Abstract
Background
The majority of experimentally verified molecular interaction and biological pathway data are present in the unstructured text of biomedical journal articles where they are inaccessible to computational methods. The Biomolecular interaction network database (BIND) seeks to capture these data in a machine-readable format. We hypothesized that the formidable task-size of backfilling the database could be reduced by using Support Vector Machine technology to first locate interaction information in the literature. We present an information extraction system that was designed to locate protein-protein interaction data in the literature and present these data to curators and the public for review and entry into BIND.
Results
Cross-validation estimated the support vector machine's test-set precision, accuracy and recall for classifying abstracts describing interaction information was 92%, 90% and 92% respectively. We estimated that the system would be able to recall up to 60% of all non-high throughput interactions present in another yeast-protein interaction database. Finally, this system was applied to a real-world curation problem and its use was found to reduce the task duration by 70% thus saving 176 days.
Conclusions
Machine learning methods are useful as tools to direct interaction and pathway database back-filling; however, this potential can only be realized if these techniques are coupled with human review and entry into a factual database such as BIND. The PreBIND system described here is available to the public at http://bind.ca. Current capabilities allow searching for human, mouse and yeast protein-interaction information.
Background
Currently, the vast majority of biomolecular interaction and pathway data are stored in printed journal articles where it is difficult to manage and to compute upon. The goal of the BIND database (Biomolecular Interaction Network Database) is to curate and archive these data from the literature using a standard data representation so that it may be effectively used for knowledge discovery http://bind.ca [1,2].
PreBIND and Textomy are two components of a literature-mining system designed to find protein-protein interaction information and present this to curators or public users for review and submission to the BIND database. Backfilling interaction data from the biomedical literature is an ongoing task that will not be completed for some time. In one sense, PreBIND represents a stopgap measure; a database where researchers can find interaction data (albeit imperfect and un-reviewed) for their molecule of interest until it has been properly indexed in BIND. Hence, the name, PreBIND. At the same time, researchers may use this resource to facilitate proper indexing of their molecules of interest by submitting data they find using PreBIND to curators at BIND. In this sense, PreBIND is complementary to BIND.
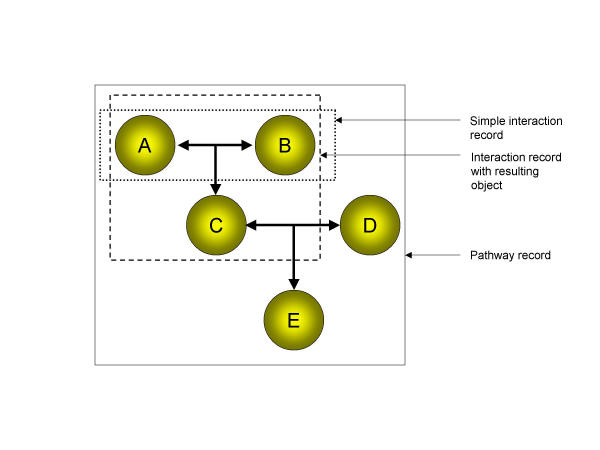
The basic unit of the BIND database is the "Interaction" record (see Figure 1). The minimum information required to define an Interaction record is a description of a molecule 'A' and a molecule 'B' and a publication supporting the interaction. Interacting molecules may include proteins, RNA, DNA, small molecules and complexes among others. Other information may be optionally added to the record such as the previous interactions that gave rise to the interactors, the set of resulting molecules/complexes of the interaction or the precise residues (for biopolymers) that mediate the interaction. A series of Interaction records may be used to describe a biomolecular pathway since the set of molecules that are created as a result of an interaction are contained within the Interaction record itself. These new molecules may have biomolecular functions not possessed by either of the two original interactors in that they may interact with a different set of molecules to yet again, create new molecules with new functionalities. In this way, one can imagine a representation of a biological pathway that consists of a set of bi-molecular interactions where the product of one interaction becomes the interactor of a subsequent bi-molecular interaction. This abstraction is the basis of the data structure for BIND (see Figure 1). For the purposes of this paper and the initial population of the BIND database, we have chosen to focus on extracting information from the literature that is sufficient for defining a simple protein-protein interaction record in BIND. Subsequent text-mining modules can be added to PreBIND in future that will help fill out other aspects of the BIND data model.
Figure 1.
Representation of a pathway using the BIND data-model. Each letter represents a molecular object such as DNA, RNA, protein, complex or small molecule. A simple interaction record (dotted box) describes an interface between two molecular objects. This paper focuses on information extraction for this type of record. Interaction records may describe a new molecule(s) created as a result of the interaction (dashed box). Molecular results of one interaction record can become the interactors in subsequent interaction records. In this way, multiple interaction records can be strung together to describe a biological pathway (solid box).
A number of other systems that also mine protein-protein interaction information and other biological relationships from the literature have been described recently: [3-14]. PreBIND and Textomy differ from these methods by a combination of five factors.
1) Support Vector Machine (SVM) technology is used to identify articles about biomolecular interactions and confirm sentences that mention specific protein-protein interactions. This method can be used to quickly train a machine learning algorithm to recognize interaction-like articles and bypasses the laborious process of building a domain-specific semantic grammar required for Natural Language Processing (NLP). Marcotte et al. [13] recently used a related method (Bayesian) to classify articles that described protein-protein interaction information.
2) Protein names and their gene-symbols are derived from a non-redundant sequence database (RefSeq) [15], and from the Saccharomyces Genome Database (SGD) [16]. Only these names are used for literature searching. This allows an explicit mapping of names to sequences and aids in the preparation of BIND interaction records for submission. Other names are detected by the Textomy module, but only for the purposes of marking-up text for review by users. This approach of using names to create a co-occurrence network of identifiable biomolecules in the literature is similar to the approach used by PubGene [11].
3) This information extraction (IE) system is coupled to a human-reviewed data-entry queue for a publicly available biomolecular interaction database (BIND). No data extraction technique to date has perfect accuracy and/or precision and even a small error rate is intolerable in a curated database that is envisioned to contain millions of records. Our guiding principle in constructing this tool has not been to completely automate information extraction but rather to make the curation task easier for expert biologist BIND users and indexers. There is no intention to apply discovery algorithms to this raw data set; rather only the curated BIND data set will be analyzed where uncertainty due to IE error will have been removed. All occurrences of protein names in abstracts are stored (regardless of the likelihood that the abstract describes an interaction). Also all co-occurrences of names within an abstract are stored as potential relationships. Each relationship is scored as to its likelihood of being a direct protein-protein interaction. This leaves viewing of search results to the discretion of the curator (even for those papers with low scores). In addition, this allows alternative scoring methods (for alternative types of papers or relationships between proteins) to be applied to the set of continuously updated search results.
4) PreBIND and Textomy allow for user feedback into the SVM training set that can constantly improve the performance of the system's ability to detect abstracts that describe biomolecular interactions.
5) PreBIND is available for use http://bind.ca by those interested in finding interaction information in the literature about their protein of interest. Users are encouraged to submit their findings to the BIND database. Once a record is submitted, it will be validated by BIND curators and by at least one other expert before it is made available in any public data release.
Results and Discussion
Part 1: Description of the Information Extraction system
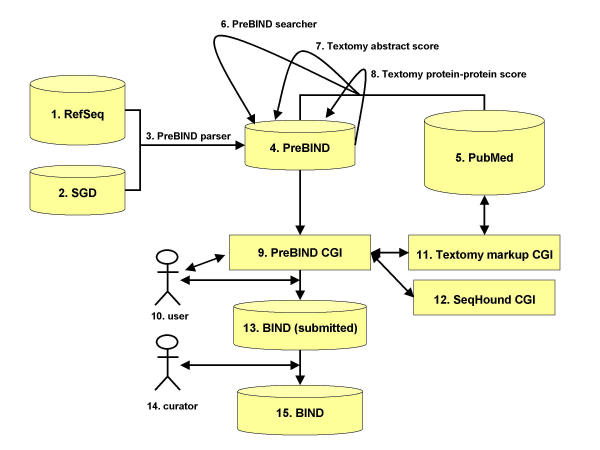
The PreBIND/Textomy information-extraction system is summarized in Figure 2. The PreBIND parser (Fig. 2, item 3) collects synonyms for proteins and their encoding loci for a non-redundant set of proteins present in the NCBI RefSeq sequence database (Fig. 2, item 1) [15]. These synonyms are stored with their corresponding RefSeq GenInfo (GI) identifier [17] in the PreBIND database (Fig. 2, item 4). Additional synonyms are collected by the parser for each unique GI from the Saccharomyces cerevisiae genome database (SGD) [16] (Fig. 2, item 2) using database cross-references found in RefSeq records. The PreBIND searcher program (Fig. 2, item 6) searches the PubMed literature database [18] (Fig. 2, item 5) for each of these synonyms in either the abstract or title fields while limited to the MESH listed taxon relevant to the protein. The PubMed Identifiers (PMIDs) for abstracts returned by these searches are stored in the PreBIND database (Fig. 2, item 4). Textomy (Fig. 2, item 7) http://www.litminer.ca/ retrieves these abstracts from PubMed and assigns a score that describes the relative likelihood that the abstract contains molecular interaction information. Textomy, or 'text anatomy', is text processing software that uses an SVM [19-21] to capture the statistical pattern of word use in papers that have previously been presented to the machine as 'papers of interest', in this case, a training set of abstracts that discuss biomolecular interactions. These SVM scores are stored in the PreBIND database (Fig. 2, item 4). Textomy is employed in a second round to score the likelihood that an interaction is described for any given pair-wise combination of proteins mentioned in an abstract (Fig. 2, item 8). A pair-wise combination of names is awarded a score of 1 for every time the two names (and only those two names) appear in a sentence that is deemed by an SVM to describe a biomolecular interaction (the same SVM that was used to classify abstracts is used here to classify the sentence). In addition, if the names correspond to yeast proteins, they both must conform to yeast-protein nomenclature rules. This avoids awarding a protein interaction score to a set of names that are described in the context of a genetic interaction. These potential interaction scores for each pair-wise combination of names appearing in an abstract are also stored in the PreBIND database (Fig. 2, item 4). The PreBIND CGI (Fig. 2, item 9) allows users (Fig 2, item 10) to search for interaction information in PreBIND that may be relevant to their protein of interest via the web.
Figure 2.
The PreBIND information extraction system. Details are provided in the text of the Results and Discussion section.
In addition, users may view PubMed abstracts that have been marked-up by Textomy (Fig. 2, item 11). The two highest scoring sentences are highlighted in the Textomy interface thus allowing a human judge to rapidly verify the SVM's decisions by only reading two sentences instead of the entire abstract. We have shown in a separate study that this technique was effective in choosing the best "interaction-sentences" in 66% of the abstracts tested [22]. Textomy also offers highlighting of protein names, interaction phrases, and organism names. This is done with a mix of three techniques: dictionary lookup, applying rules on the morphology of term names, and rules about the context of terms. The dictionary of protein names was derived from the 'short description of entries' field in SwissProt [23]. The dictionary of organism names was derived from the MeSH vocabulary with some augmentations to handle plural or adjective forms (for e.g., rats, bovine) and acronyms ("C. elegans"). Rules on term morphology were applied mostly to protein names, but also to interaction phrases. These were programmed using Perl regular expressions. For example, the regular expression \bp\ w*? [0-9A-Z]\ w*?\ b will capture strings like p47 and p56lck. The regular expression \ b [A-Z] [a-zA-Z]+? [0-9]+?(p|\ proteins?)\ b will capture strings that start with a capital letter, contain more letters and at least one digit and end with 'p' or are followed with the word 'protein' (or 'proteins'). As such, strings like 'COR14 protein' or 'Gp23p' are detected. Contextual rules allowed protein name identification from a number of contexts, including enumerations and acronym declarations. The protein name highlighting function was similar in part to the one presented by Fukuda et al. [24]. These mark-up features allow users to quickly locate interaction information and interactor names.
Once the user has finished reviewing the abstract and has confirmed the potential interactions mentioned in it, they could submit them to the BIND database (Fig. 2, item 13 and Fig. 3). A BIND record may be created using the PreBIND CGI and submitted to curators at BIND via the Web. The SeqHound database is consulted to ensure that molecule type, taxon and GI identifier are up-to-date (Fig 2.12 and [25]). A subsequent second review by BIND curators (Fig. 2, item 14) is required before the record is released to the public BIND database (Fig. 2, item 15) at http://bind.ca.
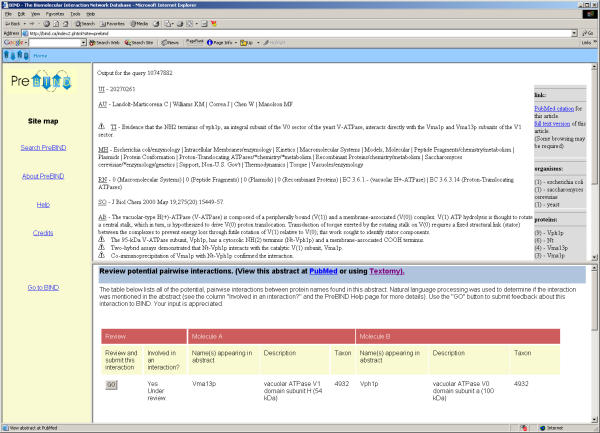
Figure 3.
The PreBIND interface. Users can freely view and submit feedback about all potential interactions present in PreBIND. Potential interactions that are judged by users to be legitimate may be submitted to the BIND database for review by curators. Information gathered in this way will be used to further train the support vector machine used in the initial search and help develop natural language analysis algorithms.
Part 2: Expected performance of the IE system
SVM training and evaluation
The heart of the PreBIND/Textomy IE system is an SVM that was trained to recognize abstracts describing biomolecular interactions. The SVM employed by the current PreBIND system available at http://bind.ca is described here. An SVM is a model that specifies a decision boundary. There are two parts to the training of an SVM for text. First, positive and negative training examples are transformed into multidimensional vectors. For example, each element in the vector may be a "1" or a "0" to represent the presence or absence of some word (corresponding to that vector element) in the abstract. Second is the discovery of a boundary that best separates positive from negative examples. This boundary is learned from the set of training samples. New text samples can be classified using this boundary; if an abstract, represented in a multidimensional space, one word per dimension, falls on one side of the boundary, it is judged as an interaction paper and as a non-interaction paper otherwise.
The training articles for the SVM were collected and judged by three different experts with slightly different sampling techniques. For the first collection, T1, the expert was asked to collect abstracts that described, a) protein-protein interactions, b) protein-protein interactions and cloning, c) DNA-protein interactions or d) just cloning. The categories, 'a' through 'c' were considered to represent abstracts about biomolecular interactions while category 'd' represented non-interaction abstracts. T1 contains 187 abstracts, does not contain any yeast papers and was biased for mammalian cell-signaling papers. The second collection, T2, was selected to approximate the manual task of adding interactions to BIND. The abstracts were chosen with a search engine and then marked as positive if the abstract described a biomolecular interaction and negative otherwise. T2 has 497 abstracts. No attempt was made to focus on yeast papers. The third collection, T3a, was selected by a third expert from a collection of yeast papers to be more representative of the papers in the yeast division of PreBIND. Each of these papers were returned by PreBIND Searcher (Fig. 2, item 6) and contained at least two yeast protein or gene names. T3a has 200 abstracts. The expert was asked to distinguish all abstracts that contain a biomolecular interaction from those that do not. The fourth collection, T3b, was selected based on an SVM trained on collections T1, T2, and T3a. Only a random sample of those abstracts that were close to the decision border were judged by the third expert. T3b contains 110 abstracts. Again the expert was asked to distinguish all abstracts that contain a biomolecular interaction from those that do not. Finally, set T3c was made up of 100 abstracts selected at random from the PreBIND search results for yeast names. These were also classified on the basis of whether or not they contained biomolecular interaction data.
All of the collections, T1, T2, and T3a-c were combined into a training set for the SVM. In total, 693 training examples described a biomolecular interaction and 401 did not. The dimensions extracted from the text were words and two-word (adjacent) phrases. Term features were filtered (thrown out) if they occurred in a standard list of 300 stop words or were shorter than two characters. The stop word list used was the set of 300 most frequent words from the SMART search engine English stop word list [26]. The longer version of this list is available from ftp://ftp.cs.cornell.edu/pub/smart/english.stop. Words were strings of alphabetic characters. They were truncated at ten characters and were converted to lower case. No other text pre-processing occurred. Digits, punctuation, and white spaces were treated as non-word characters. As a result, digits and punctuation contained within words were treated as word boundaries.
The value of each dimension for each abstract was binary, 1 if a word occurred and 0 otherwise. Of all words and phrases that met the above criteria, the (at most) 1500 with the highest positive information gain were retained. Information gain is a measure of the amount of information (measured in bits) that a word or phrase conveys about the class [27]. For example, if a person is told only that an abstract contains the word "interaction" they are better enabled to decide if the abstract describes a biomolecular interaction than if they are told only that the abstract contains the word "electrophoresis". In this case the presence or absence of the word "interaction" is said to hold more information than the presence or absence of the word "electrophoresis".
SVM training was performed using a radial-basis function kernel (RBF), meaning that the decision boundary used was more complex than a hyperplane. The RBF kernel allows enclosed boundaries such as ellipses and consistently outperformed linear, polynomial and sigmoid kernels for a collection of five other classifiers that were trained for different topics on PubMed abstracts (data not shown). The gamma parameter was set at 0.01, again, optimized after an extensive search across the space of possible values. The C parameter was set to 2. Expected SVM performance was calculated from a 10-fold cross-validation training (train on 90% and test on 10% repeated 10 times on a different 10% each time).
Accuracy was estimated at 90%. Accuracy is a percentile expression of the number of times that the SVM is correct in its classification (either interaction abstract or not). Estimated precision was 92%. Precision is the percentage of times that the SVM is correct in its classification of an abstract as describing an interaction. Recall was estimated as 92% and is the percentage of known interaction articles that the SVM would classify as being about an interaction. The cross-validation F-measure was calculated to be 92%. The F-measure is an expression of precision and recall that favors a balance between the two (see the Methods section for a more detailed description).
These expected performance values are likely to reflect the real performance when applied to abstracts not seen in the training examples to the extent that the training set reflects the overall population of abstracts in PubMed. The SVM training set included abstracts from various fields including transcription regulation and cell signaling in an attempt to generalize the concept of an "interaction paper". There is, however, no guarantee that the SVM will perform the same on interaction abstracts from other fields of study. The PreBIND/Textomy system does allow for user feedback and retraining. We expect that subsequent rounds of training may be useful in optimizing performance over the whole range of PubMed abstracts.
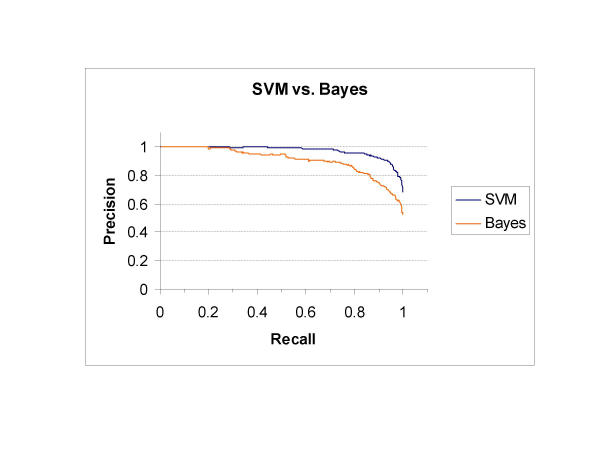
We compared the performance of this SVM classifier to a naïve-Bayes method. The same set of examples was used to train a naïve-Bayes classifier as described in the Methods section. Predicted precision was measured by 10-fold cross-validation over a range of recall settings for both methods (see Fig. 4). The SVM consistently out-performed the naïve-Bayes classifier once recall was greater than 12%. The naïve-Bayes precision and recall were balanced at an F-measure of 87% compared to 92% for the SVM. This suggests that the SVM is the better method to direct curator's attention to interaction articles in PubMed.
Figure 4.
Performance of SVM and naïve-Bayes classifiers. The performance of the SVM for identifying interaction abstracts was evaluated using 10-fold cross-validation on a set of 1094 abstracts. The performance on this task is measured in precision and recall. There is an implicit tradeoff between precision and recall that can be varied if the decision boundary is set to some value other than 0. In this evaluation, when the decision boundary for the SVM is set to 1, recall and precision are 0.57 and 0.99 respectively. When the decision boundary is set to -0.99, recall and precision are 0.997 and 0.71 respectively. Finally, if the decision boundary is set to zero then precision and recall are both 92%. In other words, when the decision boundary is set to zero and the SVM is applied to all abstracts in PubMed, it will miss approximately 8% of interaction documents (recall) and 8% of the identified interaction documents will not be interaction documents (precision). Under similar conditions, the naïve-Bayes classifier described here would only have a precision and recall of 87%.
Comparison to MIPS
In addition to classifying abstracts as either being "about an interaction" or not, the PreBIND interface presents information about the confidence in a prediction for an interaction between any two proteins mentioned in an abstract. A "potential interaction score" of one (1) is awarded to a particular pair-wise interaction for every time that the two protein names (and only those two names) appear in the same sentence and that sentence has been classified as being "about an interaction" by the same SVM used to classify the abstract. A potential interaction score of zero indicates that two protein names co-occur in the same abstract.
We assessed the ability of PreBIND/Textomy system to find interactions in yeast-related abstracts that had been independently entered into the MIPS yeast protein-protein interaction table http://mips.gsf.de/proj/yeast/tables/interaction [28]. We first removed interactions from the MIPS list that would not be detected by PreBIND regardless of its performance; these included homodimers, interactions with 'mRNA', interactions without a literature reference and interactions identified solely by a high-throughput study. We were left with 1378 non-redundant interactions from this list and compared these to interactions found by PreBIND.
Sixty percent (826) of the MIPS interactions were "found" by PreBIND solely from PubMed abstracts. In 563 cases (41%), this meant that the two names were simply in the same "interaction" abstract but they were not awarded an interaction score greater than zero. Only 263 (19%) of the MIPS interactions were found by PreBIND in "interaction" abstracts and were awarded an interaction score greater than zero. This suggests that curators might expect to index a maximum of 60% of known interactions if they were to read every abstract (and only the abstract) that was classified as containing interaction data.
Obviously, if PreBIND were able to search full-text, many more potential interactions would be found. PreBIND failed to find about 40% of the interactions in MIPS because there were no abstracts where both the protein names occurred. For example, we examined 20 abstracts used by MIPS to support interactions that were not found by PreBIND. Four of the twenty were missing both names in the abstract and sixteen were missing only one name. Seventeen of the twenty papers had two names present in the full-text articles that could have been detected by PreBIND. In at least eight cases, these two protein names appeared in the same paragraph and a biomolecular interaction was described for the two proteins in that paragraph. It is impossible to use these data to extrapolate the percentage of MIPS interactions that would be recovered if full-text searching were used. PreBIND can and does find additional references for interactions that are not listed by MIPS (see below); the full-text of these references could contain easily detectable descriptions of interactions using names from the PreBIND lexicon. Full-text searching would obviously recall more interactions than abstract searching alone, but, until full-text searching has been attempted, it not possible to tell how much better it will be.
The PreBIND system, in its current state was designed primarily to direct the attention of curators to abstracts containing interaction information for a given protein. However, PreBIND can also indicate the likelihood that any two protein names co-occurring in an interaction abstract really are interactors. If PreBIND moves to full-text searching, this ability will become increasingly important if curators are to efficiently sift through all the potential interactions that may be described in a paper. For this reason, we were interested in determining why PreBIND failed to recognize many co-occurrences as real interactions. A co-occurrence is indicated in PreBIND by a potential interaction score of zero (see above). Approximately 41 % of the MIPS interactions were found by PreBIND only as co-occurrences. To determine the reason for this, we began with a list of 20 interactions from MIPS that were found by PreBIND only as co-occurrences. We retrieved all of the supporting papers used by MIPS and compared them to those references where the interactions were found by PreBIND as co-occurrences. There were seven interactions for which PreBIND failed to find any of the same papers as MIPS to support the interaction. For five of these seven interactions, PreBIND found alternative papers that had supporting evidence for the interaction in the abstract. In fact, PreBIND found additional references not used by MIPS where the interaction was described in the abstract for 12 of the 20 interactions. These examples demonstrate that PreBIND is able to retrieve interaction abstracts used by MIPS or abstracts that contain equivalent information.
However, PreBIND never identified any of these 20 interactions as anything more than a co-occurrence in an interaction-like abstract. Why? For five of the MIPS interactions, the interaction was not described in the abstract used by MIPS nor in any of the additional abstracts identified by PreBIND. This would not be an issue if full-text searching were to be employed. For the remaining 15 interactions, PreBIND identified a total of 45 interaction papers where a co-occurrence of names appeared. PreBIND failed to identify these as interactions because both names were not in the same sentence (14 cases), because there were more than two names in a sentence (24 cases) or because the sentence where the co-occurrence happened was not identified as an interaction sentence (7 cases). If a more sophisticated method of detecting interactions was employed (for examples, see [9,10] and [12]), each of these co-occurrences may have been correctly identified as an interaction. Construction of such a natural language processing algorithm will be included in the next round of development for PreBIND.
In conclusion, the PreBIND/Textomy system is able to recall many of the interactions present in MIPS. This recall will be improved by full-text searching. Full-text searching has been attempted by few text-mining systems [10] due mainly to the difficulties involved in obtaining access to full-text articles. PreBIND is useful for focusing attention on those documents and document sections that are likely to contain interaction information. With full-text searching, the ability to differentiate interactions from co-occurrences will become more important. This ability will be facilitated by natural language processing algorithms that analyze text at the sentence level.
Part 3: Usefulness of the PreBIND IE system
Application to a high-throughput yeast study
Ultimately, IE systems are useful in their ability to reduce the time taken to locate and enter information into a factual database. The usefulness of the PreBIND/Textomy system was demonstrated by its application to the task of giving domain-knowledge context to the results of a high-throughput interaction study.
Recently, immunoprecipitation and mass spectrometry methods were used for a systematic identification of protein complexes in the yeast Saccharomyces cerevisiae [29]. In this study, 600 'bait' proteins were tagged with the Flag epitope, expressed in yeast and subsequently immunopurified from extract. Proteins that co-immunopurified with these baits (called hits) were resolved by SDS-polyacrylamide gel electrophoresis and identified by mass spectrometric analysis. The resulting collection of identified proteins included those that either interacted directly with the bait protein or with some other co-purified target. In total, 493 starting bait proteins co-immunoprecipitated 1,578 different target proteins. This corresponds to a potential 3,618 interactions if one assumes that every target interacts with its bait directly (spoke representation). If one assumes that all bait and target proteins identified in an immunoprecipitation interact with one another (matrix representation) the number of potential interactions increases significantly [30]. PreBIND was used to locate interaction information relevant to the starting bait proteins.
At the time of analysis, PreBIND contained 11,575 names that had been collected from the RefSeq database and the Saccharomyces Genome Database for 6,230 unique proteins. Each of these names was used to search the yeast literature in PubMed (42,070 papers). These searches returned 17,043 abstracts of which at least 9,631 mentioned two or more recognized protein names. The Textomy SVM classified 3981 of these 17,043 papers as containing interaction information. The training of this SVM was performed earlier and under slightly different conditions than the training of the SVM used for the rest of this study. These differences are described in the Methods section.
In order to estimate the real performance of this SVM for the purposes of curators at MDS Proteomics Inc., we reviewed 100 abstracts chosen at random that were either predicted to contain interaction data or not (50 abstracts each). Analysis of all 100 papers for performance on correctly classifying papers that contain interaction information revealed that indexers looking at papers with scores greater than zero would be assured that they contained interaction data 96% of the time (high precision). This was at the cost of missing some papers that talked about interactions (lower recall of 84%). The SVM's classification was correct approximately 9 times out of 10 (accuracy of 89%). Approximately half (4 out of 9) missed interaction papers were about protein-DNA or protein-small molecule interactions. Thus we suggest that the effective recall of protein-protein interaction data was closer to 90%. This reflects the bias in our initial training set for protein-protein interactions and against protein-DNA interactions.
BIND curators at MDS Proteomics Inc. used PreBIND to examine the literature regarding 835 yeast proteins including all of the 600 bait proteins used. A search of the yeast literature for names corresponding to these proteins returned 7021 unique abstracts. The curation team read all abstracts whose SVM scores were greater than zero. This corresponded to 2078 abstracts for the literature regarding 473 of the 835 proteins. A total of 2372 intermediate interactions were generated from this exercise including 711 genetic interaction records.
After removing redundant, genetic and protein-DNA interactions, the PreBIND curation effort resulted in approximately 644 validated protein-protein interaction BIND records covering 608 proteins and 659 publications arising from the original 835 yeast proteins.
In addition, we computed the intersection of the matrix representation of the experimental data with the entire list of putative pair wise interactions with scores greater than zero found in PreBIND. This intersection formed a list of an additional 346 non-redundant interactions that required validation by the BIND curation team. Using this method an additional 53 interactions were confirmed bringing the total to 697 literature interactions identified using PreBIND. These records represent indexed protein-protein interactions. BIND curators are in the process of reading the corresponding papers in their entirety before the records are approved and released to BIND.
As a measure of the usefulness of PreBIND, we calculated that the generation of the 644 preliminary interaction records took approximately 74 FTE (full-time equivalent) days. We estimate that the PreBIND/Textomy system saved at least 176 FTE days since curators would otherwise have had to scan 7021 abstracts instead of 2078. This does not include the time saved by other PreBIND functionalities such as collecting protein name synonyms, performing literature searches and looking up corresponding GI and PMID references for the preparation of BIND records. We estimate that bi-molecular protein interactions mentioned in the remaining 1903 abstracts classified as containing interaction information could be processed at the same level of detail in 68 FTE days. This demonstrated the timesaving usefulness of the PreBIND interface in a real world situation.
Classification of all of PubMed and reusability
We have recently completed searches for all known names for yeast, human and mouse proteins found in RefSeq. These names returned 1.88 million abstracts from PubMed at the time of writing. We used the SVM described above to find that 269,000 of these were classified as containing interaction information. We estimate that the set of interaction abstracts containing yeast, mouse and human protein names is presently comprised of 7,700, 87,000 and 204,000 abstracts respectively.
Based upon this exercise, we believe that a learned SVM can be applied to all of PubMed in a relatively rapid fashion. If the abstracts are stored locally, they can be transformed into feature vectors and be classified based on the SVM in fewer than three days on a single 2 GHz Intel processor. Furthermore, if the vectors are preprocessed (in a three day process), new SVM's can be applied to all abstracts in only six to eight hours on the same processor. This facility will allow for the rapid re-training of the SVM on an on-going basis as feedback is retrieved from PreBIND users. Furthermore, the computational efficiency afforded by this method will allow us to train additional SVM's to classify other types of abstracts and relationships between proteins mentioned in these abstracts.
Conclusions
The BIND project seeks to address, as completely as possible, the problem of encoding information about molecular interactions and their biochemical mechanisms. Proteomics discovery engines, largely emerging from industrial scale efforts, are driving interest in this information set. The system presented here, has proven useful in giving partial context to one such industrial scale effort and will be used to expand the BIND database.
The approach presented here provides a reasonable classifier for finding interaction data in the over 14 million PubMed abstracts that are available to us. The SVM method performed better than a naïve-Bayesian classifier. A natural consequence of the PreBIND interface is that more training examples can be collected on an ongoing basis, as a broader sample of the literature is examined in a feedback loop that will hopefully improve the performance of the SVM. The IE solution presented here is by no means complete. Other systems that use deeper linguistic analysis to recognize noun and verb phrases would be a better choice in extracting specific protein-protein interactions from abstracts identified as containing interaction information. This is especially the case for interactions that occur in sentences with multiple names or across sentence boundaries.
We have chosen here to focus on attaining the highest possible recall for the most general type of interaction data. Any interaction IE system is of limited use if there is no human process attached to it to reliably complete the transfer of data to a machine-readable format such as BIND. We have presented here the first such system that is maintained and publicly available over the web. The PreBIND interface was first released on the Internet in late 2000. Lastly, the PreBIND/Textomy IE system's recall of the MIPS interaction dataset demonstrates that the system must eventually be applied to full-text articles if it ever hopes to retrieve more than a fraction of the interaction knowledge base.
Tools like Textomy, integrated into the PreBIND web-based submission system, provide the BIND effort a focused queue of interaction papers to convert into BIND data records. The enabling SVM technology has allowed us to achieve high precision and recall, keys to providing BIND annotators relevant information with little noise. Currently, this system aids curators in finding and entering protein-protein interaction data. We will add similar capabilities for small-molecule and molecular complex information as the PreBIND system grows to match the requirements of the BIND database.
Methods
Collection of names
The PreBIND parser (Fig. 2, item 3) collects synonyms for proteins and their encoding loci for a non-redundant set of proteins present in the NCBI RefSeq sequence database [15]. The cumulative RefSeq database is distributed as an ASN.1 file in binary format (see the file "rscu.bna" at ftp://ftp.ncbi.nih.gov/refseq/cumulative/. The PreBIND parser program (Figure 2, item 3) used this file as its primary input. Gene loci names and synonyms were retrieved from the "locus" and "syn" fields of the Gene-ref data structure ("Gene-ref is a type of sequence feature found in "Bioseq-set" and "Bioseq" sequence records. For more information, search for "Gene-ref" at http://www.ncbi.nlm.nih.gov/IEB/ToolBox/SB/hbr.html and for an introduction to the NCBI data model see [31]).
Gene loci names present in nucleotide records were matched to their corresponding proteins by comparing location information for the Gene-Ref with location information for the protein. These synonyms were stored with their corresponding RefSeq GenInfo (GI) number in the PreBIND database (Fig. 2, item 4). Additional synonyms were collected by the parser for each unique GI from the Saccharomyces cerevisiae genome database (SGD) [16] (Fig. 2, item 2) using database cross-references found in RefSeq records.
The collected names were filtered before being used to search PubMed. The following types of names were not used: names with only one character, names with only one letter followed by numbers and systematic open reading frame names for yeast (for example YDL140C).
The complete list of names used for searching will be made available upon request until the PreBIND GI and name table have been incorporated into the SeqHound database system along with a remote API (application programming interface) that allows access to these names (see reference [25]).
Locating names in PubMed abstracts
The PreBIND searcher program (Fig. 2, item 6) searches the PubMed literature database [18] for names in either the abstract or title fields while limited to the MESH listed taxon relevant to the protein. Names are detected by searching the PubMed database remotely with the NCBI C-toolkit function "EntrezTLEvalXString(query, TYP_ML, -1, NULL, NULL)" where an example query is "rpo21 [WORD] & Saccharomyces_cerevisiae [MESH]". The results returned by this search are essentially equivalent to typing the query string into the NCBI web interface to PubMed http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=PubMed. More details on the use of this function can be found at http://www.ncbi.nlm.nih.gov/IEB/ToolBox/SB/hbr.html.
Training of the SVM for the yeast high-throughput study
The SVM used for the literature collection described for the yeast high-throughput study [29] is described here. This SVM accompanied the initial release of PreBIND in November of 2000. The training examples were essentially as described above with some minor modifications. Also, the dimensions extracted from the text were the words and two-word (adjacent) phrases. N-grams were tried with little advantage over whole words. Term features were filtered (thrown out) if they occurred in fewer than 3 articles, if they occurred in a standard list of the most common 300 stop words [26], or if they were shorter than two characters. Words were strings of alphanumeric characters that included internal apostrophes, underscores, periods, and hyphens. They were truncated at ten characters and were case sensitive. No other text pre-processing occurred. The value for each dimension for each abstract was the TFIDF score (term frequency, inverse document frequency). Term frequency is the number of times the term occurs in the abstract and the document frequency is the number of abstracts in which it appeared at least once.
SVM training was performed using a linear kernel, meaning that the decision boundary used was a hyperplane. Polynomial kernels were tried but showed no significant improvement. Ten-fold cross-validation training and testing was performed as described above.
Precision, Recall and F-measure
For a set of examples that have been labeled as either positive or negative and for an algorithm that predicts this positive or negative class, then the "recall" of the predictive algorithm is calculated as the number of correct positive predictions divided by the number of examples labeled as positive. "Precision" is calculated as the number of correct positive predictions divided by the total number of positive predictions. Precision can be increased at the cost of recall by increasing the cutoff at which the SVM score is considered to be indicative of a positive example (i.e., an interaction paper). Using zero as the cutoff often maximizes accuracy and the F-measure. As the cutoff rises above zero, the chance of a false positive decreases, but the chance of a false negative increases. The F-measure (F) is an expression of precision and recall that favors a balance between the two such that:
F = (αP-1 + (1-α)R-1)-1
where 'P' is precision 'R' is recall and 'α' is some value between 0 and 1. When an equal weighting between precision and recall is chosen (α = 0.5), the F measure becomes:
F = 2PR(P+R)-1
Naïve-Bayes Text Classification
Naïve-Bayes text classification was performed using the BOW software toolkit http://www-2.cs.cmu.edu/~mccallum/bow/). Modifications were made to allow for 10-fold cross-validation. Similar to our SVM method, term features were words and two-word (adjacent) phrases. Term features were thrown out if they occurred in a standard list of 300 most frequent stop words [26] or were shorter than two characters. Words were strings of alphabetic characters. They were truncated at ten characters and were converted to lower case. No other text pre-processing occurred. Digits, punctuation, and white spaces were treated as non-word characters. As a result, digits and punctuation contained within words were treated as word boundaries. Information gain was used to restrict the classifier to the most useful terms. The number of terms was chosen to be 100 because it optimized performance on a similar dataset. The naïve-Bayes classifier used Laplace smoothing and a word event model. Further details are available in the BOW documentation [32].
Software
PreBIND associated software was written entirely in the C programming language using the NCBI toolkit http://ncbi.nlm.nih.gov/IEB[31]. All code is cross-compilable on several platforms including Windows®, Linux, and Solaris®. The application uses a CodeBase® database backend system from Sequiter® Software Inc. http://www.sequiter.com. The PreBIND interface has been tested using fourth generation and higher Internet Explorer and Netscape clients. PreBIND data is accessible via the web interface at http://bind.ca.
Textomy software was written in a combination of Java and Perl. The Textomy tools are back ended to a MySQL database and run on a loose network of five processors connected by Condor http://www.cs.wisc.edu/condor. Textomy is accessible at http://www.litminer.ca/.
Authors Contributions
The PreBIND system was conceived of and programmed by ID based on conversations with CWVH, JM, TP and GDB. The Textomy system was conceived and programmed by JM and BdB. CW, BB and ID supplied the original examples to train the SVM. CW, VL, BT and SZ reviewed abstracts identified by PreBIND for the high-throughput yeast study. KM is the primary developer of the SeqHound system and supplied invaluable advice and guidance to ID. GDB was the primary developer of the BIND database and performed the MIPS comparison analysis. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We would like to thank Roy Walker, Lynn Wei, Brandon Smith and Maria Moreno of the Institute for Biological Sciences, NRC, for useful comments and suggestions. Funding for developing this project was provided by separate Canadian Institutes of Health Research grants to C. Hogue and T. Pawson as well as NRC funding to J. Martin and by in-kind contributions from MDS Proteomics Inc. Funding to provide the PreBIND web interface as a service is provided by Genome Canada and the Ontario R&D Challenge Fund.
Contributor Information
Ian Donaldson, Email: ian.donaldson@utoronto.ca.
Joel Martin, Email: joel.martin@nrc-cnrc.gc.ca.
Berry de Bruijn, Email: Berry.deBruijn@nrc-cnrc.gc.ca.
Cheryl Wolting, Email: wolting@mshri.on.ca.
Vicki Lay, Email: vicki@mshri.on.ca.
Brigitte Tuekam, Email: tuekam@mshri.on.ca.
Shudong Zhang, Email: szhang@mdsp.com.
Berivan Baskin, Email: berivan.baskin@sickkids.ca.
Gary D Bader, Email: gary.bader@utoronto.ca.
Katerina Michalickova, Email: katerina@mshri.on.ca.
Tony Pawson, Email: pawson@mshri.on.ca.
Christopher WV Hogue, Email: hogue@mshri.on.ca.
References
- Bader GD, Hogue CW. BIND--a data specification for storing and describing biomolecular interactions, molecular complexes and pathways. Bioinformatics. 2000;16:465–477. doi: 10.1093/bioinformatics/16.5.465. [DOI] [PubMed] [Google Scholar]
- Bader GD, Donaldson I, Wolting C, Ouellette BF, Pawson T, Hogue CW. BIND--The Biomolecular Interaction Network Database. Nucleic Acids Res. 2001;29:242–245. doi: 10.1093/nar/29.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimizu T, Park HS, Tsujii J. Identifying the Interaction between Genes and Gene Products Based on Frequently Seen Verbs in Medline Abstracts. Genome Inform Ser Workshop Genome Inform. 1998;9:62–71. [PubMed] [Google Scholar]
- Rindflesch TC, Tanabe L, Weinstein JN, Hunter L. EDGAR: extraction of drugs, genes and relations from the biomedical literature. Pac Symp Biocomput. 2000:517–528. doi: 10.1142/9789814447331_0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys K, Demetriou G, Gaizauskas R. Two applications of information extraction to biological science journal articles: enzyme interactions and protein structures. Pac Symp Biocomput. 2000:505–516. doi: 10.1142/9789814447331_0048. [DOI] [PubMed] [Google Scholar]
- Proux D, Rechenmann F, Julliard L. A pragmatic information extraction strategy for gathering data on genetic interactions. Proc Int Conf Intell Syst Mol Biol. 2000;8:279–285. [PubMed] [Google Scholar]
- Stapley BJ, Benoit G. Biobibliometrics: information retrieval and visualization from co- occurrences of gene names in Medline abstracts. Pac Symp Biocomput. 2000:529–540. doi: 10.1142/9789814447331_0050. [DOI] [PubMed] [Google Scholar]
- Thomas J, Milward D, Ouzounis C, Pulman S, Carroll M. Automatic extraction of protein interactions from scientific abstracts. Pac Symp Biocomput. 2000:541–552. doi: 10.1142/9789814447331_0051. [DOI] [PubMed] [Google Scholar]
- Blaschke C, Valencia A. The potential use of SUISEKI as a protein interaction discovery tool. Genome Inform Ser Workshop Genome Inform. 2001;12:123–134. [PubMed] [Google Scholar]
- Friedman C, Kra P, Yu H, Krauthammer M, Rzhetsky A. GENIES: a natural-language processing system for the extraction of molecular pathways from journal articles. Bioinformatics. 2001;17:S74–NaN. doi: 10.1093/bioinformatics/17.suppl_1.s74. [DOI] [PubMed] [Google Scholar]
- Jenssen TK, Laegreid A, Komorowski J, Hovig E. A literature network of human genes for high-throughput analysis of gene expression. Nat Genet. 2001;28:21–28. doi: 10.1038/88213. [DOI] [PubMed] [Google Scholar]
- Ono T, Hishigaki H, Tanigami A, Takagi T. Automated extraction of information on protein-protein interactions from the biological literature. Bioinformatics. 2001;17:155–161. doi: 10.1093/bioinformatics/17.2.155. [DOI] [PubMed] [Google Scholar]
- Marcotte EM, Xenarios I, Eisenberg D. Mining literature for protein-protein interactions. Bioinformatics. 2001;17:359–363. doi: 10.1093/bioinformatics/17.4.359. [DOI] [PubMed] [Google Scholar]
- Wong L. PIES, a protein interaction extraction system. Pac Symp Biocomput. 2001:520–531. [PubMed] [Google Scholar]
- Pruitt KD, Maglott DR. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwight SS, Harris MA, Dolinski K, Ball CA, Binkley G, Christie KR, Fisk DG, Issel-Tarver L, Schroeder M, Sherlock G, Sethuraman A, Weng S, Botstein D, Cherry JM. Saccharomyces Genome Database (SGD) provides secondary gene annotation using the Gene Ontology (GO) Nucleic Acids Res. 2002;30:69–72. doi: 10.1093/nar/30.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostell JM, Wheelan SJ, Kans JA. The NCBI data model. In: Baxevanis AD and Ouellette B F, editor. Bioinformatics. 2nd. Vol. 43. New York, John Wiley and Sons, Inc.; 2001. pp. 19–43. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Church DM, Lash AE, Leipe DD, Madden TL, Pontius JU, Schuler GD, Schriml LM, Tatusova TA, Wagner L, Rapp BA. Database resources of the National Center for Biotechnology Information: 2002 update. Nucleic Acids Res. 2002;30:13–16. doi: 10.1093/nar/30.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes C, Vapnik V. Support-Vector Networks. Machine Learning. 1995;20:273–297. doi: 10.1023/A:1022627411411. [DOI] [Google Scholar]
- Joachims T. Text categorization with Support Vector Machines: Learning with many relevant features. Machine Learning: ECML-98, Tenth European Conference on Machine Learning. 1998. pp. 137–142.
- Dumais S, Platt J, Heckerman D, Sahami M. Inductive learning algorithms and representations for text categorization. Proceedings of the International Conference on Information and Knowledge Management. 1998. pp. 148–155.
- de Bruijn B, Martin J, Wolting C, Donaldson I. Extracting sentences to justify categorization. Proceedings of the American Society for Information Science and Technology Annual Meeting ASIST. 2001. pp. 450–457.
- Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, Phan I, Pilbout S, Schneider M. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda K, Tamura A, Tsunoda T, Takagi T. Toward information extraction: identifying protein names from biological papers. Pac Symp Biocomput. 1998:707–718. [PubMed] [Google Scholar]
- Michalickova K, Bader GD, Dumontier M, Lieu HC, Betel D, Isserlin R, Hogue CW. SeqHound: biological sequence and structure database as a platform for bioinformatics research. BMC Bioinformatics. 2002;3:32. doi: 10.1186/1471-2105-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salton G. The SMART Retrieval System. Englewood Cliffs, NJ, Prentice Hall; 1971. [Google Scholar]
- Mitchell T. Machine Learning. 1st. McGraw-Hill; 1997. p. 414. [Google Scholar]
- Mewes HW, Frishman D, Guldener U, Mannhaupt G, Mayer K, Mokrejs M, Morgenstern B, Munsterkotter M, Rudd S, Weil B. MIPS: a database for genomes and protein sequences. Nucleic Acids Res. 2002;30:31–34. doi: 10.1093/nar/30.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Bader GD, Hogue CW. Analyzing yeast protein-protein interaction data obtained from different sources. Nat Biotechnol. 2002;20:991–997. doi: 10.1038/nbt1002-991. [DOI] [PubMed] [Google Scholar]
- Ostell JM, Wheelan SJ, Kans JA. The NCBI data model. Methods Biochem Anal. 2001;43:19–43. doi: 10.1002/0471223921.ch2. [DOI] [PubMed] [Google Scholar]
- McCallum Andrew Kachites. Bow: A toolkit for statistical language modeling, text retrieval, classification and clustering. http://www.cs.cmu.edu/~mccallum/bow. 1996.