Abstract
Antirrhinum majus DEFICIENS (DEF) and Arabidopsis thaliana APETALA3 (AP3) MADS box proteins are required to specify petal and stamen identity. Sampling of DEF/AP3 homologs revealed two types of DEF/AP3 proteins, euAP3 and TOMATO MADS BOX GENE6 (TM6), within core eudicots, and we show functional divergence in Petunia hybrida euAP3 and TM6 proteins. Petunia DEF (also known as GREEN PETALS [GP]) is expressed mainly in whorls 2 and 3, and its expression pattern remains unchanged in a blind (bl) mutant background, in which the cadastral C-repression function in the perianth is impaired. Petunia TM6 functions as a B-class organ identity protein only in the determination of stamen identity. Atypically, Petunia TM6 is regulated like a C-class rather than a B-class gene, is expressed mainly in whorls 3 and 4, and is repressed by BL in the perianth, thereby preventing involvement in petal development. A promoter comparison between DEF and TM6 indicates an important change in regulatory elements during or after the duplication that resulted in euAP3- and TM6-type genes. Surprisingly, although TM6 normally is not involved in petal development, 35S-driven TM6 expression can restore petal development in a def (gp) mutant background. Finally, we isolated both euAP3 and TM6 genes from seven solanaceous species, suggesting that a dual euAP3/TM6 B-function system might be the rule in the Solanaceae.
INTRODUCTION
Ever since the formulation of the classic ABC model of flower development (Coen and Meyerowitz, 1991), tremendous progress has been made in the understanding of the genetic control of flower development. The ABC model proposes the existence of three types of gene function (A, B, and C) that act in different combinations to specify the identity of the floral organs. A alone yields sepals; A in combination with B yields petals; B with C yields stamens; and C alone yields carpels. Moreover, the model implies an antagonistic relationship between the A and C functions. The analysis of floral developmental mutants in the model species Arabidopsis thaliana and Antirrhinum majus revealed the central role that MADS box transcription factors play in flower development. It turned out to be mostly members of this gene family that carry out all or part of the A, B, and C functions (Sommer et al., 1990; Yanofsky et al., 1990; Huijser et al., 1992; Jack et al., 1992; Mandel et al., 1992; Trobner et al., 1992; Bradley et al., 1993; Goto and Meyerowitz, 1994).
The synthesis of this knowledge has greatly facilitated the analysis and understanding of flower development in other species and now offers an exciting framework to understand how flower development might have evolved at the molecular level. Of the plethora of morphological innovations that have been associated with plant evolution, the invention of a petal identity program has certainly been one of the key innovations, because it is tightly associated with a revolution in pollination mechanisms through the recruitment of insects as pollen carriers. Therefore, the molecular and functional evolution of the genes encoding these organ identity programs in relation to the evolution of the flower during the radiation of the plant kingdom is of particular interest.
In the higher eudicot model species Antirrhinum and Arabidopsis, the respective DEFICIENS (DEF) and APETALA3 (AP3) MADS box proteins are required to specify petal identity in the second whorl and stamen identity in the third whorl (Sommer et al., 1990; Jack et al., 1992). As heterodimers together with the GLOBOSA (GLO) and PISTILLATA (PI) proteins, respectively, they encode the B function according to the classic ABC model of flower development (Schwarz-Sommer et al., 1992; Goto and Meyerowitz, 1994). Arabidopsis and Antirrhinum are distantly related core eudicot species; therefore, the highly similar organization of the B function in both species initially suggested that the functions identified for DEF/AP3 and GLO/PI genes might be conserved across the core eudicots.
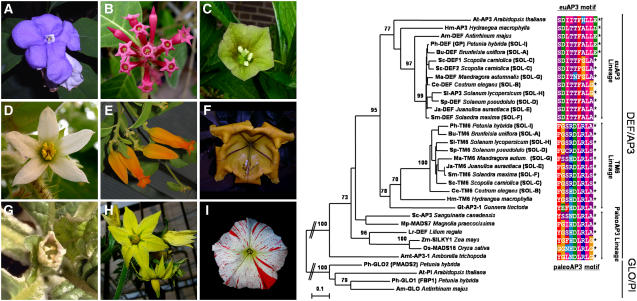
However, the isolation of DEF/AP3 homologs from a wider range of eudicot species revealed that, in fact, two paralogous DEF/AP3 lineages can be found in the core eudicots, and a phylogenetic analysis indicated that the two lineages have arisen by a gene duplication event that occurred close to or at the base of the higher eudicot radiation (Kramer et al., 1998). These two types can easily be distinguished on the basis of their fully divergent but highly conserved C-terminal motifs, called the euAP3 and paleoAP3 motifs (Kramer et al., 1998). Interestingly, euAP3-type genes (to which both DEF and AP3 belong) so far have been found exclusively in higher eudicot species. On the other hand, paleoAP3-type genes have a much broader distribution range and have been encountered in many taxa throughout the angiosperms, underscoring the antiquity and strong conservation of this motif throughout plant evolution. PaleoAP3-type genes are being identified in an accumulating number of core eudicot species (Kramer et al., 1998, 2006; Kramer and Irish, 2000; Kim et al., 2004), including the seven additional solanaceous species examined here. DEF/AP3 genes of the paleoAP3 type in core eudicot species are called TOMATO MADS BOX GENE6 (TM6) genes after the first isolated member from tomato (Solanum lycopersicum) (Pnueli et al., 1991). Comparison of sequences encoding the two different types of C-terminal motifs revealed that the origin of the novel euAP3 motif can be traced back to a frameshift mutation just upstream of the paleoAP3 motif, because second reading frame translation of a number of paleoAP3 coding sequences yields euAP3-like motifs (Vandenbussche et al., 2003a).
Therefore, both the species distribution and the C-terminal motif conservation ascribe the more ancestral B-class characteristics to the TM6 lineage, whereas euAP3 proteins seem to be a novelty restricted to the core eudicot species. EuAP3 proteins have been studied extensively. By contrast, little is known about the function of TM6 proteins in higher eudicots. Elucidating TM6 function, therefore, might help to understand the mode of B-function evolution and thereby the evolutionary development of the eudicot flower. Petunia hybrida (and a range of other species) has, unlike Arabidopsis, maintained both a euAP3 and a TM6 gene (van der Krol et al., 1993; Kramer and Irish, 2000), making it an ideal model in which to study the evolutionary development of the B function.
Previous work has provided indirect genetic evidence that in Petunia, the B function is organized differently compared with Arabidopsis and Antirrhinum. Mutations in the Petunia euAP3 homolog DEF (also called Green Petals [GP]) cause full homeotic conversion of petals to sepals in the second whorl, whereas stamen identity remains unaffected (van der Krol et al., 1993). The simplest explanation for such a phenotype is that subfunctionalization has occurred and Petunia DEF has a unique function in conferring petal identity, whereas another gene would be responsible for stamen identity, possibly in a redundant way with DEF. However, when def mutants are combined with the A-function-like blind (bl) mutant, the phenotype of the double mutants suggests that more is at stake (Tsuchimoto et al., 2000; Vandenbussche et al., 2004). In the recessive Petunia bl mutant (Vallade et al., 1987), the C-function MADS box genes PETUNIA MADS BOX GENE3 (PMADS3) and FLORAL BINDING PROTEIN6 (FBP6) are ectopically expressed in whorls 1 and 2, leading to the homeotic conversion of the corolla to antheroids in the second whorl and, occasionally, the development of carpelloid tissue in the first whorl (Tsuchimoto et al., 1993). In agreement with this, it has been demonstrated that ectopic PMADS3 expression is sufficient to phenocopy the bl mutation (Tsuchimoto et al., 1993; Kater et al., 1998). Therefore, the function of BL corresponds to the cadastral component of the A function as a repressor of C expression in the perianth.
Bearing in mind that the complete conversion of petals to sepals in the Petunia def mutant indicates the full absence of B-function activity in the second whorl, the rules of the classic ABC model would predict that def bl double mutants should develop carpelloids in the second whorl as a result of the action of the C function alone. By contrast, def bl double mutant flowers develop antheroids in the second whorl, only the tip of which is converted to stigmatic tissue (Tsuchimoto et al., 2000; Vandenbussche et al., 2004).
Together, the phenotypes of def and def bl flowers suggest the presence of an atypical B-function protein in Petunia that cannot complement DEF as a petal identity gene in the second whorl but that is capable of conferring stamen identity to the third whorl. In addition, this protein might also be responsible for the unexpected development of stamenoids in the second whorl of def bl flowers.
Here, we present evidence that the atypical function and regulation of Petunia TM6 is responsible for the unusual single and double mutant floral phenotypes, demonstrating that the euAP3- and TM6-type genes have functionally diverged in Petunia. The euAP3-type protein DEF is expressed mainly in whorls 2 and 3 (van der Krol et al., 1993), and its expression pattern remains unchanged in a bl mutant background (Tsuchimoto et al., 2000). Here, we show that in the wild-type-appearing tm6 mutant background, loss of DEF function causes the homeotic conversion of petals to sepals and of stamens to carpels, as has been described for def and ap3 mutants. Therefore, the unusual one-whorled phenotype of single Petunia def (gp) mutants can be entirely ascribed to TM6 function. Petunia DEF thus displays all typical characteristics that have been associated with normal euAP3 gene function as described for Antirrhinum DEF and Arabidopsis AP3.
On the other hand, Petunia TM6 does not exhibit the full B-function spectrum, because it is only involved in the determination of stamen organ identity. In addition, TM6 is regulated fundamentally differently than euAP3 genes: Petunia TM6 is expressed mainly in whorls 3 and 4 and is repressed directly or indirectly by the C-repression function of BL in the perianth, thereby excluding any involvement in petal development (Vandenbussche et al., 2004). A comparison between DEF and TM6 putative promoter sequences further supports the notion that an important change in regulatory elements took place during or after the duplication event that resulted in the euAP3 and TM6 lineages. These observations support the hypothesis of an ancestral role for B-type MADS box genes in distinguishing male from female reproductive structures (Mouradov et al., 1999; Sundström et al., 1999; Sundström and Engström, 2002; Winter et al., 2002).
Even though Petunia TM6 is not naturally involved in petal determination, we found that petal formation in the def mutant could be restored qualitatively by complementation with a 35S:TM6 construct. We will discuss our findings in the light of current models of flower evolution. Finally, we were able to isolate members of both the euAP3- and TM6-type gene lineages for seven additional solanaceous species from seven different genera, suggesting that a dual euAP3/TM6 B-function system, as found in Petunia, might be the rule in the Solanaceae, rather then the single euAP3 system from Arabidopsis.
RESULTS
Isolation of Petunia tm6 Knockout Mutants
To reveal the function of TM6, we screened the available Petunia dTph1 (for defective Transposon Petunia hybrida1) transposon libraries for insertions into the TM6 coding sequence. Screening ∼13,000 plants, we identified a single family of 20 individuals segregating for a dTph1 insertion in the TM6 gene. Sequence analysis of the progeny of these plants confirmed the presence of a dTph1 element in the TM6 gene, inserted 463 bp downstream of the ATG start codon, in the fourth exon at the 3′ splice junction. Because the dTph1 transposon encodes stop codons in all three reading frames in both directions (Gerats et al., 1990), the tm6-1 allele most likely represents a null allele, because the insertion results in a truncated protein lacking approximately one-third of its normal length. Nevertheless, no clear morphological aberrations were found in the flowers of homozygous mutant plants compared with wild-type flowers (Figure 1A). To investigate the effect of the transposon insertion on tm6-1 transcription, we monitored tm6 expression in the four floral organs of tm6-1 and wild-type flowers (Figure 2). Real-time PCR analysis indicates that in wild-type plants, TM6 is expressed highest in stamens and pistils and at considerably lower levels in petals and sepals, confirming previous results obtained by conventional RT-PCR and in situ hybridization (Vandenbussche et al., 2004). In all organs of tm6-1 flowers, expression decreased to very low levels or below the detection limit. These results suggest that a functional Petunia TM6 protein might be required to maintain its own expression, as has been found for the euAP3 genes DEF, AP3, and DEF (GP) in Antirrhinum, Arabidopsis, and Petunia, respectively (Schwarz-Sommer et al., 1992; Halfter et al., 1994; Jack et al., 1994). In agreement with the wild-type appearance of tm6-1 flowers, the other Petunia B-class MADS box genes, DEF, GLO1, and GLO2, remain normally expressed in tm6-1 mutants (Figure 2).
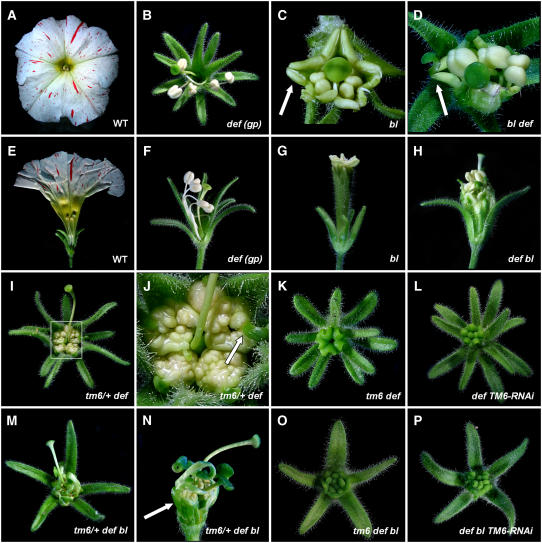
Figure 1.
Phenotypes Observed in Various P. hybrida B-Class and bl Mutant Combinations.
(A) to (D) Top views of wild-type W138 (A) and Petunia def (gp) (B) flowers and closeup top views of bl (C) and def bl (D) flowers. Arrows in (C) and (D) indicate second whorl antheroids and second whorl antheroids terminating with a short style/stigma structure, respectively.
(E) to (H) Side views of wild-type W138 (E), def (gp) (F), bl (G), and def bl (H) flowers. Some sepals were removed in (E), (F), and (H) to reveal the inner organization. Note that the front sepal in (G) has a carpelloid tip.
(I) and (J) Top view (I) and detail (J) of a Petunia tm6/+ def flower showing proliferating anther tissue in the third whorl terminating in a short style/stigma structure (arrow).
(K) Petunia tm6 def double mutant flower showing full conversion of stamens to carpels, forming a central congenitally fused chimney-like structure.
(L) Flower of a Petunia TM6-RNAi line in a def mutant background showing a similar phenotype as in tm6 def flowers.
(M) and (N) Top view (M) and detail (N) of tm6/+ def bl flowers showing proliferating anther tissue in the third whorl and carpelloid structures in the second whorl (arrow in [N]). Sepals were removed in (N) to reveal inner organs.
(O) A tm6 def bl triple mutant flower showing full conversion of petals and stamens to carpelloids, forming a central multiwhorled, congenitally fused chimney-like structure.
(P) Flower of a TM6-RNAi line in a def bl mutant background showing a similar phenotype as in tm6 def bl flowers.
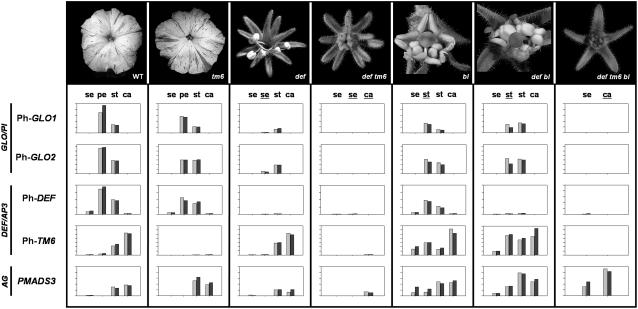
Figure 2.
Expression Analysis of P. hybrida B- and C-Class MADS Box Genes in Floral Whorls of Wild-Type and Various Mutant Flowers as Determined by Real-Time PCR.
Organ types are indicated above bars showing gene expression levels as follows: se, sepal; pe, petal; st, stamen; ca, carpel. Homeotically converted organs are shown underlined. All reactions were performed in duplicate using a biological replicate for each sample (represented by pairs of gray and black bars). The height of the bars for a given gene indicates relative differences in expression levels within the range of tissues tested. For each primer combination separately, the highest expression value encountered in the series of different tissues tested was set equal to 100, and lower values are plotted relative to this highest value on a linear y axis scale. GAPDH (for glyceraldehyde-3-phosphate dehydrogenase) expression levels were used for normalization.
We also developed transgenic Petunia lines in which TM6 is downregulated by RNA interference (RNAi). Two independent lines in which RNAi resulted in a >10-fold downregulation of steady state mRNA levels were selected for further analysis. Similar to the tm6-1 transposon insertion mutants, no obvious phenotype was observed in flowers of these lines (data not shown). These results indicate that Petunia TM6 functions redundantly with other factors, or alternatively, that the phenotype is so subtle that we have missed it.
Petunia tm6 def, tm6 glo1, and tm6 glo2 Double Mutant Analysis
The most obvious candidate to share function(s) with TM6 among the other Petunia B-class MADS box genes is DEF (GP), because both genes belong to the DEF/AP3 clade. To obtain def tm6 double mutants, homozygous tm6-1 individuals were crossed with the previously described def-1 bl double mutants (Vandenbussche et al., 2004). The resulting F1 progeny were self-fertilized to obtain tm6 def double and tm6 def bl triple mutants in the next generation (for triple mutants, see next paragraph). In a first F2 population of ∼250 individuals, only four phenotypic classes were observed, either displaying wild-type, def, bl, or def bl phenotypes, as described previously (Vandenbussche et al., 2004). When we analyzed these plants at the molecular level, all homozygous def-1 mutants appeared to be homozygous wild type for TM6, whereas homozygous tm6-1 mutants were all homozygous wild type for DEF. These results suggested a close linkage between the TM6 and DEF loci. To identify rare recombination events between the two loci, we screened a second F2 population of ∼2000 plants. In this population, homozygous def and def bl mutants were selected by phenotype and subsequently screened for the presence of the tm6-1 allele by PCR. Two plants were found to be homozygous for the def-1 mutation and heterozygous for tm6-1, and one plant was homozygous for def-1 and bl and heterozygous for tm6-1 (see below). Although stamen development was virtually unaffected in def and def bl mutants (Figures 1B to 1D), normal development of the third whorl organs in the tm6/+ def recombinants was seriously disturbed. In flowers of such plants, development of the anther filaments was strongly suppressed, whereas the loculi of the anthers developed as brain-like folded structures consisting of proliferating antheroid tissue and terminating in a short style/stigma-like structure (Figures 1I to 1J), suggesting a partial loss of determinacy in the third whorl of tm6/+ def flowers. This finding is in agreement with our previous observation of a partial loss of determinacy in the third floral whorl of Petunia glo1/+ glo2 plants (Vandenbussche et al., 2004), giving further support for the involvement of B-class MADS box genes in cell proliferation and determinacy (Bowman et al., 1992; Trobner et al., 1992; Jack et al., 1994; Sakai et al., 1995; Krizek and Meyerowitz, 1996).
Although pollen maturation and fertility in tm6/+ def plants were heavily impaired, selfing occasionally gave rise to viable seeds. In the resulting F3 progeny, besides plants displaying a def or tm6/+ def phenotype, a new phenotype was encountered: a class of plants displayed flowers exhibiting a complete homeotic conversion of the second whorl petals to sepals and of the third whorl stamens to carpels (Figure 1K). The five carpelloid organs in the third whorl were fused and formed a tubular structure. The development of the original pistil in the fourth whorl was severely reduced in most flowers. Molecular analysis confirmed all of these plants to be def-1 tm6-1 double mutants. These results demonstrate that Petunia DEF and TM6 act redundantly in the determination of stamen identity in the third whorl and in preventing premature termination of the flower meristem in the center of the flower, as has been described for DEF in Antirrhinum (Trobner et al., 1992). In accordance with the sepal and carpel identities of second and third whorl organs in tm6 def double mutant flowers, the expression of both GLO1 and GLO2 was almost undetectable in these two whorls (Figure 2), whereas in def (gp) single mutants, expression in the third whorl was maintained (cf. Angenent et al., 1992; van der Krol et al., 1993). This finding indicates that Petunia DEF and TM6 act redundantly to maintain GLO expression in the third whorl. The phenotype of tm6 def double mutant plants is very similar to that of the previously described Petunia glo1 glo2 and def glo2 double mutants (Vandenbussche et al., 2004).
When the Petunia TM6 RNAi transgene was introduced in the def mutant background, we observed a homeotic conversion of anthers to carpelloid organs very similar to that in tm6 def mutants (Figure 1L), although we observed some variation in the completeness of the homeotic conversion between different lines and between flowers of the same plants during development (data not shown), most likely as a result of differences in the degree of silencing. In strongly silenced flowers, the expression of both GLO1 and GLO2 was reduced to a similar degree as in tm6 def flowers (data not shown).
To analyze the remaining possible genetic interactions between the four Petunia B-type MADS box proteins, we also created tm6 glo1 and tm6 glo2 double mutants. Flowers of these double mutants did not show any additional phenotypes compared with glo1 (fbp1) and glo2 (pmads2) single mutants (Vandenbussche et al., 2004).
Petunia tm6 def bl Triple Mutant Analysis
Analysis of the expression of TM6 in bl mutant flowers shows that although TM6 is expressed at low levels in wild-type second whorl organs, its expression is strongly upregulated in the first two whorls of bl mutants (Figure 2). This offers an explanation for the striking phenotype of def bl double mutant flowers, which unexpectedly showed restoration of B-function activity in the second whorl compared with def single mutants (Tsuchimoto et al., 2000; Vandenbussche et al., 2004). To test whether this atypical expression profile of Petunia TM6 might be the cause of the striking phenotype of def bl mutants, we created tm6 def bl triple mutants. These plants were obtained by self-fertilizing the single tm6/+ def bl individual mentioned in the previous section. As in tm6/+ def plants, the third whorl anthers of this plant developed as brain-like structures consisting of proliferating antheroid tissue terminating in a style/stigma-like structure (Figures 1M and 1N). In addition, most of the antheroid tissue produced in the second whorl of def bl double mutants was converted to carpelloid tissue, often forming a clear pistil-like structure including an ovary at the base. Again, selfing was problematic because of strongly reduced pollen viability, although we occasionally obtained some seed set. In the next generation, we observed a new phenotype: plants exhibiting flowers in which both petal and stamen whorls were fully converted to carpelloid organs, together forming one central multiwhorled, congenitally fused structure (Figure 1O). Molecular analysis confirmed these plants to be tm6 def bl triple mutants. In agreement with this, when the TM6 RNAi transgene was crossed into a def bl mutant background, a very similar phenotype was obtained in the heavily silenced lines (Figure 1P). These results demonstrate that the unexpected development of antheroid tissue in the second whorl of def bl double mutants is entirely attributable to ectopic TM6 expression in this whorl. In agreement with the observed phenotypes, GLO1 and GLO2 expression disappeared entirely in tm6 def bl triple mutants (Figure 2) and in def bl TM6-RNAi plants (data not shown), whereas the GLO genes were normally expressed in def bl double mutants (Figure 2). This finding clearly demonstrates that Petunia TM6 is capable of maintaining GLO expression also in the second whorl, on the condition that BL is mutated.
Petunia TM6 Overexpression in the Wild Type and in a def Mutant Background
In wild-type Petunia flower development, TM6 acts redundantly with DEF to determine stamen identity but apparently plays no role in petal development. In agreement with this, although there is some TM6 expression in emerging petal primordia at early stages (Vandenbussche et al., 2004), its expression is not maintained in these regions as it is in stamens and ovary. Moreover, because TM6 expression is strongly upregulated in the perianth organs of a bl mutant flower, TM6 expression seems to be repressed directly or indirectly in the first two whorls by the BL product in wild-type flowers, thereby excluding any involvement of TM6 in petal development.
To test whether the TM6 protein is capable of determining petal identity on itself, we constitutively expressed TM6 under the control of a 35S promoter in wild-type and def genetic backgrounds. In a wild-type genetic background, several independent transformants developed flowers with an extra whorl of smaller separate petals, fused to the abaxial side of the petal tube (Figure 3A). Remarkably, all abaxial characteristics of wild-type petals, such as a dense trichome coverage on the petal tube and abaxial epidermal cell types, are found on the adaxial side of the ectopic petals, whereas the typical wild-type adaxial conical cells and the pigmentation of the inner tube are found on the abaxial side of the ectopic petals. These ectopic petals thus seem to have switched abaxial–adaxial polarity, and although they remain smaller and do not fuse with each other, they form a perfect mirror image of wild-type petals. Occasionally, even sepals acquired a partial petal identity: although the distal ends of the sepals retained their wild-type identity, the development of a clear tube-like structure was observed at their bases (Figure 3B). This petaloid tissue senesced at the same time as the petals, long before the remaining part of the sepals senesced.
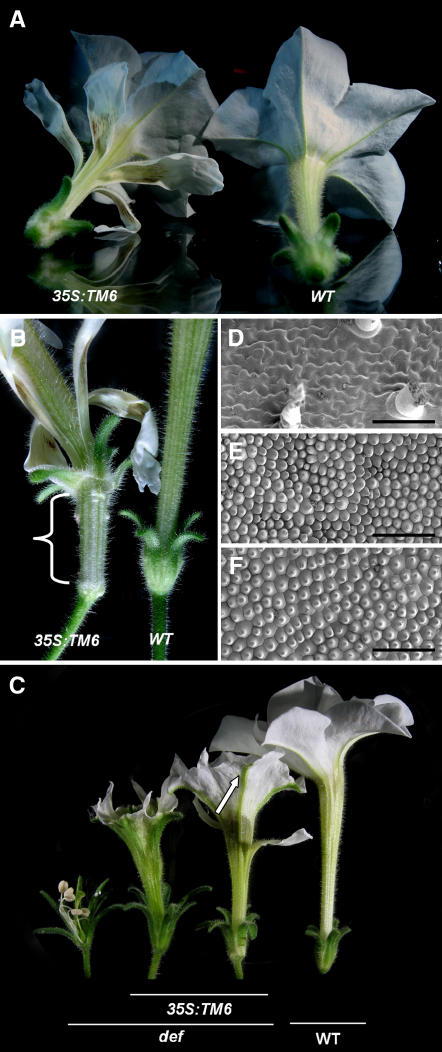
Figure 3.
Petunia TM6 Overexpression Phenotypes in Wild-Type and def Mutant Backgrounds.
(A) 35S:TM6 overexpression in wild-type Petunia showing the development of ectopic petals on the outer surface of the petal tube.
(B) Detail of first whorl organs in 35S:TM6 plants showing a partial conversion to a petal tube-like structure.
(C) 35S:TM6 overexpression in a def mutant background showing partial and more complete complementation of petal development. The arrow indicates the main petal veins retaining sepal identity in a strongly complemented overexpression line.
(D) to (F) Scanning electron microscopy images of the adaxial epidermis of def second whorl sepalloids showing the typical sepal epidermal characteristics, such as jigsaw-shaped epidermal cells interspersed by trichomes and stomata (D), wild-type petals showing the characteristic conical petal cells (E), and second whorl petals of def mutants complemented with 35S:TM6 (F). Bars = 100 μm.
When the 35S:TM6 construct was introduced in a def genetic background, a range of phenotypes was observed, from a very partial to an almost complete restoration of petal development (Figure 3C). Electron microscopy analysis of these restored petals revealed a semicomplete restoration in epidermal cell type differentiation: whereas the second whorl organs of def flowers displayed jigsaw-shaped epidermal cells covered by trichomes (Figure 3D) typical for Petunia first whorl sepals, the restored petals in def 35S:TM6 plants displayed the characteristic conical petal cells (Figure 3F) similar to wild-type petals (Figure 3E). These results indicate that although TM6 in wild-type Petunia flower development is not involved in petal development, when ectopically expressed in the second whorl the protein is clearly capable of inducing petal development in a def mutant background.
Promoter Comparison between euAP3 and TM6 Genes
To further investigate the differences in expression between Petunia DEF and TM6, we isolated putative 5′ regulatory sequences of both genes by genome walking. For comparison, we also isolated the corresponding regulatory sequences from the putative orthologs from tomato, Sl AP3 (also called TAP3) and Sl TM6, which have a similar expression pattern compared with Petunia DEF and TM6, respectively (V. Irish, personal communication). The resulting genome walks yielded putative promoter fragments for the four genes varying in size between 1.5 and 3 kb upstream of the putative ATG. To test sequence conservation within the Solanaceae, we first aligned the obtained putative promoter sequences of the two euAP3-type genes Ph DEF and Sl AP3. Over most of the alignment, only dispersed short regions of homology were found. On the other hand, a highly conserved region was identified closer to the ATG, present in both Ph DEF and Sl AP3 putative 5′ regulatory sequences, suggesting that this domain might be important for proper Ph DEF and Sl AP3 expression. Comparison of this domain with other published euAP3-type 5′ regulatory sequences showed a major part of this conserved region to correspond with a previously identified highly conserved domain in the upstream regulatory regions of euAP3-type DEF and St DEF4 genes (Schwarz-Sommer et al., 1992; Garcia-Maroto et al., 1993) and also found in a large set of Brassica AP3 homologs (Hill et al., 1998; Tilly et al., 1998; Koch et al., 2001). The alignment of this euAP3-type 5′ regulatory domain from Ph DEF and Sl AP3 (Solanaceae) is shown in Figure 4A, together with the corresponding domain from two Lamiaceae species (Am DEF and Mo DEF) and two Brassica species (At AP3 and Cp AP3).
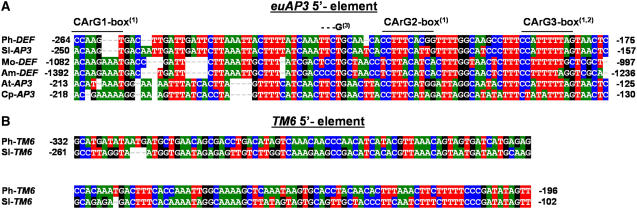
Figure 4.
Alignment of Conserved Regions Identified in euAP3- and TM6-Type 5′ Putative Regulatory Sequences.
Species names are abbreviated as follows: Ph, Petunia hybrida; Sl, Solanum lycopersicum; Mo, Misopates orontium; Am, Antirrhinum majus; At, Arabidopsis thaliana; Cp, Cochlearia pyrenaica. Accession numbers of these sequences can be found in Methods. Numbers flanking the fragment names indicate their positions relative to the ATG start codon: (1), position of the three proposed CArG boxes in the Arabidopsis AP3 promoter (Hill et al., 1998; Tilly et al., 1998); (2), position of the proposed CArG box in the Antirrhinum DEF promoter; (3), mutation site of the chlorantha def allele (Schwarz-Sommer et al., 1992).
A homology search with the obtained 2.7-kb upstream fragment of the tomato TM6 gene revealed the presence of two large elements with frequent occurrence elsewhere in the tomato genome but not in the Petunia TM6 promoter, suggesting that these represent tomato-specific repetitive elements not related to TM6 function. Notably, in the tomato TM6 promoter, a 0.7-kb element was found at position −362 relative to the ATG start codon, which shows high homology with the tomato putative XC transposon (Guyot et al., 2005). In the 1.5-kb 5′ fragment of the Petunia TM6 gene, a repetitive element was found at position −1002 bp relative to the ATG. For the alignment of Petunia TM6 and tomato TM6 5′ regulatory sequences, we removed these repetitive elements. The general picture that emerged was similar to that observed for the euAP3 sequences: most of the upstream region displays only short stretches of local homology, whereas closer to the putative ATG, a region was found that displays high sequence conservation between the Petunia and tomato TM6 (Figure 4B). This region is located in approximately the same position relative to the ATG start codon as the euAP3 5′ regulatory domain. Because euAP3 genes originate from the duplication of an ancestral paleoAP3-type gene, we compared the putative regulatory elements of both lineages, but no obvious homology between these elements could be identified (Figure 4). These results indicate that, upon or during the duplication event, an important change in the upstream regulatory sequences between the euAP3 and TM6 lineages took place.
Isolation of euAP3 and TM6 Homologs from Other Members of the Solanaceae
With Petunia, tomato, potato (Solanum tuberosum), and Nicotiana benthamiana, the Solanaceae is well represented in the group of core eudicot species for which both euAP3 and TM6 genes have been submitted to the database. This suggests that a dual euAP3/TM6 B-function system might be conserved among the Solanaceae. To further investigate this notion, we applied a degenerate PCR approach to isolate DEF/AP3 members from seven additional solanaceous species, all belonging to different genera. The result of this analysis is presented in a neighbor-joining tree (Figure 5) with the addition of some DEF/AP3 genes from other informative taxa. For a recent more complete overview of other available TM6 sequences, see Kramer et al. (2006). For all seven solanaceous species tested, we were able to isolate both a euAP3-type and a TM6-type B-class gene, whereas for one species, two euAP3 copies and one TM6 gene were found. Without exception, all newly isolated genes terminate with either a paleoAP3 motif, in the case of TM6 members, or a euAP3 motif, for the euAP3 lineage genes. The latter motif shows some variation, because a shorter variant is present in all solanaceous species tested, except for Ph DEF and Brunfelsia uniflora (Bu DEF) (Figure 5). The euAP3 motif of Ph DEF and Bu DEF more resembles that of the Antirrhinum, Hydrangea, and Arabidopsis euAP3 genes than that of the euAP3 genes of the other solanaceous species examined, suggesting that the shorter form of the euAP3 motif evolved fairly recently within a subset of the Solanaceae. This is further supported by the observation that these shorter forms in the coding sequence still show homology beyond their stop codons with the slightly longer variant of Petunia and Brunfelsia when a one-nucleotide gap is introduced (data not shown). These two variants of C-terminal motifs within the Solanaceae have been described in detail by Kramer et al. (2006).
Figure 5.
Neighbor-Joining Tree of Newly Isolated euAP3 and TM6 Homologs from Various Solanaceous Species, Including a Selection of B-Class Lineage MADS Box Genes from Other Informative Taxa.
Members of the GLO/PI subfamily were used as an outgroup, and 1000 bootstrap samples were generated to assess support for the inferred relationships. Local bootstrap probabilities of >70% are shown near the major branching points. euAP3 and TM6 putative proteins from solanaceous species included in this analysis are indicated with the extension SOL-x, of which the x corresponds to the lettering on the images of the flowers of these species at left.
DISCUSSION
Comparison between DEF and TM6 Function and Regulation in Petunia
Although the function of euAP3 proteins has been studied extensively, little is known about the function of TM6 proteins. In previous studies of the B function in Petunia, it was found that loss of function of the euAP3-type DEF (GP) protein causes homeotic conversion of petals to sepals in the second whorl (van der Krol et al., 1993), whereas stamen development remains unaffected. Because of this one-whorl phenotype, it was suggested that Petunia DEF might function differently than DEF and AP3 in Antirrhinum and Arabidopsis. However, when the def-1 null allele is introduced in the wild-type-appearing tm6-1 mutant background, a homeotic conversion of petals to sepals and stamens to carpels is observed, exactly as has been described for def and ap3 mutants in Antirrhinum and Arabidopsis. Therefore, the one-whorled phenotype of def mutants is entirely attributable to the action of the Petunia TM6 gene. Also at the level of gene expression and regulation, Petunia DEF is very similar to DEF and AP3. Petunia DEF is expressed mainly in whorls 2 and 3 (van der Krol et al., 1993), and its expression domain remains unaltered in a bl mutant background (Tsuchimoto et al., 2000; Vandenbussche et al., 2004). Furthermore, DEF expression is autoregulated (Halfter et al., 1994) and requires the presence of at least one of the two Petunia GLO proteins to maintain its own expression (Vandenbussche et al., 2004). Therefore, we conclude that Petunia DEF displays all typical characteristics that have been associated with euAP3 gene function as identified for Arabidopsis AP3 and Antirrhinum DEF.
Petunia TM6, by contrast, is expressed mainly in whorls 3 and 4 throughout flower development (Vandenbussche et al., 2004). Because stamen development is not impaired in either the def or tm6 single mutant, and stamens are fully converted to carpels in tm6 def double mutants, we conclude that TM6 functions as a stamen identity gene, fully redundant with DEF. Unlike DEF, the expression pattern of TM6 in both wild-type and bl mutant backgrounds mimics that of a C-class gene rather than a B-class gene (Figure 2). The full conversion of antheroids to carpels in tm6 def bl triple mutants and in def bl TM6-RNAi lines (Figures 1O and 1P) further demonstrates that this unusual expression pattern forms the molecular basis for why the phenotype of def bl double mutant flowers seemed to elude the rules of the classical ABC model.
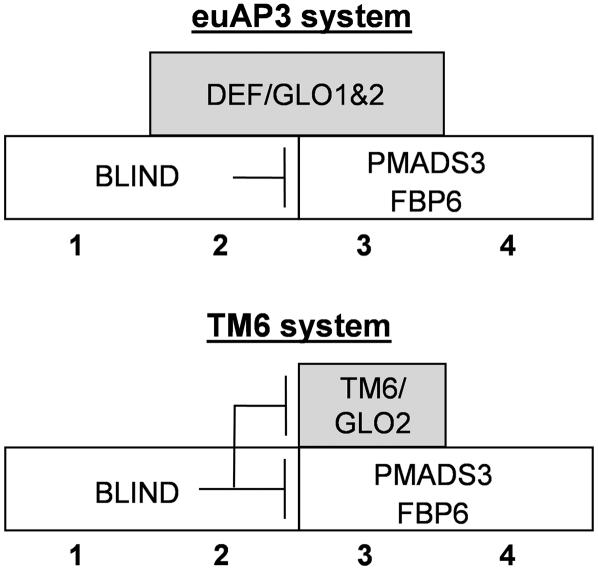
Together, these results indicate that the regulatory control of Petunia TM6 and DEF expression is fundamentally different (Figure 6). Although the expression pattern of DEF remains unchanged in a bl mutant background, TM6 expression is strongly upregulated in the second whorl and to a lesser extent in the first whorl, suggesting that TM6 expression is repressed by the BL product in the Petunia perianth organs, similar to C-class genes. Alternatively, the TM6 expression domain shift in bl mutants also could be explained as an indirect effect. If the maintenance of TM6 expression required C-class activity, then the ectopic C-class expression in bl mutants might upregulate TM6 expression in the perianth (see also last paragraph of Discussion). In addition, the strong expression levels throughout ovule development suggest additional functions for TM6 in the fourth whorl. The answer to the latter question may be found in the analysis of redundant ovule function(s).
Figure 6.
Model of B-Function Regulation in P. hybrida.
Regulatory Interactions and Redundant and Unique Functions of the B-Class Heterodimers in Petunia Flower Development
In Antirrhinum and Arabidopsis, it has been shown that DEF and AP3 function as an obligate heterodimer in combination with GLO and PI (Trobner et al., 1992; Krizek and Meyerowitz, 1996; McGonigle et al., 1996), providing a molecular explanation for the similar phenotypes observed in either the def (ap3) or glo (pi) mutant. In Petunia, the situation is more complex, because two GLO/PI genes have been identified and, within the DEF/AP3 lineage, both DEF and TM6 are present (Angenent et al., 1992; van der Krol et al., 1993; Kramer and Irish, 2000). Nevertheless, the full conversion of petals to sepals and of stamens to carpels observed in Petunia glo1 glo2 double mutants (Vandenbussche et al., 2004), and in the tm6 def double mutants described here, demonstrates that DEF/AP3 and GLO/PI functions in Petunia are also interdependent. In both double mutant combinations, no heterodimers can be formed because either both GLO/PI or both DEF/AP3 proteins have been mutated. Interestingly, in a yeast two-hybrid assay, although Petunia DEF was shown to interact with both Petunia GLO1 and GLO2 (Immink et al., 2003), Petunia TM6 displayed a strong partner preference for GLO2 (Vandenbussche et al., 2004). The phenotype of tm6 def flowers, together with the phenotypes of glo2 def and glo1 def mutant flowers (Vandenbussche et al., 2004), now provide full genetic support for this partner preference. Because no B-function activity can be performed without expression of at least one of the GLO genes, and because TM6 interacts only with GLO2, abolishing GLO2 function should result in the concomitant loss of B-class activity for TM6 as well. On the other hand, loss of GLO1 function should not influence the B-function activity of TM6. Indeed, the full conversion of stamens to carpels in tm6 def double mutants is identical to the phenotype of glo2 def mutants, whereas in glo1 def mutants, stamen development is virtually unaffected. These results, together with the complete absence of B-function activity in glo1 glo2 double mutant flowers, indicate that although TM6 interacts with only one of the GLO genes, obligate heterodimerization with a GLO/PI protein as a prerequisite for B-function activity is also conserved for TM6.
To further specify the functions of each of the three Petunia DEF/GLO heterodimers that apparently can be formed, we made all possible B-class double mutant combinations to create the different genetic backgrounds in which the capacity to specify petal and/or stamen identity of the remaining B-class MADS box pair(s) can be tested.
Previously, we proposed that petal development in Petunia is controlled by the concerted action of DEF/GLO1 and DEF/GLO2 heterodimers. In addition, the downregulation of DEF expression levels in the second whorl of glo1 glo2 double mutants and of GLO1 and GLO2 expression levels in the second whorl of def mutants demonstrates that the maintenance of DEF and GLO1/2 expression in the second whorl is interdependent (Vandenbussche et al., 2004).
In tm6 glo1, tm6 glo2, and def glo1 double mutants, stamens develop normally, indicating that either the DEF/GLO2, DEF/GLO1, or TM6/GLO2 heterodimer is sufficient to determine stamen identity. At the level of transcriptional regulation, the complete absence of Petunia GLO expression in tm6 def flowers (Figure 2) but not in def (Angenent et al., 1992; van der Krol et al., 1993) or tm6 flowers indicates that DEF and TM6 act redundantly in maintaining GLO expression in the third whorl. Summarizing, we conclude that petal development is redundantly controlled by DEF/GLO1 and DEF/GLO2 heterodimers, whereas stamen development can be accomplished by DEF/GLO1, DEF/GLO2, or TM6/GLO2 heterodimers in Petunia.
Promoter Comparison between euAP3 and TM6 Genes
The clearly divergent expression patterns of Petunia DEF and TM6 suggest that at the moment of or after the duplication that gave rise to the TM6 and euAP3 lineages, an important change in the regulatory sequences of these genes must have taken place, at least in the lineage leading to Petunia. In the Ph DEF and Sl AP3 promoter sequences, a very similar conserved domain was found that also is conserved in promoters of euAP3-type genes from more distantly related species. In Antirrhinum, the chlorantha mutation, which causes defects in the maintenance of def expression in petals and stamens (Schwarz-Sommer et al., 1992), maps to this region (Figure 4). In Arabidopsis, this region has been shown to be essential for the initiation and maintenance of AP3 expression in both petals and stamens. In addition, CArG box binding sites have been identified in this region in both Antirrhinum and Arabidopsis (Schwarz-Sommer et al., 1992; Hill et al., 1998; Tilly et al., 1998). However, only the third Arabidopsis CArG box seems to reside within the most conserved parts of this domain (Figure 4), suggesting that the positions of CArG boxes might differ between species.
Surprisingly, this highly conserved region present in euAP3-type genes is absent in the TM6 promoter sequences of Petunia and tomato. Instead, an unrelated conserved region was found, suggesting that the euAP3 5′ regulatory element may have been acquired during or soon after the duplication event that created the euAP3 gene lineage. At present, we cannot rule out the alternative scenario that the euAP3 element was originally present in the ancestral gene and that it was lost in the TM6 lineage as a result of the subcompartmentalization of initially entirely overlapping expression domains with the euAP3 lineage genes. The answer to this question will require a comparative promoter study of TM6/paleoAP3-type sequences from a wider range of species, including the basal angiosperms. The fact that both the TM6- and euAP3-type genes contain conserved but different domains in their respective promoters raises the question of how these elements might have been acquired during evolution. When the tomato genomic sequence becomes available, a synteny comparison upstream and downstream of the tomato AP3 and TM6 loci might reveal whether the TM6/euAP3 duplication was part of a larger duplicated chromosomal fragment, as has been found for C-class MADS box genes (Causier et al., 2005), or resulted from a single gene duplication event.
Speculation about the Molecular Origin of the Functional Divergence between Petunia DEF and TM6 in Relation to the Evolution of Petal Development in Higher Eudicots
At the protein level, the most obvious difference between euAP3 and paleoAP3 proteins is their completely divergent C-terminal euAP3 and paleoAP3 motifs, which are highly conserved within both lineages (Kramer et al., 1998). In Arabidopsis, the AP3 motif was shown to be essential for proper AP3 function, because truncation of the AP3 protein by removing the C-terminal region eliminates its function (Lamb and Irish, 2003). Lamb and Irish (2003) also showed that a chimeric AP3 protein in which the euAP3 motif was replaced by a paleoAP3 motif from Dicentra eximia (including the preceding PI-derived motifs) could partially complement the Arabidopsis ap3 mutant in stamen development but not in petal development. These results led to the suggestion that the paleoAP3 C terminus retains stamen-promoting activity but is unable to promote petal development in a higher eudicot genetic background.
In agreement with this, we found that in Petunia, TM6 is involved in the determination of stamen but not petal identity, whereas DEF is involved in both stamen and petal identity. By contrast, despite the presence of a paleoAP3-type motif in its C terminus, Petunia TM6 as a stamen identity gene is perfectly capable of inducing petal development when ectopically expressed, even in the first whorl (Figure 3). A very similar phenotype was described for the overexpression of DEF (GP) in Petunia (Halfter et al., 1994), including the development of additional petals fused to the abaxial side of the petal tube. Thus, the petal-promoting capacity of the paleoAP3-type Petunia TM6 protein sequence on itself seems not to differ qualitatively from that of Petunia DEF, a classical euAP3-type protein.
This is not the first example of petal rescue by overexpression of a B-class MADS box gene, which in its natural context is not involved in petal development. In maize (Zea mays) and rice (Oryza sativa), the paleoAP3 genes Silky1 (Sil) and Superwoman1 have been shown to be required to specify stamen identity in the third floral whorl and lodicule development in the second whorl, respectively (Ambrose et al., 2000; Nagasawa et al., 2003). Lodicules are small specialized organs that swell at anthesis to open the grass floret. When Si1 was expressed in an Arabidopsis ap3 mutant background under the control of the AP3 promoter, the development of stamens but also of petals was almost completely restored (Whipple et al., 2004), although this required higher levels of expression than endogenous AP3 expression. Apparently, the euAP3 motif in itself is not essential to confer a petal identity function to a B-class protein in the higher eudicots, because both Si1 and Petunia TM6 carry paleoAP3 motifs. The discrepancy between the results for both TM6 and Si1 overexpression and the outcome of the motif-swapping experiments in Arabidopsis might be attributable to the heterologous and chimeric nature of the construct used in the latter experiment. Differences in the rest of a paleoAP3 protein might normally functionally compensate for the lack of a euAP3 motif, but using chimeric constructs, this compensation would be missed. In this respect, it would be interesting to test the petal-promoting capacity of the native Dicentra paleoAP3 protein used by Lamb and Irish (2003).
In Petunia, the restoration of petal development in def mutants by TM6 overexpression indicates that the incapacity of the TM6 regulatory unit to maintain high expression levels in the second whorl is the main cause of the lack of a petal identity function of TM6. Therefore, the functional divergence between Petunia DEF and TM6 would be caused mainly by changes in expression pattern. However, the classic distinction of either changes in the protein sequence and/or changes in the expression pattern as a source of functional divergence between two paralogous genes might not be so easily made for genes whose protein products function in a positive autoregulatory loop. Arabidopsis AP3/PI and Antirrhinum DEF/GLO protein complexes are known to enhance the initially low expression levels of the genes that encode them and to maintain them at sufficiently high levels to function properly as organ identity genes (Trobner et al., 1992; Goto and Meyerowitz, 1994; Jack et al., 1994; Krizek and Meyerowitz, 1996; McGonigle et al., 1996; Riechmann et al., 1996; Yang et al., 2003a, 2003b). Mutations in the protein sequence can affect partner affinity or the specificity of protein–protein interactions in the protein complexes involved in autoregulation and, in theory, can thereby cause a shift in the floral domains in which the maintenance of expression is achieved. Therefore, it is important to consider the possibility that changes in the protein-coding sequence might have contributed to the difference in expression between Petunia TM6 and DEF.
Gomez-Mena et al. (2005) recently showed that maintaining AP3 expression requires, besides the presence of a GLO/PI protein, the presence of either AG (stamens) or AP1 (petals). Because of the antiquity of the stamen identity program, the autoregulatory loop involving AG most likely represents the ancestral regulatory mechanism of B-class MADS box proteins. In this respect, it is interesting that the expression pattern of Petunia TM6 in wild-type and bl mutant backgrounds can also be explained as a result of the incapacity of TM6 to maintain expression outside of a C expression domain context. TM6 is initially expressed in petal primordia, but at later stages it fails to get strongly upregulated, in contrast with what happens during stamen development. When using a constitutive promoter for complementation experiments, any potential effect of changes in protein sequence on the capacity to maintain expression will obviously remain obscured. Therefore, to fully understand the influence of promoter evolution and/or protein changes on the functional divergence between euAP3 and paleoAP3 genes, these two aspects should be studied both in combination and in parallel. Our future research will include complementation experiments in a tm6 def double mutant background using either the Petunia DEF or TM6 promoter fused to either wild-type DEF and TM6 cDNA sequences or modified versions of these proteins, including a frameshifted version of the TM6 coding sequence, as such mimicking the mutation that was the basis of the euAP3 lineage.
METHODS
Isolation of Petunia tm6∷dTph1 Knockout Alleles
The available W138 insertion libraries were screened as described previously (Vandenbussche et al., 2003b) with either a Petunia hybrida TM6 forward (5′-TGATTGATTTGTACCAGAGGACACT-3′) or reverse (5′-GCAAGACGTAGATCACGAGAACCA-3′) primer located at the start of exon 2 and at the C terminus, respectively, in combination with the inverted repeat primer (5′-GAATTCGCTCCGCCCCTG-3′) complementary to the terminal inverted repeats of the dTph1 transposon. These libraries consist of 20 to 25 progeny plants each, obtained upon selfing of individual W138 plants (Koes et al., 1995).
Plant Material and Genotyping
Genotyping of the tm6-1 allele was performed by PCR using a gene-specific primer pair flanking the insertion site (tm6-1-fw, 5′-GACTCAAACAGATACCTGCAGGAAG-3′; tm6-1-rv, 5′-ACCAAATCATGTACGAGGCTTCCAT-3′), and products were sized by PAGE. Genotyping for the phglo1-2 and phglo2-3 insertion alleles was done as described previously (Vandenbussche et al., 2004). Segregation analysis of the ethyl methanesulfonate–induced Petunia def-1 (gp) allele (de Vlaming et al., 1984) was initially done on a phenotypic basis but later was performed as follows. The full coding sequence of def-1 was amplified, sequenced, and compared with the wild-type DEF-coding sequence, revealing a single point mutation (A to T) in the MADS domain of the def-1 allele, resulting in the conversion of an Arg codon (AGA) to a stop codon (TGA). Translation of this sequence gives a premature termination of the DEF protein sequence after 24 amino acids. Genotyping was done by hot-start PCR using two discriminating forward primers terminating at the 3′ end with either a T or an A and displaying a 4-bp length difference mibus454 (def-1-fw, 5′-AAACAGGCAAGTGACATATTCTAAGAGAT-3′) and mibus455 (DEF-fw, 5′-AGGCAAGTGACATATTCTAAGAGAA-3′) in combination with a nondiscriminating reverse DEF primer, mibus456 (DEF-rv, 5′-TCTTGGATACGTACGTGATAGAT-3′). Resulting products were sized by PAGE. Genotypes for bl mutant alleles were scored on a phenotypic basis and, in specific cases, confirmed by backcrossing with bl. All expected double and triple mutants were obtained in the F2 populations in agreement with the expected Mendelian ratios, with the exception of the cross between tm6-1 and the double mutant def-1 bl, as described in Results.
Construction of Transgenic Petunia TM6 Overexpression and RNAi Lines in Wild-Type, def-1, and def-1 bl Backgrounds
The full-length Petunia TM6 coding sequence was amplified by PCR from a W138 stamen-derived cDNA template with TM6-ORF-fw (5′-AAAAAGCAGGCTTCGAAGGAGATAAAAAAATGGGTCGTGGTAAAATTGAG-3′) and TM6-ORF-rv (5′-AGAAAGCTGGGTTATCTTCAAGCAAGACGTAGATCAC-3′) primers containing part of the Gateway attB1 and attB2 sites. A part of the TM6 nonconserved 3′ untranslated region was chosen as a template for RNAi and was amplified by PCR using TM6-RNAi-fw (5′-AAAAAGCAGGCTTGAAGATAACAATTTGGCAATG-3′) and TM6-RNAi-rv (5′-AGAAAGCTGGGTCATCTCTTTGGTATCATTCTGATG-3′) primers containing part of the Gateway attB1 and attB2 sites. After a second PCR with universal attB adapter primers to add the full attB adapters, both PCR products were cloned in pDONR221 (Invitrogen). The full-length TM6 coding sequence was then introduced in the pK2GW7 overexpression vector, and the TM6 3′ untranslated region fragment was introduced in the binary RNAi vector pK7GWIWG2(1) (Karimi et al., 2002). The resulting destination vectors were used subsequently to transform Agrobacterium tumefaciens EHA105 cells via freeze–thaw transformation (Chen et al., 1994). Leaf discs from Petunia F1 progeny plants resulting from a cross between def-1 bl or def-1 mutants and the easily transformable Mitchell variety (W115) were transformed as described (Horsch et al., 1985). T1 kanamycin-resistant plants, which were also positive in a kanamycin gene PCR test, were self-fertilized. The resulting T2 progeny were genotyped for def-1 and phenotypically scored for the bl mutation.
Quantitative Real-Time RT-PCR Analysis
The different flower organs (sepals, petals, stamens, and pistils) were sampled separately from 1-cm flower buds. Every organ type was sampled in duplicate. Total RNA was extracted from these tissues using Trizol reagent (Life Technologies) according to the instructions of the manufacturer. RNA samples were measured and equalized, and their integrity was analyzed by gel electrophoresis. First-strand cDNA synthesis was done by combining 1 μg of total RNA diluted up to 10 μL with diethyl pyrocarbonate–water and denatured for 5 min at 65°C with a mix containing 1 μL of 50 μM oligo(dT)25 primers, 0.85 μL of diethyl pyrocarbonate–treated water, 4 μL of 5× first-strand buffer, 2 μL of 0.1 M DTT, 2 μL of 5 mM deoxynucleotide triphosphate, and 0.15 μL of Superscript III (200 units/μL; Gibco BRL) in a total volume of 20 μL. After incubation for 4 h at 42°C and inactivation of the polymerase for 10 min at 70°C, the mixture was diluted 20 times by adding 380 μL of diethyl pyrocarbonate–treated water. Quantitative real-time RT-PCR was performed on 5 μL of the diluted cDNA (50 ng of total RNA) with the addition of 10 pmol for both primers, in a total volume of 25 μL, with the iQ SYBR Green supermix (Bio-Rad) in a MyiQ single-color real-time PCR machine (Bio-Rad). Expression levels were normalized to GAPDH expression levels and were determined to be identical upon normalization with ACTIN. Specific primer pairs were designed with the help of Beacon Designer 4 software (Premier Biosoft International) in such a way that they would amplify only cDNA and no genomic DNA under the PCR conditions used. To avoid amplification of the transgene in the TM6-RNAi plants, the TM6 primer pair was chosen outside of the 3′ untranslated region used for RNAi. The primers used to quantify gene expression levels were Pet-GAPDH-fw (5′-ACCACTGTCCACTCACTTACTG-3′) and Pet-GAPDH-rv (5′-TGCTGCTAGGAATGATGTTGAATG-3′); and Pet-ACTIN-fw (5′-GTTGGACTCTGGTGATGGTGTG-3′) and Pet-ACTIN-rv (5′-CCGTTCAGCAGTGGTGGTG-3′). The primers used for the quantitative PCR of the B- and C-function genes were TM6-fw (5′-GCAGGAAGAGGGTGAGGAAC-3′) and TM6-rv (5′-GCCATAGCAGAGTTGAAATGTCC-3′); DEF-fw (5′-AGAAGAAGGTCAGGAATGTGGAAG-3′) and DEF-rv (5′-GTTGAAGGCGTAAGGCTAATATGC-3′); GLO1-fw (5′-AATGAGGTTCTGAGGATGATGAGG-3′) and GLO1-rv (5′-CTTCGCCAATTTCTCCCATATTCC-3′); GLO2-fw (5′-TGGAGGAGGAACACAAGCAAC-3′) and GLO2-rv (5′-CGAAGGGCAAATGGCATCTG-3′); and PMADS3-fw (5′-TTCTTGGTGAATCTCTTGCTG-3′) and PMADS3-rv (5′-GGTAATGGTTGTTGGTCTGC-3′).
The PCR program consisted of a first step of denaturation and Taq activation at 95°C for 3 min, followed by 40 cycles of 95°C for 15 s and 57°C for 45 s. To determine the specificity of the PCR, the amplified products were subjected to melt curve analysis using the machine's standard method and analyzed by gel electrophoresis. For all PCRs, we used a negative control that did not contain cDNA template. Each sample had a biological replicate (shown as pairs of gray and black bars). The relative expression was calculated as described by Vandesompele et al. (2002) using GAPDH expression for normalization. For each primer combination separately, the highest expression value encountered in the series of different tissues tested was set equal to 100, and lower values were plotted relative to the highest value on a linear y axis scale.
Cloning of euAP3 and TM6 Homologs from a Range of Other Solanaceous Species
Seven species flowering at the time of analysis and all belonging to different genera were selected from the extensive Nijmegen Solanaceae collection (http://www.bgard.science.ru.nl/) for further analysis. Species accession numbers are shown in parentheses after the species names: Brunfelsia uniflora (954750024); Scopolia carniolica (984750174); Cestrum fasciculatum (944750210); Juanulloa aurantiaca (A14750191); Mandragora autumnalis (934750027); Solandra maxima (944750283); and Solanum pseudolulo (A44750258).
Samples consisting of flower buds from various developmental stages were collected and subjected to RNA extraction and cDNA synthesis as described for Petunia above. euAP3- and TM6-like sequences were amplified from these templates using either a Sol-euAP3-fw degenerate primer (5′-ATGGCTCGTGGDAARATCCAGATCAAG-3′) or a Sol-TM6-fw degenerate primer (5′-ATGGGYCGTGGDAARATTGARATCAAG-3′) in combination with an oligo(dT)29-(G/A/C)-3′ reverse primer in a hot-start PCR using Platinum Taq (BD Sciences). For each PCR, the following components were mixed together: 5 μL of cDNA template, 2.5 μL of 10× PCR buffer (with 15 mM MgCl2), 1 μL of Sol-DEF-fw or Sol-TM6-fw primer (10 pmol/μL), 0.5 μL of deoxynucleotide triphosphates (10 mM), 1 μL of oligo(dT)29-(G/A/C) (10 pmol/μL), 0.08 μL of Platinum Taq DNA polymerase (Gibco BRL), and deionized water to 25 μL. The mixture was amplified in a Perkin-Elmer 9600 thermocycler with the following PCR profile: 10 cycles (94°C for 15 s, 60°C for 30 s, −1°C/cycle, and 72°C for 60 s) followed by 35 cycles (94°C for 15 s, 50°C for 30 s, and 72°C for 60 s). The samples were checked on a 1.5% agarose gel and subsequently cloned and sequenced.
Phylogenetic Analysis
For the phylogenetic analysis of the newly isolated euAP3 and TM6 homologs from the various solanaceous species, we aligned 34 protein sequences using ClustalW (Thompson et al., 1994). This alignment was checked manually and edited using BioEdit (Hall, 1999) (see supplemental data online). A neighbor-joining tree was computed using Treecon (Van de Peer and De Wachter, 1994). To assess the support for the inferred relationships, 1000 bootstrap samples were generated.
Electron Microscopy
Samples for cryoscanning electron microscopy were first frozen in slush, prepared in an Oxford Alto 2500 cryosystem (Catan), and then analyzed in a JEOL JSM-6330F field emission electron scanning microscope.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Ph DEF, DQ539416; Ph TM6, DQ539417; Sl AP3, DQ539418; Sl TM6, DQ539419; Mo DEF, AM162207; Am DEF, X62810; At AP3, U30729; Cp AP3, AF248970; Bu DEF, DQ539401; Sp DEF, DQ539402; Ma DEF, DQ539403; Ja DEF, DQ539404; Sm DEF, DQ539405; Sc DEF1, DQ539406; Sc DEF2, DQ539407; Ce DEF, DQ539408; Ja TM6, DQ539409; Ma TM6, DQ539410; Bu TM6, DQ539411; Sc TM6, DQ539412; Ce TM6, DQ539413; Sm TM6, DQ539414; Sp TM6, DQ539415.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure 1. Protein Sequence Alignment of Newly Isolated euAP3 and TM6 Homologs from Various Solanaceous Species, Including a Selection of B-Class Lineage MADS Box Genes from Other Informative Taxa.
Supplementary Material
Acknowledgments
We thank Walter Hendrickx, Yvette Evers, Harry Van Zuijlen, and Daniëlle Geerlings for assistance with the plant work, Geert-Jan Janssen for assistance with the electron microscopy pictures, and Richard Feron for general laboratory support. M.V. also thanks Suzanne Rodriques Bento for continuous support and stimulating discussions. We thank Vivian Irish for sharing unpublished results and the three anonymous reviewers for their comments and suggestions. The work of A.S.R. is funded by Netherlands Organization for Scientific Research Grant 814.02.009.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Tom Gerats (t.gerats@science.ru.nl).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.106.042937.
References
- Ambrose, B.A., Lerner, D.R., Ciceri, P., Padilla, C.M., Yanofsky, M.F., and Schmidt, R.J. (2000). Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell 5 569–579. [DOI] [PubMed] [Google Scholar]
- Angenent, G.C., Busscher, M., Franken, J., Mol, J.N.M., and van Tunen, A.J. (1992). Differential expression of two MADS box genes in wild-type and mutant petunia flowers. Plant Cell 4 983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L., Sakai, H., Jack, T., Weigel, D., Mayer, U., and Meyerowitz, E.M. (1992). SUPERMAN, a regulator of floral homeotic genes in Arabidopsis. Development 114 599–615. [DOI] [PubMed] [Google Scholar]
- Bradley, D., Carpenter, R., Sommer, H., Hartley, N., and Coen, E. (1993). Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell 72 85–95. [DOI] [PubMed] [Google Scholar]
- Causier, B., Castillo, R., Zhou, J., Ingram, R., Xue, Y., Schwarz-Sommer, Z., and Davies, B. (2005). Evolution in action: Following function in duplicated floral homeotic genes. Curr. Biol. 15 1508–1512. [DOI] [PubMed] [Google Scholar]
- Chen, H., Nelson, R.S., and Sherwood, J.L. (1994). Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 16 664–668. [PubMed] [Google Scholar]
- Coen, E.S., and Meyerowitz, E.M. (1991). The war of the whorls: Genetic interactions controlling flower development. Nature 353 31–37. [DOI] [PubMed] [Google Scholar]
- de Vlaming, P., Cornu, A., Farcy, E., Gerats, A.G., Wiering, H., and Wijsman, H.J.W. (1984). Petunia hybrida: A short description of the action of 91 genes, their origin and their map locations. Plant Mol. Biol. Rep. 2 21–42. [Google Scholar]
- Garcia-Maroto, F., Salamini, F., and Rohde, W. (1993). Molecular cloning and expression patterns of three alleles of the Deficiens-homologous gene St-Deficiens from Solanum tuberosum. Plant J. 4 771–780. [DOI] [PubMed] [Google Scholar]
- Gerats, A.G., Huits, H., Vrijlandt, E., Marana, C., Souer, E., and Beld, M. (1990). Molecular characterization of a nonautonomous transposable element (dTph1) of petunia. Plant Cell 2 1121–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Mena, C., de Folter, S., Costa, M.M., Angenent, G.C., and Sablowski, R. (2005). Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development 132 429–438. [DOI] [PubMed] [Google Scholar]
- Goto, K., and Meyerowitz, E. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8 1548–1560. [DOI] [PubMed] [Google Scholar]
- Guyot, R., Cheng, X., Su, Y., Cheng, Z., Schlagenhauf, E., Keller, B., and Ling, H.Q. (2005). Complex organization and evolution of the tomato pericentromeric region at the FER gene locus. Plant Physiol. 138 1205–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, T.A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41 95–98. [Google Scholar]
- Halfter, U., Ali, N., Stockhaus, J., Ren, L., and Chua, N. (1994). Ectopic expression of a single homeotic gene, the Petunia gene green petal, is sufficient to convert sepals to petaloid organs. EMBO J. 13 1443–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, T., Day, C., Zondlo, S., Thackeray, A., and Irish, V. (1998). Discrete spatial and temporal cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene APETALA3. Development 125 1711–1721. [DOI] [PubMed] [Google Scholar]
- Horsch, R.B., Fry, J.E., Hoffmann, N.L., Eichholtz, D., Rogers, S.G., and Fraley, R.T. (1985). A simple and general method for transferring genes into plants. Science 227 1229–1231. [DOI] [PubMed] [Google Scholar]
- Huijser, P., Klein, J., Lonnig, W.E., Meijer, H., Saedler, H., and Sommer, H. (1992). Bracteomania, an inflorescence anomaly, is caused by the loss of function of the MADS-box gene squamosa in Antirrhinum majus. EMBO J. 11 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink, R.G.H., Ferrario, S., Busscher-Lange, J., Kooiker, M., Busscher, M., and Angenent, G.C. (2003). Analysis of the petunia MADS-box transcription factor family. Mol. Genet. Genomics 268 598–606. [DOI] [PubMed] [Google Scholar]
- Jack, T., Brockman, L.L., and Meyerowitz, E.M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68 683–697. [DOI] [PubMed] [Google Scholar]
- Jack, T., Fox, G.L., and Meyerowitz, E.M. (1994). Arabidopsis homeotic gene APETALA3 ectopic expression: Transcriptional and posttranscriptional regulation determine floral organ identity. Cell 76 703–716. [DOI] [PubMed] [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY(TM) vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Kater, M.M., Colombo, L., Franken, J., Busscher, M., Masiero, S., Van Lookeren Campagne, M.M., and Angenent, G.C. (1998). Multiple AGAMOUS homologs from cucumber and petunia differ in their ability to induce reproductive organ fate. Plant Cell 10 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S., Yoo, M.-J., Albert, V.A., Farris, J.S., Soltis, P.S., and Soltis, D.E. (2004). Phylogeny and diversification of B-function MADS-box genes in angiosperms: Evolutionary and functional implications of a 260-million-year-old duplication. Am. J. Bot. 91 2102–2118. [DOI] [PubMed] [Google Scholar]
- Koch, M.A., Weisshaar, B., Kroymann, J., Haubold, B., and Mitchell-Olds, T. (2001). Comparative genomics and regulatory evolution: Conservation and function of the Chs and Apetala3 promoters. Mol. Biol. Evol. 18 1882–1891. [DOI] [PubMed] [Google Scholar]
- Koes, R., et al. (1995). Targeted gene inactivation in petunia by PCR-based selection of transposon insertion mutants. Proc. Natl. Acad. Sci. USA 92 8149–8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, E.M., Dorit, R.L., and Irish, V.F. (1998). Molecular evolution of genes controlling petal and stamen development: Duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics 149 765–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, E.M., and Irish, V.F. (2000). Evolution of petal and stamen developmental programs: Evidence from comparative studies of the lower eudicots and basal angiosperms. Int. J. Plant Sci. 161 (suppl. 6), S29–S40. [Google Scholar]
- Kramer, E.M., Su, H.J., Wu, C.C., and Hu, J.M. (2006). A simplified explanation for the frameshift mutation that created a novel C-terminal motif in the Apetala3 gene lineage. BMC Evol. Biol. 24 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek, B.A., and Meyerowitz, E.M. (1996). The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 122 11–22. [DOI] [PubMed] [Google Scholar]
- Lamb, R.S., and Irish, V.F. (2003). Functional divergence within the APETALA3/PISTILLATA floral homeotic gene lineages. Proc. Natl. Acad. Sci. USA 100 6558–6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel, M.A., Gustafson-Brown, C., Savidge, B., and Yanofsky, M.F. (1992). Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360 273–277. [DOI] [PubMed] [Google Scholar]
- McGonigle, B., Bouhidel, K., and Irish, V.F. (1996). Nuclear localization of the Arabidopsis APETALA3 and PISTILLATA homeotic gene products depends on their simultaneous expression. Genes Dev. 10, 1812–1821. Erratum. Genes Dev. 10 2235. [DOI] [PubMed] [Google Scholar]
- Mouradov, A., Hamdorf, B.A., Teasdale, R.D., Kim, J.T., Winter, K.U., and Theissen, G. (1999). A DEF/GLO-like MADS-box gene from a gymnosperm: Pinus radiata contains an ortholog of angiosperm B class floral homeotic genes. Dev. Genet. 25 245–252. [DOI] [PubMed] [Google Scholar]
- Nagasawa, N., Miyoshi, M., Sano, Y., Satoh, H., Hirano, H., Sakai, H., and Nagato, Y. (2003). SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130 705–718. [DOI] [PubMed] [Google Scholar]
- Pnueli, L., Abu-Abeid, M., Zamir, D., Nacken, W., Schwarz-Sommer, Z., and Lifschitz, E. (1991). The MADS box gene family in tomato: Temporal expression during floral development, conserved secondary structures and homology with homeotic genes from Antirrhinum and Arabidopsis. Plant J. 1 255–266. [PubMed] [Google Scholar]
- Riechmann, J., Wang, M., and Meyerowitz, E. (1996). DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 24 3134–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, H., Medrano, L.J., and Meyerowitz, E.M. (1995). Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 378 199–203. [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer, Z., Hue, I., Huijser, P., Flor, P., Hansen, R., Tetens, F., Lonnig, W., Saedler, H., and Sommer, H. (1992). Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: Evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J. 11 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer, H., Beltran, J., Huijser, P., Pape, H., Lonnig, W., Saedler, H., and Schwarz-Sommer, Z. (1990). Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: The protein shows homology to transcription factors. EMBO J. 9 605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundström, J., Carlsbecker, A., Svensson, M.E., Johanson, U., Theissen, G., and Engström, P. (1999). MADS-box genes active in developing pollen cones of Norway spruce (Picea abies) are homologous to the B-class floral homeotic genes in angiosperms. Dev. Genet. 25 253–266. [DOI] [PubMed] [Google Scholar]
- Sundström, J., and Engström, P. (2002). Conifer reproductive development involves B-type MADS-box genes with distinct and different activities in male organ primordia. Plant J. 31 161–169. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly, J., Allen, D., and Jack, T. (1998). The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development 125 1647–1657. [DOI] [PubMed] [Google Scholar]
- Trobner, W., Ramirez, L., Motte, P., Hue, I., Huijser, P., Lonnig, W., Saedler, H., Sommer, H., and Schwarz-Sommer, Z. (1992). GLOBOSA: A homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J. 11 4693–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimoto, S., Mayama, T., van der Krol, A., and Ohtsubo, E. (2000). The whorl-specific action of a petunia class B floral homeotic gene. Genes Cells 5 89–99. [DOI] [PubMed] [Google Scholar]
- Tsuchimoto, S., van der Krol, A.R., and Chua, N.H. (1993). Ectopic expression of pMADS3 in transgenic petunia phenocopies the petunia blind mutant. Plant Cell 5 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallade, J., Maizonnier, D., and Cornu, A. (1987). La morphogenese florale chez le petunia. I. Analyse d'un mutant corolle staminee. Can. J. Bot. 65 761–764. [Google Scholar]
- Vandenbussche, M., Theissen, G., Van de Peer, Y., and Gerats, T. (2003. a). Structural diversification and neo-functionalization during floral MADS-box gene evolution by C-terminal frameshift mutations. Nucleic Acids Res. 31 4401–4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche, M., Zethof, J., Royaert, S., Weterings, K., and Gerats, T. (2004). The duplicated B-class heterodimer model: Whorl-specific effects and complex genetic interactions in Petunia hybrida flower development. Plant Cell 16 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche, M., Zethof, J., Souer, E., Koes, R., Tornielli, G.B., Pezzotti, M., Ferrario, S., Angenent, G.C., and Gerats, T. (2003. b). Toward the analysis of the petunia MADS box gene family by reverse and forward transposon insertion mutagenesis approaches: B, C, and D floral organ identity functions require SEPALLATA-like MADS box genes in petunia. Plant Cell 15 2680–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer, Y., and De Wachter, R. (1994). TREECON for Windows: A software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10 569–570. [DOI] [PubMed] [Google Scholar]
- van der Krol, A., Brunelle, A., Tsuchimoto, S., and Chua, N. (1993). Functional analysis of petunia floral homeotic MADS box gene pMADS1. Genes Dev. 7 1214–1228. [DOI] [PubMed] [Google Scholar]
- Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., and Speleman, F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3 RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple, C.J., Ciceri, P., Padilla, C.M., Ambrose, B.A., Bandong, S.L., and Schmidt, R.J. (2004). Conservation of B-class floral homeotic gene function between maize and Arabidopsis. Development 131 6083–6091. [DOI] [PubMed] [Google Scholar]
- Winter, K.U., Saedler, H., and Theissen, G. (2002). On the origin of class B floral homeotic genes: Functional substitution and dominant inhibition in Arabidopsis by expression of an orthologue from the gymnosperm Gnetum. Plant J. 31 457–475. [DOI] [PubMed] [Google Scholar]
- Yang, Y., Fanning, L., and Jack, T. (2003. a). The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins, APETALA3 and PISTILLATA. Plant J. 33 47–59. [DOI] [PubMed] [Google Scholar]
- Yang, Y., Xiang, H., and Jack, T. (2003. b). pistillata-5, an Arabidopsis B class mutant with strong defects in petal but not in stamen development. Plant J. 33 177–188. [DOI] [PubMed] [Google Scholar]
- Yanofsky, M.F., Ma, H., Bowman, J.L., Drews, G.N., Feldmann, K.A., and Meyerowitz, E.M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346 35–39. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.