguelph.ca).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.081935.
References
- Adams DJ (2004) Fungal cell wall chitinases and glucanases. Microbiology 150: 2029–2035 [DOI] [PubMed] [Google Scholar]
- Akiyama T, Kaku H, Shibuya N (1996) Purification and properties of a basic endo-1,3-β-glucanase from rice (Oryza sativa L.). Plant Cell Physiol 37: 702–705 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Antikainen M, Griffith M (1997) Antifreeze accumulation in freezing tolerant cereals. Physiol Plant 99: 423–432 [Google Scholar]
- Antikainen M, Griffith M, Zhang J, Hon WC, Yang D, Pihakaski-Maunsbach K (1996) Immunolocalization of antifreeze proteins in winter rye leaves, crowns, and roots by tissue printing. Plant Physiol 110: 845–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz A, Poinssot B, Daire X, Adrian M, Bézier A, Lambert B, Joubert JM, Pugin A (2003) Laminarin elicits defense responses in grapevine and induces protection against Botrytis cinerea and Plasmopara viticola. Mol Plant Microbe Interact 16: 1118–1128 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Browse J, Xin Z (2001) Temperature sensing and cold acclimation. Curr Opin Plant Biol 4: 241–246 [DOI] [PubMed] [Google Scholar]
- Chen R, Li L, Weng Z (2003) ZDOCK: an initial-stage protein-docking algorithm. Proteins 52: 80–87 [DOI] [PubMed] [Google Scholar]
- Collins T, Gerday C, Feller G (2005) Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol Rev 29: 3–23 [DOI] [PubMed] [Google Scholar]
- Collins T, Meuwis MA, Gerday C, Feller G (2003) Activity, stability and exibility in glycosidases adapted to extreme thermal environments. J Mol Biol 328: 419–428 [DOI] [PubMed] [Google Scholar]
- Collins T, Meuwis MA, Stals I, Claeyssens M, Feller G, Gerday C (2002) A novel family 8 xylanase: functional and physico-chemical characterization. J Biol Chem 277: 35133–35139 [DOI] [PubMed] [Google Scholar]
- Coombs J, Brenchley JE (2001) Characterization of two new glycosyl hydrolases from the lactic acid bacterium Carnobacterium piscicola strain BA. Appl Environ Microbiol 67: 5094–5099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal P, Sonnichsen FD (2000) Source of the ice-binding specificity of antifreeze protein type I. J Chem Inf Comput Sci 40: 276–284 [DOI] [PubMed] [Google Scholar]
- D'Amico S, Gerday C, Feller G (2004) Temperature adaptation of proteins: engineering mesophilic-like activity and stability in a cold-adapted alpha-amylase. J Mol Biol 332: 981–988 [DOI] [PubMed] [Google Scholar]
- Davail S, Feller G, Narinx E, Gerday C (1994) Purification, characterization, and sequence of the heat-labile subtilisin from the Antarctic Psychrophile bacillus TA41. J Biol Chem 269: 17448–17453 [PubMed] [Google Scholar]
- Dayhoff MO, Schwartz RM, Orcutt BC (1978) A model of evolutionary change in proteins. Matrices for detecting distant relationships. In MO Dayhoff, ed, Atlas of Protein Sequence and Structure, Vol 5. National Biomedical Research Foundation, Washington, DC, pp 345–358
- Feller G, Gerday C (1997) Psychrophilic enzymes: molecular basis of cold adaptation. Cell Mol Life Sci 53: 830–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller G, Lonhienne T, Deroanne C, Libioulle C, Beumen JV, Gerday C (1992) Purification, characterization, and nucleotide sequence of the thermolabile α-amylase from the Antarctic psychrotroph Alteromonas haloplanctis A23. J Biol Chem 267: 5217–5221 [PubMed] [Google Scholar]
- Georlette D, Blaise V, Collins T, D'Amico S, Gratia E, Hoyoux A, Marx JC, Sonan G, Feller G, Gerday C (2004) Some like it cold: biocatalysis at low temperatures. FEMS Microbiol Rev 28: 25–42 [DOI] [PubMed] [Google Scholar]
- Georlette D, Blaise V, Dohmen C, Bouillenne F, Damien B, Depiereux E, Gerday C, Uversky VN, Feller G (2003) Cofactor binding modulates the conformational stabilities and unfolding patterns of NAD (+)-dependent DNA ligases from Escherichia coli and Thermus scotoductus. J Biol Chem 278: 49945–49953 [DOI] [PubMed] [Google Scholar]
- Grenier J, Potvin C, Trudel J, Asselin A (1999) Some thaumatin-like proteins hydrolyse polymeric β-1,3-glucans. Plant J 19: 473–480 [DOI] [PubMed] [Google Scholar]
- Griffith M, Ala P, Yang DSC, Hon WC, Moffatt BA (1992) Antifreeze protein produced endogenously in winter rye leaves. Plant Physiol 100: 593–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith M, Yaish MW (2004) Antifreeze proteins in overwintering plants: a tale of two activities. Trends Plant Sci 9: 399–405 [DOI] [PubMed] [Google Scholar]
- Guan KL, Dixon JE (1991) Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem 192: 262–267 [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18: 2714–2723 [DOI] [PubMed] [Google Scholar]
- Guy CL, Huber JLA, Huber SC (1992) Sucrose phosphate synthase and sucrose accumulation at low temperature. Plant Physiol 100: 502–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochachka PA, Somero GN (1984) Biochemical Adaptations. Princeton University Press, Princeton
- Høj PB, Fincher GB (1995) Molecular evolution of plant β-glucan endohydrolases. Plant J 7: 367–379 [DOI] [PubMed] [Google Scholar]
- Høj PB, Hartman DJ, Morrice NA, Doan DN, Fincher GB (1989) Purification of (1,3)-β-glucan endohydrolase isoenzyme II from germinated barley and determination of its primary structure from a cDNA clone. Plant Mol Biol 13: 31–42 [DOI] [PubMed] [Google Scholar]
- Hon WC, Griffith M, Chong P, Yang D (1994) Extraction and isolation of antifreeze proteins from winter rye (Secale cereale L.) leaves. Plant Physiol 104: 971–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon WC, Griffith M, Mlynarz A, Kwok YA, Yang DCS (1995) Antifreeze proteins in winter rye are similar to pathogenesis-related proteins. Plant Physiol 109: 879–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyo AH, Forsberg CW (1999) A cold-active glucanase from the ruminal bacterium Fibrobacter succinogenes S85. Appl Environ Microbiol 65: 995–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamalainen EA (1974) Resistance in winter cereals and grasses to low-temperature parasitic fungi. Annu Rev Phytopathol 12: 281–302 [Google Scholar]
- Kaufmann E, Giesler N, Weber K (1984) SDS-PAGE strongly overestimates the molecular masses of the neurofilament proteins. FEBS Lett 170: 81–84 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5: 150–163 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Livingston DP, Premakumar R (2002) Apoplastic carbohydrates do not account for differences in freezing tolerance of two winter-oat cultivars that have been second phase cold-hardened. Cereal Res Commun 30: 375–381 [Google Scholar]
- Lonhienne T, Gerday C, Feller G (2000) Psychrophilic enzymes: revisiting the thermodynamic parameters of activation may explain local flexibility. Biochim Biophys Acta 1543: 1–10 [DOI] [PubMed] [Google Scholar]
- Madura JD, Baran K, Wierzbicki A (2000) Molecular recognition and binding of thermal hysteresis proteins to ice. J Mol Recognit 13: 101–113 [DOI] [PubMed] [Google Scholar]
- Marchler–Bauer A, Anderson JB, Cherukuri PF, DeWeese–Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, et al (2005) CDD: a conserved domain database for protein classification. Nucleic Acids Res 33: D192–D196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CJ (1997) Cold-adapted enzymes. Trends Biotechnol 15: 359–364 [DOI] [PubMed] [Google Scholar]
- McFeeters RF (1980) A manual method for reducing sugar determinations with 2,2′-bicinchoninate reagent. Anal Biochem 103: 302–306 [DOI] [PubMed] [Google Scholar]
- Meins FJ, Neuhaus JM, Sperisen C, Ryals J (1992) In T Boller, F Meins, eds, Genes Involved in Plant Defense. Springer-Verlag, Berlin, pp 245–282
- Nicholas KB, Nicholas HB Jr, Deerfield DW II (1997) GeneDoc: analysis and visualization of genetic variation. EMBNET News 4: 1–4 [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10: 1–6 [DOI] [PubMed] [Google Scholar]
- Pihakaski-Maunsbach K, Griffith M, Antikainen M, Maunsbach AB (1996) Immunogold localization of glucanase-like antifreeze protein in cold acclimated winter rye. Protoplasma 191: 115–125 [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sakai A, Larcher W (1987) Frost Survival of Plants. Springer-Verlag, New York, pp 31–33
- Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31: 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siminovitch D (1981) Common and disparate elements in the processes of adaptation of herbaceous and woody plants to freezing: a perspective. Cryobiology 18: 166–185 [DOI] [PubMed] [Google Scholar]
- Simmons CR (1994) The physiology and molecular biology of plant 1,3-β-d-glucanases and 1,3;1,4-β-d-glucanases. CRC Crit Rev Plant Sci 13: 325–387 [Google Scholar]
- Snider CS, Hsiang T, Zhao GY, Griffith M (2000) Role of ice nucleation and antifreeze activities in pathogenesis and growth of snow molds. Phytopathology 90: 354–361 [DOI] [PubMed] [Google Scholar]
- Stawiski EW, Baucom AE, Lohr SC, Gregoret M (2000) Predicting protein function from structure: unique structural features of proteases. Proc Natl Acad Sci USA 97: 3954–3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung DY, Kaplan F, Lee KJ, Guy CL (2003) Acquired tolerance to temperature extremes. Trends Plant Sci 8: 179–187 [DOI] [PubMed] [Google Scholar]
- Taguchi S, Ozaki A, Momose H (1998) Engineering of a cold-adapted protease by sequential random mutagenesis and a screening system. Appl Environ Microbiol 64: 492–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi T, Nakamura M, Hayashi H, Inatsugi R, Yano R, Nishida I (2003) The leaf-order-dependent enhancement of freezing tolerance in CA Arabidopsis rosettes is not correlated with the transcript levels of the cold-inducible transcription factors of CBF/DREB1. Plant Cell Physiol 44: 922–931 [DOI] [PubMed] [Google Scholar]
- Thibaud MC, Gineste S, Nussaume L, Robaglia C (2004) Sucrose increases pathogenesis-related PR-2 gene expression in Arabidopsis thaliana through an SA-dependent but NPR1-independent signalling pathway. Plant Physiol Biochem 42: 81–88 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese JN, Garrett TP, Colman PM, Chen L, Hoj PB, Fincher GB (1994) Three-dimensional structures of two plant beta-glucan endohydrolases with distinct substrate specificities. Proc Natl Acad Sci USA 91: 2785–2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner L, Junttila O (1999) Cold-induced freezing tolerance in Arabidopsis. Plant Physiol 120: 391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh S, Moffatt BA, Griffith M, Xiong F, Yang DS, Wiseman SB, Sarhan F, Danyluk J, Xue YQ, Hew CL, et al (2000) Chitinase genes responsive to cold encode antifreeze proteins in winter cereals. Plant Physiol 124: 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XM, Griffith M (1999) Antifreeze proteins in winter rye leaves form oligomeric complexes. Plant Physiol 119: 1361–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]