Abstract
Polyamine oxidase (PAO) is a flavin adenine dinucleotide-dependent enzyme involved in polyamine catabolism. Animal PAOs oxidize spermine (Spm), spermidine (Spd), and/or their acetyl derivatives to produce H2O2, an aminoaldehyde, and Spd or putrescine, respectively, thus being involved in a polyamine back-conversion pathway. On the contrary, plant PAOs that have been characterized to date oxidize Spm and Spd to produce 1,3-diaminopropane, H2O2, and an aminoaldehyde and are therefore involved in the terminal catabolism of polyamines. A database search within the Arabidopsis (Arabidopsis thaliana) genome sequence showed the presence of a gene (AtPAO1) encoding for a putative PAO with 45% amino acid sequence identity with maize (Zea mays) PAO. The AtPAO1 cDNA was isolated and cloned in a vector for heterologous expression in Escherichia coli. The recombinant protein was purified by affinity chromatography on guazatine-Sepharose 4B and was shown to be a flavoprotein able to oxidize Spm, norspermine, and N1-acetylspermine with a pH optimum at 8.0. Analysis of the reaction products showed that AtPAO1 produces Spd from Spm and norspermidine from norspermine, demonstrating a substrate oxidation mode similar to that of animal PAOs. To our knowledge, AtPAO1 is the first plant PAO reported to be involved in a polyamine back-conversion pathway.
Polyamines putrescine (Put), spermidine (Spd), and spermine (Spm) are small aliphatic amines commonly found in both prokaryotic and eukaryotic cells with an essential role during growth and differentiation (Cohen, 1998; Bagni and Tassoni, 2001; Thomas and Thomas, 2003; Wallace et al., 2003). In higher plants as well, polyamines are key players in a range of developmental processes and are implicated in responses to various types of abiotic stress (such as potassium deficiency, osmotic shock, drought, and salt stress) and plant-pathogen interactions (both incompatible and compatible; Bouchereau et al., 1999; Walters, 2003).
Several other di- and polyamines are present in plants and microorganisms, such as the diamines 1,3-diaminopropane (Dap) and cadaverine. Furthermore, uncommon polyamines, such as norspermidine (Nor-Spd), homospermidine, norspermine (Nor-Spm), homospermine, thermospermine, caldopentamine, caldohexamine, homocaldopentamine, and homocaldohexamine, are abundant in the extreme thermophilic bacterium Thermus thermophilus and have also been detected in bacteria, algae, fungi, animals, and higher plants (Cohen, 1998). In plants, a putative role for these molecules in growth, differentiation, and stress tolerance has been hypothesized (Rodriguez-Garay et al., 1989; Bagga et al., 1997; Koc et al., 1998).
Intracellular polyamine pools appear to be sensitively regulated by various homeostatic processes that include pathways for polyamine biosynthesis, catabolism, and transport across the cell membrane (Wallace et al., 2003). Polyamine oxidase (PAO) is a flavin adenine dinucleotide (FAD)-dependent enzyme involved in the catabolic pathway of polyamines (Cona et al., 2006). It catalyzes the oxidation of Spm, Spd, and/or their acetylated derivatives at the secondary amino group (Federico and Angelini, 1991; Wang et al., 2001; Wu et al., 2003; Cona et al., 2006). The chemical identity of the products of PAO reactions depends on the enzyme source and reflects the mode of substrate oxidation. Plant and bacterial PAOs oxidize the carbon on the endo-side of the N4-nitrogen of Spd and Spm, producing 4-aminobutanal and N-(3-aminopropyl)-4-aminobutanal, respectively, in addition to Dap and H2O2 (Federico and Angelini, 1991). Animal PAOs and yeast (Saccharomyces cerevisiae) spermine oxidase (Fms1) oxidize the carbon on the exo-side of N4-nitrogen of N1-acetyl-Spm, N1-acetyl-Spd, and N1,N12-bis-acetyl-Spm to produce Spd, Put, and N1-acetyl-Spd, respectively, in addition to 3-acetamidopropanal and H2O2 (Landry and Sternglanz, 2003; Vujcic et al., 2003; Wu et al., 2003). In this catabolic pathway, polyamine acetylation is catalyzed by the tightly regulated Spd/Spm N1-acetyltransferase (SSAT), which is the rate-limiting factor (Wallace et al., 2003). Animal spermine oxidase (SMO) and Fms1 also oxidize the carbon on the exo-side of N4-nitrogen of Spm to produce Spd, 3-aminopropanal, and H2O2 (Wang et al., 2001; Vujcic et al., 2002; Cervelli et al., 2003; Landry and Sternglanz, 2003). Thus, animal PAO and SMO, as well as yeast Fms1, are all involved in a polyamine back-conversion pathway (Seiler, 2004) at variance with plant PAOs characterized thus far, which are involved in the terminal catabolism of polyamines. However, the conversion of N1-acetyl-Spd and of Spd to Put has also been hypothesized in plants (De Agazio et al., 1995; Tassoni et al., 2000), although none of the enzymes purified to date match the catalytic profile of the PAO involved in the polyamine back-conversion pathway.
Plant PAOs, which are highly expressed mainly in monocots, have been purified and partially characterized in a few species (Federico and Angelini, 1991; Šebela et al., 2001). PAO from maize (Zea mays; MPAO), the most studied member of this enzyme class, is a 53-kD monomeric glycoprotein containing one molecule of FAD (Federico and Angelini, 1991; Tavladoraki et al., 1998; Binda et al., 1999; Šebela et al., 2001). MPAO has been found mainly in the apoplast by means of biochemical and immunocytochemical approaches (Federico and Angelini, 1991; Šebela et al., 2001; Cona et al., 2005). The MPAO gene family includes three gene copies that encode for identical secretory proteins (Cervelli et al., 2000). Barley (Hordeum vulgare) PAO (BPAO) has also been characterized and two genes have been isolated (BPAO1 and BPAO2), both of which encode proteins bearing a cleavable N-terminal signal peptide (Cervelli et al., 2001). Interestingly, contrary to the extracellular localization of MPAO, symplastic localization for BPAO2 has been demonstrated. In particular, it has been suggested that a C-terminal extension of eight amino acid residues present in a BPAO2 sequence with respect to the MPAO and BPAO1 sequences is a signal for protein targeting into the plant vacuole (Cervelli et al., 2004).
Recently, most biochemical, genetic, and molecular studies have been carried out in the model plant Arabidopsis (Arabidopsis thaliana) for reasons of genome simplicity, short reproductive cycle, ease of transformation, small plant size, availability of insertional mutants of Arabidopsis plants, and the abundant amount of information available on its metabolic pathways. In particular, in the last few years, a lot of information has been gathered on polyamine metabolic pathways in Arabidopsis (Panicot et al., 2002; Illingworth et al., 2003; Imai et al., 2004; Alcázar et al., 2005). However, although several genes encoding for polyamine biosynthetic enzymes have been characterized in Arabidopsis, a gene encoding for a PAO has not been studied in this plant species. Recently, a gene encoding for a putative PAO (At5g13700; GenBank accession no. NM_121373) with a 45% amino acid identity with MPAO has been reported in Arabidopsis (Cervelli et al., 2001), but its biochemical properties are not known yet. To determine the catalytic properties and the physiological role of a PAO coming from a dicotyledonous plant, in general, and from Arabidopsis, in particular, we have undertaken a study on this Arabidopsis gene. Here, we present evidence that this gene indeed encodes for a PAO, which differs from MPAO in substrate specificity, catalytic constants, and mode of substrate oxidation. In particular, we demonstrate that this enzyme displays a substrate oxidation mode similar to that of animal PAO, producing Spd from Spm and Nor-Spd from Nor-Spm. Thus, this Arabidopsis PAO is involved in a polyamine back-conversion pathway.
RESULTS
Description of cDNA Encoding for AtPAO1
A search of the Arabidopsis database with the amino acid sequence of MPAO has revealed the presence of five cDNAs encoding for putative PAOs. These include AtPAO1 (formerly APAO [Cervelli et al., 2001]; At5g13700; GenBank accession no. NM_121373; Fig. 1), AtPAO2 (At2g43020; GenBank accession no. AF364952), AtPAO3 (At3g59050; GenBank accession no. AY143905), AtPAO4 (At1g65840; GenBank accession no. AF364953), and AtPAO5 (At4g29720; GenBank accession no. AK118203). AtPAO1 has a 45% identity (67% similarity) at the amino acid level with MPAO (Fig. 2) and only a 19% to 24% identity (42%–46% similarity) with the other four putative Arabidopsis PAOs. AtPAO2, AtPAO3, and AtPAO4 display low sequence identity (23% identity and 46% similarity) with MPAO and elevated sequence identity (58%–85% identity and 72%–88% similarity) to each other. On the contrary, AtPAO5 has low sequence identity not only with MPAO (23% sequence identity and 38% similarity), but also with AtPAO2, AtPAO3, and AtPAO4 (23% identity and 40%–42% similarity). Based on the high sequence identity with MPAO, we have chosen to characterize AtPAO1 first.
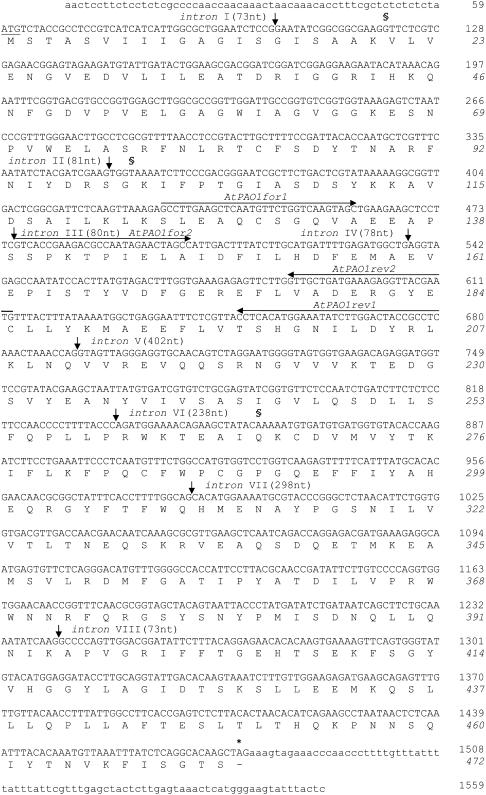
Figure 1.
Nucleotide sequence and translated protein sequence of AtPAO1 cDNA. Nucleotides and amino acids are numbered on the right. Numbering of amino acids is indicated in italics. Nucleotide sequences in capital letters correspond to the coding region. Small-lettered sequences correspond to the 5′- and 3′-untranslated regions. Horizontal arrows indicate the position, the length, and the orientation of the oligonucleotides used in experiments of RT-PCR analysis. Vertical arrows indicate the position of the introns. Numbers in parentheses indicate the length of introns. The first codon is underlined. The asterisk denotes the stop codon. Symbol § indicates the positions in the cDNA clone obtained from RIKEN (clone pda 11656) in which changes in the nucleotide sequence have been observed with respect to that reported in the database.
Figure 2.
Alignment of the amino acid sequence of animal and plant FAD-dependent amine oxidases. Multialignment was done using the program ClustalW sequence alignment (Altschul et al., 1990). Numbering of amino acid residues is shown at the right side. In MPAOs, numbering starts from the first amino acid of the mature protein. The signal peptide of MPAO is underlined. Percentage of identity refers to AtPAO1. Identical residues are indicated by gray boxes. The position of the Cys residue that is covalently linked to the FAD in MAO is indicated by the symbol #. Residues in MPAO putatively involved in the catalytic activity are labeled by the asterisk symbol (*), and the ones composing the tunnel entrance (carboxylate ring and aromatic portion; Binda et al., 2001) are labeled by the symbol ▾. MPAO, Maize PAO; mSMO, murine spermine oxidase; mPAO, murine PAO; Fms1, yeast spermine oxidase.
A database search has revealed a high amino acid sequence identity of AtPAO1 also with three putative PAOs of rice (Oryza sativa), rPAO1 (GenBank accession no. XP_470573), rPAO2 (GenBank accession no. XP_450669), and rPAO3 (GenBank accession no. XP_450667), being 40%, 43%, and 44%, respectively. These three putative PAOs from rice also have high sequence identity with MPAO (73%, 81%, and 79%, respectively). Interestingly, AtPAO1 has very high sequence identity (74% identity and 83% similarity) with a putative PAO from tobacco (Nicotiana tabacum), the sequence of which has recently been submitted to the database (nPAO; GenBank accession no. AB200262). On the contrary, the sequence identity (similarity) of AtPAO1 with murine SMO (mSMO), murine PAO (mPAO), and yeast Fms1 was found to be low, at 23% (37%), 20% (39%), and 19% (36%), respectively (Fig. 2).
To determine the possible subcellular localization of AtPAO1, the amino acid sequence has been analyzed by PSORT (www.psort.org). This analysis predicted the presence of an uncleavable signal peptide of 18 amino acid residues with possible transmembrane regions at the N terminus of the protein. However, sequence alignment of AtPAO1 with all the other PAOs that have been sequenced so far has indicated involvement of this region in FAD binding (Wu et al., 2003), thus excluding a targeting role. These considerations suggest an intracellular localization for AtPAO1, whereas in MPAO a signal peptide determines protein translocation through the secretory pathway with a final localization in the extracellular space. Furthermore, PSORT analysis of the AtPAO1 sequence did not reveal the presence of any signal peptide for protein targeting to a specific intracellular compartment, thus suggesting cytosolic localization.
The promoter region of AtPAO1 was analyzed for the presence of putative cis-acting regulatory elements. This analysis indicated the presence of several light-responsive elements, as well as putative responsive elements to ethylene, methyl jasmonate, heat, and wounding.
Cloning of cDNA Encoding AtPAO1
AtPAO1 cDNA was obtained from the expressed sequence tag (EST) bank of RIKEN BioResource Center (Seki et al., 1998, 2002) with the aim of characterizing the catalytic properties of the corresponding protein. The sequencing of this cDNA, when compared to the Arabidopsis genomic sequence, revealed the lack of one nucleotide at position 859, as well as the presence of a conservative mutation at position 356 (GGT to GGC) and a nonconservative one at position 120 (GTT to ATT; Fig. 1). Consequently, to clone the correct AtPAO1 cDNA by reverse transcription (RT)-PCR, the presence of AtPAO1 mRNA was initially verified in various plant organs (leaves, stem, inflorescences) by RT-PCR analysis using two pairs of internal gene-specific oligonucleotides (AtPAO1for1/AtPAO1rev1 and AtPAO1for2/AtPAO1rev2; Fig. 1). To obtain a detectable amount of amplification product, a second amplification step was necessary for all organs tested using the same or nested primers (data not shown). This suggests that AtPAO1 mRNA is present only in very low amounts in these organs of light-grown Arabidopsis plants. This has been confirmed by northern-blot analysis using a radiolabeled AtPAO1-specific probe, which repetitively failed to detect AtPAO1 mRNA accumulation in these organs (data not shown).
Despite low expression levels, the whole coding region of AtPAO1 was amplified by RT-PCR from Arabidopsis leaves using sequence-specific primers. These primers were designed in such a way as to allow AtPAO1 cDNA cloning in the pPICZαA vector for heterologous expression in the Pichia pastoris culture medium. The P. pastoris expression system was chosen because it has allowed us to obtain high accumulation levels of recombinant MPAO in the culture medium (Polticelli et al., 2005). Also, in this case, two PCR steps were necessary to obtain an amplification product, which was finally cloned in the pPICZαA vector (AtPAO1-pPIC construct). The culture medium and cellular extracts of transformed P. pastoris cells were examined at various time intervals for the presence of the recombinant AtPAO1 by western-blot analysis using an anti-6-His tag antibody and by enzymatic assays using various polyamines (Spd, Spm, Put) as substrates and at various pH values (6.0, 7.5, 8.5). However, these analyses did not show recombinant AtPAO1 accumulation under any of the various culture conditions tested (pH of the culture medium, growth, temperature). The lack of recombinant AtPAO1 accumulation in the P. pastoris expression system could be due to the presence of A-T-rich regions in the AtPAO1 cDNA, which may constitute polyadenylation signals resulting in the formation of incomplete mRNAs. Instability of the recombinant protein in this system cannot be ruled out.
Heterologous Expression of AtPAO1 in Escherichia coli
To determine the catalytic properties of AtPAO1, heterologous expression in Escherichia coli has also been attempted using the pET17b vector, which guides cytoplasmic expression of recombinant proteins. Soluble lysates of transformed bacteria with the AtPAO1-pET17b plasmid have been tested for PAO activity at various pH values and using various nonacetylated and acetylated polyamines as substrates. This analysis has shown the presence of PAO activity using Spm, Nor-Spm, and N1-acetyl-Spm as substrates (Vmax Nor-Spm > Vmax Spm >> Vmax N1-acetyl-Spm) in the bacterial cultures treated with 0.4 mm isopropylthio-β-galactoside (IPTG) for recombinant protein expression and not in the nontreated ones. On the contrary, PAO activity has not been detected using Put, Spd, and N1-acetyl-Spd as substrates in the IPTG-treated bacterial cultures (data not shown). The accumulation levels of recombinant AtPAO1 were higher when the expression was induced at 24°C (approximately 12 units/L of culture) than when induced at 30°C (approximately 4 units/L of culture). Furthermore, a preliminary study on the pH dependence of AtPAO1 enzyme activity in the bacterial extracts showed the highest enzymatic activity at pH 8.0 for all three substrates tested.
Purification of Recombinant AtPAO1 from E. coli Cellular Extracts
Purification of recombinant AtPAO1 from bacterial extracts has been initially attempted by affinity chromatography using a resin charged with Ni2+. However, despite the addition of a sequence encoding for a 6-His tag at the 3′ terminus of AtPAO1 cDNA, recombinant protein was not bound on the resin under any of the binding conditions tested (variations in pH and/or ionic strength). To verify whether the 6-His tag was indeed present in recombinant protein, the total bacterial extracts, the fraction of the total soluble proteins, and the inclusion bodies have been analyzed by western-blot analysis using the anti-6-His antibody (Fig. 3). This analysis proved the presence of a large amount of recombinant AtPAO1 with an intact 6-His tag in the inclusion bodies (Fig. 3). On the contrary, the anti-6-His antibody did not recognize any protein in the cellular soluble extracts, despite the presence of an elevated amount of PAO activity in these extracts (Fig. 3). These results suggest that the 6-His tag may be subjected to proteolysis in the bacterial cytoplasm while remaining intact in the inclusion bodies.
Figure 3.
Expression of AtPAO1 in E. coli. Total cellular extracts (T), total soluble proteins (S), and inclusion bodies (B) from E. coli transformed with the AtPAO1-pET17b construct have been analyzed by immunoblotting using an anti-6-His-tag antibody. Expression of recombinant protein in E. coli has been induced by 0.4 mm IPTG at 24°C for 5 h (I). A part of the same culture was allowed to grow for the same time period in the absence of IPTG (NI) as a control. The various fractions have been normalized for the volume of the initial culture. MPAO, Recombinant MPAO expressed in P. pastoris. A representative experiment, which has been repeated twice, is shown.
To purify recombinant protein from bacterial extracts, a cyanogen bromide (CNBr)-activated Sepharose on which guazatine was immobilized has also been used. Guazatine is a potent competitive inhibitor of MPAO, pig liver PAO, mSMO, and mPAO (Federico et al., 2001; Cona et al., 2004; Bianchi et al., 2006). Furthermore, preliminary studies indicated that this compound is also an efficient competitive inhibitor of recombinant AtPAO1 (see above). This resin allowed the one-step purification (3 mg/L of culture) of the recombinant protein to electrophoretic homogeneity (Fig. 4A). This protein has an apparent molecular mass of 52.5 kD, which is similar to that predicted for AtPAO1 from the amino acid sequence and to that of recombinant AtPAO1 present in the inclusion bodies (Fig. 4B). Furthermore, as expected from the analysis of the amino acid sequence of the two proteins, recombinant AtPAO1 migrates faster than recombinant MPAO expressed in P. pastoris (calculated molecular mass of 6-His-tagged MPAO is 54.5 kD; Fig. 4A).
Figure 4.
Analysis of the purified recombinant AtPAO1. A, Recombinant AtPAO1, purified from E. coli using the resin of guazatine, was analyzed by SDS-PAGE and stained by Coomassie Brilliant Blue (APAO). E, Bacterial extract before application to the guazatine resin. B, Inclusion bodies from E. coli expressing (I) or not expressing (NI) recombinant AtPAO1 have also been analyzed by SDS-PAGE and stained by Coomassie Brilliant Blue. MPAO, Purified recombinant MPAO expressed in the culture medium of P. pastoris. M, Molecular mass marker (MBI Fermentas).
The purified protein was tested for the presence of the 6-His tag through western-blot analysis, using the same amount of recombinant MPAO expressed in P. pastoris as a control. The anti-6-His antibody did not recognize any 6-His-tagged protein in the fraction of purified recombinant AtPAO1, whereas it recognized recombinant MPAO (data not shown), thus confirming that the 6-His tag in recombinant AtPAO1 is subjected to proteolysis. This proteolysis did not cause a detectable change in the molecular mass of the enzyme by SDS-PAGE analysis (Fig. 4), suggesting that the removed proteolytic fragment might be rather small.
The purified protein displayed the characteristic UV-visible spectrum of the oxidized flavoproteins with three absorbance peaks at 280, 380, and 460 nm (Fig. 5, solid line). Addition of saturating amounts of Spm converted the spectrum of the fully oxidized form of FAD to that of the fully reduced form (Fig. 5, dotted line), as indicated by the decrease in the absorbance bands in the visible range (at 365 and 450 nm). This suggests that all FAD molecules in the purified enzyme are catalytically active. Precipitation of purified AtPAO1 with TCA resulted in the release of the cofactor into the supernatant, suggesting a noncovalent linkage to the protein. This is in agreement with the presence of a Ser residue at position 367, which in MAO-A and MAO-B is involved in covalent binding to the isoalloxazine ring of the FAD through a Cys residue (Edmondson et al., 2004). His and Tyr residues have also been shown to be involved in covalent binding to the flavin ring in some flavoenzymes (Edmondson and Newton-Vinson, 2001), whereas until now such a covalent linkage has not been observed for Ser residues.
Figure 5.
Spectra of FAD in the fully oxidized and fully Spm-dependent reduced recombinant AtPAO1. Absorbance spectra were recorded in 100 mm Tris-HCl, pH 8.0, at 22°C. To obtain the spectrum of the FAD in the fully reduced AtPAO1, a saturating amount of Spm (4 mm) was added in the fully oxidized recombinant enzyme.
Partial Biochemical Characterization of Recombinant AtPAO1
Analysis of the catalytic constants of purified recombinant AtPAO1 has indicated that this enzyme oxidizes Spm (Table I) but not Spd (data not shown), in agreement with data from the analysis of the bacterial lysates. This substrate specificity is different from that of MPAO, which is active with both Spd and Spm (Polticelli et al., 2005) and similar to that of mSMO, which oxidizes only Spm and not Spd (Cervelli et al., 2003). Furthermore, analysis of the catalytic properties of the purified enzyme has determined a kcat value for Spm of 2.7 s−1, which is approximately 20-fold lower than that of the recombinant MPAO expressed in P. pastoris (54 s−1; Polticelli et al., 2005) and similar to that of the recombinant mSMO expressed in E. coli (4.5 s−1; Cervelli et al., 2003). Biochemical characterization of recombinant AtPAO1 has also determined a Km value for Spm of 0.11 mm, which is 100-fold higher than that of recombinant MPAO (1.6 μm; Polticelli et al., 2005) and similar to that of recombinant mSMO (0.09 mm; Cervelli et al., 2003) for the same substrate. These data suggest that recombinant AtPAO1 is a less efficient enzyme (kcat/Km = 24.1 s−1 mm−1) than recombinant MPAO (kcat/Km = 23.9 × 103 s−1 mm−1) and similarly efficient to mSMO (kcat/Km = 50.0 s−1 mm−1) in catalyzing Spm oxidation.
Table I.
Kinetic constants of Spm, Nor-Spm, and N1-acetyl-Spm oxidation by recombinant AtPA01 expressed in E. coli
| Substrate | kcata | Kma | kcat/Km |
|---|---|---|---|
| s−1 | mm | s−1 mm−1 | |
| Spm | 2.7 ± 0.3 | 0.11 ± 0.02 | 24.1 |
| Nor-Spm | 6.9 ± 1.3 | 0.09 ± 0.01 | 81.4 |
| N1-acetyl-Spm | 0.2 ± 0.4 | 0.47 ± 0.02 | 0.5 |
Enzymatic activity of recombinant AtPAO1 has been determined in 100 mm Tris-HCl, pH 8.0, using a constant O2 concentration at the air-saturated level and an amine substrate concentration either saturating (for apparent kcat determination) or varying between 50 and 400 μm for Spm and N1-acetyl-Spm and between 10 and 100 μm for Nor-Spm (for apparent Km determination). Data are mean ± se of at least three independent experiments.
Interestingly, recombinant AtPAO1 has a kcat/Km value for Nor-Spm that is about 4-fold higher than that for Spm, indicating that Nor-Spm is the best substrate for recombinant AtPAO1 in vitro (Table I). On the contrary, recombinant mSMO expressed in E. coli was not active with Nor-Spm and recombinant MPAO expressed in P. pastoris was able to oxidize Nor-Spm with a kcat value of 5.5 s−1, which is about 10-fold lower than that for Spm. Recombinant AtPAO1 also oxidizes N1-acetyl-Spm with a kcat/Km value for this substrate of 0.5 s−1 mm−1, which is about 150-fold lower than that for Nor-Spm and 50-fold lower than that for Spm (Table I).
The pH dependence of purified recombinant enzyme catalytic activity has also been examined using Spm, Nor-Spm (Fig. 6), or N1-acetyl-Spm (data not shown) as a substrate. The results obtained show similar pH dependence for all three substrates. In particular, the catalytic activity increases with the increase in pH, reaching a maximum at around pH 8.0, similar to what has been observed with crude bacterial extracts. At higher pH, catalytic activity of the recombinant enzyme diminishes (Fig. 6). The pH dependence of recombinant enzyme catalytic activity for all the three tested substrates can be described in terms of deprotonation of an ionizable group with a pKa value of 6.5 to 6.8 responsible for the acidic side of the bell-shaped dependence (pK1; Fig. 6) and an ionizable group with a pKa value of 9.1 to 9.2 responsible for the alkaline side (pK2; Fig. 6). A similar bell-shaped pH dependence of the catalytic activity has also been observed for MPAO (Polticelli et al., 2005) and mSMO (P. Mariottini, R. Federico, and P. Tavladoraki, unpublished data). However, whereas the pH of maximal catalytic activity of AtPAO1 is similar to that of mSMO (pH optimum 8.5), with which the AtPAO1 has a low sequence homology, it is different from that of MPAO (pH optimum 6.5), with which the AtPAO1 has a high sequence homology.
Figure 6.
Catalytic activity of recombinant AtPAO1 as a function of pH. Apparent kcat values of recombinant AtPAO1 purified from E. coli toward Spm and Nor-Spm were calculated at various pH values using saturating concentrations of amine substrate and O2. Data are expressed as percent of maximum activity. Continuous lines represent the best fit of the experimental data. Each point represents the mean value from at least three independent experiments and the bars indicate the se.
The AtPAO1 inhibition constants of a number of animal and plant PAO inhibitors have also been determined (Table II). Our data demonstrate that, similar to MPAO (Cona et al., 2004; Bianchi et al., 2006), guazatine, N-prenylagmatine, and 1,12-diaminododecane are good competitive inhibitors of AtPAO1. MDL72527, 1,8-diaminooctane, and Spd also competitively inhibit recombinant AtPAO1 activity, although with Ki values higher than those of guazatine, N-prenylagmatine, and 1,12-diaminododecane (Table II). The differences in the Ki values between 1,12-diaminododecane and 1,8-diaminooctane, which display a similar chain length to that of Spm and Spd, respectively, reflect the AtPAO1 specificity for Spm and not for Spd.
Table II.
Inhibition constants of recombinant AtPA01 by various plant and animal PAO inhibitors
| Compound | Kia |
|---|---|
| μm | |
| Guazatine | 0.7 ± 0.4 |
| N-Prenylagmatine | 4.0 ± 0.5 |
| 1,12-Diaminododecane | 11.7 ± 0.8 |
| MDL72527 | 137.3 ± 32.7 |
| Agmatine | 147.0 ± 8.2 |
| 1,8-Diaminooctane | 293.0 ± 30.5 |
| Spermidine | 546.0 ± 40.0 |
Ki values were determined using Spm as substrate in 100 mm Tris-HCl, pH 8.0, for all inhibitors except in the case of 1,12-diaminododecane, for which the Ki value was determined in 100 mm Tris-HCl, pH 7.0. The Km for Spm in 100 mm Tris-HCl, pH 7.0, was 200 μm. Data are mean ± se of three independent experiments.
Characterization of the Reaction Products of AtPAO1
The similarity of AtPAO1 substrate specificity to that of mSMO (i.e. Spm and not Spd oxidation) prompted us to determine the mode of AtPAO1 substrate oxidation by analyzing the polyamine products. In particular, it has been examined whether AtPAO1 oxidizes Spm and Nor-Spm in a similar mode to that of MPAO, producing Dap among the other reaction products, or similar to that of animal PAO and SMO producing Spd from Spm and Nor-Spd from Nor-Spm.
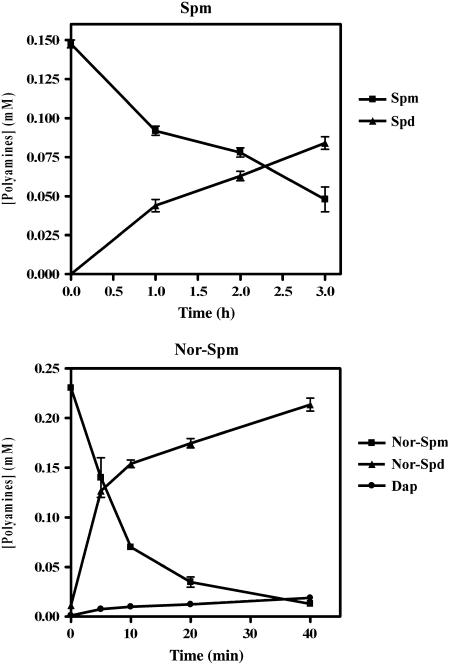
Analysis by HPLC of AtPAO1 polyamine reaction products using Spm as a substrate showed production of an increasing amount of a substance with the retention time of Spd (t = 21.3 min) in parallel with a decrease in the amount of Spm (t = 24.4 min). The formation of a product with the retention time of Dap (t = 16.1 min) was not observed (Figs. 7 and 8). Similarly, analysis of the reaction products using Nor-Spm (t = 24.1 min) as a substrate showed the formation of an increasing amount of a product with a slightly shorter retention time (t = 21.1 min) than that of Spd (Fig. 8). This product most probably corresponds to Nor-Spd, which, having a charge-to-mass ratio slightly smaller than that of Spd, should have a shorter retention time than the latter under our HPLC conditions. Nor-Spd could not be used as a standard because it is not commercially available. Interestingly, in the case of Nor-Spm, the formation of a very small amount of a product with the retention time of Dap was also observed (Figs. 7 and 8). As indicated in Figure 7, less time was necessary for the complete oxidation of Nor-Spm (about 40 min) than for the complete oxidation of Spm (about 3 h). This is in line with the calculated differences in the catalytic constants for these two substrates.
Figure 7.
Time course of polyamine oxidation by AtPAO1 and of reaction product accumulation. Purified recombinant AtPAO1 (1 nm) was incubated with 0.15 mm of Spm or 0.25 mm of Nor-Spm for up to 3 h or 40 min, respectively. Aliquots were analyzed for polyamine levels at various time intervals. Data are from a single representative experiment, which was repeated twice. Each point represents the mean value from two independent analyses of polyamine levels and the bars indicate the se.
Figure 8.
HPLC analysis of the reaction products generated when Spm or Nor-Spm is oxidized by recombinant AtPAO1. Chromatograms A and B show the analysis of the reaction products at t = 0 and t = 2 h, respectively, using 1 nm of purified recombinant AtPAO1 and 0.15 mm Spm as amine substrate. Chromatograms C and D show the analysis of the reaction products at t = 0 and t = 40 min, respectively, using 1 nm of purified recombinant AtPAO1 and 0.25 mm Nor-Spm as amine substrate. Samples were analyzed for polyamine content after addition of an equal volume of 5% (w/v) perchloric acid containing 0.12 mm Dah as an internal standard. Data are from a single representative experiment, which was repeated twice. The retention times of the various polyamines are: Dap, 16.1 min; Put, 16.6 min; Dah, 18.1 min; Spd, 21.3 min; Nor-Spm, 24.1 min; and Spm, 24.4 min.
To further confirm Spd production by AtPAO1-catalyzed Spm oxidation, an indirect enzymatic assay was performed. In detail, native MPAO was added in the AtPAO1 reaction mixture at the end of the reaction when all Spm was oxidized, as shown by the equimolar production of H2O2 and by HPLC analysis of the reaction products. Addition of a saturating amount of MPAO, which specifically oxidizes Spd and Spm, in the reaction mixture further produced an equimolar amount of H2O2 in relation to the initial amount of Spm. Moreover, HPLC analysis of the reaction products showed that addition of MPAO eliminates the AtPAO1 reaction product with the retention time of Spd, producing a substance with the retention time of Dap (data not shown).
DISCUSSION
Experimental data indicate that PAOs are involved in fundamental cellular processes, not only through their contribution to polyamine homeostasis but also through their reaction products (i.e. H2O2, Dap, and aminoaldehydes). In particular, in animal cells, H2O2 produced from polyamine degradation may cause characteristic changes in redox signaling, leading to modulation of cell proliferation or apoptosis (Ha et al., 1997; Hu and Pegg, 1997). In plants, H2O2 produced from polyamine degradation by apoplastic PAO is involved in cell wall development during plant growth and in response to biotic and abiotic stress (Cona et al., 2006). Furthermore, H2O2 derived from polyamine oxidation in plants has been shown to contribute also to hypersensitive response-induced cell death (Yoda et al., 2003). Dap produced by plant PAO is a precursor of uncommon polyamines and β-Ala (Terano and Suzuki, 1978; Koc et al., 1998), which in plants are associated with stress tolerance (Rodriguez-Garay et al., 1989; Hanson et al., 1994). β-Ala may also be produced from 3-acetamidopropanal and 3-aminopropanal, which are produced from the oxidation of Spm, N1-acetyl-Spd, and N1-acetyl-Spm by animal PAOs. Interestingly, a role in the biosynthesis of pantothenic acid (a metabolic precursor of important cofactors of several metabolic enzymes, such as CoA and acyl carrier protein) through β-Ala production has recently been proposed for yeast Fms1 (White et al., 2001). Furthermore, 4-aminobutanal also produced by oxidation of Spd in plants can be further metabolized to γ-aminobutyric acid, which is an important metabolite associated with various physiological processes (Bouché and Fromm, 2004). The nature of the reaction products and the substrate specificity of PAO depend on the source of the enzyme. The biological significance of such differences and the specific role of each distinct PAO are not understood yet. To this end, the characterization of a major number of PAOs is fundamental. In plants, PAOs from monocotyledonous plants represent the most characterized plant PAOs, whereas little is known about PAOs from dicotyledonous plants. Thus, the characterization of polyamine catabolism in Arabidopsis, a dicotyledonous plant used widely as a plant reference system for several years, will greatly contribute to understanding the structural bases for the differences in biological function between plant, yeast, and animal PAOs.
Recombinant AtPAO1 catalyzes oxidative deamination of Spm and Nor-Spm, producing Spd and Nor-Spd, respectively, thus constituting a plant PAO for which involvement in a polyamine back-conversion pathway has been shown; other plant PAOs until now characterized determine a terminal catabolism of polyamines. Recombinant AtPAO1 also oxidizes N1-acetyl-Spm, but much less efficiently than Spm. Thus, AtPAO1 might be involved in a polyamine back-conversion pathway in which the SSAT probably does not participate, this being similar to SMOs and contrary to animal PAOs, which mainly oxidize acetylated polyamines. The specific biological roles of the three catabolic pathways (i.e. terminal polyamine catabolism, SSAT-dependent back-conversion of polyamines, SSAT-independent back-conversion of polyamines) have not been determined.
Identification of the polyamine oxidation products demonstrated that, in the case of Nor-Spm, AtPAO1 catalyzes not only oxidation of the carbon on the exo-side of the N4-nitrogen but also on the endo-side, although with a low efficiency. Our biochemical and structural data do not allow us to understand why AtPAO1 oxidizes Nor-Spm in two different ways but Spm only in one. This may reflect a flexible mode of Nor-Spm binding in the AtPAO1 catalytic site. Two ways of substrate oxidation have also been observed for Fms1. In particular, whereas Fms1 oxidizes Spm, N1-acetyl-Spm, and N1-acetyl-Spd at the carbon on the exo-side of the N4-nitrogen, it oxidizes N8-acetyl-Spd at the carbon on the endo-side of the N4-nitrogen (Landry and Sternglanz, 2003). It would be interesting to know whether the double mode of polyamine oxidation has any physiological relevance or not.
Analysis of the catalytic properties of the recombinant AtPAO1 in vitro has evidenced that Nor-Spm is a better amine substrate (kcat/Km = 81.4 s−1 mm−1) than Spm (kcat/Km = 24.1 s−1 mm−1). This raises the important question as to which is the most relevant substrate in vivo. To gain further insight into this matter, it is necessary first to verify whether Nor-Spm and other uncommon polyamines are present in Arabidopsis under physiological or stress conditions and then whether AtPAO1 expression pattern correlates well with accumulation of these polyamines. AtPAO1 participation in vivo in a catabolic pathway involving uncommon polyamines may suggest a role for this enzyme in the plant response mechanisms to environmental stress.
AtPAO1 differs from the other plant PAOs characterized so far not only in the mode of substrate oxidation but also in substrate specificity. In particular, AtPAO1 oxidizes Spm, but not Spd, contrary to MPAO (Polticelli et al., 2005), BPAO1 (P. Mariottini and A. Cona, personal communication), and BPAO2 (Cervelli et al., 2001), which oxidize both of them. Furthermore, recombinant AtPAO1 has a kcat value for Nor-Spm about 3-fold higher than that for Spm, whereas recombinant MPAO has a kcat value for Nor-Spm that is about 10-fold lower than that for Spm. These differences in substrate specificity between AtPAO1 and MPAO, together with the differences in localization and mode of substrate oxidation, may reflect differences in the physiological role.
Analysis of the amino acid sequence of AtPAO1 demonstrated that, despite the differences in substrate specificity and mode of substrate oxidation from MPAO, AtPAO1 has conserved most of the residues constituting the MPAO catalytic site. In particular, residues Glu-170, Lys-300, Tyr-298, and Tyr-439 (numbering of mature MPAO) are conserved in AtPAO1 and residues Phe-403 and Tyr-169 (numbering of mature MPAO) are conservatively substituted with other aromatic residues (Phe-403Tyr and Tyr-169Phe). Only residue Glu-62 (numbering of mature MPAO) results to be remarkably different, being substituted in AtPAO1 with an Ala residue. However, because the Glu-62Gln substitution in MPAO did not significantly alter the catalytic properties of the enzyme (Polticelli et al., 2005), it seems improbable that only this substitution determines the different catalytic properties of AtPAO1 as compared to MPAO. On the other hand, although AtPAO1 and mSMO have similar catalytic properties (substrate specificity, mode of substrate oxidation, pH optimum of catalytic activity, and kcat value), the two enzymes have a low sequence similarity in the residues making up the catalytic site. In particular, both Glu-62 and Glu-170 (numbering of mature MPAO) are substituted in mSMO with His and Gln, respectively, and several of the aromatic residues present in the catalytic site are substituted with nonaromatic ones, as, for example, Tyr-439Thr, Tyr-298Asn, and Tyr-169Gln (numbering of mature MPAO). These data, together with data available from the analysis of MPAO and Fms1 crystal structure (Binda et al., 1999, 2001; Huang et al., 2005), multiple sequence alignment of various FAD-dependent PAO, and site-directed mutagenesis experiments (Polticelli et al., 2005), are not sufficient to uncover the determinants of substrate specificity and catalytic mechanisms. To this end, more information is definitely necessary. It is likely that several amino acid residues together form the architecture of the catalytic site and establish interactions with the substrate, thus determining the catalytic properties of the enzyme.
The fact that AtPAO1, SMO, and Fms1 oxidize Spm, but not Spd, is of great interest and leads to us to hypothesize a distinct role for these two polyamines in specific intracellular sites. It is possible that Spm oxidation by these enzymes is required for the production of Spd, which in turn may be necessary for other metabolic pathways, as, for example, for the biosynthesis of hypusine, a compound essential for cell growth (Park et al., 1981; Chattopadhyay et al., 2003).
The presence of a putative PAO in tobacco plants with a sequence similarity to AtPAO1 of 83% is also of great interest. Such an elevated sequence similarity between the two enzymes allows us to hypothesize similar catalytic properties and leaves open the possibility that nPAO could also be involved in a polyamine back-conversion pathway. If this proves to be the case, the biochemical characterization of more PAOs from dicotyledonous plants would be necessary to examine the possibility that the presence of a polyamine back-conversion pathway is a specific characteristic of these plants.
In conclusion, to our knowledge, AtPAO1 is the first plant PAO shown to be involved in a polyamine back-conversion pathway. Although some data on this pathway in Arabidopsis already exist (Tassoni et al., 2000), further study is required to understand its physiological role.
MATERIALS AND METHODS
Materials
Put, Spd, Spm, N1-acetyl-Spd and N1-acetyl-Spm, 1,8-diaminooctane, 1,12-diaminododecane, agmatine, 4-aminoantipyrine, 3,5-dichloro-2-hydroxybenzenesulfonic acid, and horseradish peroxidase were purchased from Sigma-Aldrich-Fluka. Guazatine was obtained from Rhone-Poulenc Agro-Italia. Restriction and DNA-modifying enzymes were purchased from New England Biolabs, Invitrogen, Stratagene, and Promega. Other chemicals were obtained from Bio-Rad and J.T. Baker. All oligonucleotides were synthesized by Invitrogen. N-prenylagmatine was a generous gift from Prof. M. Botta (University of Siena, Italy; Cona et al., 2004) and MDL72527 from Dr. M. De Agazio (Consiglio Nazionale delle Ricerche, Montelibretti, Rome).
Sequence Analysis and cDNA Acquisition
EST database searches were performed using BLAST (Altschul et al., 1990). Multiple sequence alignment of the amino acid sequences was done using the program ClustalW (Thompson et al., 1994). The cDNA encoding for the putative Arabidopsis (Arabidopsis thaliana) PAO (AtPAO1; At5g13700; GenBank accession no. NM_121373) was obtained from the EST bank of the RIKEN BioResource Center (clone pda 11656). Scans of promoter sequences for putative regulatory elements were performed using the PlantCARE database (http://sphinx.rug.ac.be:8080/PlantCARE).
RT-PCR
Total RNA was isolated from various plant organs (leaves, stems, inflorescences) of Arabidopsis (ecotype Columbia) plants using TRIZOL reagent (Invitrogen) according to the manufacturer's instructions. Poly(A+) RNA was prepared from total RNA using the Oligotex mRNA kit (Qiagen). First-strand cDNA was synthesized from total or poly(A+) RNA using the SuperScript first-strand synthesis system for RT-PCR (Invitrogen) and an oligo(dT) primer. When total RNA was used for RT-PCR, RNA samples were first treated with RNase-free DNase I (Invitrogen) to avoid amplification from genomic DNA. The cDNA encoding AtPAO1 was obtained from first-strand cDNA using gene-specific oligonucleotides. PCR amplification was carried out with the Pfu Turbo DNA polymerase (Stratagene) for cDNA cloning or the EurobioTaq DNA polymerase (Eurobio) for AtPAO1 expression studies in vivo in a DNA GeneAmp PCR System 2400 (Perkin-Elmer) with the following parameters: 5 min of denaturation at 94°C; 30 cycles of 94°C for 1 min, 58°C for 2 min, and 72°C for 2 min; and 10 min at 72°C for final extension. When necessary, for further amplification, a 10-μL aliquot of the PCR was used in a nested PCR using the same or a more internal pair of sequence-specific oligonucleotides. For AtPAO1 expression studies in various plant organs, nested PCR was performed initially using the pair of sequence-specific oligonucleotides AtPAO1for1 (5′-GCCTTGAAGCTCAATGTTCTGGTCAAGTAGC-3′) and AtPAO1rev1 (5′-GAGGCGGTAGTCCAAGATATTTCCATGTGAGG-3′), and then the pair AtPAO1for2 (5′-GTCACCGAAGACGCCAATAGAACTAGCC-3′)/AtPAO1rev2 (5′-CATTCGTAACCTCTTTCATCAGCAACCAAGAAC-3′). The forward and reverse oligonucleotides were designed to include introns in the amplification product from genomic DNA (introns III and IV in the case of the AtPAO1for1/AtPAO1rev1 pair and intron IV in the case of the AtPAO1for2/AtPAO1rev2 pair; Fig. 1) to distinguish the RT-PCR amplification products from the genomic DNA amplification products.
Preparation of the Construct for AtPAO1 Expression in Pichia pastoris
For AtPAO1 expression in the culture medium of Pichia pastoris, the sequence encoding the AtPAO1 protein was amplified from first-strand cDNA using the sequence-specific oligonucleotides AtPAO1for4 (5′-GGGTATCTCTCGAGAAAAGAGAGGCTGAAGCTATGTCTACCGCCTCCGTCATCATCATTGGC-3′) and AtPAO1rev4 (5′-CGGATGCGGATCCTCTAGACCTCAATGATGATGATGATGATGGCTTGTGCCTGAGATAAATTTAAC-3′) designed to insert restriction sites XhoI and XbaI, respectively, necessary for AtPAO1 cDNA cloning in the pPICZαA vector (Invitrogen). The underlined regions in AtPAO1for4 and AtPAO1rev4 oligonucleotides indicate XhoI and XbaI sites, respectively. In particular, the AtPAO1for4 and AtPAO1rev4 primers were designed to clone the AtPAO1 cDNA flush with the Kex2 cleavage and to insert, at the 3′ terminus of AtPAO1 cDNA, a 6-His-tag sequence prior to the stop codon. The PCR product was purified using the QIAquick gel extraction kit (Qiagen) and cloned in the pGEM-T Easy vector (Promega) yielding the AtPAO1-pGEMT-I construct. Sequence analysis of this plasmid revealed two nonconservative mutations at positions 315 and 1,351, which have been corrected through two cycles of site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene), thus obtaining the plasmid AtPAO1-pGEMT-III. Subsequently, the correct AtPAO1 cDNA was excised from plasmid AtPAO1-pGEMT-III by digestion with restriction enzymes XhoI and XbaI and ligated into the corresponding sites of the pPICZαA vector to give the AtPAO1-pPIC plasmid. This construct was then linearized with SacI and used to transform P. pastoris strain X-33 according to the Pichia EasyComp kit (Invitrogen). Transformants were plated onto YPDS medium (Invitrogen) and selected on 100 μg mL−1 zeocin.
Expression of AtPAO1 in P. pastoris
Expression in shake flasks was performed following the instructions of the EasySelect Pichia expression kit (Invitrogen) and as described by Polticelli et al. (2005). Aliquots of the culture were sampled each day to determine expression levels both in the culture medium and the cellular extracts.
Expression of AtPAO1 in Escherichia coli
The pET17b vector (Novagen) was used to construct an AtPAO1 prokaryotic expression system. To clone AtPAO1 cDNA between restriction sites NdeI and XhoI of this vector, AtPAO1 cDNA was amplified using the AtPAO1-pGEMT-III plasmid as a template and the sequence-specific oligonucleotides AtPAO1for6 (5′-GTGTATCTCATATGTCTACCGCCTCCGTCATCATCATTGG-3′)/AtPAO1rev6 (5′-CGGATGCCTCGAGCTAATGATGATGATGATGATGGCTTGTGC-3′). The underlined regions in oligonucleotides AtPAO1for6 and AtPAO1rev6 indicate NdeI and XhoI sites, respectively. The AtPAO1rev6 primer was designed to insert, at the 3′ terminus of AtPAO1 cDNA, a 6-His-tag coding sequence prior to the stop codon of the AtPAO1 cDNA. The amplified AtPAO1 cDNA was subcloned into the pGEM-T Easy vector, sequenced, and then cloned in the pET17b plasmid yielding the construct AtPAO1-pET17b. This plasmid was then used to transform E. coli BL21 (DE3) cells.
Growth of Transformed Bacteria and Expression of Recombinant Protein
A single colony of E. coli BL21 (DE3) cells transformed with the plasmid AtPAO1-pET17b was inoculated into Luria-Bertani broth containing 100 μg mL−1 ampicillin for overnight growth at 30°C to an A600 value of 0.7 to 1.0. IPTG was added to the culture to a final concentration 0.4 mm to induce recombinant protein expression. The culture was incubated at 25°C or at 30°C for various time intervals (for time-course studies) or directly for 5 h (for recombinant protein purification). The culture was centrifuged at 3,000g and the cell paste was used to determine expression levels and/or to purify the recombinant protein. To determine total expression levels by western-blot analysis, the cell paste was resuspended in loading buffer for SDS-PAGE analysis and lysed by boiling (total bacterial extracts). To determine recombinant protein accumulation in the soluble protein fraction, the cell paste was resuspended in 50 mm Tris-HCl, 0.5 m NaCl, pH 8.0, and disrupted by sonication. After centrifugation at 13,000g for 30 min at 4°C, the cleared supernatant, containing only the soluble proteins, was analyzed for recombinant protein accumulation by immunoblotting and/or enzyme activity assays. The remaining pellet was extensively washed with 50 mm Tris-HCl, 0.5 m NaCl, pH 8.0, by three to four cycles of sonication/centrifugation to eliminate the whole amount of the soluble recombinant protein and resuspended in SDS-PAGE loading buffer to determine recombinant protein accumulation in inclusion bodies.
Purification of Recombinant AtPAO1
Purification of recombinant AtPAO1 was carried out by affinity chromatography using guazatine (a good competitive PAO inhibitor; Federico et al., 2001; Cona et al., 2004; Bianchi et al., 2006) bound on CNBr-activated Sepharose 4B (Amersham Biosciences). The E. coli cell paste was resuspended in 0.05 culture volumes of 50 mm Tris-HCl, 0.5 m NaCl, pH 8.0, and disrupted by sonication. After centrifugation at 13,000g for 30 min at 4°C, the clear supernatant was applied to the guazatine-Sepharose 4B resin. After binding, the column was washed with 50 mm Tris-HCl, 0.5 m NaCl, pH 8.0, and with 50 mm Tris-HCl, pH 7.0. The recombinant AtPAO1 was eluted with 50 mm 1,12-diaminododecane in 50 mm Tris-HCl, pH 7.0, and immediately dialyzed against 50 mm Tris-HCl, pH 8.0, using centrifugal filter devices (Millipore).
Preparation of Guazatine-Sepharose 4B Resin
CNBr-activated Sepharose 4B was treated with 1 mm HCl, equilibrated in 100 mm NaHCO3, 0.5 m NaCl, pH 8.3 (binding buffer), and then incubated with guazatine (7 mg mL−1 of resin) under slow agitation at 4°C for 16 h. The resin was washed with 10 volumes of binding buffer and incubated with 100 mm Tris-HCl, pH 8.0, at 4°C for 16 h to inactivate the excess of CNBr groups. The resin was washed with three cycles of alternating pH, each cycle consisting of a wash with 100 mm sodium acetate, 0.5 m NaCl, pH 4.0, and a wash with 100 mm Tris-HCl, 0.5 m NaCl, pH 8.0. Finally, the resin was equilibrated in 50 mm sodium phosphate buffer at pH 8.0 and stored at 4°C.
Determination of AtPAO1 Catalytic Parameters
The catalytic parameters (Km and kcat) for the oxidation of Spm, N1-acetyl-Spm, and Nor-Spm by E. coli-expressed recombinant AtPAO1 were determined from purified protein by following spectrophotometrically the formation of a pink adduct (ɛ515 = 2.6 × 104 m−1 cm−1), as a result of oxidation of 4-aminoantipyrine and 3,5-dichloro-2-hydroxybenzesulfonic acid catalyzed by horseradish peroxidase in 100 mm Tris-HCl buffer, pH 8.0, at 25°C (Holt and Baker, 1995). One unit of enzyme represents the amount of enzyme catalyzing the oxidation of 1 μmol of substrate/min. kcat values were calculated using saturating concentrations of amine substrates (4 mm) and keeping O2 concentration constant at the air-saturated level (apparent kcat). Km values for recombinant AtPAO1 for Spm, Nor-Spm, and N1-acetyl-Spm were determined from Michaelis-Menten plots using 7 × 10−3 units of enzyme, a constant O2 concentration at the air-saturated level, and amine substrate concentrations varying between 50 and 400 μm for Spm and N1-acetyl-Spm and between 10 and 100 μm for Nor-Spm (apparent Km). Nonlinear least-squares fitting of data was performed using Graphpad Prism software. Ki studies were performed using Spm as a substrate in 100 mm Tris-HCl, pH 8.0, with guazatine, N-prenylagmatine, 1,8-diaminooctane, Spd, N,N1-bis(2,3-butadienyl)-1,4-butanediamine (MDL72527), and agmatine as inhibitors and in 100 mm Tris-HCl, pH 7.0, with 1,12-diaminododecane as inhibitor (this substance is scarcely soluble at pH 8.0). Ki values were determined from the plot of  as a function of inhibitor concentration and from the Dixon plot (Dixon, 1953) using Graphpad Prism software.
as a function of inhibitor concentration and from the Dixon plot (Dixon, 1953) using Graphpad Prism software.
Studies of the pH dependence of recombinant AtPAO1 activity were conducted in 100 mm Tris-HCl buffer (for pH range 7.0–9.5) or in 100 mm sodium phosphate buffer (for pH range 5.0–7.5) at 25°C using O2 concentration at the air-saturated level and 4 mm of amine substrate. Best fit of the experimental data was carried out by the steepest descent method, using an equation to simulate a 2-pK dissociation equilibrium (Eq. 1):
 |
(1) |
where A is the catalytic activity of the enzyme, Amax is the maximal catalytic activity, K1 and K2 are the equilibrium dissociation constants, %pK1 and %pK2 are the relative contributions of each pK to the activity curve, and %pK1 + %pK2 = 1.
SDS-PAGE and Western-Blot Analysis
SDS-PAGE was made according to the method of Laemmli (1970) and western-blot analysis was performed utilizing a mouse anti-6-His monoclonal antibody (Sigma-Aldrich-Fluka). An anti-mouse antibody coupled to horseradish peroxidase (Amersham Biosciences) was used as the secondary antibody and the detection of the labeled proteins was done with a chemiluminescence kit (Boehringer Mannheim).
Analysis of Polyamine Products of Spm and Nor-Spm Oxidation by Recombinant AtPAO1
A reaction mixture of 2 mL in 100 mm Tris-HCl, pH 8.0, containing purified recombinant AtPAO1 at 1 nm final concentration and either 0.15 mm Spm or 0.25 mm Nor-Spm was prepared. Aliquots of 100 μL of the reaction mixture were removed at various time intervals and analyzed for polyamine content after addition of an equal volume of 5% (w/v) perchloric acid containing 0.12 mm 1,6-diaminohexane (Dah) as an internal standard. A reference solution containing Dap, Put, Spd, Nor-Spm, and Spm was also prepared and treated as above to establish retention times and signal intensities for each compound and the internal standard during the following HPLC analysis. Polyamines were quantified after derivatization with dansyl chloride according to Smith and Davies (1985) with minor modifications. Dansylated polyamines were separated by HPLC (Spectra System P 2000; Thermo Finnigan) on a reverse-phase C18 column (Spherisorb S5 ODS2, 5-μm particle diameter, 4.6 × 250 mm) using a discontinued methanol to water gradient (40%–60% methanol in 2 min, 60%–95% methanol in 20 min, 95%–100% in 2.5 min, 100% for 1.5 min, 100%–40% in 6 min at a flow rate of 1.5 mL min−1). Eluted peaks were detected by a spectrofluorometer (Spectra System FL 3000; excitation 365 nm, emission 510 nm), recorded, and integrated by an attached computer using Thermo Finnigan Chrom-Card 32-bit software. The retention times are as follows: Dap, 16.1 min; Put, 16.6 min; Dah, 18.1 min; Spd, 21.3 min; Nor-Spm, 24.1 min; and Spm, 24.4 min.
DNA Sequencing
DNA sequencing was performed on double-stranded plasmid DNA using the automated fluorescent dye terminator technique (ABI model 373A; Perkin-Elmer).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_121373, AF364952, AY143905, AF364953, AK118203, XP_470573, XP_450669, XP_450667, and AB200262.
Acknowledgments
We thank Silvana Pranteda and Valerio Giarrizzo for technical assistance and Prof. Paolo Mariottini and Dr. Carmen Faso for critical review of the manuscript.
This work was supported by the University Roma Tre and the Italian Ministry of University and Research (project MIUR–PRIN 2005).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Paraskevi Tavladoraki (tavlador@uniroma3.it).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.080911.
References
- Alcázar R, García-Martínez JL, Cuevas JC, Tiburcio AF, Altabella T (2005) Overexpression of ADC2 in Arabidopsis induces dwarfism and late-flowering through GA deficiency. Plant J 43: 425–443 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Bagga S, Rochford J, Klaene Z, Kuehn GD, Phillips GC (1997) Putrescine aminopropyltransferase is responsible for biosynthesis of spermidine, spermine, and multiple uncommon polyamines in osmotic stress-tolerant alfalfa. Plant Physiol 114: 445–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagni N, Tassoni A (2001) Biosynthesis, oxidation and conjugation of aliphatic polyamines in higher plants. Amino Acids 20: 301–317 [DOI] [PubMed] [Google Scholar]
- Bianchi M, Polticelli F, Ascenzi P, Botta M, Federico R, Mariottini P, Cona A (2006) Inhibition of polyamine and spermine oxidases by polyamine analogues. FEBS J 273: 1115–1123 [DOI] [PubMed] [Google Scholar]
- Binda C, Angelini R, Federico R, Ascenzi P, Mattevi A (2001) Structural bases for inhibitor binding and catalysis in polyamine oxidase. Biochemistry 40: 2766–2776 [DOI] [PubMed] [Google Scholar]
- Binda C, Coda A, Angelini R, Federico R, Ascenzi P, Mattevi A (1999) A 30-angstrom-long U-shaped catalytic tunnel in the crystal structure of polyamine oxidase. Structure 7: 265–276 [DOI] [PubMed] [Google Scholar]
- Bouché N, Fromm H (2004) GABA in plants: just a metabolite? Trends Plant Sci 9: 110–115 [DOI] [PubMed] [Google Scholar]
- Bouchereau A, Aziz A, Larher F, Martin-Tanguy J (1999) Polyamines and environmental challenges: recent development. Plant Sci 140: 103–125 [Google Scholar]
- Cervelli M, Cona A, Angelini R, Polticelli F, Federico R, Mariottini P (2001) A barley polyamine oxidase isoform with distinct structural features and subcellular localization. Eur J Biochem 268: 3816–3830 [DOI] [PubMed] [Google Scholar]
- Cervelli M, Di Caro O, Di Penta A, Angelini R, Federico R, Vitale A, Mariottini P (2004) A novel C-terminal sequence from barley polyamine oxidase is a vacuolar sorting signal. Plant J 40: 410–418 [DOI] [PubMed] [Google Scholar]
- Cervelli M, Polticelli F, Federico R, Mariottini P (2003) Heterologous expression and characterization of mouse spermine oxidase. J Biol Chem 278: 5271–5276 [DOI] [PubMed] [Google Scholar]
- Cervelli M, Tavladoraki P, Di Agostino S, Angelini R, Federico R, Mariottini P (2000) Isolation and characterization of three polyamine oxidase genes from Zea mays. Plant Physiol Biochem 38: 667–677 [Google Scholar]
- Chattopadhyay MK, Tabor CW, Tabor H (2003) Spermidine but not spermine is essential for hypusine biosynthesis and growth in Saccharomyces cerevisiae: Spermine is converted to spermidine in vivo by the FMS1-amine oxidase. Proc Natl Acad Sci USA 100: 13869–13874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SS (1998) A Guide to the Polyamines. Oxford University Press, Oxford
- Cona A, Manetti F, Leone R, Corelli F, Tavladoraki P, Polticelli F, Botta M (2004) Molecular basis for the binding of competitive inhibitors of maize polyamine oxidase. Biochemistry 43: 3426–3435 [DOI] [PubMed] [Google Scholar]
- Cona A, Moreno S, Cenci F, Federico R, Angelini R (2005) Cellular re-distribution of flavin-containing polyamine oxidase in differentiating root and mesocotyl of Zea mays L. seedlings. Planta 221: 265–276 [DOI] [PubMed] [Google Scholar]
- Cona A, Rea G, Angelini R, Federico R, Tavladoraki P (2006) Functions of amine oxidases in plant development and defence. Trends Plant Sci 11: 80–88 [DOI] [PubMed] [Google Scholar]
- De Agazio M, Zacchini M, Federico R, Grego S (1995) Putrescine accumulation in maize roots treated with spermidine: evidence for spermidine to putrescine conversion. Plant Sci 111: 181–185 [Google Scholar]
- Dixon M (1953) The determination of enzyme inhibitor constants. Biochem J 55: 170–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson DE, Binda C, Mattevi A (2004) The FAD binding sites of human monoamine oxidases A and B. Neurotoxicology 25: 63–72 [DOI] [PubMed] [Google Scholar]
- Edmondson DE, Newton-Vinson P (2001) The covalent FAD of monoamine oxidase: structural and functional role and mechanism of the flavinylation reaction. Antioxid Redox Signal 3: 789–806 [DOI] [PubMed] [Google Scholar]
- Federico R, Angelini R (1991) Polyamine catabolism in plants. In RD Slocum, HE Flores, eds, Biochemistry and Physiology of Polyamines in Plants. CRC Press, Boca Raton, FL, pp 41–56
- Federico R, Leone L, Botta M, Binda C, Angelini R, Venturini G, Ascenzi P (2001) Inhibitor of pig liver and Zea mays L. polyamine oxidase: a comparative study. J Enzyme Inhib 13: 465–471 [DOI] [PubMed] [Google Scholar]
- Ha HC, Woster PM, Yager JD, Casero RA Jr (1997) The role of polyamine catabolism in polyamine analogue-induced programmed cell death. Proc Natl Acad Sci USA 94: 11557–11562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AD, Rathinasabapathi B, Rivoal J, Burnet M, Dillon MO, Gage DA (1994) Osmoprotective compounds in the Plumbaginaceae: a natural experiment in metabolic engineering of stress tolerance. Proc Natl Acad Sci USA 91: 306–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt A, Baker GB (1995) Metabolism of agmatine (clonidine-displacing substance) by diamine oxidase and the possible implications for studies of imidazoline receptors. Prog Brain Res 106: 187–197 [DOI] [PubMed] [Google Scholar]
- Hu RH, Pegg AE (1997) Rapid induction of apoptosis by deregulated uptake of polyamine analogues. Biochem J 328: 307–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Liu Q, Hao Q (2005) Crystal structures of Fms1 and its complex with spermine reveal substrate specificity. J Mol Biol 348: 951–959 [DOI] [PubMed] [Google Scholar]
- Illingworth C, Mayer MJ, Elliott K, Hanfrey C, Walton NJ, Michael AJ (2003) The diverse bacterial origins of the Arabidopsis polyamine biosynthetic pathway. FEBS Lett 549: 26–30 [DOI] [PubMed] [Google Scholar]
- Imai A, Matsuyama T, Hanzawa Y, Akiyama T, Tamaoki M, Saji H, Shirano Y, Kato T, Hayashi H, Shibata D, et al (2004) Spermidine synthase genes are essential for survival of Arabidopsis. Plant Physiol 135: 1565–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koc EC, Bagga S, Songstad DD, Betz SR, Kuehn GD, Phillips GC (1998) Occurrence of uncommon polyamines in culture tissues of maize. In Vitro Cell Dev Biol Plant 34: 252–255 [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 277: 680–685 [DOI] [PubMed] [Google Scholar]
- Landry J, Sternglanz R (2003) Yeast Fms1 is a FAD-utilizing polyamine oxidase. Biochem Biophys Res Commun 303: 771–776 [DOI] [PubMed] [Google Scholar]
- Panicot M, Minguet EG, Ferrando A, Alcázar R, Blazquez MA, Carbonell J, Altabella T, Koncz C, Tiburcio AF (2002) A polyamine metabolon involving aminopropyl transferase complexes in Arabidopsis. Plant Cell 14: 2539–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, Cooper HL, Folk JE (1981) Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proc Natl Acad Sci USA 78: 2869–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polticelli F, Basran J, Faso C, Cona A, Minervini G, Zelli M, Federico R, Angelini R, Scrutton NS, Tavladoraki P (2005) Lys300 plays a major role in the catalytic mechanism of maize polyamine oxidase. Biochemistry 44: 16108–16120 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Garay B, Phillips GC, Kuehn GD (1989) Detection of nonspermidine and norspermine in Medicago sativa L. (alfalfa). Plant Physiol 89: 525–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šebela M, Radovà A, Angelini R, Tavladoraki P, Frébort I, Pěc P (2001) FAD-containing polyamine oxidases: a timely challenge for researchers in biochemistry and physiology of plants. Plant Sci 160: 197–207 [DOI] [PubMed] [Google Scholar]
- Seiler N (2004) Catabolism of polyamines. Amino Acids 26: 217–233 [DOI] [PubMed] [Google Scholar]
- Seki M, Carninci P, Nishiyama Y, Hayashizaki Y, Shinozaki K (1998) High-efficiency cloning of Arabidopsis full-length cDNA by biotinylated CAP trapper. Plant J 15: 707–720 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y, et al (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145 [DOI] [PubMed] [Google Scholar]
- Smith TA, Davies PJ (1985) Separation and quantitation of polyamines in plant tissue by high performance liquid chromatography of their dansyl derivatives. Plant Physiol 78: 89–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassoni A, van Buuren M, Franceschetti M, Fornalè S, Bagni N (2000) Polyamine content and metabolism in Arabidopsis thaliana and effect of spermidine on plant development. Plant Physiol Biochem 38: 383–393 [Google Scholar]
- Tavladoraki P, Shininà ME, Cecconi F, Di Agostino S, Manera F, Rea G, Mariottini P, Federico R, Angelini R (1998) Maize polyamine oxidase: primary structure from protein and cDNA sequencing. FEBS Lett 426: 62–66 [DOI] [PubMed] [Google Scholar]
- Terano S, Suzuki Y (1978) Formation of β-alanine from spermine and spermidine in maize shoots. Phytochemistry 17: 148–149 [Google Scholar]
- Thomas T, Thomas TJ (2003) Polyamine metabolism and cancer. J Cell Mol Med 7: 113–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujcic S, Diegelman P, Bacchi CJ, Kramer DL, Porter CW (2002) Identification and characterization of a novel flavin-containing spermine oxidase of mammalian cell origin. Biochem J 367: 665–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujcic S, Liang P, Diegelman P, Kramer DL, Porter CW (2003) Genomic identification and biochemical characterization of the mammalian polyamine oxidase involved in polyamine back-conversion. Biochem J 370: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace HM, Fraser AV, Hughes A (2003) A perspective of polyamine metabolism. Biochem J 376: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters DR (2003) Polyamines and plant disease. Phytochemistry 64: 97–107 [DOI] [PubMed] [Google Scholar]
- Wang Y, Devereux W, Woster PM, Stewart TM, Hacker A, Casero RA Jr (2001) Cloning and characterization of a human polyamine oxidase that is inducible by polyamine analogue exposure. Cancer Res 61: 5370–5373 [PubMed] [Google Scholar]
- White WH, Gunyuzlu PL, Toyn JH (2001) Saccharomyces cerevisiae is capable of de novo pantothenic acid biosynthesis involving a novel pathway of beta-alanine production from spermine. J Biol Chem 276: 10794–10800 [DOI] [PubMed] [Google Scholar]
- Wu T, Yankovskaya V, McIntire WS (2003) Cloning, sequencing, and heterologous expression of the murine peroxisomal flavoprotein, N1-acetylated polyamine oxidase. J Biol Chem 278: 20514–20525 [DOI] [PubMed] [Google Scholar]
- Yoda H, Yamaguchi Y, Sano H (2003) Induction of hypersensitive cell death by hydrogen peroxide produced through polyamine degradation in tobacco plants. Plant Physiol 132: 1973–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]