Abstract
Regulation of seed germination requires coordinate action by the embryo and surrounding endosperm. We used Arabidopsis thaliana to establish the relative roles of embryo and endosperm in the control of seed germination and seedling establishment. We previously showed that endospermic oil reserves are used postgerminatively via gluconeogenesis to fuel seedling establishment and that lipid breakdown is repressed by abscisic acid (ABA) in embryo but not endosperm tissues. Here, we use RNA amplification to describe the transcriptome of the endosperm and compare the hormone responses of endosperm and embryo tissues. We show that the endosperm responds to both ABA and gibberellin but that ABA in particular regulates nuclear but not plastid-encoded photosynthetic gene expression in the embryo. We also show that ABA INSENSITIVE4 (ABI4) expression is confined to the embryo, accounts for the major differences in embryo response to ABA, and defines a role for ABI4 as a repressor of lipid breakdown. Furthermore, ABI5 expression in the endosperm defines a second region of altered ABA signaling in the micropylar endosperm cap. Finally, embryo and endosperm ABA signaling mutants demonstrate the spatial specificity of ABA action in seed germination. We conclude that the single cell endosperm layer plays an active role in the regulation of seed germination in Arabidopsis.
INTRODUCTION
Arabidopsis thaliana seeds exhibit a common form of dormancy known as coat induced or coat enhanced. These terms describe the important role of tissues surrounding the seed in the maintenance of dormancy. In Arabidopsis, the availability of mutants has focused attention on the role of the testa in coat-enhanced dormancy, but in general, the term covers all tissues surrounding the embryo, which include the endosperm and pericarp where present (Bewley and Black, 1994). The key role of these embryo-surrounding tissues in the control of germination is neatly demonstrated by the fact that damaging or removing them breaks dormancy and stimulates embryo growth in many species.
The endosperm in the mature Arabidopsis seed consists of a single cell layer resembling the cereal aleurone. Until recently it was believed to be of little importance once the seed has reached maturity (Berger, 1999). However, we have shown that carbon stored as triacylglycerol in the Arabidopsis endosperm is used via gluconeogenesis to fuel seedling establishment (Penfield et al., 2004). Although abscisic acid (ABA) can inhibit lipid breakdown in the embryo, endosperm storage reserve mobilization occurs independently of ABA levels, demonstrating tissue-specific differences in ABA signaling in the seed. This differential sensitivity of lipid mobilization in the embryo and endosperm to ABA has subsequently been confirmed in tobacco (Nicotiana tabacum) seeds (Manz et al., 2005), suggesting wide conservation among seed plants and important functional significance of the process. However, the molecular basis of this effect remains unclear.
In tobacco and tomato (Solanum lycopersicum), both of which contain comparatively large endosperms in the mature seed, the endosperm is a major focus in efforts to understand the control of germination. In addition to its function as a storage tissue, in these species and others, the endosperm has been shown to exert control over germination by secreting cell wall loosening enzymes that weaken the mechanical resistance of the micropylar endosperm cap to radicle protrusion (reviewed in Bewley, 1997b). Hence, understanding the role of the endosperm is vital to our knowledge of the control of germination in Arabidopsis and other seed plants. To this end, many screens have been performed to identify endosperm-expressed genes during seed germination (Dubreucq et al., 2003; Liu et al., 2005).
Seed dormancy and germination are strongly under the control of phytohormones and their responses. Genetic analysis of seed germination in Arabidopsis has revealed that ABA and gibberellins (GAs) are crucial regulators (Bentsink and Koornneef, 2002). Both ABA-deficient and several aba insensitive (abi) mutants show reduced dormancy phenotypes. abi3 mutants also show defects in seed maturation and desiccation tolerance (Nambara et al., 1992). Two further loci, ABI4 and ABI5, are not required for dormancy but are necessary for the ABA inhibition of germination and have additive effects on seed maturation when combined with abi3, leafy cotyledon1, or fusca3, suggesting a function in the maturing seed (Brocard-Gifford et al., 2003). ABI3, ABI4, and ABI5 encode B3-, AP2-, and bZIP-type transcription factors, respectively, suggesting that regulation at the level of transcription is important for the ABA response in seeds (Giraudat et al., 1992; Finkelstein et al., 1998; Finkelstein and Lynch, 2000). Despite the central role of ABA in the imposition of seed dormancy, mature seeds treated even with high concentrations of exogenous ABA do not mimic dormant seeds but arrest with the seed coat split and the radicle held within the micropylar endosperm (Koornneef et al., 1982; Penfield et al., 2004). Importantly, the activity of endosperm-expressed cell wall loosening enzymes is also controlled by both ABA and GA (Groot et al., 1988; Toorop et al., 2000).
We have shown previously that embryo and endosperm tissues can be successfully isolated from mature Arabidopsis seeds after the seed coat has undergone programmed cell death (Penfield et al., 2004). To further probe the role of the endosperm in germination control in Arabidopsis, and ABA signaling during germination, we performed microarray expression profiling of isolated embryo and endosperm tissues using RNA amplification and the ATH1 chip. The results reveal the transcriptome of the endosperm and describe in detail the embryo and endosperm response to ABA. We demonstrate that the expression domain of the ABI4 gene is the crucial determinant of the sensitivity of lipid mobilization to ABA during seed germination and that ABI5 expression is a marker for the critical micropylar region of the endosperm. Finally, using mutants disrupted in either the embryo or endosperm ABA responses, we show the control of Arabidopsis seed germination by the endosperm.
RESULTS
Differential Gene Expression in the Embryo and Endosperm during Germination
To probe in detail the phytohormonal responses of the embryo and endosperm during germination, we undertook global expression profiling using the ATH1 gene chip 1 d after transfer to 22°C after 3 d of cold stratification. At this time point, shortly after radicle emergence, the percentage decrease in storage lipid is significantly more advanced in endosperm compared with embryo tissues, whereas transcriptional activity of various storage lipid–related genes as determined by promoter-reporter gene activity is detectable in both (Penfield et al., 2004, 2005). Additional transcriptome data sets were generated from seeds that were stratified for 3 d and transferred to 22°C for 1 d, having been maintained throughout on media containing either 20 μM ABA or 20 μM paclobutrazol (PAC). We harvested embryo and endosperm tissue in triplicate samples and compared gene expression.
First, we noticed that the number of probe sets (12,190) found to be expressed in the endosperm using the ATH1 chip was very similar to that in the embryo (13,121). These numbers closely resemble the number of genes found to be expressed in other Arabidopsis tissues, excluding pollen (Schmid et al., 2005). Significance analysis of microarray (SAM) analysis (Tusher et al., 2001) found that the expression of 9650 probe sets did not differ significantly between embryo and endosperm tissues (Figure 1A), with ∼4000 genes expressed differentially in either the embryo or endosperm. Plotting embryo and endosperm transcript levels supported this view, showing a broad correlation in gene expression levels between the two tissues (R2 = 0.75; Figure 1B). Hence, we concluded that patterns of gene expression are broadly similar between the two organs at this time point, perhaps reflecting the similar postgerminative metabolism occurring in these tissues.
Figure 1.
Comparison of the Embryo and Endosperm Transcriptome 24 h after Transfer to 22°C.
(A) Number of detected expressed genes in embryo, endosperm, or both tissues.
(B) Plot of Z-score transformed embryo versus endosperm expression data shows similar expression of most genes in both tissues.
(C) and (D) Transmission electron micrograph of representative embryo and endosperm cells 1 d after transfer to 22°C showing cells densely packed with ribosomes in the embryo (C) compared with the endosperm (D). Numbers at the bottom left of the panels indicate the mean ±se of ribosomes per μm2 of cytoplasm observable in a 100-μm-thin section. CW, cell wall; RB, ribosomes; LB, lipid body; PX, peroxisome. Bars = 1 μM.
Using highly stringent criteria, we identified embryo- and endosperm-specific transcripts during germination. To qualify, genes must be scored as present in all three replicate samples in the expressed tissue and must be called absent in the nonexpressed tissue in at least two of the three replicate experiments. Genes whose function has been previously addressed in other published work and fulfill these criteria are shown in Supplemental Table 1 online. The Arabidopsis extensin-like gene EPR1 is known to be expressed solely in the endosperm during germination and was used previously as a marker for endosperm identity (Dubreucq et al., 2003; Penfield et al., 2004). This control gene was found to be expressed highly in endosperm samples yet was absent in the embryo (3749.8 ± 122.6 versus 25.2 ± 4.5; see Supplemental Table 1 online), indicating the high fidelity of the microarray data. The highest expressed genes were consistent with known endosperm functions and included various putative proteases, transporters, and cell wall–modifying enzymes. One of these, EXPANSIN2 (EXP2), has been described as an early GA-induced transcript during seed germination (Ogawa et al., 2003). An EXP2:GUS (for β-glucuronidase) fusion confirmed that its expression was indeed endosperm specific (data not shown). Comparison with online data sets suggested that the expression of this gene is not detected in any other tissues except for low expression in seeds after imbibition (www.genevestigator.ethz.ch). Given the high endosperm expression of EXP2 in our data sets, we concluded that EXP2 is a marker for endosperm tissue in nondormant imbibed or germinating seeds. One notable gene, MYB101, was found to be highly expressed in endosperm tissue yet absent from the embryo. MYB101 is a homolog of the cereal GAMYB transcription factor required for the strong induction of amylase expression in the endosperm during germination (Gubler et al., 1995). Endosperm expression of MYB101 has been shown previously (Gocal et al., 2001); however, this study also detected expression in the embryo. Embryo-specific transcripts included known chloroplast-associated genes and those associated with shoot apical meristem function (see Supplemental Table 1 online).
To understand the functional specialization of embryo and endosperm tissues, we compared the two transcriptomes using Metabolic MapMan (Thimm et al., 2004; see Supplemental Figure 1 online). MapMan allows the visualization of changes in gene expression in a format such that genes are organized and displayed in discrete bins organized according to gene function or predicted gene function. In addition, the Wilcoxon rank sum test was used to identify major bins of genes with a common function whose expression was significantly higher in one tissue versus the other compared with the data set as a whole (Usadel et al., 2005). Several MapMan bins were found to be highly expressed in the endosperm relative to the embryo. Strikingly, these included those dedicated to storage reserve mobilization, including the entire path of primary carbon metabolism through the glyoxylate cycle, the trichloroacetic acid (TCA) cycle, glycolysis/gluconeogenesis, and sugar transport (Table 1). All the major characterized fatty acid β-oxidation transcripts were also more highly expressed in the endosperm versus the embryo (see Supplemental Table 2 online). This is consistent with our previous observation that the endosperm is supremely dedicated to reserve mobilization at this early time point when compared with the embryo (Penfield et al., 2005) and supports previous conclusions that endospermic reserves are used by the embryo during seedling establishment (Penfield et al., 2004). The higher expression of transcripts relating to lipid reserve mobilization in the embryo may reflect the relative lack of transcripts required for reserve use in the endosperm, or it may be that the peak of lipid reserve mobilization gene expression occurs at a later time point in the embryo. The latter hypothesis is supported by observations that endosperm lipid reserve mobilization is completed earlier in the endosperm than in the embryo (Penfield et al., 2005). Importantly, another group of bins highly expressed in the endosperm relative to the embryo were those involved in protein degradation and amino acid transport. This suggests that storage protein breakdown is another important function of the endosperm.
Table 1.
MapMan Bins of Genes of Common Function Identified as Significantly More Highly Expressed in the Embryo Relative to Endosperm or Endosperm Relative to Embryo
| Embryo
|
Endosperm
|
||||||
|---|---|---|---|---|---|---|---|
| Bin Code | Bin Name | No. Genes | P Value | Bin Code | Bin Name | No. Genes | P Value |
| 29.2 | Protein synthesis | 427 | <1e-20 | 29.5 | Protein degradation | 654 | <1e-20 |
| 29.2.2 | Ribosomal proteins | 233 | <1e-20 | 29.5.11.20 | Proteasome | 53 | 6.17e-09 |
| 29.2.1 | Plastid/mitochondrial ribosomal proteins | 72 | 1.75e-19 | 8.1 | TCA cycle | 33 | 2.98e-06 |
| 1.1 | Photosynthesis light reactions | 114 | 3.02e-12 | 29.4 | Posttranslational modification | 611 | 3.30e-05 |
| 28.1 | DNA synthesis/chromatin structure | 218 | 5.32e-06 | 34.3 | Amino acid transport | 40 | 1.08e-03 |
| 34.19.1 | Major intrinsic proteins PIP | 9 | 3.10e-05 | 4.0 | Glycolysis | 48 | 1.73e-03 |
| 19.0 | Tetrapyrrole biosynthesis | 27 | 3.55e-05 | 9.0 | Mitochondrial electron transport | 86 | 2.96e-03 |
| 11.6 | Lipid transfer proteins | 9 | 1.16e-04 | 31.4 | Vesicle transport | 69 | 4.69e-03 |
| 10.6.3 | Pectate lyases | 37 | 4.55e-04 | 29.11.4.3.2 | F-box proteins | 163 | 8.69e-03 |
| 16.2.1 | Lignin biosynthesis | 22 | 9.04e-04 | 2.1.1 | Sucrose synthesis | 9 | 1.00e-02 |
| 1.3 | Calvin cycle | 29 | 1.32e-03 | 5.0 | Fermentation | 10 | 1.36e-02 |
| 29.1 | Amino acid activation | 67 | 1.32e-03 | 34.9 | Metabolite transporters at the mitochondrial membrane | 51 | 1.47e-02 |
| 10.8 | Pectin esterases | 34 | 1.49e-03 | 34.14 | Unspecified cation transporters | 27 | 1.77e-02 |
| 10.5.1 | Arabinogalactan proteins | 26 | 1.60e-03 | 34.2 | Sugar transporters | 52 | 1.85e-02 |
| 11.1 | Fatty acid synthesis | 69 | 1.72e-03 | 34.12 | Metal transporters | 49 | 1.85e-02 |
| 31.3 | Cell cycle | 81 | 2.32e-03 | 6.0 | Glyoxylate cycle, gluconeogenesis | 8 | 3.24e-02 |
| 16.3 | Phenylpropanoid metabolism | 59 | 2.96e-03 | ||||
| 27.2 | Transcription | 54 | 9.43e-03 | ||||
| 2.1.2 | Starch synthesis | 23 | 1.17e-02 | ||||
MapMan bins of genes were identified by comparing the regulation of each bin to the ATH1 array as a whole using the Wilcoxon rank sum test.
Prominent among bins highly expressed in the embryo relative to the endosperm are those for transcription, amino acid activation, and protein synthesis (Table 1). As the amino acids used for early postgerminative protein synthesis are likely derived from seed storage proteins, this suggests an embryo tissue more specialized for reserve use than the endosperm. The number of observable ribosomes in embryo and endosperm cells correlates strikingly with protein synthesis–related gene expression, with embryo cells containing approximately five times more ribosomes than the endosperm (Figures 1C and 1D).
Transcriptomic Response of Embryo and Endosperm Tissues to ABA
To further probe the ABA responses of the endosperm and embryo, we compared gene expression in control and ABA-treated tissues 24 h after imbibition. The total number of genes regulated by ABA in the embryo and endosperm was compared using SAM (Tusher et al., 2001) with a stringent 1% false discovery rate (Figures 2A and 2B). Using this analysis, 1971 genes were found to be ABA downregulated in the embryo, while 1197 genes were upregulated. This represents a total of ∼22% of expressed genes regulated by ABA in the embryo. This result contrasts strikingly with the endosperm, where only 422 ABA downregulated and 49 ABA upregulated genes were found, despite a similar number of expressed genes in both tissues (Figures 2A and 2B). Hence, a large number of genes regulated by ABA in the embryo are either not regulated by ABA in the endosperm or show only a weak effect of ABA on their transcript abundance such that they fall short of the 1% significance level. Therefore, these results strongly support previous observations that the endosperm is markedly less sensitive to ABA than the embryo (Penfield et al., 2004) and show that this phenomenon is not limited to the regulation of lipid mobilization.
Figure 2.
Transcriptomic Response of Embryo and Endosperm Tissues to ABA.
(A) and (B) Number of probe sets found to be downregulated or upregulated by ABA in the embryo or endosperm by SAM analysis.
(C) and (D) Regulation of the plastid transcriptome by ABA in the embryo (C) and endosperm (D).
To understand which processes were under embryo-specific ABA regulation, we used MapMan to identify functional categories of genes that were ABA regulated in the embryo but not the endosperm (see Supplemental Figure 2 online) and using the Wilcoxon rank sum test identified bins of genes regulated by ABA in the embryo, endosperm, or both (Table 2). Several bins were found to be significantly changed in gene expression by ABA treatment in the embryo but not the endosperm. Strikingly, this group of bins was dominated by genes involved in major plastid functions: these included the photosynthetic light reactions, the Calvin cycle, tetrapyrolle synthesis, and fatty acid synthesis. In fact, genes encoding components of the photosynthetic reaction centers were downregulated by ABA in the embryo yet actually upregulated by ABA in the endosperm. A number of bins representing secondary metabolism were also ABA downregulated in the embryo only. The role of ABA in the inhibition of chloroplast development in young seedlings is well known; however, ABA had no significant affect on the expression of chloroplast-encoded gene expression in either embryo or endosperm (Figures 2C and 2D). Therefore, ABA influences chloroplast metabolism exclusively through the regulation of the nuclear genome.
Table 2.
The MapMan Wilcoxon Rank Sum Test Identifies Groups of Genes Differentially Regulated by ABA in the Embryo and Endosperm 1 d after Imbibition
| MapMan Bin Code | MapMan Bin Name | MapMan Bin Size | Embryo | P Value | Endosperm | P Value |
|---|---|---|---|---|---|---|
| Bins repressed by ABA in the embryo only | ||||||
| 1.1 | Photosynthetic light reactions | 115 | Down | 1.96e-15 | Up | 4.59e-02 |
| 10.2 | Cellulose synthesis | 27 | Down | 1.86e-07 | No change | 1.80e-01 |
| 11.1 | Fatty acid synthesis | 67 | Down | 2.63e-07 | No change | 1.90e-01 |
| 19 | Tetrapyrolle synthesis | 27 | Down | 6.18e-06 | Up | 4.17e-02 |
| 1.3 | Calvin cycle | 29 | Down | 3.31e-05 | No change | 1.20e-01 |
| 16.5 | Glucosinolates | 10 | Down | 6.46e-04 | No change | 4.30e-01 |
| 16.2 | Phenylpropanoids | 60 | Down | 8.63e-04 | No change | 2.60e-01 |
| 16.1 | Isoprenoids | 61 | Down | 1.16e-03 | No change | 9.30e-01 |
| 16.7 | Waxes | 12 | Down | 2.54e-03 | No change | 3.10e-01 |
| 23.3 | Nucleotide salvage | 21 | Down | 8.29e-03 | No change | 4.90e-01 |
| ABA-regulated bins in embryo and endosperm | ||||||
| 10.8 | Pectin esterases | 32 | Down | 2.95e-08 | Down | 5.24e-03 |
| 10.5.1 | Arabinogalactan proteins | 25 | Down | 7.16e-08 | Down | 9.32e-03 |
| 10.6.3 | Pectate lyases | 34 | Down | 7.02e-07 | Down | 2.12e-03 |
| 11.4 | Oleosins | 9 | Up | 9.20e-07 | Up | 2.01e-06 |
| 10.7 | Cell wall modification | 40 | Down | 1.82e-06 | Down | 1.53e-03 |
| 21.1 | Ascorbate and glutathione | 45 | Down | 3.35e-05 | Down | 2.10e-03 |
| 10.1 | Cell wall precursor synthesis | 36 | Down | 1.04e-04 | Down | 9.56e-06 |
| 13.2.3.2 | Amino acid degradation Asp family | 15 | Up | 6.85e-03 | Up | 3.16e-03 |
| ABA-regulated bins in endosperm only | ||||||
| 13.1.6.5 | Aromatic amino acid synthesis | 12 | No change | 9.20e-01 | Up | 7.04e-04 |
| 2.2.2 | Starch synthesis | 24 | No change | 8.70e-01 | Up | 1.71e-03 |
Several MapMan bins were also found to be significantly affected by ABA in both tissues. Prominent among these were groups of cell wall–related enzymes, such as pectinesterases, pectate lyases, and cell wall–modifying enzymes (expansins and xylogucan endotransglycosylases; Table 2). This suggests that the cell wall is a major target of ABA signaling in both the embryo and endosperm and that this role of ABA is mediated by signal transduction components expressed in both tissues. ABA-upregulated bins in both embryo and endosperm include those representing oleosins and aspartate degradation. We found no major targets of ABA repression that were unique to the endosperm. Hence, the endosperm is both qualitatively and quantitatively different in its ABA response compared with the embryo. Previously, we used reporter gene studies to show that ABA affects the transcription from a number of promoters of key storage reserve mobilization-related genes (Pritchard et al., 2002; Penfield et al., 2004). In the latter study, the temporal induction of reporter gene activity at 1 d after imbibition correlated strongly with endogenous enzyme activities. However, other published data on steady state transcript levels reported that these transcripts were already elevated in imbibed seeds (Rylott et al., 2001). Our study also shows that reserve mobilization is not dramatically regulated at the level of steady state transcript levels in either the embryo or endosperm by ABA and that transcripts are already accumulated at high levels in nongerminating PAC-treated seeds (see Supplemental Table 2 online). This agrees with previous published array work that showed that the transcription of key enzymes in β-oxidation and the glyoxylate cycle is already induced in late seed development (Schmid et al., 2005).
Comparing this data set to the AtGenExpress ABA treatment experimental data (http://www.weigelworld.org/research/projects/resources/microarray/AtGenExpress/; 24 h imbibed seeds with and without 30 μM ABA and 7-d-old seedlings with or without 3 μM ABA for 3 h), we were able to identify 52 ABA-induced probe sets representing 51 genes whose expression was affected twofold or greater by ABA treatment in all four experiments (embryo, endosperm, seed, and seedling; see Supplemental Table 3 online). This same analysis revealed eight probe sets twofold or greater repressed by ABA in each experiment. This included many known stress- or desiccation-responsive genes and a number of transcription factors. These can be considered a core set of genes that respond to ABA independently of seed tissue type. Comparing this core set with the dry seed data sets of the wild type, abi4-1, and abi5-1 of Nakabayashi et al. (2005) and imbibed seed data sets of the wild type and abi3-4 (http://affymetrix.arabidopsis.info/narrays/experimentbrowse.pl), it is apparent that the regulation of many of these genes is dependent on ABI3 and ABI4.
ABA and GA Synthesis and Response in the Embryo and Endosperm
We have shown that the ABA response differs between the embryo and endosperm. This response is potentially antagonized by GA in the seed, so GA signaling could also in principle modulate the ABA response in the endosperm. Furthermore, in situ hybridization has shown that GIBBERELLIC ACID 3-OXIDASE1 (GA3OX1), the last step in GA biosynthesis, is expressed in endosperm tissues before germination (Yamauchi et al., 2004). This has led to speculation that the endosperm could be an important site of GA biosynthesis in the seed because this pathway is divided between at least two tissues in the embryo (Yamaguchi et al., 2001). To begin to understand the differences in ABA and GA signaling between the embryo and endosperm, we compared the expression of the ABA and GA metabolic and signaling components in the microarray data (Table 3). Although levels of transcript abundance do not necessarily correlate with protein levels, it is possible that this data provides a guide to the differences in hormone synthesis and signaling between the two tissues. GA biosynthetic genes were found to be expressed at approximately equal levels in both tissues, suggesting that the endosperm could be capable of synthesizing active GAs 1 d after germination. GA biosynthetic genes are also expressed at a similar level in embryo and endosperm in the samples where germination is inhibited by ABA or PAC. Instructively, multiple isoforms of both GA20 oxidase and GA3 oxidase were subject to known feedback upregulation by PAC treatment in the endosperm, demonstrating that GA signal transduction functions normally in these cells. That GA signal transduction operates in both the embryo and endosperm is also suggested by the expression of all five DELLA protein regulators of the GA response in both tissues. Most of these are actually more highly expressed in the endosperm than the embryo.
Table 3.
GA and ABA Metabolic and Response Gene Expression in the Embryo and Endosperm during Germination
| Embryo
|
Endosperm
|
||||||
|---|---|---|---|---|---|---|---|
| Affymetrix ID | Name | PAC | ABA | CON | PAC | ABA | CON |
| GA metabolism | |||||||
| 255461_at | CPS (GA1) | 58.5 ± 7.2 | 15.4 ± 2.5* | 10.1 ± 1.8* | 12.0 ± 3.3* | 9.2 ± 3.9* | 6.3 ± 2.8* |
| 262891_at | KS (GA2) | 21.5 ± 2.5 | 27.6 ± 3.8 | 25.0 ± 5.1 | 22.8 ± 1.8 | 23.1 ± 0.5 | 20.2 ± 2.2 |
| 246864_at | KO1 (GA3) | 1053.8 ± 83.2 | 487.3 ± 33.1 | 400.8 ± 7.0 | 1231.8 ± 140.3 | 572.8 ± 55.3 | 291.3 ± 42.7 |
| 264586_at | KAO1 | 30.2 ± 1.9 | 51.2 ± 1.2 | 25.8 ± 0.6 | 25.8 ± 5.1 | 29.5 ± 4.1 | 20.7 ± 1.5 |
| 254065_at | GA20OX1 | 335.1 ± 14.1 | 28.1 ± 1.4 | 37.4 ± 4.5 | 204.1 ± 16.6 | 23.7 ± 2.9 | 14.0 ± 6.0 |
| 248371_at | GA20OX2 | 306.1 ± 4.2 | 7.7 ± 1.0* | 21.5 ± 3.4 | 203.9 ± 12.6 | 8.5 ± 0.7* | 5.8 ± 3.5* |
| 250611_at | GA20OX3 | 775.9 ± 22.3 | 24.1 ± 3.7 | 3.4 ± 1.5* | 798.3 ± 29.0 | 19.2 ± 1.2* | 10.7 ± 4.8* |
| 261768_at | GA3OX1 (GA4) | 1984.1 ± 103.4 | 282.8 ± 28.1 | 597.7 ± 89.6 | 642.6 ± 103.5 | 86.9 ± 32.8 | 102.8 ± 21.9 |
| 260300_at | GA3OX2 | 339.4 ± 38.3 | 105.2 ± 6.3 | 376.1 ± 15.0 | 107.8 ± 28.0 | 37.3 ± 9.4 | 75.5 ± 31.1 |
| 260023_at | GA2OX2 | 10.4 ± 4.2* | 26.4 ± 6.8 | 24.2 ± 10.0 | 5.7 ± 3.1* | 22.9 ± 4.1 | 18.8 ± 2.9* |
| GA signaling | |||||||
| 262850_at | GAI | 137.0 ± 4.1 | 286.5 ± 19.7 | 73.1 ± 2.1 | 158.1 ± 15.3 | 311.0 ± 30.9 | 218.0 ± 18.2 |
| 266331_at | RGA | 87.3 ± 2.7 | 90.3 ± 5.3 | 74.2 ± 6.9 | 88.3 ± 11.9 | 69.2 ± 11.6 | 47.3 ± 8.6 |
| 260141_at | RGL1 | 13.0 ± 2.6* | 14.1 ± 2.4* | 24.6 ± 1.8 | 43.5 ± 3.5 | 19.3 ± 2.3* | 37.5 ± 5.3 |
| 259042_at | RGL2 | 403.9 ± 16.3 | 488.1 ± 43.0 | 167.2 ± 7.9 | 561 ± 58.9 | 715.0 ± 13.9 | 519.2 ± 49.0 |
| 246432_at | RGL3 | 84.2 ± 6.6 | 150.3 ± 9.5 | 42.9 ± 1.6 | 81.4 ± 5.2 | 100.0 ± 11.2 | 75.5 ± 5.4 |
| 254160_at | SLY1 | 137.2 ± 28.9 | 86.4 ± 10.7 | 80.8 ± 6.2 | 187.2 ± 2.8 | 73.5 ± 10.1 | 102.8 ± 25.3 |
| 251200_at | ATGID1a | 4.3 ± 1.5* | 2.7 ± 1.6* | 6.5 ± 0.6* | 7.6 ± 2.6* | 4.5 ± 2.8* | 11.1 ± 6.3* |
| 259302_at | ATGID2 | 124.1 ± 11.1 | 80.3 ± 6.1 | 37.4 ± 2.6 | 102.7 ± 8.7 | 85.4 ± 11.6 | 53.0 ± 10.8 |
| 246782_at | ATGID3 | 24.2 ± 3.3 | 27.3 ± 4.3 | 10.9 ± 1.4* | 22.2 ± 4.3* | 15.6 ± 5.7* | 16.6 ± 2.2* |
| ABA metabolism | |||||||
| 247025_at | ABA1 | 397.8 ± 5.1 | 365.2 ± 14.8 | 138.2 ± 2.0 | 260.4 ± 10.6 | 320.0 ± 54.4 | 213.8 ± 14.3 |
| 257280_at | NCED3 | 4.4 ± 2.9* | 2.1 ± 1.3* | 2.0 ± 1.5* | 2.3 ± 1.4* | 1.5 ± 0.4* | 1.2 ± 0.3* |
| 257242_at | NCED6 | 0.9 ± 0.2* | 1.3 ± 0.3* | 0.9 ± 1.2* | 2.1 ± 0.5* | 1.6 ± 0.5* | 1.9 ± 0.4* |
| 260797_at | NCED9 | 7.7 ± 0.7* | 2.8 ± 1.0* | 3.3 ± 1.7* | 21.8 ± 2.0 | 26.7 ± 2.5 | 24.7 ± 3.7 |
| 259669_at | ABA2 | 82.0 ± 5.4 | 76.3 ± 4.3 | 78.9 ± 1.7 | 116.6 ± 6.6 | 136.0 ± 5.0 | 121.3 ± 2.7 |
| 246325_at | ABA3 | 52.5 ± 4.9 | 51.5 ± 7.1 | 15.7 ± 3.0* | 36.3 ± 1.2 | 36.5 ± 1.9 | 33.1 ± 5.6 |
| 263570_at | AAO3 | 74.4 ± 4.1 | 73.3 ± 6.8 | 42.2 ± 5.7 | 96.7 ± 28.6 | 94.2 ± 4.0 | 103.0 ± 3.8 |
| 266778_at | CYP707A2 | 53.0 ± 1.5 | 82.9 ± 12.3 | 25.6 ± 2.9 | 19.3 ± 3.5* | 176.9 ± 17.5 | 21.2 ± 10.9 |
| ABA signalingb | |||||||
| 253994_at | ABI1 | 373.9 ± 24.5 | 390.5 ± 53.5 | 436.9 ± 20.3 | 282.6 ± 21.6 | 429.5 ± 143.1 | 640.6 ± 112.3 |
| 247957_at | ABI2 | 11.2 ± 5.1* | 42.6 ± 1.7 | 17.1 ± 3.6* | 3.2 ± 1.7* | 39.0 ± 4.2 | 7.1 ± 2.8* |
| 256898_at | ABI3 | 264.8 ± 22.4 | 137.3 ± 11.4 | 30.6 ± 2.9 | 342.7 ± 29.9 | 190.2 ± 14.1 | 131.7 ± 31.6 |
| 263377_at | ABI4 | 69.5 ± 8.8 | 115.3 ± 22.2 | 61.6 ± 14.7 | 13.9 ± 2.6* | 42.3 ± 12.9c | 24.5 ± 6.2* |
| 263907_at | ABI5 | 182.4 ± 4.8 | 133.7 ± 18.3 | 23.7 ± 1.8* | 108.2 ± 8.8 | 121.2 ± 66.1 | 52.2 ± 20.3 |
| 258666_at | ABI8 | 132.6 ± 4.6 | 161.9 ± 12.9 | 122.1 ± 11.6 | 104.9 ± 13.4 | 109.1 ± 16.2 | 112.7 ± 7.0 |
| 248719_at | rbohD | 78.3 ± 9.2 | 79.8 ± 7.1 | 27.5 ± 1.4 | 649.3 ± 7.8 | 501.3 ± 78.9 | 869.9 ± 56.1 |
| 262344_at | rbohF | 7.9 ± 3.0* | 6.2 ± 1.1* | 17.0 ± 4.3* | 3.5 ± 1.1* | 6.1 ± 2.3* | 21.4 ± 3.6* |
| Light signaling | |||||||
| 264508_at | PHYA | 1385.7 ± 52.3 | 842.1 ± 50.2 | 1036.0 ± 53.5 | 2516.9 ± 190.1 | 1475.2 ± 164.2 | 2545.5 ± 145.9 |
| 266065_at | PHYB | 331.5 ± 25.5 | 261.2 ± 5.7 | 188.9 ± 1.6 | 299.0 ± 6.0 | 206.6 ± 6.1 | 198.1 ± 8.4 |
| 245487_at | PHYD | 137.8 ± 1.9 | 159.9 ± 19.8 | 46.0 ± 1.9 | 58.8 ± 4.6 | 67.9 ± 1.8 | 25.7 ± 5.8 |
| 254680_at | PHYE | 161.6 ± 8.2 | 180.2 ± 3.4 | 100.5 ± 2.9 | 143.1 ± 19.3 | 176.0 ± 4.2 | 82.1 ± 14.9 |
| 265584_at | PIL5/PIF1 | 373.7 ± 22.6 | 337.2 ± 6.0 | 207.2 ± 11.0 | 383.5 ± 41.5 | 320.4 ± 10.6 | 213.8 ± 29.6 |
| 246212_at | SPATULA | 54.5 ± 4.3 | 54.7 ± 3.5 | 207.1 ± 4.4 | 187.2 ± 21.4 | 170.7 ± 48.1 | 483.6 ± 121.5 |
Asterisk indicates an absent call in at least two of three replicates. CON, control.
The Arabidopsis homologues of the recently identified rice GA receptor GIBBERELLIN INDEPENDENT DWARF1.
Major and relevant ABA response genes shown only.
One absent, one present, and one marginal call.
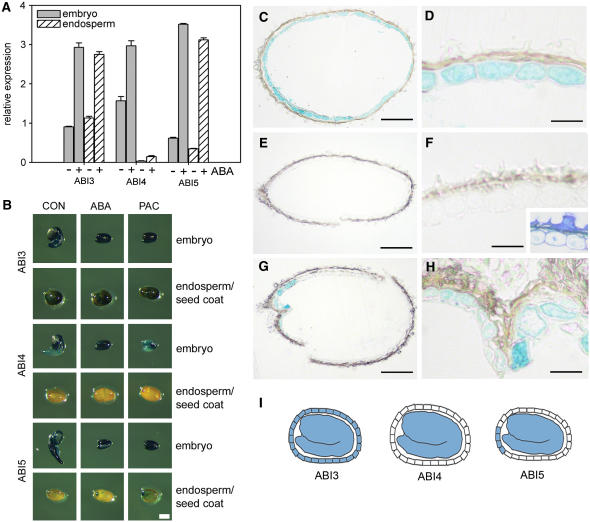
At the transcript level, the endosperm also expresses many genes required for ABA synthesis and catabolism, suggesting that the endosperm might also be able to make and degrade ABA. This has been further demonstrated by a recent report that two carotenoid cleavage dioxygenase isoforms are expressed in the endosperm of maturing seeds and are required for dormancy (Lefebvre et al., 2006). One of these, NCED9, was still expressed in an endosperm-specific manner in germinating seeds (Table 3). Multiple genes have been shown to control and affect the ABA response of young seedlings. We analyzed the expression of major ABA response genes in our data set and noted differences between the embryo and endosperm. The expression level of ABI1, ABI2, ABI3, and ABI5 was similar in the embryo and endosperm. However, although expressed at low levels in both tissues, ABI4 transcripts were much less abundant in the endosperm and were classified as absent in eight out of nine array experiments by MAS5. We used real-time RT-PCR to clarify ABI4 expression in the seed. This clearly showed that while ABI3 and ABI5 are indeed expressed in both the embryo and endosperm, ABI4 was undetectable in the endosperm (Figure 3A). Furthermore while ABI4 was highly induced at the transcriptional level by ABA in the embryo, this effect could not be observed in the endosperm. To further analyze the expression of the ABI genes in the seed, we examined the activity of the GUS reporter fused to the ABI3, ABI4, or ABI5 promoters (Parcy et al., 1994; Soderman et al., 2000; Brocard et al., 2002; Figures 3B to 3I). ABI3 was expressed ubiquitously throughout the seed, including the endosperm (Figures 3B to 3D). ABI3 is required for the regulation of oleosin gene expression by ABA (Crowe et al., 2000), and it is interesting to note that transcripts for oleosins are upregulated by ABA in both the embryo and endosperm, in agreement with the expression pattern of ABI3 observed in seeds (Table 2; see Supplemental Figure 2 online). In agreement with the real-time RT-PCR data, the ABI4 promoter directs GUS expression in the embryo only (Figures 3B, 3E, and 3F). Therefore, this result is confirmed by three independent methods of expression analysis. One further surprise was the expression of ABI5:GUS in the endosperm. Unlike ABI3, ABI5 expression was confined to the micropylar region of the endosperm (Figures 3B, 3G, and 3H). This region is important for the regulation of seed germination in many species and is characterized by differential gene regulation and cell wall composition compared with the lateral endosperm in many species (Bewley, 1997b). So ABI5 expression defines two domains of altered ABA sensitivity in the endosperm.
Figure 3.
Expression of ABI3, ABI4, and ABI5 in the Embryo and Endosperm 1 d after Transfer to 22°C.
(A) Detection by real-time RT-PCR. Data represent mean and sd of three replicate determinations.
(B) GUS expression from the ABI3, ABI4, and ABI5 promoters in dissected embryos or endosperm/seed coats in control (CON) or ABA- or PAC-treated seeds.
(C) to (H) Sections to show endosperm expression of the ABI:GUS fusions in ABA-treated seeds.
(C) and (D) ABI3:GUS.
(E) and (F) ABI4:GUS. Inset shows toluidene blue–stained ABI4:GUS section confirming the presence of the endosperm cell layer barely visible in (F) due to lack of expression.
(G) and (H) ABI5:GUS.
(I) Illustration of the expression pattern of ABI3, ABI4, and ABI5 in seeds.
ABI4 Determines the Sensitivity of Seed Lipid Breakdown to ABA
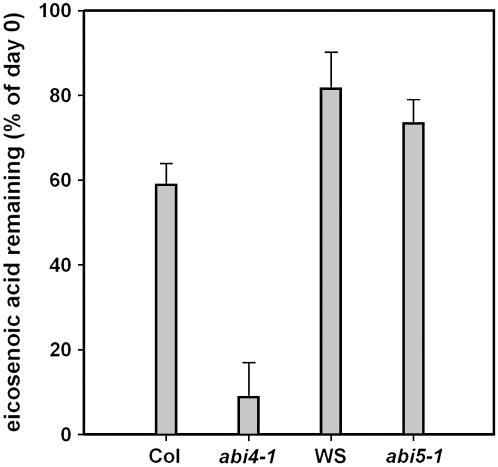
Given that both ABI4 and ABI5 are not expressed in the majority of endosperm tissues, we reasoned that the lack of either could in principle be responsible for the ABA insensitivity of lipid catabolism in the endosperm. To examine whether ABI4, ABI5, or both are required for the ABA inhibition of triacylglycerol (TAG) breakdown, eicosenoic acid levels were measured in embryos of the wild type and abi4-1 and abi5-1 mutants in the presence of ABA 5 d after imbibition (Figure 4). In wild-type embryos (Columbia-0 and Wassilewskija), TAG breakdown was estimated at 20 to 30% after 5 d of ABA treatment. Both the abi4-1 and abi5-1 mutants germinated at high frequency in the presence of ABA, yet the level of eicosenoic acid in 5-d-old abi5-1 mutants was similar to that in nongerminating wild-type seeds, indicating that ABI5 does not play a role in the ABA regulation of TAG breakdown. However, the abi4-1 mutant could break down virtually all its embryo-stored TAG in the presence of ABA. Therefore, we concluded that ABI4 expression determines the sensitivity of lipid reserve mobilization to ABA in the seed.
Figure 4.
Eicosenoic Acid Levels in Wild Type, abi4-1, and abi5-1 Embryos 5 d after Imbibition in the Presence of 20 μM ABA.
Bars represent the mean and sd of four independent determinations.
The Endosperm Is Required for the ABA and GA Regulation of Seed Germination in Arabidopsis
Germination in many endospermic seeds, including tobacco, tomato, and lettuce (Lactuca sativa), is characterized by controlled loosening of the micropylar endosperm cell walls to facilitate radicle emergence (Bewley, 1997b). This is achieved by the activity of multiple categories of cell wall–modifying enzymes, including β-mannanase, β-1,4-glucanase, expansins, xyloglucan endotransglycosidases, and polygalacturonases. Our expression data reveal that this group of genes is highly expressed in the Arabidopsis endosperm during germination (see Supplemental Table 4 online).
As ABA and GA signal transduction appeared to operate normally in the Arabidopsis endosperm, and the expression of multiple cell wall–modifying enzymes in the Arabidopsis endosperm was confirmed, we reasoned that the endosperm might play an active role in the phytohormonal regulation of Arabidopsis seed germination. Among the genes involved in ABA signaling during seed germination, we noted that the NADPH oxidase rbohD was highly expressed in the endosperm (Table 3). It has previously been shown that the atrbohd atrbohf double mutant exhibits reduced ABA inhibition of seed germination (Kwak et al., 2003). Our array data showed that rbohD was expressed in the endosperm during germination, while rbohF was not expressed in either tissue. This expression profile was confirmed by real-time RT-PCR (Figure 5A). This suggests that the described rbohd rbohf double mutant germination phenotype was a consequence of the loss of rbohD function alone. Accordingly, we found that the rbohd single mutant germination exhibited a reduced ABA sensitivity phenotype similar to that of the double mutant (Figure 5B; Kwak et al., 2003). Hence, the NADPH oxidase rbohD is an endosperm-expressed gene required for the ABA inhibition of seed germination in Arabidopsis, clearly demonstrating a role for the endosperm in this process. Given the fact that other abi mutants show a much greater level of resistance to exogenous ABA during seed germination than the rbohd mutant, it appears that either the endosperm plays a relatively minor role in the ABA control of seed germination in Arabidopsis or that loss of AtrbohD does not completely block the endosperm response to ABA.
Figure 5.
The Endosperm Is Required for the ABA Regulation of Seed Germination.
(A) Real-time RT-PCR to show the expression of rbohD and rbohF in germinating seeds.
(B) The rbohd single mutant germination shows decreased sensitivity to ABA.
(C) and (D) abi4-1 mutant seed 5 d after transfer to 22°C growing in the presence of 10 μM ABA (C) and 10 μM PAC (D). Bars = 100 μM.
We also analyzed the germination and establishment of abi4-1 mutant seeds in the presence of ABA and PAC. As ABI4 expression is confined to the embryo, we reasoned that the response of the abi4-1 endosperm to both ABA and PAC should be similar to the wild type. The abi4-1 mutant shows significant PAC- and ABA-resistant germination (data not shown). We observed that abi4-1 mutants germinating on concentrations of PAC or ABA inhibitory to wild-type seeds became entangled in the endosperm despite the completion of radicle emergence. In many cases, a ring of endosperm tissue remained attached to abi4-1 hypocotyls, and occasionally seedling development proceeded with the seedling encased in the endosperm (Figures 5C and 5D). Although we cannot completely rule out an ABI4-independent ABA- and PAC-responsive mechanism in the embryo that could account for these observations, in the context of our analysis, it seems likely that an ABA-repressible and GA requiring endosperm loosening is responsible. This is not surprising given that ABI3 and ABI5 are both expressed in the endosperm and presumably involved in mediating subsets of ABA responses. Therefore, these results show that in common with other seeds, the Arabidopsis endosperm plays an active role in the regulation of seed germination and seedling establishment.
DISCUSSION
We have used RNA amplification to facilitate tissue-specific transcript profiling and dissection of the ABA response in seeds. This type of approach adds extra value and resolution to transcriptome studies compared with the analysis of more complex groups of tissues (Lee et al., 2005). This investigation has enabled the description of the endosperm transcriptome and has been successful in understanding the molecular basis of differential ABA signaling in the seed. Furthermore, using the new findings, we have been able to demonstrate a previously unknown role for the Arabidopsis endosperm in the control of seed germination by ABA.
ABI4 Is an Embryo-Specific Regulator of TAG Breakdown
Three independent analyses have shown that ABI4 is expressed in embryo but not endosperm tissues. ABI4 is required for the ABA regulation of seed germination and plant responses to sugars (Finkelstein, 1994; Arenas-Huertero et al., 2000; Huijser et al., 2000; Laby et al., 2000; Rook et al., 2001). As ABI4 is not expressed in the endosperm, we hypothesized that ABI4 expression determines the sensitivity of lipid breakdown to exogenous ABA. This conclusion is strongly supported by the observation that loss of ABI4 results in ABA-independent TAG breakdown in the embryo (Figure 4). Hence, the embryo-specific expression of ABI4 is the crucial determinant of the ABA sensitivity of lipid mobilization in the seed. Although seeds are unlikely to encounter high ABA in the environment, ABI4 is also required for the inhibition of seed germination under osmotic stress (Quesada et al., 2000). Under these conditions, lipid breakdown also occurs in the endosperm only (see Supplemental Figure 3 online), suggesting that this differential response does have functional significance for the seeds germinating in suboptimal circumstances. In this case, the embryo-specific expression of ABI4 adds an unexpected and important plasticity to the germination process, allowing energy stored in the endosperm to be mobilized without commitment to the final phase of seedling establishment. This phased process of reserve mobilization could serve to prolong the integrity of the embryo under suboptimal conditions.
The second major difference in ABA signaling between the embryo and endosperm was the ABA sensitivity of photosynthesis-associated nuclear-encoded gene (PhANG) expression. ABA had a strong inhibitory effect on the expression of these genes in the embryo. This effect of ABA on plastid function was found to be specific to the nuclear genome, as ABA had no effect on chloroplast-encoded gene expression (Figures 2C and 2D). PhANGs with embryo-specific ABA-sensitive expression included those encoding the photosynthetic apparatus, the Calvin cycle, and tetrapyrrole biosynthesis. Interestingly, MapMan analysis of PhANG expression in the endosperm suggests that ABA treatment actually results in a small upregulation of the transcript levels of these genes in the endosperm, rather than the inhibition seen in the embryo (see Supplemental Figure 2 online; P = 0.046). Interestingly, PhANG expression is also a known key target of ABI4. For instance, ABI4 regulates the sugar and ABA responsiveness of the plastocyanin and chlorophyll a/b binding protein 2 (CAB2) promoters (Dijkwel et al., 1997; Huijser et al., 2000; Oswald et al., 2001). Furthermore, ABI4 binds to a conserved element present in the promoters of multiple genes encoding subunits of ribulose-1,5-bis-phosphate carboxylase/oxygenase and various CABs found throughout the plant kingdom and regulates their expression according to sugar and ABA levels (Acevedo-Hernández et al., 2005). In a striking correlation, Oswald et al. (2001) found that in the ABI4-deficient sucrose uncoupled6 (sun6) mutant, CAB2:LUC expression is actually induced by sugar treatment, rather than repressed as in the wild type. This situation is apparently mirrored in the ABA induction of PhANG expression in the ABI4-deficient endosperm. However, we cannot rule out the possibility that further unknown proteins also regulate the differential ABA response of PhANG gene expression in germinating seeds.
In the embryo, the glyoxylate cycle and gluconeogenesis are induced 1 d after imbibition, but here, we report that this is not accompanied by an increase in steady state transcript levels (see Supplemental Table 2 online), although we cannot rule out that these do not increase at later time points. The level of expression of reserve mobilization genes in our analysis strongly resembles that observed during late seed development and in dry seeds (Nakabayashi et al., 2005; Schmid et al., 2005), suggesting that much of the mRNA present shortly after germination was stored during desiccation. However, further evidence suggests these pathways are actually regulated at the level of transcription in the seed. For instance, the induction of isocitrate lyase, malate synthase, and PCK1 enzyme activities correlates with the onset of expression of GUS or luciferase fused to their respective promoters (Penfield et al., 2004). Furthermore, the GA-dependent accumulation of several gluconeogenic proteins, including malate synthase and PCK1, could be blocked in germinating seeds by the transcriptional inhibitor α-amantin (Rajjou et al., 2004). One possibility is that these apparently unused transcripts have been stored during quiescence, broken down after germination, and actively excluded from postgerminative translation. A study of the postgerminative induction of the TCA cycle enzyme isocitrate dehydrogenase also concluded that transcripts present before or shortly after germination were not translated (Falk et al., 1998), suggesting that this could be a general feature of the regulation of metabolism in germinating seeds. We hypothesize that the unused stored transcripts may themselves be a stored seed reserve that is broken down and the nucleotide components recycled for rapid de novo transcription during early postgerminative growth.
The Endosperm Is Required for ABA and GA Regulation of Seed Germination
Our analysis shows that the single cell endosperm layer plays an active role in the phytohormonal regulation of Arabidopsis seed germination. The endosperm is also vital for normal seed dormancy and may be the principal site for the synthesis of dormancy inducing ABA during seed development (Lefebvre et al., 2006). It is possible that the reduced ABA response observed in the endosperm has evolved as a consequence of higher endogenous levels of ABA in that tissue. We also show that the endosperm possesses a normal GA response and expresses GA biosynthetic genes at a similar level to that observed in the embryo. Together, these results demonstrate a fundamental role for the endosperm in the control of phase transition in the seed. The Arabidopsis endosperm also exhibits gene regulation specific to the micropylar endosperm, and we have discovered that this region is marked by the expression of the ABI5 transcription factor. Therefore, this study clearly highlights the importance of tissue- and cell-specific hormone responses in germination control.
METHODS
Plant Material
Arrays were performed on isolated embryo and endosperm tissue from the Landsberg erecta ecotype. The ABI3:GUS, ABI4:GUS, and ABI5:GUS lines were gifts from Ruth Finkelstein. The abi4-1 and abi5-1 mutants were obtained from the Nottingham Arabidopsis Stock Centre. The rbohd mutant was a gift from Jonathan Jones.
Affymetrix Gene Chip Experiments and Data Analysis
Tissue was harvested from wild-type (Landsberg erecta) seeds plated on Murashige and Skoog medium supplemented by 20 μM ABA (mixed isomers; Sigma-Aldrich) or 20 μM PAC (Greyhound Chromatography) where indicated, stratified for 3 d at 4°C, and placed under continuous white light at 70 μM m−2 s−1 at 22°C for 24 h. Arabidopsis thaliana embryo and endosperm samples were collected by manual dissection as described (Penfield et al., 2004). These were stored in RNAlater solution prior to RNA extraction (Qiagen). Approximately 50 embryos/endosperms were pooled for each RNA isolation. RNA was isolated using an RNA Nanoprep kit (Stratagene) following manufacturer's instructions. Isolated RNA was subjected to one round of linear amplification with the MessageAmp Kit (Ambion). Biotin labeling of amplified RNA was performed according to the Gene Chip Eukaryotic Small Sample Target Labeling Assay Version II protocol, supplied by Affymetrix, using random primers (Invitrogen) and 1 μg of amplified RNA for the first-strand synthesis reaction. Three biological replicates per treatment were hybridized independently to the Affymetrix ATH1 array, washed, stained, and scanned following the procedures described in the Affymetrix technical manual. The expression levels of genes were measured by detection calls and signal intensities using the Micro Array Suite 5.0 software with a target signal of 100. Sixty-four Affymetrix controls and 5623 Arabidopsis genes that are detected as absent in all 18 chips were removed from the 22,810 probe sets. All pairwise differentially expressed genes were identified using SAM software (Tusher et al., 2001) using the data of all remaining 17,123 Arabidopsis probe sets. A false discovery rate parameter of 1% was used for the SAM analysis. Following SAM analyses, genes that were called absent more than twice among three replicas in both control and treatment arrays were then regarded as not expressed in both conditions and removed from the above list. Z-score transformation was performed as described (Cheadle et al., 2003). This transformation normalizes the data according to the distance of each log10 value from the mean log10 value, expressed in terms of number of standard deviations. For the MapMan analysis, input files were created by calculating the natural log ratio of the mean detection of the three control samples to the mean detection in the treatment samples. Genes called absent in two out of the three replicates were regarded as not expressed under that particular experimental condition. Final analyses were performed with MapMan version 1.6.1, including automatic application of the Wilcoxon rank sum test (Usadel et al., 2005).
Comparison with public domain Affymetrix ATH1 data sets was achieved by downloading entire data sets from NascArrays (http://affymetrix.arabidopsis.info/narrays/experimentbrowse.pl) and from Nakabayashi et al. (2005). Probe sets were identified that exhibited twofold or greater change in expression in response to ABA treatment.
Real-Time RT-PCR and GUS Expression Analysis
Real-time RT-PCR on endosperm and embryo RNA was conducted as described (Penfield et al., 2004) from amplified mRNA. Three replicate quantifications were routinely used for each determination, which was compared with a standard curve. Primers for 18S (Penfield et al., 2004) were used as a standard. Further primers used were as follows: ABI3F, 5′-ACAGTAACTTTCAGTCGGGTTGATC-3′; ABI3R, 5′-CCCTGCATGTCTCCAGCAT-3′; ABI4F, 5′-TTCTGACATCGAGCTCACTGAT-3′; ABI4R, 5′-CCCTAACGCCACCTCATGAT-3′; ABI5F, 5′-CGATGCTTGACTGGGTCCTT-3′; ABI5R, 5′-CTGCAACTAGGTGGCCTTCCT-3′; RBOHDF, 5′-CGAATGGCATCCTTTCTCAATC-3′; RBOHDR, 5′-GTCACCGAGAGTGCGGATATG-3′; RBOHFF, 5′-GATGGTTTAGGCGTAACCTAGTCAA-3′; RBOHFR, 5′-AACAAATGATGCGAATACCAAAAG-3′. GUS expression assays were performed on dissected seeds as described (Jefferson et al., 1987). Seeds were dissected into embryo and endosperm/seed coat before staining (this was found to be essential because intact seeds do not permit proper access of the GUS substrate to the internal tissues) in a buffer containing ferricyanide and ferrocyanide to ensure precipitation of the reaction product. Seeds were photographed using a Leica MP6 dissecting microscope fitted with a SPOT RT image capture system (Diagnostic Instruments). Images were manipulated into a composite using Adobe Photoshop. For sectioning, seeds were dissected and stained overnight for GUS activity as described above, then fixed in 2.5% gluteraldehyde in sodium phosphate buffer before dehydration in an ethanol series and embedding in LR white resin (London Resin Company). Sectioning was performed using a Leica ultramicrotome and sections viewed and photographed as described (Penfield et al., 2004).
Transmission Electron Microscopy
This was performed on imbibed Arabidopsis seeds stratified for 3 d and then transferred to a growth chamber for 24 h under continuous white light at 70 μM m−2 s−1 at 20°C. Seeds were fixed with 2.5% gluteraldehyde in sodium phosphate buffer, postfixed in osmium tetroxide, dehydrated, and embedded in Spurr's resin. Ultrathin sections were viewed under an FEI Tecnai G2 transmission electron microscope. Using captured images, areas of cytoplasm from 10 representative embryo and endosperm cells were determined using the open source ImageJ software (rsb.info.nih.gov/ij) and the number of observable ribosomes present within each area counted.
Fatty Acid Determinations
TAG breakdown was followed by measuring the abundance of the Arabidopsis TAG-specific fatty acid eicosenoic acid (20:1) by gas chromatography as described previously (Penfield et al., 2004).
rbohd Germination Assay
Approximately 50 seeds from five independent seed batches of simultaneously grown wild type (Columbia-0) and rbohd mutant were sown on water agar plates supplemented with ABA (mixed isomers; Sigma-Aldrich) as indicated. Plates were stratified for 3 d and placed in continuous white light at 70 μM m−2 s−1 at 20°C. Germination was scored after 5 d as radicle emergence from the seed coat and endosperm.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: ABI3, At3g24650; ABI4, At2g40220; ABI5, At2g36270; rbohD, At5g47910; and EPR1, At2g27380. Affymetrix data have been deposited in the Nottingham Arabidopsis Stock Centre public repository under the accession number 386.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. MapMan Visualization Tool's Overview of the Differences in Metabolism-Related Gene Expression between Embryo and Endosperm 1 d after Imbibition.
Supplemental Figure 2. MapMan Metabolism Visualization Tool's Overview of the Differential Affect of Applied ABA on the Transcriptome of the Embryo and Endosperm.
Supplemental Figure 3. Embryo and Endosperm Eicosenoic Acid Content 5 d after Imbibition on Control Media, 20 μM ABA, or 400 mM Mannitol.
Supplemental Table 1. Genes Whose Function Has Been Previously Experimentally Addressed and Are Expressed Specifically in the Embryo or Endosperm 1 d after Imbibition.
Supplemental Table 2. Expression of Genes Involved in Lipid Storage Reserve Mobilization in Arabidopsis.
Supplemental Table 3. The Identification of a Core Set of ABA-Responsive Probe Sets in Embryo, Endosperm, 24-h-Imbibed Seeds, and 7-d-Old Seedlings.
Supplemental Table 4. Cell Wall–Modifying Genes Expressed in the Endosperm during Germination and Their Regulation by PAC and ABA.
Supplementary Material
Acknowledgments
We thank Ruth Finkelstein and Jonathan Jones for sharing plant material. This work was supported by the Garfield-Weston Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ian A. Graham (iag1@york.ac.uk).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.106.041277.
References
- Acevedo-Hernández, G.J., Leon, P., and Herrera-Estrella, L.R. (2005). Sugar and ABA responsiveness of a minimal RBCS light-responsive unit is mediated by direct binding of ABI4. Plant J. 43 506–519. [DOI] [PubMed] [Google Scholar]
- Arenas-Huertero, F., Arroyo, A., Zhou, L., Sheen, J., and Leon, P. (2000). Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev. 14 2085–2096. [PMC free article] [PubMed] [Google Scholar]
- Bentsink, L., and Koornneef, M. (2002). Seed dormancy and germination. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0050, http://www.aspb.org/publications/arabidopsis/. [DOI] [PMC free article] [PubMed]
- Berger, F. (1999). Endosperm development. Curr. Opin. Plant Biol. 2 28–32. [DOI] [PubMed] [Google Scholar]
- Bewley, J.D. (1997. b). Breaking down the walls – A role for endo-β-mannanase in release from seed dormancy? Trends Plant Sci. 2 464–469. [Google Scholar]
- Bewley, J.D., and Black, M. (1994). Seeds: Physiology of Development and Germination. (New York: Plenum Press).
- Brocard, I.M., Lynch, T.J., and Finkelstein, R.R. (2002). Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol. 129 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard-Gifford, I.M., Lynch, T.J., and Finkelstein, R.R. (2003). Regulatory networks in seeds integrating developmental, abscisic acid, sugar, and light signaling. Plant Physiol. 131 78–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheadle, C., Vawter, M.P., Freed, W.J., and Becker, K.G. (2003). Analysis of microarray data using Z score transformation. J. Mol. Diagn. 5 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe, A.J., Abenes, M., Plant, A., and Moloney, M.M. (2000). The seed-specific transactivator, ABI3, induces oleosin gene expression. Plant Sci. 151 171–181. [DOI] [PubMed] [Google Scholar]
- Dijkwel, P.P., Huijser, C., Weisbeek, P.J., Chua, N.-H., and Smeekens, S.C.M. (1997). Sucrose control of phytochrome A signaling in Arabidopsis. Plant Cell 9 583–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreucq, B., Berger, N., Vincent, E., Boisson, M., Pelletier, G., Caboche, M., and Lepiniec, L. (2003). The Arabidopsis AtEPR1 extensin-like gene is specifically expressed in endosperm during seed germination. Plant J. 23 643–652. [DOI] [PubMed] [Google Scholar]
- Falk, K.L., Behal, R.H., Xiang, C., and Oliver, D.J. (1998). Metabolic bypass of the tricarboxylic acid cycle during lipid mobilization in germinating oilseeds. Regulation of nad+-dependent isocitrate dehydrogenase versus fumarase. Plant Physiol. 117 473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R. (1994). Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 5 765–771. [Google Scholar]
- Finkelstein, R.R., and Lynch, T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R., Wang, M.L., Lynch, T.J., Rao, S., and Goodman, H.M. (1998). The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat, J., Hauge, B.M., Valon, C., Smalle, J., Parcy, F., and Goodman, H.M. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocal, G.F., Sheldon, C.C., Gubler, F., Moritz, T., Bagnall, D.J., MacMillan, C.P., Li, S.F., Parish, R.W., Dennis, E.S., Weigel, D., and King, R.W. (2001). GAMYB-like genes, flowering, and gibberellin signaling in Arabidopsis. Plant Physiol. 127 1682–1693. [PMC free article] [PubMed] [Google Scholar]
- Groot, S.P.C., Kieliszewka-Rokicka, B., Vermeer, E., and Karssen, C.M. (1988). Gibberellin-induced hydrolysis of endosperm cell walls in gibberellin-deficient tomato seeds prior to radicle protrusion. Planta 174 500–504. [DOI] [PubMed] [Google Scholar]
- Gubler, F., Kalla, R., Roberts, J.K., and Jacobsen, J.V. (1995). Gibberellin-regulated expression of a myb gene in barley aleurone cells: Evidence for Myb transactivation of a high-pI alpha-amylase gene promoter. Plant Cell 7 1879–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser, C., Kortstee, A., Pego, J., Weisbeek, P., Wisman, E., and Smeekens, S. (2000). The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: Involvement of abscisic acid in sugar responses. Plant J. 23 577–585. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Jorna, M.L., Brinkhorst-van der Swan, D.L.C., and Karssen, C.M. (1982). The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana L. Heynh. Theor. Appl. Genet. 61 385–393. [DOI] [PubMed] [Google Scholar]
- Kwak, J.M., Mori, I.C., Pei, Z.M., Leonhardt, N., Torres, M.A., Dangl, J.L., Bloom, R.E., Bodde, S., Jones, J.D., and Schroeder, J.I. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22 2623–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laby, R.J., Kincaid, M.S., Kim, D., and Gibson, S.I. (2000). The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J. 23 587–596. [DOI] [PubMed] [Google Scholar]
- Lee, J.Y., Levesque, M., and Benfey, P.N. (2005). High-throughput RNA isolation technologies. New tools for high-resolution gene expression profiling in plant systems. Plant Physiol. 138 585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre, V., North, H., Frey, A., Sotta, B., Seo, M., Okamoto, M., Nambara, E., and Marion-Poll, A. (2006). Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 45 309–319. [DOI] [PubMed] [Google Scholar]
- Liu, P.P., Koizuka, N., Homrichhausen, T.M., Hewitt, J.R., Martin, R.C., and Nonogaki, H. (2005). Large-scale screening of Arabidopsis enhancer-trap lines for seed germination-associated genes. Plant J. 41 936–944. [DOI] [PubMed] [Google Scholar]
- Manz, B., Muller, K., Kucera, B., Volke, F., and Leubner-Metzger, G. (2005). Water uptake and distribution in germinating tobacco seeds investigated in vivo by nuclear magnetic resonance imaging. Plant Physiol. 138 1538–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi, K., Okamoto, M., Koshiba, T., Kamiya, Y., and Nambara, E. (2005). Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: Epigenetic and genetic regulation of transcription in seed. Plant J. 41 697–709. [DOI] [PubMed] [Google Scholar]
- Nambara, E., Naito, S., and McCourt, P. (1992). A mutant of Arabidopsis which is defective in seed development and storage protein accumulation is a new abi3 allele. Plant J. 2 435–441. [Google Scholar]
- Ogawa, M., Hanada, A., Yamauchi, Y., Kuwahara, A., Kamiya, Y., and Yamaguchi, S. (2003). Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald, O., Martin, T., Dominy, J.P., and Graham, I.A. (2001). Plastid redox state and sugars: Interactive regulators of nuclear encoded photosynthetic gene expression. Proc. Natl. Acad. Sci. USA 98 2047–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy, F., Valon, C., Raynal, M., Gaubier-Comella, P., Delseny, M., and Giraudat, J. (1994). Regulation of gene expression programs during Arabidopsis seed development: Roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6 1567–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield, S., Graham, S., and Graham, I.A. (2005). Storage reserve mobilization in germinating oilseeds: Arabidopsis as a model system. Biochem. Soc. Trans. 33 380–383. [DOI] [PubMed] [Google Scholar]
- Penfield, S., Rylott, E.L., Gilday, A.D., Graham, S., Larson, T.R., and Graham, I.A. (2004). Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid, and requires PHOSPHOENOLPYRUVATE CARBOXYKINASE1. Plant Cell 16 2705–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, S.L., Charlton, W.L., Baker, A., and Graham, I.A. (2002). Germination and storage reserve mobilization are regulated independently in Arabidopsis. Plant J. 31 639–647. [DOI] [PubMed] [Google Scholar]
- Quesada, V., Ponce, M.R., and Micol, J.L. (2000). Genetic analysis of salt-tolerant mutants in Arabidopsis thaliana. Genetics 154 421–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou, L., Gallardo, K., Debeaujon, I., Vandekerckhove, J., Job, C., and Job, D. (2004). The effect of alpha-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol. 134 1598–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook, F., Corke, F., Card, R., Munz, G., Smith, C., and Bevan, M.W. (2001). Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J. 26 421–433. [DOI] [PubMed] [Google Scholar]
- Rylott, E.L., Hooks, M.A., and Graham, I.A. (2001). Co-ordinate regulation of genes involved in storage lipid mobilization in Arabidopsis thaliana. Biochem. Soc. Trans. 29 283–287. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37 501–506. [DOI] [PubMed] [Google Scholar]
- Soderman, E.M., Brocard, I.M., Lynch, T.J., and Finkelstein, R.R. (2000). Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol. 124 752–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm, O., Blaesing, O., Gibon, Y., Nagel, A., Meyer, S., Kruger, P., Selbig, J., Muller, L.A., Rhee, S.Y., and Stitt, M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37 914–939. [DOI] [PubMed] [Google Scholar]
- Toorop, P.E., van Aelst, A.C., and Hilhorst, H.W. (2000). The second step of the biphasic endosperm cap weakening that mediates tomato (Lycopersicon esculentum) seed germination is under control of ABA. J. Exp. Bot. 51 1371–1379. [PubMed] [Google Scholar]
- Tusher, V.G., Tibshirani, R., and Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel, B., et al. (2005). Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol. 138 1195–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, S., Kamiya, Y., and Sun, T. (2001). Distinct cell-specific expression patterns of early and late gibberellin biosynthetic genes during Arabidopsis seed germination. Plant J. 28 443–453. [DOI] [PubMed] [Google Scholar]
- Yamauchi, Y., Ogawa, M., Kuwahara, A., Hanada, A., Kamiya, Y., and Yamaguchi, S. (2004). Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.