Abstract
During plant reproduction, the central cell of the female gametophyte becomes fertilized to produce the endosperm, a storage tissue that nourishes the developing embryo within the seed. The molecular mechanisms controlling the specification and differentiation of the central cell are poorly understood. We identified a female gametophyte mutant in Arabidopsis thaliana, fem111, that is affected in central cell development. In fem111 female gametophytes, the central cell's nucleolus and vacuole fail to mature properly. In addition, endosperm development is not initiated after fertilization of fem111 female gametophytes. fem111 contains a T-DNA insertion in AGAMOUS-LIKE80 (AGL80). FEM111/AGL80 is a member of the MADS box family of genes that likely encode transcription factors. An AGL80–green fluorescent protein fusion protein is localized to the nucleus. Within the ovule and seed, FEM111/AGL80 is expressed exclusively in the central cell and uncellularized endosperm. FEM111/AGL80 expression is also detected in roots, leaves, floral stems, anthers, and young flowers by real-time RT-PCR. FEM111/AGL80 is required for the expression of two central cell–expressed genes, DEMETER and DD46, but not for a third central cell–expressed gene, FERTILIZATION-INDEPENDENT SEED2. Together, these data suggest that FEM111/AGL80 functions as a transcription factor within the central cell gene regulatory network and controls the expression of downstream genes required for central cell development and function.
INTRODUCTION
The central cell within the female gametophyte (Figure 1A) is essential for seed formation in angiosperms. Angiosperms undergo double fertilization, in which the female gametophyte's haploid egg cell and diploid central cell both become fertilized. After fertilization, the egg cell and central cell give rise to the seed's diploid embryo and triploid endosperm, respectively. Endosperm is important for the seed because it provides nutrients and other factors to the developing embryo during seed development and, in some species, to developing seedlings after germination (Olsen, 2004).
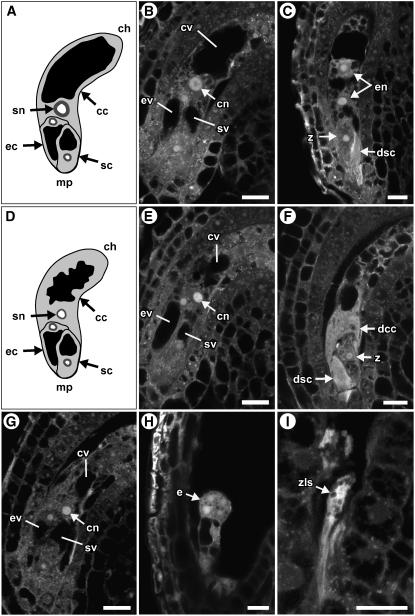
Figure 1.
Microscopic Analysis of Wild-Type and fem111 Female Gametophytes.
(A) and (D) Depictions of the Arabidopsis female gametophyte at the terminal developmental stage (stage FG7) in the wild type (A) and fem111 (D). Modified from Drews et al. (1998). In these panels, cytoplasm is light gray, vacuoles are black, nuclei are dark gray, and nucleoli are white. Only one of the two synergid cells is depicted in this orientation.
(B), (C), and (E) to (I) CLSM images. In these images, cytoplasm is gray, vacuoles are black, and nucleoli are white.
(B), (E), and (G) Wild type (B) and fem111 ([E] and [G]) female gametophytes at the terminal developmental stage (stage FG7). The wild-type female gametophyte in (B) and the fem111 female gametophyte in (E) are from ovules at 24 h after emasculation of a stage 12c flower. The fem111 female gametophyte in (G) is from an ovule at 42 h after emasculation of a stage 12c flower. The vacuole and nucleolus are smaller in fem111 ([E] and [G]) compared with the wild type (B). In the wild type (B), the egg cell is elongated along the micropylar/chalazal axis and has a single vacuole at the micropylar end and its nucleus at the chalazal end.
(C) and (F) Wild-type (C) and fem111 (F) seeds at 18 h after pollination. In (C), only two of the four endosperm nuclei can be seen. The zygote, in contrast with the egg cell, is rounded, has the nucleus in the center of the cell, and contains many smaller vacuoles that surround the nucleus on all sides. In (F), the zygote, degenerating synergid cell, and degenerating central cell are in focus and endosperm nuclei are not observed.
(H) and (I) Wild-type (H) and fem111 (I) seeds at 40 h after pollination. Wild-type seeds (H) contain embryos at the globular stage. fem111 seeds (I) contain zygote-like structures.
cc, central cell; ch, chalazal pole; cn, central cell nucleolus; cv, central cell vacuole; dcc, degenerating central cell; dsc, degenerating synergid cell; e, embryo; ec, egg cell; ev, egg cell vacuole; mp, micropylar pole; sc, synergid cell; sn, secondary nucleus; sv, synergid cell vacuole; z, zygote; zls, zygote-like structure. Bars = 10 μm.
During female gametophyte development, a haploid megaspore undergoes three rounds of mitosis without cellularization to produce an eight-nucleate structure. Cellularization results in a seven-celled gametophyte containing three antipodal cells at the chalazal pole, one egg cell and two synergid cells at the micropylar pole, and a central cell in the center. The largest of these cells is the central cell, which inherits two nuclei referred to as the polar nuclei. In Arabidopsis thaliana and many other species, the polar nuclei fuse to form the diploid central cell nucleus (secondary nucleus) and the antipodal cells degenerate before fertilization. Thus, in the mature female gametophyte of Arabidopsis, the central cell occupies most of the volume of the embryo sac, is located at the chalazal pole, contains a large vacuole at its chalazal pole, and has a relatively large nucleus at its micropylar pole (Figure 1A) (Willemse and van Went, 1984; Huang and Russell, 1992; Yadegari and Drews, 2004).
Little is known about the molecular processes controlling the specification and differentiation of the central cell during female gametophyte development. During the last 10 years, many genes required for female gametophyte development have been identified and analyzed, including FERTILIZATION-INDEPENDENT ENDOSPERM (FIE) (Ohad et al., 1996, 1999), MEDEA (MEA) (Chaudhury et al., 1997; Grossniklaus et al., 1998; Kiyosue et al., 1999), FERTILIZATION-INDEPENDENT SEED2 (FIS2) (Chaudhury et al., 1997; Luo et al., 1999), GFA2 (Christensen et al., 2002), CYTOKININ-INDEPENDENT1 (Pischke et al., 2002; Hejatko et al., 2003), DYAD (Siddiqi et al., 2000; Agashe et al., 2002), MULTICOPY SUPPRESSOR OF IRA (MSI1) (Kohler et al., 2003a; Guitton et al., 2004), NOMEGA (Kwee and Sundaresan, 2003), RETINOBLASTOMA RELATED1 (Ebel et al., 2004), ARABINOGALACTAN PROTEIN18 (Acosta-Garcia and Vielle-Calzada, 2004), SLOW WALKER1 (Shi et al., 2005), DEMETER (DME) (Choi et al., 2002), LYSOPHOSPHATIDYL ACYLTRANSFERASE2 (Kim et al., 2005), GLUCOSE 6-PHOSPHATE/PHOSPHATE TRANSLOCATOR1 (Niewiadomski et al., 2005), CHROMATIN REMODELLING PROTEIN11 (Huanca-Mamani et al., 2005), and MYB98 (Kasahara et al., 2005). Of these, only FIE, FIS2, MEA, MSI1, and DME are known to function in the central cell specifically. Loss-of-function mutations in the FIE, FIS2, MEA, and MSI1 genes result in autonomous endosperm development in the absence of fertilization. Based on this phenotype and on similarity to polycomb group proteins in Drosophila and mammals, it has been proposed that the FIE, FIS2, MEA, and MSI1 proteins form a complex that represses genes involved in endosperm development (Grossniklaus et al., 1998; Luo et al., 2000; Spillane et al., 2000; Guitton et al., 2004). DME is a regulatory molecule required for MEA expression in the central cell and endosperm (Choi et al., 2002; Gehring et al., 2006).
Expression-based screens have identified a battery of central cell–expressed genes, including Zea mays MADS1 (Heuer et al., 2001), Zm EBE-1 and Zm EBE-2 (Magnard et al., 2003), C053 to C195 (Le et al., 2005), and EC-52, EC-57, and EC-217 (Sprunck et al., 2005). At present, the functions of these genes are unknown. Other aspects of the central cell gene regulatory network have not been determined.
Here, we report the identification of a mutant, fem111, that has defects in central cell and endosperm development. We show that the fem111 mutant has a lesion in the AGAMOUS-LIKE80 (AGL80) gene. FEM111/AGL80 encodes a type I MADS domain–containing protein, which likely functions as a transcription factor. In the context of the ovule, FEM111/AGL80 is expressed exclusively in the central cell. We also show that FEM111/AGL80 is required for the expression of DME, indicating that FEM111/AGL80 is upstream of this gene in the central cell gene regulatory network. Thus, FEM111/AGL80 appears to encode a regulatory molecule controlling central cell differentiation during female gametophyte development.
RESULTS
fem111 Affects the Female Gametophyte but Not the Male Gametophyte
We previously identified a large collection of female gametophyte mutants using the criteria of segregation distortion and reduced seed set (Yadegari and Drews, 2004). One of these mutants, fem111, is described here.
As summarized in Table 1, fem111 exhibited segregation distortion in self-crosses (P < 0.001), suggesting that the gametophyte generation is affected. To determine whether the fem111 mutation affects the female gametophyte, we crossed fem111/FEM111 females with wild-type males and scored the number of fem111/FEM111 and FEM111/FEM111 progeny. As shown in Table 1, transmission of the fem111 mutation was significantly reduced compared with that of the wild-type allele (P < 0.001). In addition, siliques resulting from this cross exhibited reduced seed set (47% aborted ovules; n = 300) (see Supplemental Figure 1 online). These observations indicate that fem111 affects the female gametophyte.
Table 1.
Segregation of the fem111 Mutation
| Parental Genotypes
|
Progeny Genotypes
|
|||
|---|---|---|---|---|
| Male | Female | FEM111/FEM111 | fem111/FEM111 | fem111/fem111 |
| fem111/FEM111 | fem111/FEM111 | 137 | 149 | 0 |
| FEM111/FEM111 | fem111/FEM111 | 343 | 0 | – |
| fem111/FEM111 | FEM111/FEM111 | 218 | 211 | – |
To determine whether the fem111 mutation also affects the male gametophyte, we crossed fem111/FEM111 males with wild-type females and scored the number of fem111/FEM111 and FEM111/FEM111 progeny. As shown in Table 1, transmission of the fem111 mutation through the male gametophyte was not significantly different from that of the wild-type allele (P > 0.5), indicating that the male gametophyte is not affected.
As shown in Table 1, the penetrance of the fem111 mutation is 100% in the female gametophyte. As a consequence, we were not able to isolate homozygous mutants and, thus, were not able to evaluate whether this mutation affects the sporophyte generation.
fem111 Affects Central Cell Development
To determine whether fem111 affects megagametogenesis, we analyzed female gametophytes at the terminal developmental stage (stage FG7) using confocal laser-scanning microscopy (CLSM) (Christensen et al., 1997). We emasculated flowers at stage 12c (Christensen et al., 1997), waited 24 h, and fixed ovule tissue for confocal analysis. Wild-type female gametophytes at this stage have one egg cell, one central cell, and two synergid cells (Figures 1A and 1B); the three antipodal cells undergo cell death during the transition from stage FG6 to FG7 (Christensen et al., 1997). The nucleus and nucleolus of the central cell are larger than those of the other cells as a result of fusion of the two polar nuclei during central cell development (Figures 1A and 1B).
In fem111 female gametophytes, all four cells were present (the antipodal cells degenerated normally), and the egg cell and synergid cells appeared indistinguishable from those of the wild type (Figures 1D and 1E). By contrast, the central cell in fem111 female gametophytes exhibited two defects: the nucleolus and vacuole both were smaller than those of the wild type (Figures 1D and 1E).
We analyzed female gametophytes at earlier developmental stages to determine why the nucleolus and vacuole are small in fem111 central cells. We first analyzed these structures in the wild type. The central cell is first formed during stage FG5. During this stage, the central cell contains the two polar nuclei that fuse to form a single nucleus (secondary nucleus) and a large vacuole that occupies most of the volume of this cell. The central cell's nucleolus is relatively small just after fusion (stage FG6) and enlarges as the female gametophyte matures (stages FG6 and FG7), until it reaches a diameter of ∼6 μm at the terminal developmental stage (stage FG7) (Figure 2). During these stages, the vacuole continues to occupy most of the volume of the central cell (Christensen et al., 1997).
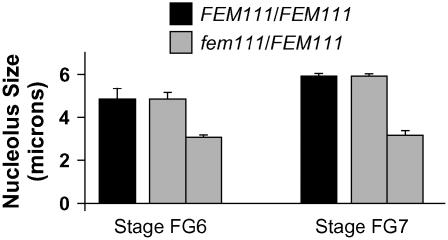
Figure 2.
Size of the Central Cell Nucleolus in Wild-Type and fem111 Female Gametophytes.
Black bars, nucleolus size in wild-type plants; gray bars, nucleolus size in fem111/FEM111 plants. The two sets of bars for fem111/FEM111 plants represent two distinct classes of female gametophytes within the pistil. For the wild type at stages FG6 and FG7, n = 100 for each stage. For fem111/FEM111 at stage FG6, n = 235; for fem111/FEM111 at stage FG7, n = 250. Error bars indicate sd.
To analyze the maturation of the central cell's nucleolus and vacuole in fem111, we used CLSM to observe the female gametophytes within fem111/FEM111 pistils from flower stage 12c, which contain female gametophytes at stages FG4 to FG6 (Christensen et al., 1997). Female gametophytes at stages FG4 and FG5 exhibited no phenotypic variation (n = 113 female gametophytes), indicating that migration and fusion of the polar nuclei occur normally in fem111 female gametophytes. By contrast, two distinct populations of female gametophytes were observed at stage FG6. One population (54%) was indistinguishable from the wild type in all features, including the sizes of the central cell's vacuole and nucleolus (Figure 2). In the second population (46%), the central cell's vacuole and nucleolus had reduced size (Figure 2). Together, these data indicate that the phenotype conferred by fem111 first becomes apparent soon after fusion of the polar nuclei and includes defects in nuclear maturation and the maintenance of vacuole size.
fem111 Affects Endosperm Development
In the wild type, the central cell becomes fertilized and gives rise to the seed's endosperm. To determine whether the central cell of fem111 female gametophytes is competent to give rise to endosperm, we emasculated fem111/FEM111 flowers at stage 12c (Christensen et al., 1997), waited 24 h, pollinated with wild-type pollen, waited 18 h, and fixed ovule tissue for confocal analysis. In the wild type at 18 h after pollination, one of the synergid cells is degenerated and both the egg cell and the central cell have become fertilized. At this time point, the embryo is a single-celled zygote and the endosperm consists of four to eight nuclei (Figure 1C).
As with the wild type, fem111 female gametophytes at 18 h after pollination contained a degenerated synergid cell and a single-celled zygote (Figure 1F). In the wild type, egg cells and zygotes are distinguished by several structural features (see the legends of Figures 1B and 1C) (Faure et al., 2002). fem111 zygotes exhibited all of these structural features, strongly suggesting that fem111 egg cells become fertilized (Figure 1F). In contrast with the wild type, endosperm nuclei were not present (Figure 1F). Instead, the central cell cavity was filled with highly fluorescent material. Within the fluorescent material, the secondary nucleus and the nucleolus could sometimes be observed but were difficult to distinguish from the surrounding cytoplasm (Figure 1F).
The absence of endosperm at 18 h after pollination could be attributable to either a failure to form endosperm or a delay in endosperm development. To distinguish between these possibilities, we analyzed fem111 seeds at 40 h after pollination. In wild-type seeds at this time point, endosperm consists of >50 nuclei and the embryo is at the globular stage (Figure 1H). By contrast, in fem111/FEM111 siliques, 45% of seeds at this time point did not contain endosperm nuclei (n = 100 seeds). Of those seeds lacking endosperm, 76% had a zygote-like structure (Figure 1I); in the remaining 24%, it could not be determined whether a zygote-like structure was present because the embryo sacs were collapsed. This result suggests that the failure to form endosperm is not the result of a delay in endosperm development.
The data presented above suggest that the structural changes associated with the central cell are induced by pollination. To explore this possibility further, we emasculated stage 12c flowers, waited 42 h (corresponds to 18 h after manual pollination), and fixed ovule tissue for confocal analysis. The autofluorescence associated with the central cell in fertilized fem111 ovules was not observed in fem111 female gametophytes of unpollinated pistils at this time point (Figure 1G), indicating that these structural changes require pollination.
Together, these observations indicate that the central cell in fem111 female gametophytes is not competent to give rise to endosperm, even when fertilized with wild-type pollen.
FEM111 Encodes the AGL80 MADS Domain Protein
The fem111 mutant was identified in a screen of T-DNA–mutagenized lines. To identify the affected gene, we used thermal asymmetric interlaced PCR (Liu et al., 1995) to determine the T-DNA insertion site in the fem111 mutant. The T-DNA in fem111 is inserted into gene At5g48670. To determine whether the female gametophyte defect is attributable to disruption of At5g48670, we introduced a wild-type copy of this gene into the fem111 mutant. We generated plants homozygous for both the fem111 allele and the rescue construct; these plants had full seed set, indicating that disruption of At5g48670 is responsible for the female gametophyte defect in fem111 mutants.
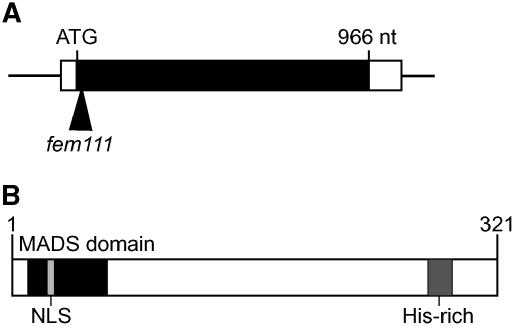
This gene corresponds to AGL80, which encodes a type I MADS domain protein (Parenicova et al., 2003) that likely functions as a transcription factor. To determine the structure of the AGL80 gene, we isolated a cDNA clone and compared its sequence with that of the genomic sequence. We also performed 5′ and 3′ rapid amplification of cDNA ends (RACE) to characterize the 5′ and 3′ untranslated regions of this gene. As shown in Figure 3A, FEM111/AGL80 comprises a single exon that contains a 966-nucleotide open reading frame. As shown in Figure 3B, FEM111/AGL80 is predicted to encode a protein of 321 amino acids that contains a MADS domain near the N terminus of the protein and a His-rich region near the C terminus of the protein.
Figure 3.
Structures of the FEM111/AGL80 Gene and FEM111/AGL80 Protein.
(A) FEM111/AGL80 gene structure. The black box represents coding sequence, and the white boxes represent the 5′ (51 nucleotides [nt]) and 3′ (109 nucleotides) untranslated regions. The insertion site of the T-DNA in the fem111 mutant is marked by an arrowhead.
(B) FEM111/AGL80 protein structure. FEM111/AGL80 contains a MADS domain (amino acids 9 to 60; black box), a predicted nuclear localization signal (NLS) (amino acids 23 to 26; light gray box), and a His-rich domain (amino acids 275 to 291; dark gray box).
The T-DNA is inserted immediately after the second nucleotide in the predicted open reading frame and is associated with a 6-bp deletion in the coding sequence. This observation suggests that fem111 is a null allele. However, we were not able to confirm this prediction because we were unable to obtain a homozygous line (Table 1), which precluded the analysis of FEM111/AGL80 expression in fem111 mutants.
FEM111/AGL80 Is Expressed in the Central Cell during Female Gametophyte Development
To characterize the expression of FEM111/AGL80 during female gametophyte development, we analyzed transgenic Arabidopsis plants containing a protein fusion construct in which the FEM111/AGL80 promoter and the entire FEM111/AGL80 coding sequence were fused to green fluorescent protein (AGL80-GFP). This construct was sufficient to rescue the mutant phenotype in the fem111 background, suggesting that expression of this construct mimics that of the endogenous gene.
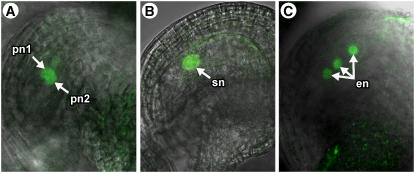
As shown in Figure 4B, at the terminal developmental stage (stage FG7), AGL80-GFP expression was detected exclusively in the central cell. Figure 4B also shows that the AGL80-GFP fusion protein was localized to the nucleus, consistent with its predicted function as a transcriptional regulator. To determine whether FEM111/AGL80 is expressed at earlier developmental stages, we analyzed expression in flowers at stage 12c, which contain both uncellularized (stages FG4 and early FG5) and cellularized (late stage FG5 and stage FG6) female gametophytes (Christensen et al., 1997). Within flowers at this stage, we detected AGL80-GFP expression in the polar nuclei before fusion (late stage FG5) (Figure 4A). In all cases in which expression in the polar nuclei was detected, the two nuclei were immediately adjacent to one another. Expression was never observed in polar nuclei that were separated. Together, these data indicate that FEM111/AGL80 is expressed specifically in the central cell and that its expression is first detected in female gametophytes soon after cellularization and before the polar nuclei fuse (late stage FG5).
Figure 4.
AGL80-GFP Expression.
(A) Expression of AGL80-GFP in female gametophytes before fusion of the polar nuclei (late stage FG5). Expression is detected only in the two polar nuclei.
(B) Expression of AGL80-GFP in female gametophytes at the terminal developmental stage (stage FG7). Expression is detected only in the secondary nucleus.
(C) Expression of AGL80-GFP in a fertilized female gametophyte at 18 h after pollination. Expression is detected only in the endosperm nuclei. Only three of the four endosperm nuclei can be seen in this focal plane.
All images are composites of CLSM micrographs of AGL80-GFP expression merged with bright-field images of ovules. en, endosperm nuclei; pn1 and pn2, the two polar nuclei before fusion; sn, secondary nucleus of the central cell.
To determine whether FEM111/AGL80 is also expressed in developing seeds, we analyzed AGL80-GFP expression at 8 to 72 h after pollination. During this period, AGL80-GFP expression was detected exclusively in the endosperm (Figure 4C). During endosperm development, AGL80-GFP expression was strongest in young seeds and diminished gradually at progressively older stages; expression was not detected in seeds older than 3 d after pollination. In reciprocal crosses with plants homozygous for the AGL80-GFP construct and the wild type, expression was detected only when the reporter construct was present in the female parent.
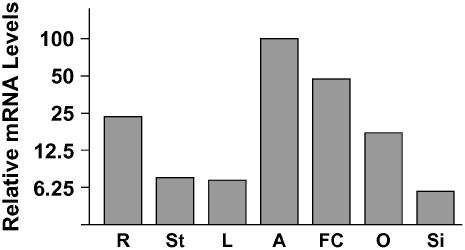
To analyze FEM111/AGL80 expression elsewhere in the plant, we performed real-time RT-PCR with RNA from various organs. Consistent with the expression of AGL80-GFP in the female gametophyte and endosperm, we detected FEM111/AGL80 expression in ovaries and young siliques (Figure 5). In addition, expression was detected in roots, leaves, stems, young flowers, and anthers (Figure 5). Our results are consistent with previous studies showing expression of AGL80 in leaves, stems, siliques, and anthers (Parenicova et al., 2003).
Figure 5.
Real-Time RT-PCR Analysis of FEM111/AGL80 Expression.
AGL80 transcript levels were normalized to ACTIN2 levels using the formula ΔCT = CT(AGL80) – CT(ACTIN2). Real-time RT-PCR was performed with cDNAs from anthers harvested from flowers at stages 10 to 13 (A; ΔCT = 7.3 ± 1.5), flowers at stages 1 to 10 (FC; ΔCT = 8.4 ± 0.3), leaves (L; ΔCT = 11.1 ± 0.6), ovaries from unpollinated flowers (O; ΔCT = 9.8 ± 0.4), roots (R; ΔCT = 9.4 ± 0.3), siliques at 1 to 3 d after pollination (Si; ΔCT = 11.4 ± 0.8), and stems (St; ΔCT = 11.0 ± 0.8). Each bar represents an average of four independent reactions.
FEM111/AGL80 Is Required for the Expression of DME and DD46 during Central Cell Development
The data presented above suggest that FEM111/AGL80 functions as a transcription factor within the central cell and controls the expression of downstream genes required for central cell development and function. To test this further, we analyzed the expression of the DME, FIS2, and DD46 central cell–expressed genes in fem111 mutants. We analyzed the expression of DME–β-glucuronidase (GUS), FIS2-GFP, and ProDD46:GFP in fem111/FEM111 and wild-type plants.
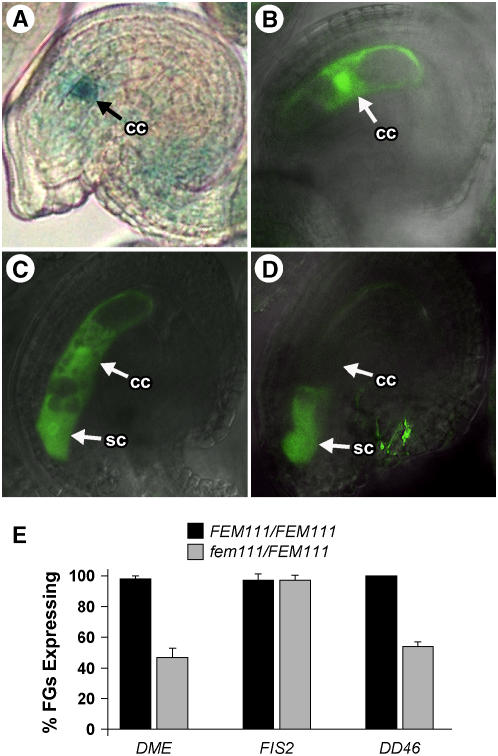
In wild-type female gametophytes, DME-GUS (Choi et al., 2002) and FIS2-GFP are expressed in the central cell (Figures 6A and 6B). As shown in Figure 6E, the number of female gametophytes expressing DME-GUS in fem111/FEM111 plants was ∼50% that in wild-type plants. By contrast, the number of female gametophytes expressing FIS2-GFP was approximately equal in fem111/FEM111 and wild-type plants (Figure 6E).
Figure 6.
Expression of DME, FIS2, and DD46 in Wild-Type and fem111 Female Gametophytes.
(A) Expression of DME-GUS in a wild-type female gametophyte. Expression is detected in the central cell nucleus. The weak signal in the integuments is an artifact of the staining protocol and is not associated with DME-GUS expression.
(B) Expression of FIS2-GFP in a wild-type female gametophyte. Expression is detected only in the central cell (cc).
(C) and (D) Expression of ProDD46:GFP in wild-type (C) and fem111 (D) female gametophytes. In the wild type (C), expression is detected in the central cell (cc) and synergid cells (sc). In fem111 (D), expression is detected only in the synergid cells.
(E) Bar graph of the percentage of ovules expressing DME-GUS, FIS2-GFP, and ProDD46:GFP in wild-type (black bars) and fem111/FEM111 (gray bars) plants. Percentage expression is based on an average of the number of ovules expressing the reporter construct per plant analyzed. For each reporter construct, a minimum of three plants and two siliques per plant were analyzed. Error bars indicate sd. The reporter construct was homozygous for all plants scored. FGs, female gametophytes.
In wild-type female gametophytes, ProDD46:GFP (DD46 corresponds to gene At1g22015) is expressed in both the central cell and the synergid cells (Figure 6C) (J.G. Steffen, J. Macfarlane, and G.N. Drews, unpublished data). By contrast, in fem111/FEM111 plants, the reporter was expressed in the synergid cells but not in the central cell (Figure 6D) in approximately half of the ovules (Figure 6E). Together, these data indicate that FEM111/AGL80 is required for the expression of DME and DD46 but not FIS2 during central cell development.
DISCUSSION
FEM111/AGL80 Encodes a Type I MADS Domain Protein
FEM111/AGL80 is a member of the MADS box family of genes. MADS box genes have been identified in a diverse array of organisms, including plants, animals, slime molds, algae, and fungi (Robles and Pelaz, 2005; Tanabe et al., 2005). Genes in this family encode proteins that contain a structurally conserved MADS domain through which they dimerize (Pellegrini et al., 1995) and bind DNA (Hayes et al., 1988; Treisman, 1992; Tilly et al., 1998). MADS domain–containing proteins function as transcriptional regulators that control many developmental processes in plants, including flowering time, specification of floral meristem identity, fruit development, specification of floral organ identity, and seed coat development (Irish, 2003; Robles and Pelaz, 2005).
Two groups of MADS box genes, referred to as type I and type II (including the MIKC type), are present in plants, animals, and fungi and are probably the result of a duplication event that occurred in a common ancestor (Alvarez-Buylla et al., 2000). Type I and type II MADS box genes are most similar to Serum Response Factor and Myocyte Enhancer Factor2, respectively, in animals. In Arabidopsis, the type I and type II clusters comprise ∼62 and 46 members, respectively (Parenicova et al., 2003). The type II cluster contains many well-characterized genes, including AGAMOUS, APETALA1, APETALA3, PISTILLATA, SEPALLATA1, FLOWERING LOCUS C, FRUITFULL, and SHATTERPROOF1 (Parenicova et al., 2003).
In contrast with the type II genes, little functional characterization has been performed with the type I genes in plants. Functional information is available for only one type I gene, PHERES1 (AGL37), which is a target of the FIE/FIS2/MEA/MSI1 polycomb complex and is thought to play a role in endosperm development (Kohler et al., 2003b). The type I MADS box genes have been subdivided into three clades, Mα, Mβ, and Mγ (Parenicova et al., 2003). FEM111/AGL80 and PHERES1 are both members of the Mγ clade, which includes 14 other genes (Parenicova et al., 2003). RT-PCR analysis showed that 14 of the 16 Mγ genes are expressed within siliques (Kohler et al., 2003b; Parenicova et al., 2003). Together, these observations raise the possibility that other members of this clade may also function during female gametophyte and/or seed development.
FEM111/AGL80 Is Required for Central Cell and Endosperm Development
In the context of the ovule and seed, FEM111/AGL80 is expressed exclusively in the central cell and endosperm (Figure 4). During central cell development, FEM111/AGL80 expression is first detected just before the fusion of the polar nuclei (Figure 4A). This expression pattern is consistent with the phenotype of fem111 mutants, which are affected in central cell development. The central cell defects include failure of maturation of both the nucleolus and the vacuole and are first detected just after the fusion of the polar nuclei. FEM111/AGL80 most likely regulates the expression of genes required for these processes. The nucleolus is the center of ribosome biogenesis within the cell (Hernandez-Verdun, 2005). FEM111/AGL80, therefore, may regulate genes involved in ribosome biogenesis, perhaps to prepare the cell for rapid endosperm development after fertilization. The smaller vacuole in fem111 female gametophytes suggests a loss of osmotic strength in this structure. Osmotic pressure in the vacuole is maintained by proton pumps and other ion transporters (Gaxiola et al., 2002; Horie and Schroeder, 2004). Thus, FEM111/AGL80 may regulate genes required for the proper maintenance of ionic gradients across the tonoplast.
After pollination, endosperm fails to form from fem111 female gametophytes. Instead, the central cell cavity becomes filled with highly fluorescent material that resembles a degenerating synergid cell (Figure 1F), suggesting that fertilization triggers a cell death process in fem111 central cells. Degeneration of fem111 central cells does not occur in the absence of pollination (Figure 1G), suggesting that it may be induced by the fertilization process. At present, we do not know whether the central cell actually becomes fertilized, because the degeneration process was extremely rapid and prevented the observation of sperm nuclei in the central cell (Faure et al., 2002). However, the egg cell does appear to become fertilized based on known structural differences between egg cells (Figures 1B and 1E) and zygotes (Figures 1C and 1F) and on the presence of zygote-like structures in seeds resulting from fertilized fem111 female gametophytes (Figure 1I). It recently was shown that fertilization of the egg cell generates a positive signal for endosperm development (Nowack et al., 2005). Thus, the structural changes in the central cell could possibly be triggered by fertilization of the egg cell.
We also detected AGL80-GFP expression in the endosperm (Figure 4C). Consistent with this observation, AGL80 expression was previously detected within the endosperm by in situ hybridization analysis (Parenicova et al., 2003). These observations suggest that FEM111/AGL80 plays a role during endosperm development. However, at present, whether FEM111/AGL80 plays a role in the endosperm is unclear, because fem111 female gametophytes fail to initiate endosperm development (Figure 1F). Future studies involving FEM111/AGL80 overexpression, directed RNA interference, or mosaic analysis may allow the analysis of FEM111/AGL80 function in the endosperm.
Fertilized fem111 egg cells give rise to zygote-like structures (Figure 1I). FEM111/AGL80 is not expressed in the egg cell (Figure 4B) or the developing embryo (Figure 4C). These observations suggest that the embryo development defect results from the lack of endosperm. Zygote-like structures were also observed in autonomously developing mea (fis1) and fis2 seeds (Chaudhury et al., 1997).
Using real-time RT-PCR, we detected FEM111/AGL80 RNA in roots, stems, leaves, young flowers, and anthers (Figure 5). With the exception of the root expression, these results are consistent with those of a prior analysis of this gene (Parenicova et al., 2003; de Folter et al., 2004). This ubiquitous expression pattern suggests that FEM111/AGL80 functions throughout the plant. However, we were not able to determine whether this mutation affects the sporophyte generation because, as a result of high penetrance, we were unable to isolate homozygous mutants. Within young flowers, FEM111/AGL80 expression was detected by in situ hybridization exclusively in microspores (Parenicova et al., 2003). Thus, the RT-PCR signal in young flowers and anthers is probably attributable to expression in the pollen. This expression pattern suggests a possible function in pollen. The fem111 mutation does not exhibit reduced transmission through the male gametophyte (Table 1), suggesting that FEM111/AGL80 functions redundantly with other pollen-expressed genes, the mutation produces a subtle defect that does not affect transmission, or FEM111/AGL80 does not function during pollen development.
Models for FEM111/AGL80 Function
We show that DME (Figure 6E) and DD46 (Figures 6C and 6E) are not expressed in the central cell in fem111 female gametophytes, suggesting that FEM111/AGL80 is upstream of these two genes in the central cell gene regulatory network. The requirement of FEM111/AGL80 for DME and DD46 expression may be direct or indirect. Many members of the MADS box transcription factor family bind to a conserved DNA regulatory motif termed the CArG box [CC(A/T)6GG]. We did not identify a CArG box within 2.0 kb of sequence upstream of the translational start site of either DME or DD46, suggesting an indirect mechanism of activation. By contrast, FIS2 appears to be part of an independent pathway, because its expression does not require FEM111/AGL80 during central cell development (Figure 6E). These data suggest that FEM111/AGL80 functions as a transcriptional regulator required for specific aspects of central cell differentiation, including vacuole maintenance or growth, maturation of the nucleolus, and expression of DME and DD46. In addition, because fem111 mutants fail to initiate endosperm development, FEM111/AGL80 may regulate genes required for the initiation of endosperm development and/or genes required for endosperm viability after fertilization. Identification of additional central cell–expressed genes regulated by FEM111/AGL80 will be required to fully dissect the regulatory pathways downstream of FEM111/AGL80.
METHODS
Growth Conditions
Arabidopsis thaliana seeds were sterilized in chlorine gas and germinated on plates containing 0.5× Murashige and Skoog salts (Sigma-Aldrich M-9274), 0.05% MES, 0.5% sucrose, and 0.8% Phytagar (Life Technologies). Ten-day-old seedlings were transferred to Scott's Redi-Earth and grown under 24-h illumination.
Plant Transformation
T-DNA constructs were introduced into Agrobacterium tumefaciens strain LBA4404 by electroporation. Arabidopsis plants (ecotype Columbia) were transformed using a modified floral dip procedure (Clough and Bent, 1998). Transformed progeny were selected by germinating surface-sterilized T1 seeds on growth medium containing antibiotics. Resistant seedlings were transplanted to soil after 10 d of growth.
Segregation Analysis
For reciprocal cross analysis, fem111/FEM111 plants were crossed with wild-type plants as outlined in Table 1. F1 seeds were germinated on growth medium containing 50 μg/mL kanamycin, and the number of resistant and sensitive plants was scored. Resistant and sensitive plants were scored as genotypes fem111/FEM111 and FEM111/FEM111, respectively. Statistical analyses of these data were performed using the χ2 test.
For self-cross analysis, fem111/FEM111 plants were allowed to self-pollinate, progeny seeds were collected, and the number of resistant and sensitive progeny was scored. Resistant progeny from the self-cross could be either heterozygous or homozygous for the fem111 T-DNA insertion. To distinguish between these possibilities, resistant plants were grown and seed set was scored; all resistant plants exhibited ∼50% seed set, indicating that they were heterozygous. Statistical analyses of these data were performed using the χ2 test.
Confocal Analysis
CLSM of ovules and seeds was performed as described previously (Christensen et al., 1997, 1998, 2002), with the modification that we used a Zeiss LSM510 microscope. Measurements of nucleoli diameter were obtained using the Zeiss LSM software package.
Analysis of GFP Expression Patterns
Tissue from plants containing GFP constructs was analyzed using a Zeiss LSM510 confocal microscope. GFP was excited with an argon laser at a wavelength of 488 nm, and emission was detected between 500 and 530 nm. For analysis of the terminal developmental stage, we emasculated flowers at stage 12c, waited 24 h, removed the flowers from the plants, removed the sepals, petals, and stamens, and dissected off the carpel walls using a 30-gauge syringe needle. The pistils were then mounted on a slide in 1× Murashige and Skoog salts, pH 7.0, for CLSM analysis. For analysis of different developmental stages, we used flowers of the appropriate stage (Christensen et al., 1997) and processed them as described above.
Gene Identification
The fem111 mutant was isolated in a screen of T-DNA–mutagenized lines generated with the pCGN1547 vector (McBride and Summerfelt, 1990). We used thermal asymmetric interlaced PCR (Liu et al., 1995) to identify the sequences flanking the T-DNA. We used primers oligo-L1 (5′-CTCAATGCAAAAGGGGAACG-3′) and FEM111CL1 (5′-AGCTACCGCAACAGAAGAAGAT-3′) to characterize the T-DNA insertion of the left border and primers RBMP2 (5′-GCGTTACCCAACTTAATCGC-3′) and FEM111CR1 (5′-ATCTTATAATACCAAATCCTACT-3′) to characterize the T-DNA insertion of the right border. The PCR products from these reactions were then sequenced to determine the nature of the insertion site. The T-DNA right border inserts between the first and second nucleotides of the open reading frame and faces the 5′ region of the gene. The T-DNA left border inserts after the seventh nucleotide of the open reading frame and faces the 3′ region of the gene.
Molecular Complementation
Molecular complementation was performed using a 2600-bp DNA fragment containing the FEM111/AGL80 genomic coding sequence (966 bp) along with 1332 bp of sequence upstream of the translational start codon and 366 bp of sequence downstream of the stop codon. This DNA fragment was amplified by PCR using the primers FEM111-R2 (5′-AAGACGGATTTGGAGTG-3′) and FEM111F2 (5′-CAGCGTTGAGTATGACA-3′) and cloned into the TOPO II PCR cloning vector (Invitrogen) to generate clone FEM111-2.6/TOPO. The insert was released from clone FEM111-2.6/TOPO with BamHI and XhoI, and the fragment was subcloned into pCAMBIA1300 at BamHI and SalI, producing clone FEM111-2.6/pCAMBIA. FEM111-2.6/pCAMBIA was introduced into Arabidopsis plants heterozygous for the fem111 mutation as described above. The pCAMBIA1300 T-DNA contains a marker gene conferring resistance to hygromycin, and the pCGN1547 T-DNA contains a marker gene conferring resistance to kanamycin. Transformed plants were selected by germinating T1 seeds on growth medium containing 15 μg/mL hygromycin and 50 μg/mL kanamycin to select for plants containing both the rescue transgene and the fem111 mutation. The presence of the fem111 mutation was verified by PCR with primers oligo-L1 and FEM111CL1.
Plants heterozygous for the fem111 allele and hemizygous for the rescue construct at a single locus were allowed to self-cross. The progeny seeds were germinated on growth medium containing 50 μg/mL kanamycin, and the number of resistant and sensitive plants was scored. We observed 77 kanamycin-resistant progeny and 43 kanamycin-sensitive progeny. Plants homozygous for the fem111 allele and homozygous for the rescue construct were identified by having progeny seeds that were 100% kanamycin-resistant and 100% hygromycin-resistant. These plants had seed set indistinguishable from the wild type.
Cloning the FEM111/AGL80 cDNA
To amplify a cDNA encompassing the entire open reading frame of FEM111/AGL80, we performed RT-PCR on RNA extracted from pistils harvested from flowers at stage 13 (Smyth et al., 1990). RNA was extracted using the Qiagen RNeasy kit according to the manufacturer's instructions (www.qiagen.com). Aliquots of RNA (1 μg) were reverse-transcribed using the RETROscript kit (Ambion) according to the manufacturer's instructions. PCR was performed with primers FEM111-cDNA-F (5′-CAATGACAAGAAAGAAAGTG-3′) and FEM111-cDNA-R (5′-ACCTAATGGAACCATGTTT-3′). The cDNA was cloned into the TOPO II PCR vector using the TOPO TA cloning kit (Invitrogen), resulting in plasmid pCRII-FEM111.
We identified the 5′ and 3′ untranslated sequences with RACE using the First Choice RLM-RACE kit (Ambion). For 5′ RACE, the gene-specific outer primer was 5′RACE-Outer#2 (5′-AAGAGCGAGAGTTCGTGTACCTTC-3′) and the gene-specific inner primer was 5′RACE-Inner#2 (5′-AAGAATCGTTGGAAATGTAAGCAAG-3′). For 3′ RACE, the gene-specific outer primer was AGLSEQp4 (5′-AGCGACAGAAACTTTGAGGAGA-3′) and the gene-specific inner primer was AGLSEQp6 (5′-TCGCAGGATTGAGATTTTAC-3′). This analysis showed that FEM111/AGL80 contains 5′ and 3′ untranslated regions of 51 and 109 bp, respectively.
Sequence Analysis
We used pfam 17.0 (http://pfam.wustl.edu/hmmsearch.shtml) to identify the MADS domain. We used psort (http://psort.nibb.ac.jp/form.html) to identify the putative nuclear localization signal. We identified the His-rich motif at the C terminus of the protein using prosite (http://ca.expasy.org/prosite/).
Generating AGL80-GFP
The AGL80-GFP fusion construct has GFP inserted 33 bp upstream of the stop codon of the FEM111/AGL80 gene. As a first step in cloning this gene, we generated three PCR products. An upstream fragment was amplified using primers FEM111GFPF1 (5′-CTCGCCCTTGCTCACCATAGTAACAGGGATGATGCT-3′; contains 18 bp of sequence found in GFP) and FEM111GFPR1 (5′-ATTTAAATCAGAACTATAATC-3′; includes a native SwaI site). A downstream fragment was amplified using primers FEM111GFPF2 (5′-CCTCGAGCAGCGTTGAGTATGACAAAG-3′; contains an XhoI site) and FEM111GFPR2 (5′-ATGGACGAGCTGTACAAGAGTTCAAGTATTACCAAT-3′; contains 18 bp of sequence found in GFP). The GFP insert was amplified from pBI-GFP(S65T) (Choi et al., 2002) using primers GFPfuseF1 (5′-ATGGTGAGCAAGGGCGAG-3′) and GFPfuseR1 (5′-CTTGTACAGCTCGTCCAT-3′). The GFP fragment contained the entire GFP open reading frame, excluding the stop codon. The three PCR fragments were gel-purified using QIAquick spin columns and then used as the template for a PCR using primers FEM111GFPF2 and FEM111GFPR1, which resulted in a 1.2-kb fusion product containing GFP and FEM111 sequence. This product was TOPO-cloned and sequenced to verify that no PCR errors existed. The TOPO clone was then cut with SwaI and XhoI and inserted into FEM111-2.6/TOPO at XhoI/SwaI to create FEM111-GFP/TOPO. Finally, FEM111-GFP/TOPO was cut with BamHI and XhoI and was subcloned into pCAMBIA1300 at BamHI/SalI to create FEM111-GFP/pCAMBIA.
This construct was introduced into Arabidopsis plants as described above, and transformed plants were selected by germinating seeds on growth medium containing 50 μg/mL hygromycin. The expression patterns reported in Results are derived from the analysis of 10 single-insert AGL80-GFP lines. Plants homozygous for both the AGL80-GFP construct and the fem111 T-DNA insert (as determined by kanamycin and hygromycin selection of progeny) had 100% seed set.
Real-Time RT-PCR
Tissue was harvested from plants and placed immediately into liquid nitrogen. Ovaries were harvested from the oldest stage 12 flower (Smyth et al., 1990) on each plant as well as the next two older flowers from plants harboring the male sterile1 mutation (Wilson et al., 2001; Ito and Shinozaki, 2002) to prevent self-pollination. Floral cluster tissue includes the inflorescence meristem and flowers at stages 1 to 10 (Smyth et al., 1990). Silique tissue includes siliques at 1 to 2 d after pollination. Leaf tissue includes leaves of 5 to 12 mm. Roots were harvested from seedlings at 11 d after germination. Floral stem tissue includes internodes from 4-week-old plants.
RNA was extracted from these tissues using the Qiagen RNeasy kit according to the manufacturer's instructions (www.qiagen.com). DNA contamination was removed from samples using the Ambion TURBO DNA-free DNase kit according to the manufacturer's instructions. After DNase treatment, RNA samples were repurified using the Qiagen RNeasy kit according to the manufacturer's instructions. Aliquots of RNA (1 μg) were reverse-transcribed using the RETROscript kit (Ambion) according to the manufacturer's instructions.
For real-time RT-PCR, PCR was performed using the Roche FastStart DNA Master SYBR Green I master mix (www.roche.com) in a volume of 10 μL on a Roche LightCycler system. The PCR mixture consisted of 0.5 μL of cDNA, 0.5 μM primers, and 1× master mix. In every real-time RT-PCR run, ACTIN2 was used as an internal control to normalize for variation in the amount of cDNA template. The PCR primers used were MP1-1F (5′-GACTCTTTGTGGCATCACTGC-3′) and MP1-1R (5′-CCTCAAAGTTTCTGTCGCTTTC-3′). The PCR program consisted of a first step of denaturation and Taq activation (95°C for 5 min) followed by 45 cycles of denaturation (95°C for 15 s), annealing (60°C for 15 s), and extension (72°C for 10 s). To determine the specificity of the PCR, the amplified products were subjected to melt curve analysis using the machine's standard method. The reported values are averages of four independent trials (two biological replicates and two technical replicates). We calculated relative expression levels as follows. We first normalized AGL80 transcript levels relative to a standard (ACTIN2) using the formula ΔCT = CT(AGL80) − CT(ACTIN2). We next calculated an average ΔCT value for each tissue. Anther, the tissue with the highest relative expression (lowest ΔCT value), was used as the standard for the comparison of expression levels. We then calculated relative expression levels using the equation 100 × 2−[average ΔCT (tissue) − average ΔCT (anther)].
Analysis of DME-GUS, FIS2-GFP, and ProDD46:GFP in fem111 Female Gametophytes
The DME-GUS construct contains 2.3 kb of promoter and 1.9 kb of the DME gene (including a nuclear localization signal) fused to GUS (Choi et al., 2002). The FIS2-GFP construct contains 2.5 kb of promoter and the entire genomic coding region fused to GFP. The ProDD46:GFP reporter construct contains 1 kb of 5′ flanking sequence of the gene At1g22015 fused to GFP in the vector pBI-GFP(S65T).
To analyze promoter:reporter constructs in the fem111 mutant background, we crossed fem111/FEM111 plants as males with plants homozygous for the reporter construct. To identify F1 plants containing the fem111 T-DNA allele, PCR was performed with the primers RBMP2 (5′-GCGTTACCCAACTTAATCGC-3′) and FEMCR1 (5′-ATCTTATAATACCAAATCCTACT-3′). Plants heterozygous for the fem111 T-DNA insertion and hemizygous for the reporter construct were allowed to self-cross. Progeny from the self-cross were then scored for the fem111 T-DNA insertion by PCR, as described above. One-quarter of these plants should also be homozygous for the reporter construct. To identify these plants, we made use of the fact that all reporter constructs were inserted into vectors conferring kanamycin resistance. Offspring containing the fem111 T-DNA allele were then allowed to self-cross. Seeds from this cross were plated on medium containing 50 μg/mL kanamycin, and the ratio of kanamycin-sensitive to kanamycin-resistant seedlings was scored. Plants that produced 100% kanamycin-resistant progeny were determined to be homozygous for the promoter:reporter construct.
Analysis of reporter expression was performed at stage FG7. Flowers were emasculated at stage 12c (Christensen et al., 1997), and pistils were collected at 24 h after emasculation. Pistils from plants containing DME-GUS were stained for GUS activity in a solution containing 50 mM sodium phosphate buffer, pH 7.0, 0.2% Triton X-100, 2.5 mM potassium ferrocyanide, 2.5 mM potassium ferricyanide, and 1 mM 5-bromo-4-chloro-3-indolyl-β-glucuronic acid. Tissue was placed into this solution on ice, vacuum-infiltrated for 15 min, and incubated for 4 h at 37°C. The ovules were then analyzed on a Zeiss Axioplan upright microscope with ×20 and ×40 objectives. Analysis of the GFP lines was performed as described above.
Accession Numbers
The GenBank accession number for the AGL80 nucleotide sequence is DQ406752. The Arabidopsis Genome Initiative number for the protein sequence of AGL80 is At5g48670.1.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure 1. Silique Phenotype of fem111 Mutants.
Supplementary Material
Acknowledgments
We thank Ramin Yadegari and members of the Drews laboratory for critical review of the manuscript. We thank Yeonhee Choi and Ramin Yadegari for the DME-GUS and FIS2-GFP lines, respectively. We thank Ed King and the Department of Biology Microscopy Facility for guidance with the microscopic analysis. This work was supported by Department of Energy Grant DE-FG02-04ER15620 to G.N.D. and USDA Cooperative State Research, Education, and Extension Services Fellowship 2004-35304-14931 to M.F.P.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Gary N. Drews (drews@bioscience.utah.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.106.040824.
References
- Acosta-Garcia, G., and Vielle-Calzada, J.P. (2004). A classical arabinogalactan protein is essential for the initiation of female gametogenesis in Arabidopsis. Plant Cell 16 2614–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agashe, B., Prasad, C.K., and Siddiqi, I. (2002). Identification and analysis of DYAD: A gene required for meiotic chromosome organisation and female meiotic progression in Arabidopsis. Development 129 3935–3943. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla, E.R., Pelaz, S., Liljegren, S.J., Gold, S.E., Burgeff, C., Ditta, G.S., Ribas de Pouplana, L., Martinez-Castilla, L., and Yanofsky, M.F. (2000). An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 97 5328–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury, A.M., Ming, L., Miller, C., Craig, S., Dennis, E.S., and Peacock, W.J. (1997). Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94 4223–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Y., Gehring, M., Johnson, L., Hannon, M., Harada, J.J., Goldberg, R.B., Jacobsen, S.E., and Fischer, R.L. (2002). DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110 33–42. [DOI] [PubMed] [Google Scholar]
- Christensen, C.A., Gorsich, S.W., Brown, R.H., Jones, L.G., Brown, J., Shaw, J.M., and Drews, G.N. (2002). Mitochondrial GFA2 is required for synergid cell death in Arabidopsis. Plant Cell 14 2215–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, C.A., King, E.J., Jordan, J.R., and Drews, G.N. (1997). Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex. Plant Reprod. 10 49–64. [Google Scholar]
- Christensen, C.A., Subramanian, S., and Drews, G.N. (1998). Identification of gametophytic mutations affecting female gametophyte development in Arabidopsis. Dev. Biol. 202 136–151. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- de Folter, S., Busscher, J., Colombo, L., Losa, A., and Angenent, G.C. (2004). Transcript profiling of transcription factor genes during silique development in Arabidopsis. Plant Mol. Biol. 56 351–366. [DOI] [PubMed] [Google Scholar]
- Drews, G.N., Lee, D., and Christensen, C.A. (1998). Genetic analysis of female gametophyte development and function. Plant Cell 10 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel, C., Mariconti, L., and Gruissem, W. (2004). Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429 776–780. [DOI] [PubMed] [Google Scholar]
- Faure, J.E., Rotman, N., Fortune, P., and Dumas, C. (2002). Fertilization in Arabidopsis thaliana wild type: Developmental stages and time course. Plant J. 30 481–488. [DOI] [PubMed] [Google Scholar]
- Gaxiola, R.A., Fink, G.R., and Hirschi, K.D. (2002). Genetic manipulation of vacuolar proton pumps and transporters. Plant Physiol. 129 967–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring, M., Huh, J.H., Hsieh, T.F., Penterman, J., Choi, Y., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (2006). DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus, U., Vielle-Calzada, J.P., Hoeppner, M.A., and Gagliano, W.B. (1998). Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280 446–450. [DOI] [PubMed] [Google Scholar]
- Guitton, A.E., Page, D.R., Chambrier, P., Lionnet, C., Faure, J.E., Grossniklaus, U., and Berger, F. (2004). Identification of new members of Fertilisation Independent Seed Polycomb Group pathway involved in the control of seed development in Arabidopsis thaliana. Development 131 2971–2981. [DOI] [PubMed] [Google Scholar]
- Hayes, T.E., Sengupta, P., and Cochran, B.H. (1988). The human c-fos serum response factor and the yeast factors GRM/PRTF have related DNA-binding specificities. Genes Dev. 2 1713–1722. [DOI] [PubMed] [Google Scholar]
- Hejatko, J., Pernisova, M., Eneva, T., Palme, K., and Brzobohaty, B. (2003). The putative sensor histidine kinase CKI1 is involved in female gametophyte development in Arabidopsis. Mol. Genet. Genomics 269 443–453. [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun, D. (2005). Nucleolus in the spotlight. Cell Cycle 4 106–108. [DOI] [PubMed] [Google Scholar]
- Heuer, S., Hansen, S., Bantin, J., Brettschneider, R., Kranz, E., Lorz, H., and Dresselhaus, T. (2001). The maize MADS box gene ZmMADS3 affects node number and spikelet development and is co-expressed with ZmMADS1 during flower development, in egg cells, and early embryogenesis. Plant Physiol. 127 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie, T., and Schroeder, J.I. (2004). Sodium transporters in plants. Diverse genes and physiological functions. Plant Physiol. 136 2457–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huanca-Mamani, W., Garcia-Aguilar, M., Leon-Martinez, G., Grossniklaus, U., and Vielle-Calzada, J.P. (2005). CHR11, a chromatin-remodeling factor essential for nuclear proliferation during female gametogenesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102 17231–17236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, B.-Q., and Russell, S.D. (1992). Female germ unit: Organization, isolation, and function. Int. Rev. Cytol. 140 233–292. [Google Scholar]
- Irish, V.F. (2003). The evolution of floral homeotic gene function. Bioessays 25 637–646. [DOI] [PubMed] [Google Scholar]
- Ito, T., and Shinozaki, K. (2002). The MALE STERILITY1 gene of Arabidopsis, encoding a nuclear protein with a PHD-finger motif, is expressed in tapetal cells and is required for pollen maturation. Plant Cell Physiol. 43 1285–1292. [DOI] [PubMed] [Google Scholar]
- Kasahara, R.D., Portereiko, M.F., Sandaklie-Nikolova, L., Rabiger, D.S., and Drews, G.N. (2005). MYB98 is required for pollen tube guidance and synergid cell differentiation in Arabidopsis. Plant Cell 17 2981–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.U., Li, Y., and Huang, A.H. (2005). Ubiquitous and endoplasmic reticulum-located lysophosphatidyl acyltransferase, LPAT2, is essential for female but not male gametophyte development in Arabidopsis. Plant Cell 17 1073–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue, T., Ohad, N., Yadegari, R., Hannon, M., Dinneny, J., Wells, D., Katz, A., Margossian, L., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (1999). Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc. Natl. Acad. Sci. USA 96 4186–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler, C., Hennig, L., Bouveret, R., Gheyselinck, J., Grossniklaus, U., and Gruissem, W. (2003. a). Arabidopsis MSI1 is a component of the MEA/FIE polycomb group complex and required for seed development. EMBO J. 22 4804–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler, C., Hennig, L., Spillane, C., Pien, S., Gruissem, W., and Grossniklaus, U. (2003. b). The polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 17 1540–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwee, H.S., and Sundaresan, V. (2003). The NOMEGA gene required for female gametophyte development encodes the putative APC6/CDC16 component of the anaphase promoting complex in Arabidopsis. Plant J. 36 853–866. [DOI] [PubMed] [Google Scholar]
- Le, Q., Gutierrez-Marcos, J.F., Costa, L.M., Meyer, S., Dickinson, H.G., Lorz, H., Kranz, E., and Scholten, S. (2005). Construction and screening of subtracted cDNA libraries from limited populations of plant cells: A comparative analysis of gene expression between maize egg cells and central cells. Plant J. 44 167–178. [DOI] [PubMed] [Google Scholar]
- Liu, Y.G., Mitsukawa, N., Oosumi, T., and Whittier, R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8 457–463. [DOI] [PubMed] [Google Scholar]
- Luo, M., Bilodeau, P., Dennis, E.S., Peacock, W.J., and Chaudhury, A. (2000). Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 97 10637–10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M., Bilodeau, P., Koltunow, A., Dennis, E.S., Peacock, W.J., and Chaudhury, A.M. (1999). Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnard, J.L., Lehouque, G., Massonneau, A., Frangne, N., Heckel, T., Gutierrez-Marcos, J.F., Perez, P., Dumas, C., and Rogowsky, P.M. (2003). ZmEBE genes show a novel, continuous expression pattern in the central cell before fertilization and in specific domains of the resulting endosperm after fertilization. Plant Mol. Biol. 53 821–836. [DOI] [PubMed] [Google Scholar]
- McBride, K.E., and Summerfelt, K.R. (1990). Improved binary vectors for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 14 269–276. [DOI] [PubMed] [Google Scholar]
- Niewiadomski, P., Knappe, S., Geimer, S., Fischer, K., Schulz, B., Unte, U.S., Rosso, M.G., Ache, P., Flugge, U.I., and Schneider, A. (2005). The Arabidopsis plastidic glucose 6-phosphate/phosphate translocator GPT1 is essential for pollen maturation and embryo sac development. Plant Cell 17 760–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack, M.K., Grini, P.E., Jakoby, M.J., Lafos, M., Koncz, C., and Schnittger, A. (2006). A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat Genet. 38 63–67. [DOI] [PubMed] [Google Scholar]
- Ohad, N., Margossian, L., Hsu, Y.C., Williams, C., Repetti, P., and Fischer, R.L. (1996). A mutation that allows endosperm development without fertilization. Proc. Natl. Acad. Sci. USA 93 5319–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad, N., Yadegari, R., Margossian, L., Hannon, M., Michaeli, D., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (1999). Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, O.-A. (2004). Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell 16 (suppl.), S214–S227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenicova, L., de Folter, S., Kieffer, M., Horner, D.S., Favalli, C., Busscher, J., Cook, H.E., Ingram, R.M., Kater, M.M., Davies, B., Angenent, G.C., and Colombo, L. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 15 1538–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini, L., Tan, S., and Richmond, T.J. (1995). Structure of serum response factor core bound to DNA. Nature 376 490–498. [DOI] [PubMed] [Google Scholar]
- Pischke, M.S., Jones, L.G., Otsuga, D., Fernandez, D.E., Drews, G.N., and Sussman, M.R. (2002). An Arabidopsis histidine kinase is essential for megagametogenesis. Proc. Natl. Acad. Sci. USA 99 15800–15805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles, P., and Pelaz, S. (2005). Flower and fruit development in Arabidopsis thaliana. Int. J. Dev. Biol. 49 633–643. [DOI] [PubMed] [Google Scholar]
- Shi, D.Q., Liu, J., Xiang, Y.H., Ye, D., Sundaresan, V., and Yang, W.C. (2005). SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell 17 2340–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi, I., Ganesh, G., Grossniklaus, U., and Subbiah, V. (2000). The dyad gene is required for progression through female meiosis in Arabidopsis. Development 127 197–207. [DOI] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillane, C., MacDougall, C., Stock, C., Kohler, C., Vielle-Calzada, J.P., Nunes, S.M., Grossniklaus, U., and Goodrich, J. (2000). Interaction of the Arabidopsis polycomb group proteins FIE and MEA mediates their common phenotypes. Curr. Biol. 10 1535–1538. [DOI] [PubMed] [Google Scholar]
- Sprunck, S., Baumann, U., Edwards, K., Langridge, P., and Dresselhaus, T. (2005). The transcript composition of egg cells changes significantly following fertilization in wheat (Triticum aestivum L.). Plant J. 41 660–672. [DOI] [PubMed] [Google Scholar]
- Tanabe, Y., Hasebe, M., Sekimoto, H., Nishiyama, T., Kitani, M., Henschel, K., Munster, T., Theissen, G., Nozaki, H., and Ito, M. (2005). Characterization of MADS-box genes in charophycean green algae and its implication for the evolution of MADS-box genes. Proc. Natl. Acad. Sci. USA 102 2436–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly, J.J., Allen, D.W., and Jack, T. (1998). The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development 125 1647–1657. [DOI] [PubMed] [Google Scholar]
- Treisman, R. (1992). The serum response element. Trends Biochem. Sci. 17 423–426. [DOI] [PubMed] [Google Scholar]
- Willemse, M.T.M., and van Went, J.L. (1984). The female gametophyte. In Embryology of Angiosperms, B.M. Johri, ed (Berlin: Springer-Verlag), pp. 159–196.
- Wilson, Z.A., Morroll, S.M., Dawson, J., Swarup, R., and Tighe, P.J. (2001). The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 28 27–39. [DOI] [PubMed] [Google Scholar]
- Yadegari, R., and Drews, G.N. (2004). Female gametophyte development. Plant Cell 16 (suppl.), S133–S141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.