Abstract
To increase our understanding of imprinting and epigenetic gene regulation, we undertook a search for new imprinted genes. We identified Gatm, a gene that encodes l-arginine:glycine amidinotransferase, which catalyzes the rate-limiting step in the synthesis of creatine. In mouse, Gatm is expressed during development and is imprinted in the placenta and yolk sac, but not in embryonic tissues. The Gatm gene maps to mouse chromosome 2 in a region not previously shown to contain imprinted genes. To determine whether Gatm is located in a cluster of imprinted genes, we investigated the expression pattern of genes located near Gatm: Duox1-2, Slc28a2, Slc30a4 and a transcript corresponding to LOC214616. We found no evidence that any of these genes is imprinted in placenta. We show that a CpG island associated with Gatm is unmethylated, as is a large CpG island associated with a neighboring gene. This genomic screen for novel imprinted genes has elucidated a new connection between imprinting and creatine metabolism during embryonic development in mammals.
Of the 30,000–40,000 genes in a mammalian genome, ≈0.1–0.2% are expressed differently depending on whether they were inherited from the mother or father, a phenomenon known as genomic imprinting. Many of these genes are expressed during embryonic development and influence the growth and nutritional demands of an embryo on the mother (1–3). Imprinted gene expression is most often observed in the placenta and yolk sac, the extraembryonic tissues of a developing embryo. This tissue distribution is consistent with the parent–offspring conflict hypothesis that has been proposed to explain the evolution and maintenance of imprinting in mammals (4). According to this model, imprinting is a manifestation of a conflict between the parental genomes over the allocation of maternal resources. Genes that play a role in regulating the exchange of energy and nutrients between mother and embryo are predicted to be likely candidates for imprinting control. In general, this prediction is well supported by experimental evidence.
Imprinted genes share some unusual characteristics, which presumably reflect aspects of the mechanism of this epigenetic regulation. For example, imprinted genes are often found in clusters of both maternally and paternally expressed genes (ref. 5 and www.mgu.har.mrc.ac.uk/imprinting/implink.html). Such clusters of imprinted genes usually contain one or more CpG islands that are differentially methylated between the maternal and paternal alleles of the gene (1, 6). In a few instances, these differentially methylated regions have been shown to control the imprinting of multiple genes in a cluster (7–14) by binding methylation-sensitive regulatory proteins (15, 16).
To investigate the mechanism and function of genomic imprinting, we used a differential allelic display strategy to identify new imprinted genes (17, 18). In this report we show that Gatm, the gene encoding the rate-limiting enzyme in creatine synthesis (19), is imprinted, being expressed exclusively from the maternal allele in extraembryonic tissues of mouse. Analysis of the allelic expression pattern of genes near Gatm failed to identify any other gene whose expression was imprinted. The first exon of the Gatm gene is located in a CpG island that is unmethylated on both alleles, as is a large CpG island associated with the nearest neighboring transcript. Thus, we have identified a new imprinted gene, Gatm, which resides in apparent isolation from other imprinted genes.
Materials and Methods
Peromyscus-Imprinting Assay.
RNA samples were isolated from embryonic day 14.5 (E14.5) placenta and embryo of Peromyscus polionotus (PO) and Peromyscus maniculatus (BW) parental strains and reciprocal F1 crosses. RNA samples were treated with DNase (Stratagene) for 30 min at 37°C. RT-PCRs were performed by using an RNA amplification Core kit (Applied Biosystems) supplemented with [33P]dCTP to label the products. Samples were denatured at 94°C for 5 min in 95% formamide/10 mM NaOH. The primers used to amplify the noncoding transcript in Peromyscus were T11C for the reverse transcriptase reaction and 5′-TATAAGCAAGTGACTTCGGTGTGG-3′ and 5′-GAAATGGATCATCCTGGTCCTTG-3′ for the PCR. Radiolabeled RT-PCR products were separated on gels made of MDE gel solution (Cambrex, East Rutherford, NJ) to visualize differences based on single-strand conformational polymorphisms. Parallel amplification reactions were performed with and without reverse transcriptase, to control for DNA contamination.
Mus-Imprinting Assays.
RNA samples were isolated from C57BL/6J and CAST/Ei parental strains and reciprocal F1 crosses by extraction with Trizol (GIBCO/BRL). cDNA synthesis was performed by using an oligo(dT) primer and reverse transcriptase. RT-PCR amplification was performed by using an RNA amplification Core kit (Applied Biosystems), and the resulting products were separated on 7% acrylamide gels and visualized by ethidium bromide staining. To test imprinting, oligo(dT)-primed cDNA was PCR amplified by using the following primers: for Gatm, 5′-GACCATAGTTACTCCTCCAACAC-3′ and 5′-GAAGGATAACCCACTTGAGATG-3′, which generates a 1,007-bp product that spans exons 7–9; for Duox1-2, 5′-AAGTTTGAGGTGTCAGTGCTGGTAG-3′ and 5′-GCAGAGAGTTGAAAAAAGGGCTC-3′, which amplifies a product ≈400 bp in length that spans three or more exons in one or both of the Duox transcripts; for Slc28a2, 5′-TCCATAGGAATCACACTGGGAGG-3′ and 5′-CCATACTTTGGCTCAAGAGGGTC-3′, which amplifies a 585-bp product that spans the last three exons of the Slc28a2 gene; for LOC214616, 5′-GGGAGCGAGTTCTTTCAGTTCTG-3′ and 5′-GGATGGCTTCACAGTCTTGAG-3′, which amplifies a 493-bp product that spans three introns; and for Slc30a4, 5′-GGAGACATAGTGAGTGGCACAACC-3′ and 5′-CCAACGATGAACTGAACCAAATGG-3′, which produces a 994-bp product. Because the Slc30a4 RT-PCR product does not span an intron, Slc30a4 reactions were performed with and without reverse transcriptase, as a control for exclusive amplification of RNA (data not shown). Because the computationally predicted reference sequence for the mouse LOC214616 transcript, XM141336, appeared to be chimeric by comparison with the known and predicted human homologs, we opted to base our design of primers for the LOC214616 transcript on the mouse EST AV141009. Because there are gaps in the mouse genomic sequence for this region, genomic structure for EST AV141009 transcript was deduced from comparison with human genomic sequence.
For the Gatm gene, an allele-specific size polymorphism was visualized directly by running the RT-PCR products on acrylamide gels. For Duox1-2, Slc28a2, and Slc30a4, RT-PCR products were digested with polymorphic restriction enzymes to reveal the allele specificity of the expressed RNA template. The polymorphic restriction enzymes used were: Duox1-2, HhaI; Slc28a2, HaeIII; and Slc30a4, ApaLI. The LOC214616 product was sequenced directly.
Gatm Expression Analysis.
The mouse Northern blots (Seegene, Del Mar, CA) were probed with a 353-bp PCR-generated fragment that spanned exon 7 to exon 9 of the Gatm cDNA. The probe was radiolabeled with [32P]dCTP by a random primer reaction, and hybridization was carried out in ExpressHyb (CLONTECH) at 68°C.
DNA Methylation Analysis.
Genomic DNAs were isolated by using a DNeasy kit (Qiagen, Valencia, CA), digested with restriction enzymes, separated by using 1× 90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3/0.8% agarose gels, and transferred to Hybond N+ (Amersham Biosciences). The Gatm CpG island probe was a 799-bp Tth111I–NcoI fragment that includes flanking DNA at the 5′ end of the Gatm gene. The LOC214616 CpG island probe was generated from a PCR product amplified by the following primers: 5′-ACAGGGAGTTTGTGGTTGTGGG-3′ and 5′-AAAGACGCTAAAAAAGCCAGCC-3′. This primer pair produces a 394-bp product that extends from exon 1 into intron 1 of the predicted LOC214616 transcript.
Results
Identification of a Maternally Expressed Transcript in Peromyscus.
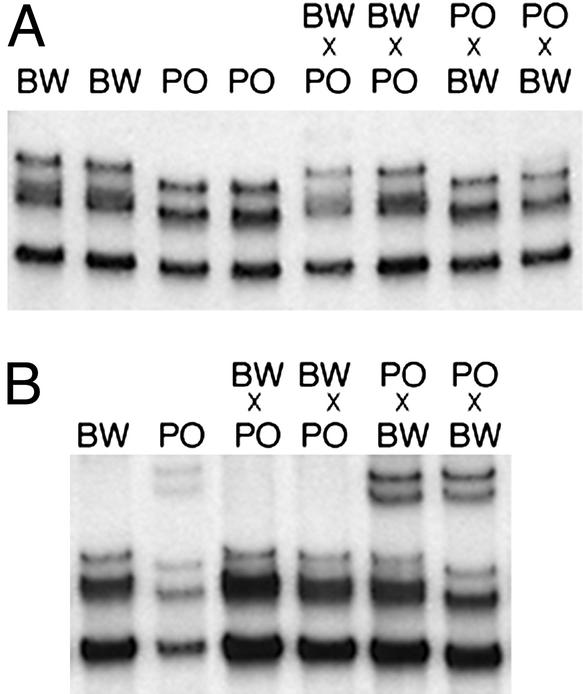
To identify new imprinted genes, we used a differential display screen that took advantage of the high degree of polymorphism between two species of deer mice of the genus Peromyscus (18). RNAs from placental tissue of reciprocal F1 crosses between PO and BW were amplified with PCR, and the resulting products were subjected to differential display by gel electrophoresis. Comparison of the patterns for the BW × PO and the PO × BW progeny revealed cDNA products that derived from individual parental alleles. From this screen, we identified a placental transcript whose expression derived exclusively from the maternal allele (Fig. 1A). Furthermore, we determined that the transcript exhibited imprinted expression in the embryo (Fig. 1B), demonstrating that the maternal-specific expression was not a result of maternal tissue in the placenta.
Figure 1.
A polymorphic transcript is maternally expressed in Peromyscus placenta and embryo. RNA was isolated from BW and PO parental strains and from reciprocal F1 hybrids. For the hybrids, the maternal species is listed first. Single strand conformation polymorphism RT-PCR analysis detected a difference between the two parental strains. (A) E14.5 placental RNA. (B) E14.5 whole embryo RNA.
The Imprinted Transcript in Mus Overlaps the Gatm Gene.
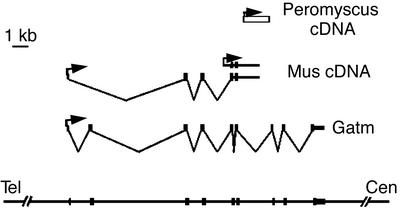
The sequence of the Peromyscus transcript did not match known sequences in the EST and whole genome databases. To further investigate its identity, we cloned two cDNAs that shared homology with the Peromyscus transcript from a Mus domesticus adult brain library (Fig. 2). Portions of the Mus cDNAs contained exons of a previously identified gene, l-arginine:glycine amidinotransferase (Gatm, also known as AGAT in human). This enzyme catalyzes the rate-limiting step in the formation of creatine, an important molecule in energy metabolism (19). The shorter of the two Mus cDNAs was contiguous with genomic DNA and contained exons 5 and 6 of the Gatm gene embedded within intronic sequence, indicating that the transcript was unspliced. The other cDNA contained exons 1, 3, and 4 of the Gatm gene spliced correctly into exon 5, followed by unspliced genomic sequence. The homology with the original Peromyscus clone was contained within intron 6 of the Gatm gene.
Figure 2.
Mus Gatm gene and splice variants. The inferred genomic organization of Peromyscus and Mus transcripts overlapping the Gatm gene are depicted. The position of the Peromyscus cDNA clone is represented by the open rectangle. Filled rectangles indicate the positions of Gatm exons in Mus brain cDNA splice variants and in the actual Gatm transcript. Arrows represent direction of transcription. Orientation of the Mus transcripts was determined by RT-PCR analysis (data not shown). The heavy horizontal lines represent genomic sequence.
Gatm Expression Is Imprinted in Extraembryonic Tissue in Mouse.
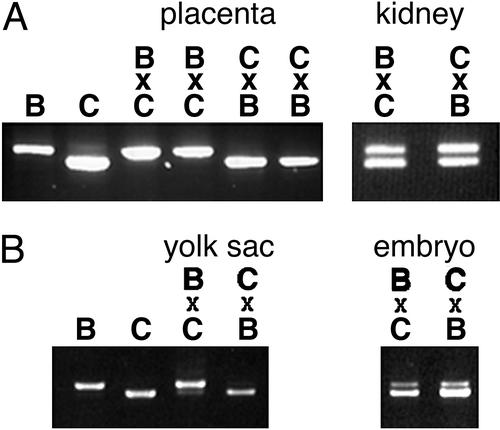
Because the unspliced and partially spliced transcripts were contained within the Gatm gene, we sought to test the imprinting status of Gatm itself. By sequencing Gatm cDNAs from C57BL/6J and CAST/Ei mice, we identified a duplicated region in the C57BL/6J allele that was represented only once in the CAST/Ei allele, a difference readily detectable as a size polymorphism in RT-PCR products (Fig. 3A).
Figure 3.
Gatm is imprinted in extraembryonic tissue in Mus. Gatm-imprinting assays were performed by RT-PCR amplification of Gatm RNA. Imprinting was assessed by visualization of RT-PCR products from C57BL/6J (B) and CAST/Ei (C) parental strains and reciprocal F1 hybrids. For the hybrids, the maternal genotype is indicated first. (A) E17.5 placenta and kidney. (B) E9.5 yolk sac and whole embryo.
Using the size polymorphism RT-PCR assay, we examined the allelic expression of the Gatm gene in tissues from reciprocal F1 crosses between C57BL/6J and CAST/Ei mice. At E17.5 the larger RT-PCR product indicative of the C57BL/6J allele was observed in crosses in which the mother was C57BL/6J. Similarly, the smaller CAST/Ei product was observed in crosses in which the mother was CAST/Ei (Fig. 3A). In contrast, products of both parental alleles are observed in kidney samples from hybrid offspring (Fig. 3A) as well as brain and gonad (data not shown). These data indicate the Gatm gene is maternally expressed in placenta but biallelic in other tissues. At E9.5, the Gatm gene is predominantly maternally expressed in the yolk sac but biallelic in the embryo (Fig. 3B). Thus, unlike the Peromyscus intronic transcript, the Mus Gatm gene is imprinted only in extraembryonic tissues during mouse development.
In mouse, Gatm maps to the central region of chromosome 2. We refined its position to 69.0 centimorgans (cM) on mouse chromosome 2 by using The Jackson Laboratory T31 radiation hybrid mapping panel, and we confirmed the map position by identification of yeast artificial chromosomes (YAC73.C3 and YAC142.B4) that contained Gatm and the markers D2Mit17 and D2Mit396 (data not shown). Mouse chromosome 2 harbors imprinted genes at both termini, with the closest imprinted gene, Nnat, a maternally expressed gene, mapping at 88 cM (ref. 5 and www.mgu.har.mrc.ac.uk/imprinting/implink.html). However, no imprinted genes have been mapped to the central region of chromosome 2 in the vicinity of D2Mit17.
The autosomal mapping of the transcripts also excluded the possibility that the maternal expression of Gatm derived from either the mitochondrial genome or the X chromosome, two potential sources of maternal-specific gene expression in the placenta.
Gatm Is Expressed During Embryonic Development.
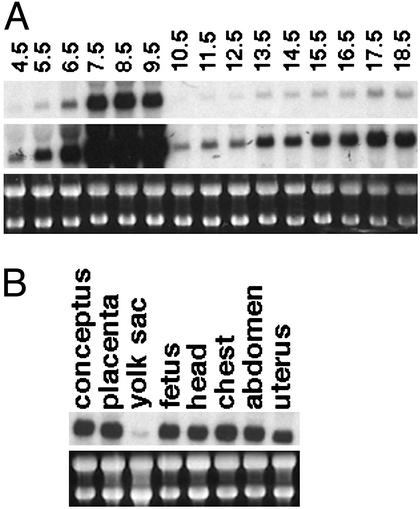
The tissue distribution of Gatm expression has been studied primarily in adult animals (19), and, aside from the observation that Gatm expression is high in decidua of rat (20), relatively little is known about its expression profile during embryonic development. To investigate the expression of Gatm during mouse embryonic development, we performed Northern blot analysis, using the 3′ end of the gene as a probe (Fig. 4A). The highest levels of Gatm expression were observed in RNA samples prepared from E7.5 to E9.5 conceptuses, which contained a mixture of embryonic and extraembryonic tissue. Analysis of the samples from E10.5 to E18.5 embryos, which contain exclusively embryonic tissue, indicates that Gatm is expressed at relatively low levels in the mid-gestation stage embryo, with expression gradually increasing throughout development. At E17.5, expression of Gatm is relatively uniform in embryo and placenta but low in the yolk sac and amnion (Fig. 4B).
Figure 4.
Gatm is expressed during embryonic development in mouse. (A) Northern blot analysis of Gatm expression throughout embryonic development is shown. Samples from E4.5 to E6.5 include embryo plus extraembryonic plus uterus RNA, samples from E7.5 to E9.5 include embryo plus extraembryonic RNA, and samples from E10.5 to E18.5 include RNA from embryos only. To allow visualization of dramatically different transcript levels, two exposures of the same Northern blot are shown. (Top) A short exposure. (Middle) A longer exposure. (Bottom) Ethidium bromide-stained ribosomal RNA as a control for RNA loading. (B) Northern blot analysis of Gatm expression in various tissues of E17.5 embryos is shown. Yolk sac sample contains yolk sac plus amnion.
Imprinting Status of Neighboring Genes.
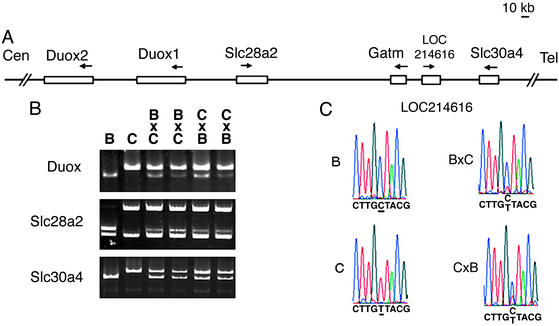
Because many imprinted genes are found in clusters, we sought to test the imprinting status of genes near Gatm. Using the University of California, Santa Cruz (21), and National Center for Biotechnology Information genome maps, we identified several genes near Gatm (Fig. 5A). Those include Slc28a2, a sodium-coupled purine transporter, and Slc30a4, a zinc transporter in which a mutation is responsible for the recessive mouse mutant lethal milk. Other extraembryonically expressed genes near Gatm included a pair of transcripts (herein referred to as Duox1-2) that are similar to the human dual oxidase genes DUOX1 and DUOX2, whose products are part of a peroxide-generating system (22). Also near Gatm is a transcript corresponding to LOC214616, which shares homology with the putative human protein MGC5347, whose predicted domains are similar to ATPases that have a variety of cellular activities. We verified that each of these genes was closely linked to Gatm in mouse by showing that genomic fragments could be amplified from Gatm-containing yeast artificial chromosomes (data not shown).
Figure 5.
Analysis of biallelic expression of neighboring genes. (A) Genomic organization within ≈450 kb surrounding the Gatm gene is depicted, with open boxes representing genes assayed for imprinting status. Arrows represent direction of transcription. (B) RT-PCR imprinting assays of Duox1-2, Slc28a2, and Slc30a4 are shown. In each case, the parental restriction fragment length polymorphism fragments are shown in the first two lanes: B, C57BL/6J; C, CAST/Ei. Fragments produced from hybrid offspring, which indicate both parental alleles and thus biallelic expression, are shown in the following lanes indicated as B × C or C × B (maternal genotype shown first). For Duox1-2, cDNA was digested with HhaI, which selectively digests the C57BL/6J product to produce a 337-bp fragment. For Slc28a2, cDNA was digested with HaeIII, which digests both C57BL/6J and CAST/Ei RT-PCR products to generate ≈240- and ≈340-bp fragments, respectively. For Slc30a4, cDNA was digested with ApaLI, which specifically cuts the C57BL/6J product, reducing its the size by 70 bp. (C) Direct sequencing assay of the LOC214616 transcript.
To identify polymorphisms for imprinting assays, fragments from each of these genes were cloned and sequenced from C57BL/6J and CAST/Ei cDNA. Restriction polymorphisms were identified for Slc30a4, Slc28a2, and Duox 1-2. Owing to the high degree of similarity between the Duox genes, the two transcripts were not distinguishable in the allele-specific imprinting assay described here. For the mouse gene corresponding to LOC214616, imprinting status was assessed by a direct sequence assay. All four genes were determined to be biallelically expressed in E17.5 placenta (Fig. 5 B and C).
Although we assayed the imprinting status of four genes, our search for imprinted genes in the vicinity of Gatm was not exhaustive. EST evidence, as well as gene prediction analysis, suggests that there may be as many as three other genes within the 450 kb surrounding Gatm.
Gatm CpG Island and Neighboring Island Are Unmethylated.
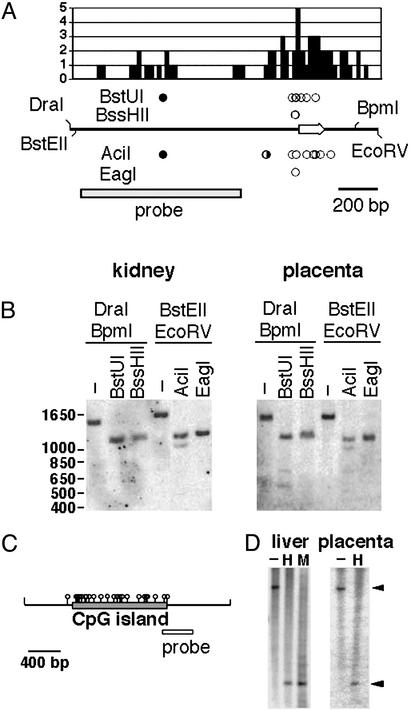
To investigate the methylation pattern of Gatm, we cloned and sequenced genomic DNA corresponding to the first exon and an associated CpG island (Fig. 6A). The island is relatively small (406 bp), but it is dense in CpG dinucleotides, having an observed/expected CpG ratio of 0.76. The methylation status of the Gatm CpG island was assessed by Southern blot analysis by using a variety of methylation-sensitive restriction enzymes. Genomic DNA samples from C57BL/6J embryonic kidney and placenta were digested with the methylation-sensitive enzymes BstUI, BssHII, EagI, and AciI (Fig. 6 A and B). The BstUI and AciI recognition sequences outside the CpG island were protected from digestion by methylation. For the kidney samples, this nonisland methylation is complete and for the placenta samples the nonisland methylation appeared slightly less, in keeping with the general undermethylation of extraembryonic tissues (23). In contrast to the nonisland CpG restriction sites, one or more of the AciI and BstUI sites within the Gatm CpG island were digested to completion for both the kidney and the placental DNA samples. Similarly, the single EagI site and one or both of the BssHII sites digested to completion. Thus, within the Gatm CpG island, one or more recognition sites for each of the four methylation-sentive enzymes were unmethylated and digested to completion for both the kidney and the placental DNA samples. Taken together, these data suggest that the Gatm CpG island is unmethylated in both kidney and placenta DNA.
Figure 6.
Methylation analysis of CpG islands associated with Gatm and LOC214616. (A) The frequency distribution of CpG dinucleotides within a 1.5-kb fragment containing the Gatm CpG island is shown. Each value represents the number of CpG dinucleotides per 20 bp. Genomic DNA is depicted below the histogram. Arrow indicates position of exon 1 as well as direction of transcription. Circles represent recognition sites for methylation-sensitive enzymes; filled circles represent sites that failed to digest owing to methylation, half-filled circles represent sites that were partially protected by methylation, and open circles indicate sites of which one or more were unmethylated and digested to completion. The position of the probe is indicated by the rectangle. (B) Methylation Southern blots of C57BL/6J DNAs prepared from E18.5 kidney and placenta. Digestion with the methylation-sensitive enzymes BstUI and BssHII was assessed in combination with DraI and BpmI, which together define an ≈1.4-kb restriction fragment. Digestion with the methylation-sensitive enzymes AciI and EagI was assessed in combination with BstEII and EcoRV, which together define an ≈1.5-kb restriction fragment. Mr positions are indicated in base pairs. (C) The structure of an ≈2.5-kb DraI fragment containing the LOC214616 CpG island. (D) DNAs prepared from E17.5 liver and E18.5 placenta from C57BL/6J were digested with DraI (−), DraI + HpaII (H), or DraI +MspI (M). Arrowheads indicate the position of the ≈2.5-kb DraI restriction fragment and the ≈0.7-kb fragment produced by complete digestion with either DraI + HpaII or DraI + MspI.
Because the Gatm CpG island appeared unmethylated on both alleles and thus unlikely to control the imprinted expression of Gatm, we looked for evidence of a differentially methylated CpG island in the vicinity that might function as an imprinting control element for the region. To that end, we investigated the methylation pattern of the CpG island of the nearest neighboring gene to Gatm. Based on the National Center for Biotechnology Information and University of California, Santa Cruz, genomic databases, the nearest neighboring gene to Gatm, which corresponds to LOC214616, is associated with a large CpG island located ≈18 kb distal to the Gatm CpG island (21). The LOC214616 CpG island is 1,160 bp and has a CpG observed/expected ratio of 0.79. Southern blot methylation analysis of embryonic liver and placenta DNA samples was performed by using HpaII as the methylation-sensitive enzyme and its methylation-insensitive isoschizomer, MspI (Fig. 6 C and D). Hybridization with a probe adjacent to the LOC214616 CpG island revealed that the HpaII and MspI digestion patterns were indistinguishable, indicating that CpG dinucleotides within both alleles of the island were unmethylated. The unmethylated state of the LOC214616 CpG island is consistent with the biallelic expression of the associated transcript. The LOC214616 island was unmethylated in both placenta and embryonic liver DNA. Thus, we have found no evidence of a differentially methylated CpG in the vicinity of Gatm.
Discussion
This study was undertaken to identify new genes that are regulated by genomic imprinting. We have determined that Gatm, a gene that encodes a metabolic enzyme involved in creatine synthesis, is imprinted throughout embryonic and extraembryonic tissues in Peromyscus and in extraembryonic tissues in Mus. These data are the first to link creatine metabolism with imprinting and the parental “tug-of-war” for energy resources during development.
Phosphocreatine, the phosphorylated form of creatine, serves as a reservoir for high-energy phosphate in ATP synthesis and, thus, is critically involved in energy balance (19, 24). The rate-limiting step in creatine synthesis is the conversion of l-arginine to guanidinoacetate by the product of the Gatm gene, l-arginine:glycine amidinotransferase. The GATM gene is important for brain development in humans, because deficiencies in creatine synthesis and transport have been implicated in certain forms of mental retardation in humans (25, 26). These include mutations in the GATM gene itself (27), as well as other genes required for synthesis or transport of creatine (28–31).
Our finding that a gene involved in brain development in humans is imprinted during embryogenesis in mice fits with a previously established pattern of imprinted genes that play a role in embryonic and fetal growth as well as brain function. Mutations in two paternally expressed genes, Peg1 and Peg3, have been shown to affect maternal behavior in mice, possibly because of a deficit of oxytocin-producing neurons in the hypothalamus of the mutant mice (32, 33). Moreover, deletions and translocations in the Prader–Willi/Angelman syndrome region, which involve disruption of imprinted genes, lead to mental retardation in humans (34).
Maternally expressed genes in the placenta are thought to act to restrain the allocation of maternal resources to the embryo and thereby counteract paternally expressed genes that promote placental or embryonic growth at the expense of the mother. For example, the maternally expressed Igf2r gene encodes a negative regulator of the paternally expressed growth factor IGFII (35–37). Given this paradigm, it is unclear what might be the evolutionary advantage of the two parental genomes differently regulating Gatm expression in the placenta. Perhaps the high level of Gatm expression in the placenta protects the mother from dramatic spikes in demand for energy by the embryo or reduces the embryonic drain on maternal resources by increasing the efficiency of energy generation within the extraembryonic tissues. To explain paternal genome silencing of placental Gatm, however, we would also have to imagine that the function of creatine in the placenta restrains the growth of the embryo. Regardless of the imprinted expression of the gene, the high level of Gatm expression in the extraembryonic tissues may point to an unrecognized role for the creatine/phosphocreatine high-energy phosphate-buffering system during embryonic development.
In previous studies of the distribution of Gatm activity in adult animals, Gatm activity was found predominantly in kidney and pancreas and at lower levels in other tissues such as liver, heart, lung, muscle, spleen, brain, testis, and thymus (19). In at least some tissues, the gene is known to be transcriptionally regulated by feedback inhibition of the final product, creatine. It is difficult to reconcile a role for imprinted regulation, which implies an effect of gene dosage, with a feedback-loop mechanism of transcriptional regulation. However, until more is known about the role of Gatm during development, it remains possible that Gatm levels, as differentially regulated by the two parental alleles, are critical for normal energy metabolism in a developing embryo.
Because most imprinted genes are found clustered with other imprinted genes in the genome, we examined several genes near Gatm to determine whether they were imprinted. We found no evidence that any of the neighboring genes were imprinted or that Gatm was part of a cluster. Our search included genes located ≈300 kb centromeric and ≈150 kb telomeric of the Gatm gene. Of course, failure to identify imprinted genes in the vicinity of Gatm does not prove that Gatm is not part of a cluster. One of the other putative genes within the region may be expressed and imprinted in placenta. Alternatively, one or more of the genes we examined may be imprinted in different tissues or at different developmental stages from Gatm.
Whereas most CpG islands within the mammalian genome are unmethylated (38), many CpG islands associated with imprinted or X-linked genes are differentially methylated (6, 39–41). A subset of these differentially methylated regions, such as the CpG island at Igf2, acquires differential methylation postzygotically (42–45) and is thought to be a consequence and not a cause of imprinting. Other differentially methylated regions, such as the imprinting control region at H19 and the intronic CpG islands in the Igf2r and KCNQ1 genes, are inherited from gametes (7–9, 11–14, 46, 47). Although many imprinted genes are associated with differentially methylated CpG islands, there are also imprinted genes that contain CpG islands unmethylated on both alleles. Examples of imprinted genes associated with unmethylated CpG islands include a paternally expressed gene, Dio3 (48), and the linked maternally expressed imprinted genes Ipl, Mash2, and Cdkn1c/p57Kip2 (data not shown). The lack of methylation at the Gatm CpG island places Gatm with this class of unmethylated imprinted gene.
For the unmethylated imprinted genes Ipl, Mash2, and Cdkn1c/p57Kip2, imprinted expression is regulated by a differentially methylated control region located some distance from the unmethylated genes (14). The mechanism of imprinting of the unmethylated Dio3 gene is not known. However, it located within a megabase of the “imprinting cluster” containing Gtl2 and Dlk and an associated intergenic CpG island (49), which is marked by germ-line differential methylation and is thus a likely candidate to function as an imprinting control center.
We found no evidence of a differentially methylated CpG island that might regulate the imprinting of Gatm. The island associated with Gatm is unmethylated, as is a large CpG island of the adjacent biallelic gene, LOC214616. Both islands are unmethylated in placenta, the tissue in which the Gatm gene is imprinted, and are thus almost certainly not responsible for regulating the imprinted expression of Gatm in that tissue.
In summary, we have determined that the gene encoding Gatm is expressed during mouse development and imprinted in extraembryonic tissues. These data suggest that creatine synthesis plays an important role in embryonic growth control. The Gatm gene resides in the middle of mouse chromosome 2 in apparent isolation from other imprinted genes and is associated with an unmethylated CpG island.
Acknowledgments
We thank Dr. Paul Matteson for generating the Peromyscus differential display cDNAs. This work was supported by a grant from the National Institute of General Medical Sciences. L.L.S. was supported by a grant from the American Cancer Society. S.M.T. was an investigator of the Howard Hughes Medical Institute.
Abbreviations
- En
embryonic day n
- PO
Peromyscus polionotus
- BW
Peromyscus maniculatus
References
- 1.Reik W, Walter J. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 2.Sleutels F, Barlow D P. Adv Genet. 2002;46:119–163. doi: 10.1016/s0065-2660(02)46006-3. [DOI] [PubMed] [Google Scholar]
- 3.Tilghman S. Cell. 1999;96:185–193. doi: 10.1016/s0092-8674(00)80559-0. [DOI] [PubMed] [Google Scholar]
- 4.Moore T, Haig D. Trends Genet. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- 5.Beechy C V. In: Results and Problems in Cell Differentiation. Ohlsson R, editor. Vol. 25. Berlin: Springer; 1999. pp. 303–323. [PubMed] [Google Scholar]
- 6.Razin A, Cedar H. Cell. 1994;77:473–476. doi: 10.1016/0092-8674(94)90208-9. [DOI] [PubMed] [Google Scholar]
- 7.Leighton P A, Ingram R S, Eggenschwiler J, Efstratiadis A, Tilghman S M. Nature. 1995;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 8.Thorvaldson J L, Duran K L, Bartolomei M S. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wutz A, Smrzka O W, Schweifer N, Schellander K, Wagner E F, Barlow D P. Nature. 1997;389:745–749. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]
- 10.Birger Y, Shemer R, Perk J, Razin A. Nature. 1999;397:84–88. doi: 10.1038/16291. [DOI] [PubMed] [Google Scholar]
- 11.Yoon B J, Herman H, Sikora A, Smith L T, Plass C, Soloway P D. Nat Genet. 2002;30:92–96. doi: 10.1038/ng795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chamberlain S J, Brannan C I. Genomics. 2001;73:316–322. doi: 10.1006/geno.2001.6543. [DOI] [PubMed] [Google Scholar]
- 13.Yang T, Adamson T E, Resnick J L, Leff S, Wevrick R, Francke U, Jenkins N A, Copeland N G, Brannan C I. Nat Genet. 1998;19:25–31. doi: 10.1038/ng0598-25. [DOI] [PubMed] [Google Scholar]
- 14.Fitzpatrick G V, Soloway P D, Higgins M J. Nat Genet. 2002;32:426–431. doi: 10.1038/ng988. [DOI] [PubMed] [Google Scholar]
- 15.Bell A C, Felsenfeld G. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 16.Hark A T, Schoenherr C J, Katz D J, Ingram R S, Levorse J M, Tilghman S M. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 17.Hagiwara Y, Hirai M, Nishiyama K, Kanazawa I, Ueda T, Sakaki Y, Ito T. Proc Natl Acad Sci USA. 1997;94:9249–9254. doi: 10.1073/pnas.94.17.9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt J V, Matteson P G, Jones B K, Guan X J, Tilghman S M. Genes Dev. 2000;14:1997–2002. [PMC free article] [PubMed] [Google Scholar]
- 19.Wyss M, Kaddurah-Daouk R. Physiol Rev. 2000;80:1107–1213. doi: 10.1152/physrev.2000.80.3.1107. [DOI] [PubMed] [Google Scholar]
- 20.Walker J B. Adv Enzymol Relat Areas Mol Biol. 1979;50:177–242. doi: 10.1002/9780470122952.ch4. [DOI] [PubMed] [Google Scholar]
- 21.Kent W J, Sugnet C W, Furey T S, Roskin K M, Pringle T H, Zahler A M, Haussler D. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Deken X, Wang D, Many M C, Costagliola S, Libert F, Vassart G, Dumont J E, Miot F. J Biol Chem. 2000;275:23227–23233. doi: 10.1074/jbc.M000916200. [DOI] [PubMed] [Google Scholar]
- 23.Razin A, Webb C, Szyf M, Yisraeli J, Rosenthal A, Naveh-Many T, Sciaky-Gallili N, Cedar H. Proc Natl Acad Sci USA. 1984;81:2275–2279. doi: 10.1073/pnas.81.8.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellington W R. Annu Rev Physiol. 2001;63:289–325. doi: 10.1146/annurev.physiol.63.1.289. [DOI] [PubMed] [Google Scholar]
- 25.Bianchi M C, Tosetti M, Fornai F, Alessandri M G, Cipriani P, De Vito G, Canapicchi R. Ann Neurol. 2000;47:511–513. [PubMed] [Google Scholar]
- 26.van der Knaap M S, Verhoeven N M, Maaswinkel-Mooij P, Pouwels P J, Onkenhout W, Peeters E A, Stockler-Ipsiroglu S, Jakobs C. Ann Neurol. 2000;47:540–543. doi: 10.1002/1531-8249(200004)47:4<540::aid-ana23>3.3.co;2-b. [DOI] [PubMed] [Google Scholar]
- 27.Item C B, Stockler-Ipsiroglu S, Stromberger C, Muhl A, Alessandri M G, Bianchi M C, Tosetti M, Fornai F, Cioni G. Am J Hum Genet. 2001;69:1127–1133. doi: 10.1086/323765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carducci C, Leuzzi V, Prudente S, Mercuri L, Antonozzi I. Mol Genet Metab. 2000;71:633–638. doi: 10.1006/mgme.2000.3108. [DOI] [PubMed] [Google Scholar]
- 29.Leuzzi V, Bianchi M C, Tosetti M, Carducci C, Cerquiglini C A, Cioni G, Antonozzi I. Neurology. 2000;55:1407–1410. doi: 10.1212/wnl.55.9.1407. [DOI] [PubMed] [Google Scholar]
- 30.Salomons G S, van Dooren S J, Verhoeven N M, Cecil K M, Ball W S, Degrauw T J, Jakobs C. Am J Hum Genet. 2001;68:1497–1500. doi: 10.1086/320595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stockler S, Isbrandt D, Hanefeld F, Schmidt B, von Figura K. Am J Hum Genet. 1996;58:914–922. [PMC free article] [PubMed] [Google Scholar]
- 32.Lefebvre L, Viville S, Barton S C, Ishino F, Keverne E B, Surani M A. Nat Genet. 1998;20:163–169. doi: 10.1038/2464. [DOI] [PubMed] [Google Scholar]
- 33.Li L, Keverne E B, Aparicio S A, Ishino F, Barton S C, Surani M A. Science. 1999;284:330–333. doi: 10.1126/science.284.5412.330. [DOI] [PubMed] [Google Scholar]
- 34.Nicholls R D, Knepper J L. Annu Rev Genomics Hum Genet. 2001;2:153–175. doi: 10.1146/annurev.genom.2.1.153. [DOI] [PubMed] [Google Scholar]
- 35.Filson A, Louvi A, Efstradiatis A, Roberston E J. Development (Cambridge, UK) 1993;118:731–736. doi: 10.1242/dev.118.3.731. [DOI] [PubMed] [Google Scholar]
- 36.Lau M M H, Stewart C E H, Liu Z, Bhatt H, Rotwein P, Stewart C L. Genes Dev. 1994;8:2953–2963. doi: 10.1101/gad.8.24.2953. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z-Q, Fung M R, Barlow D P, Wagner E F. Nature. 1994;372:464–467. doi: 10.1038/372464a0. [DOI] [PubMed] [Google Scholar]
- 38.Cross S H, Bird A P. Curr Opin Genet Dev. 1995;5:309–314. doi: 10.1016/0959-437x(95)80044-1. [DOI] [PubMed] [Google Scholar]
- 39.Barlow D P. Science. 1995;270:1610–1613. doi: 10.1126/science.270.5242.1610. [DOI] [PubMed] [Google Scholar]
- 40.Ferguson-Smith A C, Sasaki H, Cattanach B M, Surani M A. Nature. 1993;362:751–755. doi: 10.1038/362751a0. [DOI] [PubMed] [Google Scholar]
- 41.Yen P H, Patel P, Chinault A C, Mohandas T, Shapiro L J. Proc Natl Acad Sci USA. 1984;81:1759–1763. doi: 10.1073/pnas.81.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsieh C L. Curr Opin Genet Dev. 2000;10:224–228. doi: 10.1016/s0959-437x(00)00064-2. [DOI] [PubMed] [Google Scholar]
- 43.Reik W, Walter J. Curr Opin Genet Dev. 1998;8:154–164. doi: 10.1016/s0959-437x(98)80136-6. [DOI] [PubMed] [Google Scholar]
- 44.Reik W, Murrell A. Nature. 2000;405:408–409. doi: 10.1038/35013178. [DOI] [PubMed] [Google Scholar]
- 45.Shemer R, Birger Y, Dean W L, Reik W, Riggs A D, Razin A. Proc Natl Acad Sci USA. 1996;93:6371–6376. doi: 10.1073/pnas.93.13.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tremblay K D, Saam J R, Ingram R S, Tilghman S M, Bartolomei M S. Nat Genet. 1995;9:407–413. doi: 10.1038/ng0495-407. [DOI] [PubMed] [Google Scholar]
- 47.Stoger R, Kubicka P, Liu C-G, Kafri T, Razin A, Cedar H, Barlow D P. Cell. 1993;73:61–71. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- 48.Tsai C, Lin S, Ito M, Takagi N, Takada S, Ferguson-Smith A. Curr Biol. 2002;12:1221–1226. doi: 10.1016/s0960-9822(02)00951-x. [DOI] [PubMed] [Google Scholar]
- 49.Takada S, Paulsen M, Tevendale M, Tsai C E, Kelsey G, Cattanach B M, Ferguson-Smith A C. Hum Mol Genet. 2002;11:77–86. doi: 10.1093/hmg/11.1.77. [DOI] [PubMed] [Google Scholar]